The expression of cell surface molecule, IGSF4, on human and mouse T cells associate with the TCR ζ-chain and colocalizes to the c-SMAC to enhance cytokine production and adhesion to APCs.
Abstract
Immunoglobulin superfamily member 4 (IGSF4) is a known ligand of CRTAM, a receptor expressed in activated NKT and CD8+ T cells, but its function in T cell immunity has not been elucidated. In this study, we show that IGSF4 directly interacts with the T cell receptor (TCR) ζ-chain and enhances TCR signaling by enhancing ζ-chain phosphorylation. Ectopic overexpression of IGSF4 enhances TCR-mediated T cell activation. In contrast, IGSF4 knockdown shows a dramatic decrease in markers associated with T cell activation compared with those in control small interfering RNA. The transmembrane domain is essential for TCR ζ-chain association and clustering to the immunological synapse, and the ectodomain is associated with T cell interaction with antigen-presenting cells (APCs). IGSF4-deficient mice have impaired TCR-mediated thymocyte selection and maturation. Furthermore, these mice reveal attenuated effector T cell functions accompanied by defective TCR signaling. Collectively, the results indicate that IGSF4 plays a central role in T cell functioning by dual independent mechanisms, control of TCR signaling and control of T cell–APC interaction.
Immunoglobulin superfamily member 4 (IGSF4) is a member of the intercellular adhesion molecule family (Shingai et al., 2003). It was first characterized as a tumor suppressor in non-small cell lung cancer and termed TSLC1 (Murakami, 2005). Later, it was found to have roles in the adhesion of spermatogenic cells to Sertoli cells (van der Weyden et al., 2006; Yamada et al., 2006) and mast cells to fibroblasts (Ito et al., 2003, 2004; Koma et al., 2005) and was termed SgIGSF. Other researchers revealed that IGSF4 drives the synaptic formation of neural cells and termed it SynCAM (Ohta et al., 2005). Three extracellular domains of IGSF4 mediate homophilic or heterophilic interactions independently of Ca2+ (Watabe et al., 2003). The cytoplasmic (CT) domain contains the binding motifs connecting to actin fibers, but the function of the transmembrane (TM) domain is not known. Furthermore, on APCs, IGSF4 serves as a ligand of CRTAM (MHC class I–restricted T cell–associated molecule), a receptor primarily expressed on activated cytotoxic T cells (Arase et al., 2005), and regulates IFN-γ and IL-22 expression by activated CD8+ T cells (Yeh et al., 2008). However, its function with regard to T cells has not been addressed.
The TCR is the key structure recognizing its cognate peptide in an MHC molecule. However, it is not sufficient to induce intracellular signaling cascades and the subsequent T cell activation. Therefore, various integral membrane proteins are associated with or functionally involved in TCR-mediated signal transduction. For example, invariant chains such as CD3γ, CD3δ, CD3ε, and TCR ζ-chains, constituting the TCR complex, initiate a series of intracellular signaling cascades (Lin and Weiss, 2001; Le Deist et al., 2007). Surface antigens such as CD2, CD4, or CD8 and CD5, which are physically associated with the TCR complex (Beyers et al., 1992), are required for efficient signal transduction (Horejsí et al., 2004). Although co-stimulatory molecules such as B7/CD28, TNFR/TNF, CD58/CD2, and ICAM-1/LFA-1 are not directly associated with the TCR complex, they provide secondary signals for T cell activation (Wingren et al., 1995). Lastly, membrane proteins providing multiple docking sites for cytosolic signaling and effector molecules have roles in the regulation of T cell functions (Zhang and Samelson, 2000). These proteins, termed TRAPs (TM adaptor proteins), include LAT (linker for activation of T cells; Martelli et al., 2000; Wange, 2000), TRIM (TCR-interacting molecule; Bruyns et al., 1998; Kirchgessner et al., 2001), PAG (protein associated with GEMs; Davidson et al., 2003; Maksumova et al., 2005), NTAL (non–T cell activation linker; Brdicka et al., 2002), LIME (LCK-interacting membrane protein; Hur et al., 2003), and SIT (SH2-domain–containing protein tyrosine phosphatase [SHP2]–interacting TRAP; Marie-Cardine et al., 1999; Posevitz et al., 2008). However, no member of the intercellular adhesion molecule family that physically associates with the subunits of the TCR complex and modulates TCR signaling has been reported so far.
Interestingly, in this study, we found that IGSF4 messenger RNA (mRNA) and its protein product are expressed in all human and mouse T cells. Furthermore, we found that IGSF4, even without the ectodomain, localizes at the central supramolecular activation cluster (SMAC [c-SMAC]) in the immunological synapse during T cell–APC interaction. This finding raised the question whether IGSF4 serves as a physical partner with the TCR complex or at least modulates TCR signaling in an adhesion-independent way. Because IGSF4 is an intercellular adhesion molecule expressed on both T cells and APCs, we also addressed whether IGSF4 has an adhesion-dependent co-stimulatory function through homotypic or heterotypic interactions with ligands on APCs. In this study, we provide evidence that IGSF4 is an important molecule for T cell functioning.
RESULTS
IGSF4 is expressed in human and mouse T cells and positively regulates T cell responses
IGSF4 is reportedly not detectable in normal CD4+ T cells and some human T cell lines (Sasaki et al., 2005). However, we detected IGSF4 not only in immune function–related tissues (spleen, thymus, and lymph node) but also in human T cell lines (Fig. 1 A, top and bottom left) and all human and mouse T cell subsets including CD4+ and CD8+ T cells, B cells (CD19+), and DCs (CD11c+; Fig. 1 A, bottom right). In addition, anti-CD3/28 or PMA/A23187 stimulation slightly increased both the mRNA and protein levels of IGSF4 in Jurkat T cells (Fig. 1 B, top), human PBLs (Fig. 1 B, bottom left), and mouse splenic CD3+ T cells (Fig. 1 B, bottom right). These results clearly demonstrate that IGSF4 is expressed in T cells, and its expression is controlled during T cell activation.
Figure 1.
IGSF4 is expressed in human and mouse T cells and positively regulates T cell responses. (A) Western blot and RT-PCR analyses. (top) Tissue distribution of IGSF4 in 8-wk-old mice. IGSF4 expression in T cell lines (bottom left) and purified mouse or human T cell subsets (bottom right). #, clone number; SP, splenic CD3+ T cells. (B) Jurkat T cells (top), human PBLs (bottom left), and mouse splenic CD3+ T cells (bottom right) were stimulated for the indicated times with anti-CD3/28, and the expression of IGSF4 or IL-2 was assessed by quantitative PCR and flow cytometric analyses (top), RT-PCR analysis (bottom left), and Western blot analysis (bottom right). (A and B) The data are representative of three independent experiments. (C) Jurkat T cells (left) and human PBLs (middle) were transfected with either 70 µM of scrambled (SC) siRNA or siRNA targeting IGSF4. IGSF4 expression was measured after 48 h of transfection (top). Stimulation of cells and IL-2 mRNA measurements were performed as described in B. The results are the mean ± SD of triplicate experiments. (right) Purified splenic CD3+ T cells from TCR transgenic DO11.10 mice were transfected with 1 µg/100 µl shRNA targeting IGSF4. The IGSF4 knockdown efficiency was analyzed after 48 h of transfection (top). The cells were coincubated with 50 ng/ml OVA–pulsed A20 mouse B cells, and the IL-2 mRNA levels were measured. The results are the mean ± SD of triplicate experiments. (D, top) Establishment of Jurkat T cells overexpressing GFP or WT-IGSF4_GFP by lentiviral transduction. The efficiency of viral infection was determined by Western blot and flow cytometric analyses. IB, immunoblot. (bottom) The cells were then stimulated with SEE-pulsed Raji B cells, anti-CD3/28, or PMA/A23187 for the indicated time. IL-2 mRNA levels were assessed by RT-PCR (blots) and real-time quantitative PCR (graphs) analyses. The results are the mean ± SD of triplicate experiments. Molecular mass (M) is indicated in kilodaltons. EV, empty vector.
We next determined whether IGSF4 knockdown influences T cell functions. Compared with control small interfering RNA (siRNA), siRNA targeting IGSF4 significantly reduced IGSF4 expression in Jurkat T cells (Fig. 1 C, left). Interestingly, a dramatic reduction in IL-2 mRNA levels was observed in Jurkat T cells transfected with IGSF4 siRNA after stimulation with anti-CD3/28 or incubation with Staphylococcus enterotoxin E (SEE)–pulsed Raji B cells (Fig. 1 C, left). A significant decrease in IL-2 expression was also observed in human PBLs (Fig. 1 C, middle). To test the antigen-specific T cell response, we also used splenic CD3+ T cells from TCR transgenic DO11.10 mice and found a dramatic reduction in mouse IL-2 mRNA levels in the IGSF4 short hairpin RNA (shRNA)–transfected cells incubated with OVA peptide–loaded A20B cells (Fig. 1 C, right).
We also established Jurkat T cells overexpressing either GFP (J-GFP) or WT IGSF4_GFP (J-IGSF4_GFP) by lentiviral transduction (Fig. 1 D, top). J-IGSF4_GFP cells had a significant increase in IL-2 mRNA levels compared with J-GFP cells after stimulation with anti-CD3/28 or incubation with SEE-pulsed Raji B cells. In contrast, no significant difference was observed between J-GFP and J-IGSF4_GFP cells stimulated with PMA/A23187 (Fig. 1 D, bottom). These results strongly suggest a positive role of IGSF4 in TCR-mediated T cell activation.
IGSF4 localizes at the c-SMAC in the immunological synapse
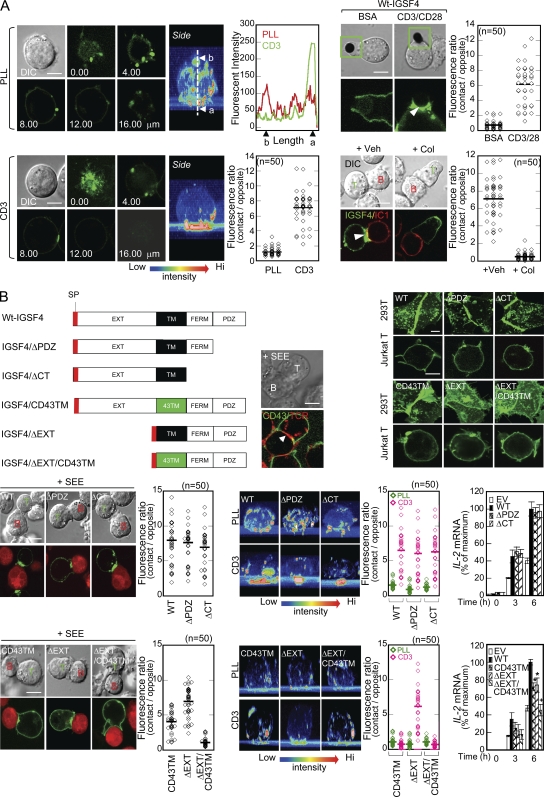
Many T cell–regulating molecules are located in a specialized junctional structure referred to as the immunological synapse (Monks et al., 1998; Grakoui et al., 1999) during T cell contact with APCs. We next explored the localization of IGSF4 during Jurkat T cell interaction with SEE-pulsed Raji B cells. In the absence of SEE, IGSF4 was most prominent on the membrane surface and was strongly relocated at the T cell–T cell contact sites (>95%; Fig. 2 A), suggesting that IGSF4 on one T cell induces homotypic trans-interaction with the same molecule on the neighboring T cell. Interestingly, relocation of IGSF4 was also observed at immature immunological synapses in the absence of SEE (28 ± 7%; Fig. 2 A). The relocation and concentration of IGSF4 was more obvious during maturation of the immunological synapse in the presence of SEE (73 ± 5%; Video 1). These results demonstrate that IGSF4 has a potential role in the early to late phase of T cell activation.
Figure 2.
IGSF4 localizes at the c-SMAC in the immunological synapse. (A) J-IGSF4_GFP cells were incubated with Raji B cells stained with orange CMRA in the presence or absence of SEE. Arrowheads indicate the polarized IGSF4 (green) at the cell–cell contact sites. The percentage of T–T or T–B conjugates with surface IGSF4 relocation at the contact zone relative to the total number of conjugates in the absence or presence of SEE was analyzed from a total of 150 conjugates of each category. The results are the mean ± SD of triplicate experiments. The boxed areas (blue and orange) are represented as zoomed images in the right panels. (B) J-IGSF4_GFP cells were stained with anti-CD3 (cy5), anti-CD45 (cy5), or anti–LFA-1 (cy5) Fabs and then incubated with SEE-loaded Raji B cells stained with anti–ICAM-1 (cy3) Fab. See also Video 1. The data are representative of four independent experiments. The panels on the right represent zoomed images of the boxed areas (yellow) indicated in the differential interference contrast images. Bars, 10 µm.
To identify its specific location, we performed a colocalization study of IGSF4 with other immunological synapse molecules such as CD3, CD45, LFA-1, and ICAM-1 in T cell–APC conjugates. IGSF4 predominantly colocalized with CD3, a key molecule of the c-SMAC, and was clearly distinguishable from LFA-1, a peripheral SMAC molecule. On the basis of these observations, we considered the potential mechanisms of IGSF4 in T cell function. Given that IGSF4 is an intercellular adhesion molecule that is also expressed in APCs including B cells and DCs (Fig. 1 A), an adhesion-dependent co-stimulatory function through homotypic or heterotypic interactions with ligands on APCs is possible. Because IGSF4 localized at the c-SMAC and highly colocalized with CD3, it is possible that IGSF4 performs an enhancer function in association with the molecules in the TCR signaling platform. Alternatively, these two mechanisms could coexist independently.
IGSF4 enhances T cell–T cell or T cell–APC adhesion, and its ectodomain mutation reduces T cell activation
We first tested the adhesion-dependent co-stimulatory function. As expected, J-GFP cells did not prominently aggregate under the normal culture condition. In contrast, J-IGSF4_GFP cells aggregated (Fig. 3 A), with intense aggregation after TCR engagement with anti-CD3/28 (Fig. 3 A). T cell adhesion to APCs was also examined in the absence or presence of the superantigen. Quantitative analysis revealed that J-IGSF4_GFP cells had significantly increased conjugate formation regardless of SEE (Fig. 3 B), demonstrating that IGSF4 mediates T cell adhesion to APCs.
Figure 3.
IGSF4 enhances T cell–T cell and T cell–APC adhesions by ectodomain interaction. (A) J-GFP or J-IGSF4_GFP cells (2 × 105 cells/well) were cultured in the presence or absence of anti-CD3/28 for 3 h and photographed. Quantitation of cell aggregation was determined as described in Materials and methods. The results are the mean ± SD of six experiments. *, P < 0.05 versus J-GFP cells without stimulation. **, P < 0.05 versus J-GFP cells with anti-CD3/28 stimulation. DIC, differential interference contrast; EV, empty vector. (B) A representative conjugate formation profile with T cells and Raji B cells and the percentage of DP cells (blue) are shown in the flow cytometric plot and bar graph, respectively. The results are the mean ± SD of triplicate experiments. *, P < 0.05 versus J-GFP cells without stimulation. **, P < 0.05 versus J-GFP cells with SEE-pulsed Raji B cells. (C and D) Domain swapping from the IGSF4 ectodomain to ICAM-1 D3–5 in IGSF4-mediated T cell activation. (C, top) Arrows indicate the relocation of IGSF4 at the cell–cell contact regions in Jurkat T cells expressing WT-IGSF4_GFP but not IC1_IGSF4_GFP. (bottom) Schematic illustration showing swapped region of IGSF4/EXTD with ICAM1/D3-5. Quantitation of IGSF4 or IC1_IGSF4 relocation was performed as described in Fig. 2 A. The results are the mean ± SD of four experiments. IG4, IGSF4; IC1, ICAM-1 D3–5. (D) These cells were stimulated with SEE-pulsed Raji B cells, and the IL-2 mRNA levels were assessed by real-time quantitative PCR. The results are the mean ± SD of triplicate experiments. *, P < 0.05, as compared with cells expressing WT-IGSF4. Bars: (A) 100 µm; (C) 10 µm.
We next questioned whether the increased IL-2 expression in J-IGSF4_GFP cells corresponds to the increased T cell adhesion to APCs. To this end, three Ig domains of IGSF4 were exchanged with domains 3–5 (D3–5) of ICAM-1 and expressed in Jurkat T cells (Fig. 3 C). Because D3–5 of ICAM-1 have no ligand-binding site on B cells, this ICAM-1–IGSF4 chimera (IC1_IGSF4) is functionally null. Polarization of IGSF4 at the cell–cell contact region (arrows) disappeared in J-IC1_IGSF4_GFP cells (95 ± 4% of IGSF4 vs. 3 ± 2% of IC1_IGSF4; Fig. 3 C). Accordingly, IL-2 expression reduced in these cells compared with J-IGSF4_GFP cells, suggesting that an adhesion-dependent function is an important mechanism for IGSF4-mediated T cell activation. Interestingly, however, we observed that the levels of IL-2 mRNA in J-IC1_IGSF4_GFP cells did not reduce to those in J-GFP cells (Fig. 3 D). This result suggests the existence of another mechanism for T cell activation by IGSF4.
IGSF4 accumulation in the c-SMAC depends on the TM domain
To completely exclude the adhesion-dependent mechanism by the IGSF4 ectodomain, J-IGSF4_GFP cells were placed on coverslips coated with poly-l-lysine (PLL) or anti-CD3. Confocal microscopy revealed that WT IGSF4_GFP significantly accumulated at the bottom of only the anti-CD3–coated coverslips (Fig. 4 A, left). The same results were obtained with anti-CD3/28–coated microbeads (Fig. 4 A, right top). These results suggest that IGSF4 relocation to the c-SMAC is independent off receptor–ligand interaction but requires other domains. Supporting this view, colchicine, a microtubule inhibitor, almost completely inhibited IGSF4 relocation to the c-SMAC (Fig. 4 A, right bottom), suggesting the involvement of the CT or TM domain rather than receptor–ligand interaction.
Figure 4.
IGSF4 recruitment to the immunological synapse is independent off receptor–ligand interaction but is mediated by the TM domain. (A, left) J-IGSF4_GFP cells were placed on chambered coverslips coated with PLL or anti-CD3. Confocal images were obtained and reconstituted to the three-dimensional images by the FLUOVIEW program. The fluorescent intensity caused by the accumulation at the PLL or anti-CD3–coated surface was quantified. Note, a = contact region and b = opposite region. Each dot represents a single measurement, and at least 50 cells were examined. The data are representative of two independent experiments. (right) J-IGSF4_GFP cells were incubated with either BSA or anti-CD3/28–coated beads (top) or SEE-pulsed Raji B cells in the presence or absence of 1 µg/ml colchicine (Col) for 30 min (bottom), and live-cell imaging was performed. Arrowheads indicate IGSF4 accumulation at the synapse sites. Boxed areas (green) are shown as magnified images in the micrographs below. The fluorescent intensity caused by the accumulation at the T cell–APC contact site was quantified. Data analysis was performed as described in A (left). The data are representative of two independent experiments. DIC, differential interference contrast. (B, top left) Schematic diagram showing deletion and swapping mutants of IGSF4 (top). T cell–B cell conjugates were stained with anti-CD43 (FITC) and anti-CD3 (cy3). The arrowhead reveals the exclusion of CD43 from the immunological synapse (bottom). (top right) Localization pattern of each mutant of IGSF4 in HEK293T and Jurkat T cells. (bottom left and middle) Jurkat T cells expressing IGSF4_GFP or mutants (IGSF4ΔPDZ, IGSF4ΔCT, IGSF4/CD43TM, IGSF4/ΔEXT, or IGSF4/ΔEXT/CD43TM) were either incubated with SEE-loaded Raji B cells (bottom left) or placed on coverslips coated with PLL or anti-CD3 (bottom middle), and confocal analysis was performed. At least 20 z-stack images were reconstituted into a three-dimensional image for the bottom middle panel. Intensity represents accumulation from low (blue) to high (red). Quantitation and data analysis were performed as described in A (left). The data are representative of four independent experiments. (bottom right) Real-time quantitative PCR analysis of IL-2 mRNA expression in response to anti-CD3/28. *, P < 0.05; and +, P < 0.01, as compared with cells expressing WT-IGSF4. The results are the mean ± SD of triplicate experiments. EV, empty vector. (A and B) Horizontal bars indicate the mean. Bars, 10 µm.
To define the functional domain of IGSF4, several deletion mutants were constructed and expressed in Jurkat T cells. GFP was fused at the end of each construct to facilitate visualization by confocal microscopy. Interestingly, a partial deletion of CT (ΔPDZ) or deletion of the entire CT domain (ΔCT) of IGSF4 (Fig. 4 B, top left) had little effect on its subcellular localization on the membrane (Fig. 4 B, top right), accumulation at the cell–cell contact region (Fig. 4 B, bottom left) and anti-CD3–coated coverslips (Fig. 4 B, bottom middle), or IL-2 expression in response to anti-CD3/28 (Fig. 4 B, bottom right). These results demonstrate that the CT domain of IGSF4 is not required for localization of IGSF4 on the membrane and is not the functional domain.
We therefore examined the function of the TM domain. We used the domain-swapping strategy to understand the apparent role of the TM domain in IGSF4-mediated T cell activation. CD43 was selected as this protein is known to be excluded from the c-SMAC during immunological synapse formation (Fig. 4 B, top left; Delon et al., 2001). Exchange of the TM domain of IGSF4 to the TM domain of CD43 generated a mutant termed IGSF4/CD43TM. Despite the similar cellular localization pattern, interestingly, this mutant revealed less accumulation at the T cell–APC contact sites (Fig. 4 B, top right and bottom left) and anti-CD3–coated surfaces (Fig. 4 B, bottom middle). In addition, IL-2 expression in response to anti-CD3/28 was significantly reduced in Jurkat T cells expressing IGSF4/CD43TM compared with WT IGSF4 (Fig. 4 B, bottom right).
The obtained result suggested that the TM domain may have its own role in T cell activation. Therefore, we deleted the ectodomain and the signal sequence was directly linked to the TM domain, thereby enabling the expression of only the TM and CT domains (IGSF4/ΔEXT) in 293T and Jurkat T cells. The membrane localization was similar to the WT (Fig. 4 B, top right). In addition, this mutant accumulated at the T cell–APC contact sites and anti-CD3–coated surfaces (Fig. 4 B, bottom left and middle). Consequently, J-IGSF4/ΔEXT_GFP cells had higher levels of IL-2 expression than J-GFP cells, but the expression levels were lower than those in J-IGSF4_GFP cells (Fig. 4 B, bottom right). In contrast, despite a similar cellular localization pattern (Fig. 4 B, top right), the mutant with CD43TM (IGSF4/ΔEXT/CD43TM) was not recruited to the c-SMAC and anti-CD3–coated surfaces (Fig. 4 B, bottom left and middle). Finally, no difference in IL-2 expression was observed compared with that in J-GFP cells (Fig. 4 B, bottom right). Together, these results strongly suggest that the TM domain of IGSF4 is essential and sufficient for recruitment of IGSF4 to the c-SMAC and is therefore important for T cell activation.
IGSF4 associates with the TCR ζ-chain through the TM domain and boosts ζ-chain–linked signaling cascades
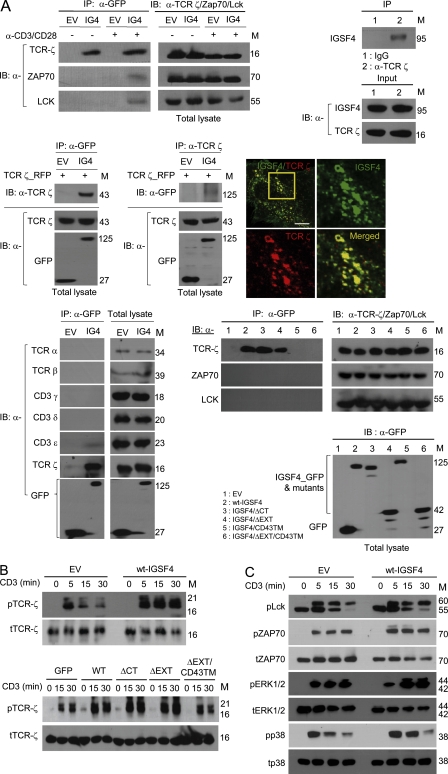
The finding that the TM domain is essential for IGSF4 relocation to the c-SMAC is intriguing because IGSF4 may interact with molecules in the c-SMAC through TM–TM interactions. To identify the physically interacting partners of IGSF4 in the c-SMAC, coimmunoprecipitation assay was performed with anti-GFP (for IGSF4), and immunoblotting was performed with antibodies against potential candidate proteins, including TCR α and β, CD3γ, CD3δ, CD3ε, TCR ζ, Zap70, and Lck. IGSF4 strongly interacted with the ζ-chain under both resting and activated conditions. The interaction was also confirmed by reverse configuration: pull-down of the ζ-chain and immunoblotting of endogenous IGSF4 (Fig. 5 A, top left and right). In parallel experiments, however, IGSF4 failed to interact with other components of the TCR, including TCR α and β, CD3γ, and CD3δ (Fig. 5 A, bottom left), suggesting that the interaction of IGSF4 is ζ-chain specific among the components of the TCR complex. Only a trace amount of CD3ε was recovered in the IGSF4 immunoprecipitates under the mild detergent (0.3% CHAPS) condition (Fig. 5 A, bottom left). To further test whether IGSF4 binding to ζ-chain occurs in the absence of other components of the TCR complex, cDNA constructs encoding WT-IGSF4_GFP and ζ_RFP were cotransfected in 293T cells, and then a coimmunoprecipitation assay was performed with anti-GFP (for IGSF4) and anti-TCR ζ-chain. The results strongly demonstrated that IGSF4 directly associates with the TCR ζ-chain (Fig. 5 A, middle left). In supporting this view, WT-IGSF4 was highly colocalized with the TCR ζ_RFP (Fig. 5 A, middle right). Low amounts of Zap70 and Lck were also detectable in the IGSF4 immunoprecipitates under the mild-detergent condition only after stimulation with anti-CD3/28, suggesting recruitments of Zap70 and Lck to the phosphorylated ITAMs in the ζ-chain (Fig. 5 A, top left). Interestingly, WT IGSF4 and mutant IGSF4 containing the TM domain could interact only with the ζ-chain (Fig. 5 A, bottom right), demonstrating that the TM domain is essential for the association with the ζ-chain and therefore localization to the c-SMAC. In addition, these results strongly demonstrate that IGSF4 is involved in TCR/CD3-mediated first signals.
Figure 5.
IGSF4 associates with the TCR ζ-chain through the TM domain and boosts TCR-mediated signal transduction. (A, top) J-GFP or J-IGSF4_GFP cells were incubated for 30 min with or without anti-CD3/28. Cell lysates were immunoprecipitated with anti-GFP (top left) or anti–TCR ζ-chain (top right) and then immunoblotted with antibodies against the indicated molecules (ζ-chain, Lck, Zap70, or IGSF4). (middle) HEK293T cells were cotransfected with WT-IGSF4_GFP and TCR ζ_RFP, and then immunoprecipitation (IP) and immunoblotting (IB) were performed as in the top left blots. Confocal images show surface localization of IGSF4_GFP and TCR ζ_RFP. The boxed area (yellow) is represented as zoomed images in the right and bottom micrographs. Bar, 10 µm. (bottom left) J-GFP or J-IGSF4_GFP cell pellets were lysed, immunoprecipitated with anti-GFP, and then immunoblotted with antibodies against the indicated molecules (TCR α and β, CD3γ, CD3δ, CD3ε, TCR ζ, and GFP). Total lysates were also immunoblotted with the same antibodies indicated above. The data are representative of two independent experiments. (bottom right) Immunoprecipitation and immunoblotting of Jurkat T cells expressing GFP, WT-IGSF4_GFP, or the indicated mutants (ΔCT, ΔEXT, CD43TM, and ΔEXT/CD43TM) as in A (top left). The data are representative of at least three independent experiments. EV, empty vector; IG4, IGSF4. (B) Jurkat T cells expressing GFP, WT-IGSF4_GFP, or mutant IGSF4 (ΔCT, ΔEXT, and ΔEXT/CD43TM) were stimulated with plate-bound anti-CD3 for the indicated time. The cells lysates were immunoblotted by using antibodies against phosphorylated (p) and total (t) ζ-chain. The data are representative of three independent experiments. (C) J-GFP or J-IGSF4_GFP cells were stimulated with plate-bound anti-CD3 for the indicated time. The cell lysates were immunoblotted for the phosphorylated and total forms of Lck, Zap70, ERK, and p38 kinase. The data are representative of at least three independent experiments. Molecular mass (M) is indicated in kilodaltons.
Next, we wondered whether the association of IGSF4 with the ζ-chain influences the phosphorylation of downstream signaling molecules upon TCR/CD3 stimulation. J-IGSF4_GFP cells showed increased levels of phospho–ζ-chain (p– ζ-chain) compared with J-GFP cells (Fig. 5 B, top). Consistently, downstream proximal kinases such as Lck and Zap70 and distal MAP kinases such as ERK and p38 kinase were highly phosphorylated in J-IGSF4_GFP cells compared with J-GFP cells (Fig. 5 C). Interestingly, the level of p–ζ-chain was significantly increased in J-IGSF4/ΔEXT_GFP cells compared with the levels in J-GFP and J-ΔEXT/CD43TM_GFP cells (Fig. 5 B, bottom). These results demonstrate that binding of IGSF4 to the ζ-chain through the TM domain is critical for enhancing ζ-chain activity and that of its following proximal and distal kinases.
IGSF4-deficient mice have defective T cell functioning
To obtain insight into the physiological significance of IGSF4 in T cell immunity, we generated mice deficient in IGSF4 by using gene trap methods (strain name: B6;CB-Cadm1Gt(pU-21W)34Imeg). Insertion of the trap vector in the first intron region of IGSF4 was confirmed by PCR analysis using genomic DNA from ear tissues (Fig. 6, A and B). However, we found a small amount of IGSF4 protein in T cells from IGSF4-deficient mice (see Fig. 8 A), suggesting inefficient use of the splice acceptor site by which a small amount of normal mRNA can be produced, as described previously (Li et al., 2010). Therefore, the insertion was considered a hypomorphic allele and homozygous mice were designated GT/GT. Interestingly, the homozygous mice were smaller and lighter than the WT mice (Fig. 6 C), implying that IGSF4 may also be involved in the developmental process or bone development.
Figure 6.
Generation of IGSF4-deficient mice and characterization of phenotype. (A) The trap vector, pU-21W, was inserted into the first intron of IGSF4. P1-2 (a primer set for the trap vector insertion point in the first intron; red arrows) and T1-2 (a primer set for the trap vector–specific sequence; orange arrows) were used for genotyping, and E1-2 (a primer set for the exon 1– and exon 2–specific sequence; blue arrows) was used for RT-PCR to detect endogenous IGSF4 mRNA. (B) PCR genotyping. Genomic DNA was extracted from mouse ear tissue. The WT (+/+) and trap (GT) alleles were detected with P1-2, but the homozygote (GT/GT) allele was not detected with this primer set (top). Total RNA was also extracted from mouse ear tissue, and the levels of IGSF4 mRNA were determined by using E1-2 (bottom). (C) Phenotypic comparison of IGSF4+/+ and IGSF4GT/GT mice. 4-wk-old and E18.5 WT and knockout littermates of size differences are shown. The organs from the IGSF4GT/GT mice are slightly smaller than those of the WT littermates (He, heart; Ki, kidney; Sp, spleen; Th, thymus). Body weight was measured weekly. The data represent the mean ± SD (n = 8).
Figure 8.
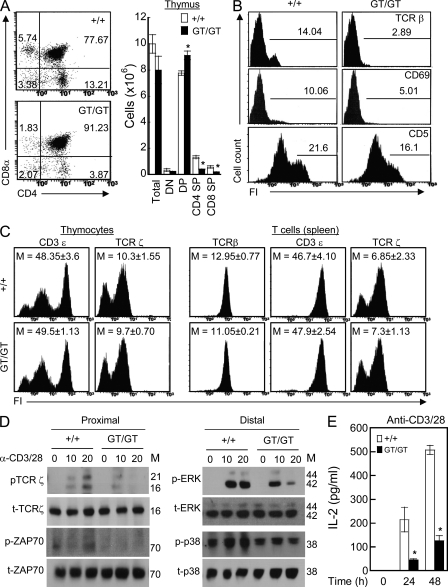
IGSF4GT/GT mice show defective T cell function. (A) IGSF4 expression in purified CD3+ T cells from the spleens and lymph nodes of WT and IGSF4GT/GT mice. Molecular mass (M) is indicated in kilodaltons. (B) The number of each cell type in the lymph node and spleen. The results are the mean ± SD of triplicate experiments. *, P < 0.05, as compared with IGSF4+/+ mice. (C) T cells purified from the spleen were stimulated with anti-CD3/28 for the indicated time. The phosphorylated (p) and total (t) forms of Lck, Zap70, ERK, and p38 kinase were determined by Western blot analysis. Data are representative of three independent experiments. (D) Allogeneic DCs and CD3+ T cells from WT or IGSF4GT/GT mice were incubated in SEB-containing medium for 3 h to allow conjugation. The cells were fixed and stained with anti–p-tyrosine (4G10; arrows). Polarization of p-tyrosine in T cells was quantified by cell counting (n > 50). Data are representative of at least three independent experiments. DIC, differential interference contrast. (E) CD3+ T cells were mixed with SEB-pulsed CD11C+ DCs (red) or CD19+ B cells from WT mice, and then conjugate formation was determined by confocal microscopy (left) and flow cytometry (bar graphs). (F) Purified splenic CD3+ T cells from WT and IGSF4GT/GT mice were incubated with anti-CD3/28 for 72 h. Cell proliferation was assessed by [3H]thymidine incorporation. (G) CD4+ or CD8+ T cells were stimulated with anti-CD3/28 or PMA/A23187 for 24 h, and the cytokine productions (IL-2, IL-4, and IFN-γ) were measured by ELISA. (E–G) The results are the mean ± SD of triplicate experiments. *, P < 0.05, as compared with cells from IGSF4+/+ mice. Bars: (D) 5 µm; (E) 10 µm.
As TCR ζ-chain signaling is critical not only for effector functions but also for T cell development (Shores and Love, 1997), we first observed whether IGSF4-deficient mice have developmental defects in terms of thymocyte selection. Intrathymic T cell maturation involves sequential differentiation stages that can be distinguished on the basis of CD4 and CD8 coreceptor expression (Shortman, 1992). Interestingly, the number and percentage of CD4+ and CD8+ single-positive cells were much lower in the thymus of IGSF4GT/GT mice than IGSF4+/+ mice (Fig. 7 A). However, the double-positive (DP) cells were higher in the thymus of IGSF4GT/GT mice than IGSF4+/+ mice (Fig. 7 A). Because we found no significant difference in the double-negative population, these data collectively suggested that the deletion of IGSF4 results in a substantial block in thymocyte development at the DP stage.
Figure 7.
IGSF4GT/GT mice show substantial blockade of intrathymic T cell development. (A) Thymocytes were stained with FITC-conjugated anti-CD4 and Cy5.5-conjugated anti-CD8 mAbs, and then the cells were analyzed by flow cytometry. On dot plots, the percentage of cells in each quadrant is indicated. Mean cell numbers of thymocyte subsets are shown in the bar graph (right). The results are the mean ± SD of triplicate experiments. *, P < 0.05, as compared with cells from IGSF4+/+ mice. (B) Flow cytometric analysis of TCR-β, CD5, and CD69 in IGSF4+/+ and IGSF4GT/GT thymocytes. Numbers above bracketed lines indicate the percentage of TCR-βhi, CD69hi, and CD5hi cells. (C) Surface expression of CD3ε, TCR ζ-chain, and TCR-β on thymocytes or lymphocytes from IGSF4+/+ and IGSF4GT/GT. (A–C) Data are representative of at least five to six independent experiments. (D) Thymocytes from IGSF4+/+ and IGSF4GT/GT mice were stimulated with anti-CD3/28 for the indicated time. The phosphorylated (p) and total (t) forms of Lck, Zap70, ERK, and p38 kinase were determined by Western blot analysis. Data are representative of two independent experiments. Molecular mass (M) is indicated in kilodaltons. (E) Thymocytes were stimulated with anti-CD3/28 for the indicated time, and the cytokine productions (IL-2) were measured by ELISA. The results are the mean ± SD of triplicate experiments. *, P < 0.05, as compared with cells from IGSF4+/+ mice.
TCR β-chain gene rearrangement and surface expression are critical for thymocyte development (Mombaerts et al., 1992). Flow cytometry showed that IGSF4GT/GT thymocytes included fewer TCR-βhi cells than did WT thymocytes (Fig. 7 B). In contrast, the proportion of TCR-βlo cells in IGSF4GT/GT thymuses was normal (Fig. 7 B). In addition to influencing TCR β-chain expression, TCR engagement initiates intracellular signals that induce the expression of thymocyte positive selection and maturation markers, such as CD69 and CD5 (Fowlkes et al., 1985). The surface expression of CD5 and CD69 was much lower on IGSF4GT/GT thymocytes than WT thymocytes (Fig. 7 B), suggesting that TCR-mediated positive selection might be defective in IGSF4GT/GT thymocytes.
In contrast, the surface expression of other TCR components, such as CD3ε and TCR ζ-chain, was similar in WT and IGSF4GT/GT thymocytes and splenic T cells (Fig. 7 C). To further determine whether thymocytes present in IGSF4GT/GT mice are competent for TCR signaling, thymocytes were stimulated with anti-mCD3/28 and assessed for phosphorylation of the TCR ζ-chain and its down-stream signaling pathways. The signaling pathways were dramatically attenuated in IGSF4GT/GT thymocytes compared with those of IGSF4+/+ mice (Fig. 7 D). Consequently, IL-2 production was also reduced in IGSF4GT/GT thymocytes (Fig. 7 E).
In accordance with the perturbed thymocyte development, the number of CD4+ and CD8+ cells in the spleens and lymph nodes of IGSF4GT/GT mice slightly decreased compared with those of IGSF4+/+ mice (Fig. 8 B). Because surface expression of TCR-β was similar in WT and IGSF4GT/GT spleen T cells (mean fluorescent intensity of TCR-β, WT vs. IGSF4GT/GT: 12.95 ± 0.77 vs. 11.05 ± 0.21; Fig. 7 C), we further determined the TCR signaling in splenic T cells of IGSF4GT/GT mice and compared it with WT. Splenic CD3+ T cells from IGSF4GT/GT mice had dramatically reduced phosphorylation of the ζ-chain, downstream kinases Lck and Zap70, and the foregoing signaling pathways of ERK and p38 kinase upon TCR stimulation with anti-mCD3/28 (Fig. 8 C). We also examined the membrane proximal signal during T cell contact with APCs. To this end, allogeneic DC (IGSF4+/+) mice were isolated, pulsed with Staphylococcus enterotoxin B (SEB), and incubated with splenic CD3+ T cells isolated from IGSF4+/+ or IGSF4GT/GT mice. As shown in Fig. 8 D, p-tyrosine staining at the T cell–APC contact regions was significantly reduced in T cells from IGSF4GT/GT mice compared with those from IGSF4+/+ mice. These results suggest that, in the absence of IGSF4, TCR cross-linking does not fully stimulate the most immediate as well as downstream distal signaling pathways.
Because the ectodomain is also important for T cell–APC interaction (Fig. 3), we performed a conjugation assay of mouse T cells with SEB-loaded DCs or B cells. Multiple WT T cells formed immune synapses with DCs and B cells (Fig. 8 E). In contrast, conjugate formation was hardly found in IGSF4-deficient T cells (Fig. 8 E, left).
Finally, we assessed the effector T cell functions by measuring cytokine expressions in CD4+ and CD8+ T cells. As the proliferation of purified T cells from IGSF4GT/GT mice was significantly impaired under the condition with anti-mCD3/28 (Fig. 8 F), presumably by defective TCR signaling, we directly stimulated purified CD4+ and CD8+ T cells with anti-mCD3/28 or PMA/A23187. ELISA demonstrated a severe decrease in IL-2, IFN-γ, and IL-4 levels in CD4+ and CD8+ T cells of IGSF4GT/GT mice stimulated with anti-mCD3/28 (Fig. 8 G).
DISCUSSION
In this study, we found that IGSF4 is important for T cell functioning both in vitro and in vivo. It is expressed in all T cell subsets including CD4+ and CD8+ cells as well as in other types of cells including B cells and DCs. These facts led us to focus on the function of IGSF4 in T cells rather than in APCs. We found that IGSF4 enhances T cell activation through dual independent mechanisms: control of TCR signaling and control of T cell–APC interaction.
The extracellular region of IGSF4 mediates intercellular adhesion through homophilic or heterophilic trans-interaction in neighboring cells. Because IGSF4 is expressed on both T cells and APCs and has a low affinity for binding between identical ectodomains (Shingai et al., 2003), we suggest that IGSF4 serves as an initial scanning receptor facilitating the exploration of the APC surface by T cells. Montoya et al. (2002) suggested a role of ICAM-3 in the early adhesive interactions between T cells and APCs; the clustering of endogenous ICAM-3 at cell–cell contacts occurred in both antigen-independent and -dependent T cell–APC interactions in their study. Similarly, IGSF4 clustering occurred rapidly after the initial cell–cell contact in our study, suggesting its role in early adhesive events. The view is supported by the finding that IGSF4-overexpressing cells show increased antigen-nonspecific conjugate formation (Fig. 3 B).
The CT domain of IGSF4 includes a couple of important motifs that interact with other proteins. One of these is a protein 4.1–binding motif in the juxtamembrane portion, which directly interacts with DAL-1/4.1b, a known spectrin-actin–binding protein (Murakami, 2005). The other is a PDZ-binding motif, through which a group of proteins belonging to membrane-associated guanylate kinase homologues (MAGuK) can interact. Given that these interacting molecules are important for cytoskeleton rearrangement, the CT domain may transmit the signal of cell adhesion toward cytoskeleton organization by modulating actin networks or cellular polarity. However, the CT domain had little function at least in synapse accumulation and T cell activation, although it appears to be multifunctional in other cell types, especially epithelial cells (Shingai et al., 2003). There are two possible explanations for the lack of CT domain function. First, IGSF4 could interact with the TCR ζ-chain through only the TM domain, which regulates its phosphorylation; in this case, the CT domain is not required for this signaling cascade. Second, although the CT domain can bind to DAL-1, CASK, MPP6 (Pals2), and MPP3 (Biederer et al., 2002; Fukuhara et al., 2003; Shingai et al., 2003), no specific function of these molecules has been reported in T cells since they were discovered; these molecules are mainly involved in suppressing tumorigenesis (Fukuhara et al., 2003) and maintaining epithelial morphology (Shingai et al., 2003).
Surprisingly, we observed that IGSF4 interacts with the TCR ζ-chain and this interaction is mediated by the TM domain. Indeed, replacement of the IGSF4 TM domain by the CD43 TM domain resulted in loss of ζ-chain association and the subsequent phosphorylation of the ζ-chain and downstream signaling pathways. However, it is unclear how the IGSF4 TM domain associates with the ζ-chain and controls its phosphorylation. As we observed no association of IGSF4 with cytosolic tyrosine kinases such as Lck and Zap70 in unstimulated conditions, the interaction with the ζ-chain may be extremely dependent on a TM–TM interaction. It is well characterized that the ζ-chain contains the homodimeric motif (LxxxxGVxxT) in the TM domain that participates in the TCR/CD3 complex assembly by interacting with CD3γδ TM domains (Bolliger and Johansson, 1999). This motif may also mediate the interaction of the ζ-chain with the specific region of IGSF4 TM domain. Interestingly, IGSF4 contains a GxxVx sequence, which is similar to the dimerization motifs in the TM domain of tyrosine kinase growth factor receptors such as Ltk (mouse leukocyte tyrosine kinase; GxxVx) and EGFR (epidermal growth factor receptor; SxxVx; Sternberg and Gullick, 1990). Therefore, it will be interesting to investigate whether the GxxVx motif mediates the heterodimeric interaction with the ζ-chain and thus enhances TCR signaling. The interaction may be important in TCR microcluster formation (Yokosuka and Saito, 2010). The signaling complexes including Lck, Zap70, and LAT are known to be recruited rapidly to the microclusters (Yokosuka and Saito, 2010). Interestingly, in a related study, we found that Lck and Zap70 were highly concentrated at the region that IGSF4 was polarized in IGSF4-overexpressing cells, suggesting that IGSF4 is involved in clustering of the TCR complex by interacting with the ζ-chain and, consequently, that the size and number of microclusters increase.
Our genetic experiments in mice revealed that IGSF4 is involved in thymocyte positive selection and maturation. The developmental defect was also associated with less expression of CD5, CD69, and TCR-β in IGSF4GT/GT thymocytes, which is regulated by TCR signaling (Azzam et al., 1998). Impaired TCR signaling in IGSF4GT/GT T cells was further demonstrated by less proliferation and less activation of kinase signaling cascades in response to TCR engagement. In addition, a dramatic reduction of effector T cell functions in terms of cytokine production strongly suggested that IGSF4 serves as a positive regulator for thymocyte development and TCR signaling. These results are particularly interesting because functions of IGSF4 are strikingly distinguishable from those of CRTAM, an adhesion molecule transiently expressed on activated CD4+ T and CD8+ T cells (Abbas et al., 2005). CRTAM−/− mice showed no signs of thymic developmental defects, and the positive selection was normal (Yeh et al., 2008). In addition, CRTAM mediated only the maintenance of late-phase T cell polarity corresponding to the selective production of IFN-γ and IL-22. In contrast to CRTAM−/− mice, however, the defect of early T cell development in IGSF4GT/GT thymocytes strongly suggests that the physiological action of IGSF4 in T cells does not overlap with that of CRTAM. Although it has been suggested that IGSF4 serves only as a ligand for CRTAM in APCs, our present results unambiguously demonstrate that IGSF4 plays a unique role in T cell immunity.
The thymus provides a specialized microenvironment for T cell development and many components are involved in thymocyte development, including ECM integrin and laminin in thymic epithelial cells (Magner et al., 2000; Schmeissner et al., 2001). Thus, it will be of interest to address whether IGSF4 has another binding partner rather than a homophilic interaction with IGSF4 on thymic epithelial cells or DCs. Recently, Garay et al. (2010) demonstrated that CRTAM is also detected in epithelial cells along the lateral membrane and is important for early cell–cell contacts and cell–substrate interactions. Therefore, reverse configuration, i.e., IGSF4-CRTAM rather than CRTAM-IGSF4, may correspond to early T cell development and positive selection in the thymus.
In conclusion, IGSF4 is a novel ζ-chain–associating protein and a key regulator of T cell responses from early to late phases. Two independent mechanisms of its function coexist. As an intercellular adhesion molecule, IGSF4 has an adhesion-dependent co-stimulatory function through homophilic or heterophilic interactions with ligands on APCs. This mechanism may be important for the initiation of T cell contact with epithelial cells in the thymus or DCs in the lymphoid tissues as well as for the formation and maintenance of the immunological synapse. As a binding partner of the ζ-chain, it enhances the transmission of outside signals to the inside of cells by modulating phosphorylation of the ζ-chain, downstream proximal kinases such as Lck and Zap70, and distal MAP kinases such as ERK and p38 kinase. In this study, we evidenced the role of IGSF4 in T cell development and activation. IGSF4 may also have roles in a variety of effector functions including Th1/Th2/T-reg responses and cytotoxic T cell or NKT cell responses. In addition, it may be involved in immune function–related diseases such as cancer, infectious diseases, shock, and autoimmune diseases as well as immediate and delayed hypersensitivity reactions through its action in activating T cell immunity.
MATERIALS AND METHODS
Mice, reagents, and antibodies.
C57BL/6 WT, TCR transgenic DO11.10, and C57BL/6 IGSF4+/GT heterozygous mice were obtained from Damul Science, the Jackson Laboratory, and Kumamoto University, respectively. The mice were housed in specific pathogen–free conditions, and all experiments were approved by the Animal Care and Use Committee of the School of Life Sciences, Gwangju Institute of Science and Technology. OmicsLink shRNA expression clone against IGSF4 was obtained from GeneCopoeia, and siRNA targeting IGSF4 and a scrambled siRNA were obtained from Thermo Fisher Scientific. SEE and SEB were obtained from Toxin Technology. OVA peptide fragments (323–339) were purchased from GeneScript Corp. Colchicine, PMA, A23187, and PLL were purchased from Sigma-Aldrich. Anti-CD3/28 Dynabeads and fluorescent dyes for cell labeling, green CMFDA (5-chloromethylfluorescein diacetate) and orange CMRA, were obtained from Invitrogen. [3H]thymidine was obtained from PerkinElmer. Welprep Total RNA isolation reagent, reverse transcript PCR premix, conventional PCR premix, and ECL Western blotting detection reagents were purchased from iNtRON Biotechnology. ECL advanced detection kit (Lumigen TMA-6) was purchased from GE Healthcare. PrimeSTAR HS DNA polymerase was purchased from Takara Bio Inc. ImmunoPure Fab preparation kit was obtained from Thermo Fisher Scientific, and Cy-3 bisfunctional dye kit was obtained from GE Healthcare. Duo Set mouse IL-2, IL-4, and IFN-γ ELISA kits and a mouse T cell enrichment column were obtained from R&D Systems. Anti–human CD28 was purchased from R&D Systems, and anti–mouse CD28 was purchased from BD. OKT3 (human anti-CD3; CRL-8001) and 145-2C11 (mouse anti-CD3; CRL-1975) hybridoma cell lines were purchased from the American Type Culture Collection. TS1/18 (anti–human LFA-1; HB-203) and R6.5 (anti–human ICAM1) hybridoma cell lines were a gift from T.A. Springer (Harvard Medical School, Boston, MA). Rabbit polyclonal anti-GFP was developed in rabbit by using purified recombinant full-length GFP protein (AbFrontier). Anti–human IGSF4, anti–TCR ζ-chain (6B10.2), anti–CD3γ chain (C-20), anti–CD3δ chain (M-20), anti-CD3ε (M-20), anti–TCR-α (H-142), anti–TCR-β (H-197), FITC-conjugated anti-CD43 (6D269), mouse polyclonal anti-GFP, and goat polyclonal anti–β-actin were purchased from Santa Cruz Biotechnology, Inc. Rabbit polyclonal anti–p-Zap70, rabbit anti-Zap70 (99F2), rabbit polyclonal anti-Lck, rabbit polyclonal anti–p-Lck, rabbit anti-p44/42 MAPK (137F5), rabbit anti–p-p44/42 (197G2), rabbit polyclonal anti-p38, rabbit anti–p-p38 (3D7), horseradish peroxidase–conjugated anti–mouse IgG, and anti–rabbit IgG were purchased from Cell Signaling Technology. Phosphorylated tyrosine (4G10) was purchased from Millipore. Mouse anti-p–CD3-ζ (K25-407.69) and mouse anti-CD45 (2D1) were purchased from BD. FITC-conjugated anti–mouse CD4, PerCP Cy5.5–conjugated CD8α, FITC-conjugated anti–mouse TCR-β, FITC-conjugated anti–mouse CD5, PE-conjugated CD3ε, PE-conjugated TCR ζ, and PE-conjugated anti-CD69 were purchased from eBioscience. Secondary antibodies including FITC-conjugated anti–rabbit IgG, anti–goat IgG, and anti–hamster IgG were purchased from Sigma-Aldrich.
Cells.
Jurkat T cells (TIB-152; American Type Culture Collection), HuT78 (TIB-161; American Type Culture Collection), Molt4 (CRL-1582; American Type Culture Collection), J77 (TIB-67; American Type Culture Collection), HEK293T (CRL-1573; American Type Culture Collection), and Raji B cells (gift from F. Sánchez-Madrid, Universidad Autónoma de Madrid, Madrid, Spain) were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% (vol/vol) FBS (Invitrogen). After written informed consent, human primary PBLs were isolated from healthy donors by dextran sedimentation and centrifugation through a discontinuous Ficoll gradient (GE Healthcare). Human PBLs and mouse splenocytes were dispersed and purified to CD4+, CD8+, CD19+, and CD11C+ populations by MACS cell separation (Miltenyi Biotec). All mouse CD3+ T cells were purified from the mouse spleen and lymph nodes on a T cell enrichment column (BD). The purity of each population was confirmed to be >95% by flow cytometry.
Cell transfection and lentiviral infection.
Transient transfection to Jurkat T cells was mostly performed by Amaxa technology using Nucleofector kitV (Lonza). Transfection to 293T cells was performed by using Lipofectamine 2000 (Invitrogen). To establish stable cell lines, cDNAs in pHJ-1 lentiviral vector were cotransfected with lentiviral packaging vectors (pHDM-Hgpm2, 1 pRC/CMV-Rev1b, and pHDM.G) into 293T cells. The supernatants were collected and spin-infected into Jurkat T cells by centrifugation at 800 g for 30 min in the presence of 8 µg/ml polybrene. For IGSF4 knockdown, 70 µM siRNAs or 2 µg/100 µl shRNAs were introduced into the target cells and cultured for 48 h before use.
cDNA constructs.
DNA fragments encoding human WT IGSF4 or deletion or swapping mutants were generated by PCR from a full-length IGSF4 ORF clone (imaGenes GmbH) and inserted into pEGFP-N1 (Takara Bio Inc.) or pHJ-1 lentiviral vector. The expression vectors for various mutants of IGSF4 were as follows: IGSF4/ΔCT (deletion of the entire CT domain, Δ398–442), IGSF4/ΔPDZ (deletion of the PDZ binding motif, Δ432–442), IGSF4/ΔEXT (deletion of the entire extracellular domain, Δ1–373), IC1_IGSF4 (the extracellular domain was substituted with D3–5 of ICAM-1), IGSF4/CD43TM (the TM domain was substituted with the TM domain of CD43), and IGSF4/ΔEXT/CD43TM (IGSF4/ΔEXT + IGSF4/CD43TM). All the IGSF4 deletion and chimeric mutants were verified by sequencing analysis. TCR ζ-chain cDNA was amplified from the ζ-chain mRNA by RT-PCR using primers 5′-ATGAAGTGGAAGGCGCTTTTCACCGCGGCC-3′ and 5′-TTAGCGAGGGGGCAGGGCCTGCATGTGAAG-3′, and then the PCR product was inserted into pERFP_N1 to generate TCR ζ_RFP.
RT-PCR and real-time quantitative RT-PCR.
Total RNA was isolated from cells or homogenized tissues of C57BL/6 mice with TRIZOL reagent and reverse transcribed by using RT-PreMix (iNtRON Biotechnology). PCR was performed with the following primers (the respective forward and reverse pairs are indicated): human IL-2, 5′-CACGTCTTGCACTTGTCAC-3′ and 5′-CCTTCTTGGGCATGTAAAACT-3′; mouse IL-2, 5′-TGAGCAGGATGGAGAATTACAGG-3′and 5′-GTCCAAGTTCATCTTCTAGGCAC-3′; human GAPDH, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′; mouse GAPDH, 5′-GCACAGTCAAGGCCGAGAAT-3′ and 5′-GCCTTCTCCATGGTGGTGAA-3′; human IGSF4, 5′-AAGTAGTCCTGAAGGACAGAAACT-3′ and 5′-ATAAATCAGCATAAGTTTTCCACA-3′; mouse IGSF4, 5′-CAGTATAAACCGCAAGTGCA-3′ and 5′-GCGGTAAGTACCGTTATCTG-3′; human Necl1, 5′-AGCCCATCATCACTGGTTATAAAT-3′ and 5′-TTTACCATTGGGATCTTCCTGTAT-3′; human nectin3, 5′-GTTCAAGGAGAATATCAGGGAAGA-3′ and 5′-TAAGAACTGCCCTTTTCAG-3′; and human CRTAM, 5′-GTAATACTACCAGCACTCTCATAATCCAC-3′ and 5′-GATGTACTAGAATCTTCCGTTACTGAGAC-3′. The amplification profile was composed of denaturation at 94°C for 30 s, annealing at 60°C for 20 s, and extension at 72°C for 40 s. The 30 cycles were preceded by denaturation at 72°C for 7 min.
Total RNA was isolated, and cDNA was synthesized. PCR amplification was performed in DNA Engine Opticon1 (MJ Research) for continuous fluorescence detection in a total volume of 10 µl containing 1 µl cDNA/control and gene-specific primers by using SYBR Premix Ex Taq (Takara Bio Inc.). The mRNA levels of the target genes, relative to GAPDH, were normalized by using the following formula: relative mRNA expression = 2−(ΔCt of target gene − ΔCt of GAPDH), where Ct is the threshold cycle value. In each sample, the expression of the gene being analyzed was normalized to that of GAPDH and described as the relative mRNA levels to GAPDH or percentage of maximum.
T cell stimulation.
Jurkat T cells (including IGSF4-overexpressing or knockdown cells) and mouse CD3+ T cells (including CD4+ or CD8+ cells) were stimulated with either plate-bound anti-CD3 (10 µg/ml OKT3 for human; 10 µg/ml 145-2C11 for mouse)/2 µg/ml CD28 or 200 nM PMA/1 µM A23187. For superantigen stimulation, Jurkat T cells were incubated with 1 µg/ml SEE–pulsed Raji B cells. Mouse T cells were incubated with 1 µg/ml SEB–pulsed mouse DCs (CD11C+) or B cells (CD19+). Splenic CD3+ T cells from TCR transgenic DO11.10 mice were stimulated with 50 ng/ml OVA–pulsed A20 B cells.
Immunofluorescence and confocal imaging analyses.
For live-time colocalization analysis, Jurkat T cells expressing GFP or WT IGSF4_GFP were stained with anti–CD3-Fab-cy3, anti–CD45-Fab-cy3, or LFA-1–Fab-cy3 for 1 h at 4°C and then incubated for 15–30 min with SEE-pulsed Raji B cells stained with Cell Tracker orange CMRA (Invitrogen) or anti–ICAM-1–Fab-cy5 in a live chamber device. CD43 was stained with anti–CD43-Fab-FITC. 1 µg/ml colchicine (or vehicle [DMSO]) was treated before T cell–B cell conjugation. For translocation analysis, Jurkat T cells expressing WT IGSF4_GFP or other mutants were incubated for 30 min at 37°C on PLL- or anti-CD3–coated coverslips or with anti-CD3/28–coated Dynabeads (Invitrogen). IGSF4 accumulation at the T cell–APC contact site or anti-CD3–coated surface was calculated as the ratio of fluorescence intensity at the contact region (Fcon = a) to the fluorescence intensity at the opposite site (Foppo = b). In some cases, the percentage of T–T or T–B conjugates with surface IGSF4 relocation at the contact zone relative to the total number of conjugates in the absence or presence of SEE was also analyzed from a total of 150 conjugates of each category (Figs. 2 A and 3 C).
In the mouse experiment, purified mouse CD3+ T cells were co-cultured with SEB-pulsed allogeneic DCs or B cells on PLL-coated glass, fixed, and stained with anti–p-tyrosine. The data were obtained, processed, and analyzed by FLUOVIEW software (Olympus). The z-section cutting area through the apical surface was chosen for the colocalization analysis. Fluorescence was represented by an intensity profile with blue indicating the lowest intensity and red indicating the highest intensity.
Quantitation of cell aggregation.
J-GFP or J-IGSF4_GFP cells were cultured at the concentration of 2 × 105 cells/well in the presence or absence with anti-CD3/28 for 3 h. Aggregate formation was quantitated by phase-contrast microscopy using a calibrated ocular grid as described previously (Dang and Rock, 1991). The percentage of cells forming aggregates was determined by counting free cells within the grid in six randomly selected grids within one well and then applying the following equation: percent aggregation = (1 − number of free cells/number of total cells) × 100.
Conjugation assay.
Raji B cells, mouse DCs (CD11C+), or mouse B cells (CD19+) were stained with Cell Tracker orange CMRA according to the manufacturer’s directions, incubated in the presence or absence of superantigen (1 µg/ml SEE for human and 1 µg/ml SEB for mouse) for 30 min, washed, and resuspended at a density of 106 cells/ml in RPMI. Jurkat or mouse CD3+ T cells were stained with Cell Tracker green CMFDA (Invitrogen) and resuspended at a density of 106 cells/ml in RPMI. For T cell–APC conjugation, equal volumes of T cells and APCs were mixed together and incubated at 37°C for 30 min. The relative proportion of red, green, and red-green populations was determined by an EPICS XL flow cytometer (Beckman Coulter). The number of gated events counted per sample was at least 10,000.
Immunoprecipitation.
For immunoprecipitation, Jurkat T cells (including WT IGSF4– or IGSF4 mutant–overexpressing cells) were incubated at 2 × 107 cells/ml in the presence or absence of anti-CD3/28 for 30 min. The cells were washed quickly once in cold PBS and lysed in 1% Triton X-100 lysis buffer containing 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, one tablet of Complete protease inhibitors (Roche), and phosphatase inhibitors (cocktails I and II; Sigma-Aldrich). The lysates of equivalent protein content were precleared on Sepharose 4B (GE Healthcare) for 1 h at 4°C. GFP-fused IGSF4 and mutant proteins or TCR ζ-chain was immunoprecipitated with anti-GFP–conjugated Sepharose 4B or anti–TCR ζ-chain–conjugated protein A/G agarose. Immunoprecipitates were washed twice with the corresponding 1% Triton X-100 lysis buffer and twice with lysis buffer without detergent. Proteins were resolved by 10–12% SDS-PAGE, and then Western blot analysis was performed as described in the next section. To analyze the direct binding of TCR ζ-chain with IGSF4, HEK293T cells were cotransfected with TCR ζ_RFP and IGSF4_GFP. The cells were lysed as above, and immunoprecipitation was performed using antibodies against GFP (for IGSF4) and TCR ζ-chain. The amount of TCR ζ-chain or IGSF4 present in the immunoprecipitates was analyzed by Western blot analysis as described in the next section. For detection of the TCR complex (α and β, CD3γ, CD3δ, CD3ε, and TCR ζ) and Zap70 and Lck in IGSF4 immunoprecipitates, cells were lysed in 0.3% CHAPS lysis buffer instead of 1% Triton X-100.
Western blotting.
Cells or tissue samples were lysed in Triton X-100 lysis buffer containing 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, one tablet of Complete protease inhibitors, and phosphatase inhibitors (cocktails I and II). The lysates were centrifuged at 14,000 rpm for 25 min at 4°C, and the supernatant was eluted with SDS sample buffer (100 mM Tri-HCl, pH 6.8, 4% SDS, and 20% glycerol with bromophenol blue) and heated for 5 min. The proteins were separated through 10% SDS-PAGE gels and transferred onto a nitrocellulose membrane (GE Healthcare) by means of Trans-Blot SD semidry transfer cell (Bio-Rad Laboratories). The membrane was blocked in 5% skim milk for 1 h at room temperature, rinsed, and incubated with the intended antibodies in TBS containing 0.1% Tween 20 (TBS-T) and 3% skim milk for 2 h at room temperature. Excess primary antibody was then removed by washing the membrane four times in TBS-T. The membrane was then incubated with 0.1 µg/ml peroxidase-labeled secondary antibody (anti–rabbit or anti–mouse) for 2 h at room temperature. After three washes in TBS-T, bands were visualized by using ECL Western blotting detection reagents (iNtRON Biotechnology) and exposed onto x-ray film. In some cases, the detection sensitivity was improved by using the ECL advanced detection kit (Lumigen TMA-6; GE Healthcare).
ELISA.
Splenic CD3+ T cells, CD4+ or CD8+ T cells, or thymocytes (5 × 104 cells/sample) were stimulated as described in the section T cell stimulation. After 24–48 h, the amounts of IL-2, IL-4, and IFN-γ in supernatants from three replicas for each condition were determined by ELISA with Duo Set Mouse ELISA kits for IL-2, IL-4, and IFN-γ (R&D Systems).
Flow cytometry and determination of cell number.
Thymus, spleen, and lymph nodes were excised from mice, and single-cell suspensions were obtained by mincing the organs through a nylon mesh. Isolated cells were stained for flow cytometry with antibodies against CD4, CD8, CD69, CD5, TCR-β, TCR ζ-chain, and CD3ε for 30 min at 4°C. Total cellularity was determined by counting the live cells. Absolute cell numbers were calculated on the basis of the percentage of each population and represented as the mean ± SD. The mice in these experiments were 4 wk of age.
Mice strains and genotyping.
A C57BL/6 IGSF4+/GT heterozygous mouse was generated by a group at Kumamoto University. In brief, exchangeable gene trap pU-21W vector was used for random gene trap mutagenesis. The mutant construct contains a splice acceptor sequence linked to the β-geo reporter gene, and its integration site is concentrated in the 5′ end of the trapped gene. The pU-21W vector (20–40 µg) was transfected by electroporation (800 V, 3 µF with Gene Pulser; Bio-Rad Laboratories) into feeder-free embryonic stem (ES) cell line KTPU8 (F1 of B6 and CBA). Thereafter, G418-resistant clones were selected and expanded. Genomic DNA was prepared from the clones and examined by PCR and Southern blotting for single-copy integration and for the existence of lox71–lox2272 sites, which are indispensable for site-specific recombination. To identify the trapped gene, 5′-RACE was performed. We found that pU-21W was inserted at 212,677 bp downstream of IGSF4 exon 1. Clones that had the trapped gene insertion in IGSF4 were used to generate chimeric mice. ES cells were aggregated with morulae from ICR (imprinting control region) mice; 125 morulae per line were used and transferred into five foster mothers. The chimeric mice were mated with C57BL/6 females. Genomic DNAs of the F1 progenies and original ES cells were subjected to Southern blotting to confirm identical integration of the vector in the mouse line and the original ES clone. The strain name is depicted as B6;CB-Cadm1Gt(pU-21W)34Imeg, and the address of the Database for the Exchange of Gene Trap Clones is http://egtc.jp/action/access/clone_detail?id=21-W34.
T cell proliferation assay.
Mouse splenocytes were enriched by using a T cell enrichment column (BD), and 5 × 104 cells/sample were incubated for 72 h on 10 µg/ml anti-CD3–coated 96-well plates or 5 µg/ml PHA in RPMI 1640 medium containing 10% FBS. [3H]thymidine was added at 1 µCi/well 14 h before measurement. The cells were harvested, and [3H] radiation was measured by liquid scintillation.
Body weight measurement.
4-wk-old mice were sacrificed, and their heart, kidney, spleen, thymus, and lymph node were removed and displayed. Heterozygous female mice crossed with a heterozygous male were sacrificed at day 18.5 after checking the vaginal plug. Embryonic day 18.5 (E18.5) embryos were isolated and photographed. All embryos were used for genotyping. All mice were weighed at 1-wk intervals from 4 to 8 wk of age.
Statistical analysis.
Data represent the mean ± SD of three or more independent experiments conducted on separate days. Unpaired Student’s t test and one-way analysis of variance were used for statistical analysis (P < 0.05).
Online supplemental material.
Video 1 reveals the c-SMAC localization of IGSF4_GFP in the T cell–B cell conjugates. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110853/DC1.
Acknowledgments
We thank Dr. F. Sánchez-Madrid for Raji B cells and Dr. T.A. Springer for TS1/18 (anti–human LFA-1; HB-203) and R6.5 (anti–human ICAM1) hybridoma cell lines.
This work was supported by the Cell Dynamics Research Center (2011-0001159), Korea Health Care Technology R&D Project of the Ministry for Health, Welfare and Family Affairs (A100159 and A090252), Bio and Medical Technology Development Program of the National Research Foundation (20110030157), and the BioImaging Research Center at Gwangju Institute of Science and Technology.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- c-SMAC
- central SMAC
- CT
- cytoplasmic
- DP
- double positive
- ES
- embryonic stem
- mRNA
- messenger RNA
- PLL
- poly-l-lysine
- SEB
- Staphylococcus enterotoxin B
- SEE
- Staphylococcus enterotoxin E
- shRNA
- short hairpin RNA
- siRNA
- small interfering RNA
- SMAC
- supramolecular activation cluster
- TM
- transmembrane
References
- Abbas A.R., Baldwin D., Ma Y., Ouyang W., Gurney A., Martin F., Fong S., van Lookeren Campagne M., Godowski P., Williams P.M., et al. 2005. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 6:319–331 10.1038/sj.gene.6364173 [DOI] [PubMed] [Google Scholar]
- Arase N., Takeuchi A., Unno M., Hirano S., Yokosuka T., Arase H., Saito T. 2005. Heterotypic interaction of CRTAM with Necl2 induces cell adhesion on activated NK cells and CD8+ T cells. Int. Immunol. 17:1227–1237 10.1093/intimm/dxh299 [DOI] [PubMed] [Google Scholar]
- Azzam H.S., Grinberg A., Lui K., Shen H., Shores E.W., Love P.E. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188:2301–2311 10.1084/jem.188.12.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyers A.D., Spruyt L.L., Williams A.F. 1992. Molecular associations between the T-lymphocyte antigen receptor complex and the surface antigens CD2, CD4, or CD8 and CD5. Proc. Natl. Acad. Sci. USA. 89:2945–2949 10.1073/pnas.89.7.2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T., Sara Y., Mozhayeva M., Atasoy D., Liu X., Kavalali E.T., Südhof T.C. 2002. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 297:1525–1531 10.1126/science.1072356 [DOI] [PubMed] [Google Scholar]
- Bolliger L., Johansson B. 1999. Identification and functional characterization of the zeta-chain dimerization motif for TCR surface expression. J. Immunol. 163:3867–3876 [PubMed] [Google Scholar]
- Brdicka T., Imrich M., Angelisová P., Brdicková N., Horváth O., Spicka J., Hilgert I., Lusková P., Dráber P., Novák P., et al. 2002. Non–T cell activation linker (NTAL): A transmembrane adaptor protein involved in immunoreceptor signaling. J. Exp. Med. 196:1617–1626 10.1084/jem.20021405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruyns E., Marie-Cardine A., Kirchgessner H., Sagolla K., Shevchenko A., Mann M., Autschbach F., Bensussan A., Meuer S., Schraven B. 1998. T cell receptor (TCR) interacting molecule (TRIM), a novel disulfide-linked dimer associated with the TCR–CD3-ζ complex, recruits intracellular signaling proteins to the plasma membrane. J. Exp. Med. 188:561–575 10.1084/jem.188.3.561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L.H., Rock K.L. 1991. Stimulation of B lymphocytes through surface Ig receptors induces LFA-1 and ICAM-1-dependent adhesion. J. Immunol. 146:3273–3279 [PubMed] [Google Scholar]
- Davidson D., Bakinowski M., Thomas M.L., Horejsi V., Veillette A. 2003. Phosphorylation-dependent regulation of T-cell activation by PAG/Cbp, a lipid raft-associated transmembrane adaptor. Mol. Cell. Biol. 23:2017–2028 10.1128/MCB.23.6.2017-2028.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delon J., Kaibuchi K., Germain R.N. 2001. Exclusion of CD43 from the immunological synapse is mediated by phosphorylation-regulated relocation of the cytoskeletal adaptor moesin. Immunity. 15:691–701 10.1016/S1074-7613(01)00231-X [DOI] [PubMed] [Google Scholar]
- Fowlkes B.J., Edison L., Mathieson B.J., Chused T.M. 1985. Early T lymphocytes. Differentiation in vivo of adult intrathymic precursor cells. J. Exp. Med. 162:802–822 10.1084/jem.162.3.802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara H., Masuda M., Yageta M., Fukami T., Kuramochi M., Maruyama T., Kitamura T., Murakami Y. 2003. Association of a lung tumor suppressor TSLC1 with MPP3, a human homologue of Drosophila tumor suppressor Dlg. Oncogene. 22:6160–6165 (published erratum appears in Oncogene. 2004. 23:629) 10.1038/sj.onc.1206744 [DOI] [PubMed] [Google Scholar]
- Garay E., Patiño-López G., Islas S., Alarcón L., Canche-Pool E., Valle-Rios R., Medina-Contreras O., Granados G., Chávez-Munguía B., Juaristi E., et al. 2010. CRTAM: A molecule involved in epithelial cell adhesion. J. Cell. Biochem. 111:111–122 10.1002/jcb.22673 [DOI] [PubMed] [Google Scholar]
- Grakoui A., Bromley S.K., Sumen C., Davis M.M., Shaw A.S., Allen P.M., Dustin M.L. 1999. The immunological synapse: a molecular machine controlling T cell activation. Science. 285:221–227 10.1126/science.285.5425.221 [DOI] [PubMed] [Google Scholar]
- Horejsí V., Zhang W., Schraven B. 2004. Transmembrane adaptor proteins: organizers of immunoreceptor signalling. Nat. Rev. Immunol. 4:603–616 10.1038/nri1414 [DOI] [PubMed] [Google Scholar]
- Hur E.M., Son M., Lee O.H., Choi Y.B., Park C., Lee H., Yun Y. 2003. LIME, a novel transmembrane adaptor protein, associates with p56lck and mediates T cell activation. J. Exp. Med. 198:1463–1473 10.1084/jem.20030232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A., Jippo T., Wakayama T., Morii E., Koma Y., Onda H., Nojima H., Iseki S., Kitamura Y. 2003. SgIGSF: a new mast-cell adhesion molecule used for attachment to fibroblasts and transcriptionally regulated by MITF. Blood. 101:2601–2608 10.1182/blood-2002-07-2265 [DOI] [PubMed] [Google Scholar]
- Ito A., Koma Y., Watabe K., Jippo T., Wakayama T., Iseki S., Kitamura Y. 2004. Contribution of the SgIGSF adhesion molecule to survival of cultured mast cells in vivo. Biochem. Biophys. Res. Commun. 319:200–206 10.1016/j.bbrc.2004.04.172 [DOI] [PubMed] [Google Scholar]
- Kirchgessner H., Dietrich J., Scherer J., Isomäki P., Korinek V., Hilgert I., Bruyns E., Leo A., Cope A.P., Schraven B. 2001. The transmembrane adaptor protein TRIM regulates T cell receptor (TCR) expression and TCR-mediated signaling via an association with the TCR ζ chain. J. Exp. Med. 193:1269–1284 10.1084/jem.193.11.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koma Y., Ito A., Watabe K., Hirata T., Mizuki M., Yokozaki H., Kitamura T., Kanakura Y., Kitamura Y. 2005. Distinct role for c-kit receptor tyrosine kinase and SgIGSF adhesion molecule in attachment of mast cells to fibroblasts. Lab. Invest. 85:426–435 10.1038/labinvest.3700231 [DOI] [PubMed] [Google Scholar]
- Le Deist F., de Saint Basile G., Rieux-Laucat F., Hivroz C., Fischer A. 2007. [Expression anomalies of the CD3-TCR complex expression and immunodeficiencies]. Med. Sci. (Paris). 23:161–166 10.1051/medsci/2007232161 [DOI] [PubMed] [Google Scholar]
- Li Y., Hu J., Höfer K., Wong A.M., Cooper J.D., Birnbaum S.G., Hammer R.E., Hofmann S.L. 2010. DHHC5 interacts with PDZ domain 3 of post-synaptic density-95 (PSD-95) protein and plays a role in learning and memory. J. Biol. Chem. 285:13022–13031 10.1074/jbc.M109.079426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Weiss A. 2001. T cell receptor signalling. J. Cell Sci. 114:243–244 [DOI] [PubMed] [Google Scholar]
- Magner W.J., Chang A.C., Owens J., Hong M.J., Brooks A., Coligan J.E. 2000. Aberrant development of thymocytes in mice lacking laminin-2. Dev. Immunol. 7:179–193 10.1155/2000/90943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksumova L., Le H.T., Muratkhodjaev F., Davidson D., Veillette A., Pallen C.J. 2005. Protein tyrosine phosphatase alpha regulates Fyn activity and Cbp/PAG phosphorylation in thymocyte lipid rafts. J. Immunol. 175:7947–7956 [DOI] [PubMed] [Google Scholar]
- Marie-Cardine A., Kirchgessner H., Bruyns E., Shevchenko A., Mann M., Autschbach F., Ratnofsky S., Meuer S., Schraven B. 1999. SHP2-interacting transmembrane adaptor protein (SIT), a novel disulfide-linked dimer regulating human T cell activation. J. Exp. Med. 189:1181–1194 10.1084/jem.189.8.1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli M.P., Lin H., Zhang W., Samelson L.E., Bierer B.E. 2000. Signaling via LAT (linker for T-cell activation) and Syk/ZAP70 is required for ERK activation and NFAT transcriptional activation following CD2 stimulation. Blood. 96:2181–2190 [PubMed] [Google Scholar]
- Mombaerts P., Clarke A.R., Rudnicki M.A., Iacomini J., Itohara S., Lafaille J.J., Wang L., Ichikawa Y., Jaenisch R., Hooper M.L., et al. 1992. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 360:225–231 10.1038/360225a0 [DOI] [PubMed] [Google Scholar]
- Monks C.R., Freiberg B.A., Kupfer H., Sciaky N., Kupfer A. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 395:82–86 10.1038/25764 [DOI] [PubMed] [Google Scholar]
- Montoya M.C., Sancho D., Bonello G., Collette Y., Langlet C., He H.T., Aparicio P., Alcover A., Olive D., Sánchez-Madrid F. 2002. Role of ICAM-3 in the initial interaction of T lymphocytes and APCs. Nat. Immunol. 3:159–168 10.1038/ni753 [DOI] [PubMed] [Google Scholar]
- Murakami Y. 2005. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci. 96:543–552 10.1111/j.1349-7006.2005.00089.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y., Itoh K., Yaoi T., Tando S., Fukui K., Fushiki S. 2005. Spatiotemporal patterns of expression of IGSF4 in developing mouse nervous system. Brain Res. Dev. Brain Res. 156:23–31 10.1016/j.devbrainres.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Posevitz V., Arndt B., Krieger T., Warnecke N., Schraven B., Simeoni L. 2008. Regulation of T cell homeostasis by the transmembrane adaptor protein SIT. J. Immunol. 180:1634–1642 [DOI] [PubMed] [Google Scholar]
- Sasaki H., Nishikata I., Shiraga T., Akamatsu E., Fukami T., Hidaka T., Kubuki Y., Okayama A., Hamada K., Okabe H., et al. 2005. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood. 105:1204–1213 10.1182/blood-2004-03-1222 [DOI] [PubMed] [Google Scholar]
- Schmeissner P.J., Xie H., Smilenov L.B., Shu F., Marcantonio E.E. 2001. Integrin functions play a key role in the differentiation of thymocytes in vivo. J. Immunol. 167:3715–3724 [DOI] [PubMed] [Google Scholar]
- Shingai T., Ikeda W., Kakunaga S., Morimoto K., Takekuni K., Itoh S., Satoh K., Takeuchi M., Imai T., Monden M., Takai Y. 2003. Implications of nectin-like molecule-2/IGSF4/RA175/SgIGSF/TSLC1/SynCAM1 in cell-cell adhesion and transmembrane protein localization in epithelial cells. J. Biol. Chem. 278:35421–35427 10.1074/jbc.M305387200 [DOI] [PubMed] [Google Scholar]
- Shores E.W., Love P.E. 1997. TCR zeta chain in T cell development and selection. Curr. Opin. Immunol. 9:380–389 10.1016/S0952-7915(97)80085-4 [DOI] [PubMed] [Google Scholar]
- Shortman K. 1992. Cellular aspects of early T-cell development. Curr. Opin. Immunol. 4:140–146 10.1016/0952-7915(92)90003-W [DOI] [PubMed] [Google Scholar]
- Sternberg M.J., Gullick W.J. 1990. A sequence motif in the transmembrane region of growth factor receptors with tyrosine kinase activity mediates dimerization. Protein Eng. 3:245–248 10.1093/protein/3.4.245 [DOI] [PubMed] [Google Scholar]
- van der Weyden L., Arends M.J., Chausiaux O.E., Ellis P.J., Lange U.C., Surani M.A., Affara N., Murakami Y., Adams D.J., Bradley A. 2006. Loss of TSLC1 causes male infertility due to a defect at the spermatid stage of spermatogenesis. Mol. Cell. Biol. 26:3595–3609 10.1128/MCB.26.9.3595-3609.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wange R.L. 2000. LAT, the linker for activation of T cells: a bridge between T cell-specific and general signaling pathways. Sci. STKE. 2000:re1 10.1126/stke.2000.63.re1 [DOI] [PubMed] [Google Scholar]
- Watabe K., Ito A., Koma Y.I., Kitamura Y. 2003. IGSF4: a new intercellular adhesion molecule that is called by three names, TSLC1, SgIGSF and SynCAM, by virtue of its diverse function. Histol. Histopathol. 18:1321–1329 [DOI] [PubMed] [Google Scholar]
- Wingren A.G., Parra E., Varga M., Kalland T., Sjögren H.O., Hedlund G., Dohlsten M. 1995. T cell activation pathways: B7, LFA-3, and ICAM-1 shape unique T cell profiles. Crit. Rev. Immunol. 15:235–253 [DOI] [PubMed] [Google Scholar]
- Yamada D., Yoshida M., Williams Y.N., Fukami T., Kikuchi S., Masuda M., Maruyama T., Ohta T., Nakae D., Maekawa A., et al. 2006. Disruption of spermatogenic cell adhesion and male infertility in mice lacking TSLC1/IGSF4, an immunoglobulin superfamily cell adhesion molecule. Mol. Cell. Biol. 26:3610–3624 10.1128/MCB.26.9.3610-3624.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh J.H., Sidhu S.S., Chan A.C. 2008. Regulation of a late phase of T cell polarity and effector functions by Crtam. Cell. 132:846–859 10.1016/j.cell.2008.01.013 [DOI] [PubMed] [Google Scholar]
- Yokosuka T., Saito T. 2010. The immunological synapse, TCR microclusters, and T cell activation. Curr. Top. Microbiol. Immunol. 340:81–107 10.1007/978-3-642-03858-7_5 [DOI] [PubMed] [Google Scholar]
- Zhang W., Samelson L.E. 2000. The role of membrane-associated adaptors in T cell receptor signalling. Semin. Immunol. 12:35–41 10.1006/smim.2000.0205 [DOI] [PubMed] [Google Scholar]