As shown by transcriptional analysis of blood samples from human volunteers, injection with synthetic dsRNA (an agonist of the TLR3 and MDA5 pattern recognition receptors) triggered up-regulation of genes involved in innate immune pathways, similar to those induced by vaccination with the efficacious yellow fever vaccine.
Abstract
Adjuvants are critical for the success of vaccines. Agonists of microbial pattern recognition receptors (PRRs) are promising new adjuvant candidates. A mechanism through which adjuvants enhance immune responses is to stimulate innate immunity. We studied the innate immune response in humans to synthetic double-stranded RNA (polyinosinic:polycytidylic acid [poly IC] stabilized with poly-l-lysine [poly ICLC]), an agonist for toll-like receptor (TLR) 3, and the cytosolic RNA helicase MDA-5. Transcriptional analysis of blood samples from eight volunteers, after subcutaneous administration of poly ICLC, showed up-regulation of genes involved in multiple innate immune pathways in all subjects, including interferon (IFN) and inflammasome signaling. Blocking type I IFN receptor ex vivo significantly dampened the response to poly IC. Comparative transcriptional analysis showed that several innate immune pathways were similarly induced in volunteers immunized with the highly efficacious yellow fever vaccine. Therefore, a chemically defined PRR agonist like poly ICLC can be a reliable and authentic microbial mimic for inducing innate immune responses in humans.
Vaccines prevent many infectious diseases. However, significant challenges remain to develop effective and safe vaccines against important diseases such as HIV, malaria, and tuberculosis (Plotkin, 2008; Germain, 2010). Most effective vaccines are live attenuated variants of the pathogens, but for many pathogens, live attenuated vaccines have not been successfully developed or are not considered safe in humans. Therefore, there is renewed interest in the identification of vaccine adjuvants that can potentiate the immunogenicity of subunit vaccines to prevent viral infections and mimic the intact pathogen.
An important mechanism for adjuvant action is to activate innate immunity, thereby leading to adaptive immunity (Pulendran and Ahmed, 2006; Reed et al., 2009; Coffman et al., 2010). Agonists for defined pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), induce innate immunity and are a potential new class of adjuvants (Beutler, 2004; Kawai and Akira, 2010). Synthetic double-stranded RNA, polyinosinic:polycytidylic acid (poly IC), and its more RNase-resistant analogue stabilized with poly-L-lysine (poly ICLC) and available for investigation in humans, are superior vaccine adjuvants in mice and in nonhuman primates (Longhi et al., 2009; Stahl-Hennig et al., 2009). The double-stranded RNA is a pathogen-associated molecular pattern and activates innate immunity (Alexopoulou et al., 2001). Both poly IC and poly ICLC are recognized by the cytosolic RNA helicase MDA-5 and by endosomal TLR3 (Meylan and Tschopp, 2006). A major mechanism underlying the strong adjuvant function of poly IC in mice is that it is superior at inducing systemic type I IFN (Longhi et al., 2009), which has many immune stimulatory roles for both T and B lymphocytes (Kolumam et al., 2005; Le Bon et al., 2006) and DCs (Le Bon et al., 2001; Longhi et al., 2009). Although poly IC and poly ICLC have been used for immunotherapy of cancer (Morse et al., 2011; Okada et al., 2011), antigen-specific responses are variable and it is not yet clear if humans exhibit a reliable innate response to poly ICLC.
Gene expression profiling has been successfully used to provide a systems-wide appreciation of the early, presumably innate molecular signatures to the yellow fever vaccine (YFV) YF17D in healthy volunteers. These results revealed an integrated innate response that includes the complement system, inflammasomes, and IFNs. This is followed by a broad polyfunctional and persistent adaptive immune response (Gaucher et al., 2008; Querec et al., 2009). Querec et al. (2006) reported that the YF17D vaccine activated many TLRs in vitro and surmised that multiple PRRs were required for the activity of powerful microbial vaccines.
Similarly, systems biology can be used to study the mechanism whereby specific PRR ligands act as vaccine adjuvants and exert differential control of innate immune responses. For example, gene expression profiling of mouse DCs in response to different PRR ligands in vitro showed two distinct transcriptional programs: a TLR2-like inflammatory response, which was induced by PAM3CSK4, and a TLR3/MDA5-like antiviral response, induced by poly IC, which was enriched for IFN-regulatory factors and for viral- and IFN-responsive genes (Amit et al., 2009). Analysis of global gene expression changes in human PBMCs showed that poly IC elicits transcriptional changes that are similar to changes after acute viral infection. At 3 h, poly IC induced changes in genes related to TLR3 signaling, NF-κB–dependent pathway, and IFN-stimulated pathway, whereas later on, at 24 h, gene expression changes were mostly cell-mediated immune responses involving activation of cell adhesion, cell mobility, and phagocytosis (Huang, et al., 2006). These ex vivo signatures have provided insights into the mechanism whereby poly IC and other PRR ligands perturb the immune system. Yet it is not clear if the in vivo innate response to synthetic microbial agonists in any species is reliable and is an authentic mimic to a microbial vaccine, which is an assumption to the use of these agonists as microbial adjuvants.
In this paper, we have gained a systems-wide appreciation of the innate immune responses induced in the blood of healthy volunteers after s.c. injection of poly ICLC. We find that poly ICLC is a reliable inducer of many arms of innate immunity in humans and that many of the triggered pathways mimic what is seen with YF17D, a successful live attenuated viral vaccine.
RESULTS AND DISCUSSION
Poly ICLC injected s.c. is tolerated in healthy volunteers
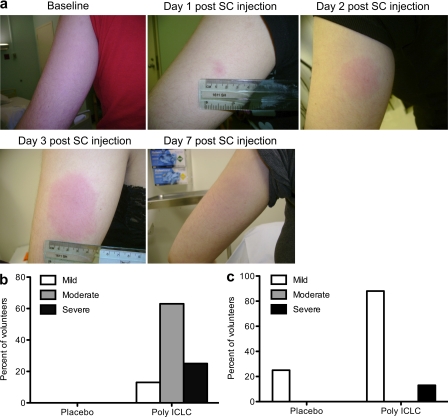
Preclinical studies in mice showed that s.c. administration of poly ICLC in combination with DC-targeted vaccines, a vaccine platform which is being developed in our laboratory, was superior to the i.m. route in inducing adaptive immune responses (unpublished data). To determine if human subjects tolerate poly ICLC administered s.c., because prior studies have mainly used the i.m. route (Rosenfeld et al., 2010; Okada et al., 2011), a total of 12 volunteers were enrolled. Eight were randomly assigned to receive a single dose of 1.6 mg poly ICLC, a dose which has often been used in prior phase I studies, and four to receive placebo (sterile saline; Table S1 and Fig. S1). No treatment-related, grade 4, or serious adverse events were reported. Nonetheless, volunteers receiving poly ICLC developed erythema and induration at the site of injection (Fig. 1 A), something which had not previously been reported in phase I studies of poly ICLC. We tested poly ICLC s.c., which might explain the higher frequency of local adverse events. Systemic reactogenicity included transient flu-like symptoms, such as malaise, headache, fever, and chills, which were generally mild to moderate in severity (Fig. 1, B and C). In addition, there were no clinically significant changes in complete blood cell counts and serum chemistries, including liver function tests, 3 and 7 d after poly ICLC administration. Thus, in this small cohort of young healthy volunteers, poly ICLC was generally safe with tolerable side effects when administered as a single dose of 1.6 mg s.c. In our ongoing studies in which 1.6 mg poly ICLC is being used as a vaccine adjuvant, the reactogenicity was similar after s.c. injections at 0, 1, and 3 mo (unpublished data).
Figure 1.
s.c. poly ICLC induces transient local and systemic reactogenicity. (a) Typical local erythematosus skin reaction in the area of poly ICLC s.c. administration (representative subject). (b and c) Frequency of local (pain/tenderness at injection site, erythema, and induration; b) and systemic (fever, headache, myalgia, and malaise; c) adverse events in the first week after poly ICLC (n = 8) or placebo administration (n = 4).
Poly ICLC reliably induces innate immunity with regulation of many IFN-regulated genes
To assess the global innate response to poly ICLC, transcriptional profiling was performed on whole blood samples, collected just before poly ICLC or placebo administration (day 0) and at multiple time points thereafter (6 and 12 h and 1, 2, 3, 7, 14, and 28 d). RNA was extracted, amplified, and hybridized onto Illumina Expression BeadChips for analysis as previously described (Gaucher et al., 2008). Multidimensional scaling (MDS) and principal component analysis (PCA) showed similar gene expression responses to poly ICLC among volunteers, with a peak response at day 1 for five out of eight individuals, at 12 h for the remaining three, and a return to baseline at day 7 for all individuals (Fig. S2). Differential expression analysis between poly ICLC and placebo groups at each time point resulted in 31, 212, and 52 genes showing more than a twofold change at 5% false discovery rate (FDR) for 12 h, day 1, and day 2, respectively (Table S2). No significant differential expression was detected in the placebo group relative to the day 0 baseline, even at a very permissive FDR (>80%).
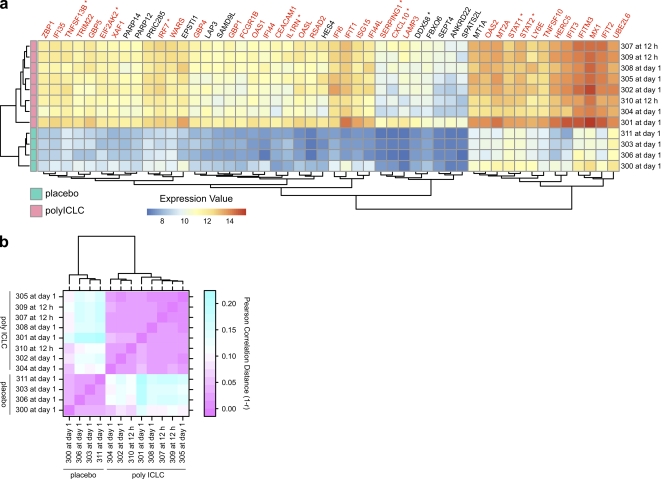
Importantly, the expression profiles of individual subjects after poly ICLC administration were comparable, both in quantity and quality, as illustrated by a heat map of the top 50 responsive genes (Fig. 2 A). As a further measure of the reliability of the response among healthy volunteers, there was a high degree of correlation in expression levels of IFN-regulated genes (IRGs) between the subjects after poly ICLC administration (Fig. 2 B).
Figure 2.
Poly ICLC reliably induces IFN signaling in healthy volunteers. (a) Heat map representation of the normalized expression values. Genes shown are the top 50 differentially expressed at day 1 (FC > 2 at 5% FDR), the group-level peak time point (poly ICLC n = 8 or placebo n = 4). Each volunteer who received poly ICLC (horizontal rows) is represented at the volunteer-specific peak time (right labels), as in Fig. S2. IRGs are highlighted in red. *, genes discussed in the text. (b) Heat map representation of the between-subject correlation of the expression values for IRGs at subject-specific peak times after poly ICLC administration. The color gradient extends from pink, representing perfect correlation (or a correlation distance r of 0, i.e., 1 − r = 0), to cyan for low correlation.
TLR-associated genes (TLR7 and TLR4), other genes involved in viral recognition (DDX58 and IFIH1), and transcription factor genes (IRF7, IRF5, and IRF1) were up-regulated by poly ICLC, all features of an IFN response leading to activation of transcription factors STAT1, STAT2, and STAT3. Many of the top regulated genes (92 out of 212 genes with fold change [FC] >2 at 5% FDR) by poly ICLC were IRGs (Waddell et al., 2010; IRGs listed in Table S3). Poly ICLC induced a broad array of IRGs, both genes associated with control of replication of several important animal and human viruses (IRF1, RIG-I, MDA-5, and IFITM3) and genes recently associated with enhanced replication of certain viruses in vitro (ADAR and LY6E; Schoggins et al., 2011).
Also, total PBMC counts and frequencies of different blood cell subsets were not significantly regulated by poly ICLC (unpublished data). Therefore, the observed transcriptional changes in PBMCs were not a result of redistribution of different cell subsets but rather of a rapid and reliable induction of gene expression.
Poly ICLC induces many specific genes associated with different innate pathways
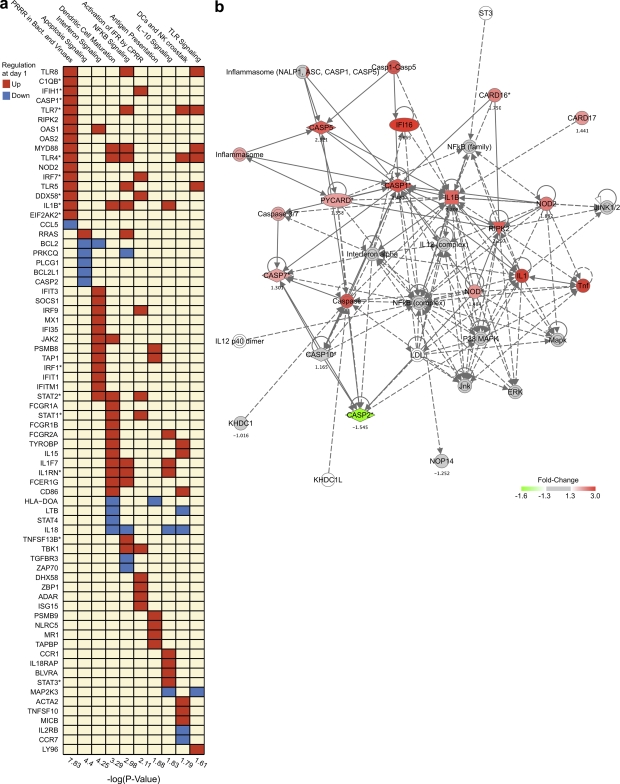
The gene-by-gene analysis approach identifies individual genes that exhibit differences between the poly ICLC and placebo groups. To detect molecular signatures that are included within a co-expressed or co-regulated set of genes, we performed canonical pathway analysis using Ingenuity Pathway Analysis software. We tested whether a co-regulated set of genes (pathway) was enriched in the list of genes that were significantly regulated by poly ICLC at day 1. Many pathways for innate immunity were induced. In addition to IFN signaling, poly ICLC stimulated other canonical innate pathways such as NF-κB signaling, DC maturation, and antigen presentation (Fig. 3 A, red blocks for up-regulated genes; and Table S4).
Figure 3.
Poly ICLC induces multiple genes of innate immunity including inflammasome components. (a) Each column is an up-regulated canonical pathway for innate immunity (Ingenuity software); each row represents an up-regulated (red) or down-regulated (blue) gene included in one or more regulated pathways. The over-representation test was performed using Fisher’s Exact Test, and the significance, displayed on the right, is achieved for P < 0.05 (−log(p) > 1.3). *, genes discussed in the text. CPRR, cytosolic PRR; PRRR, PRR recognition. (b) Network analysis of inflammasome gene signature induced 1 d after poly ICLC, generated with the Ingenuity software.
To further describe the scope of the innate pathways induced by poly ICLC, we analyzed the expression of selected genes from the regulated canonical pathways. We noted that in addition to the IRGs in Fig. 2 A, poly ICLC also up-regulated inflammasome-associated genes (Guarda and So, 2010), for example, IL-1β, IL1RN, CASP1, and CASP5 (Fig. 3 B), where inflammasomes are macromolecular complexes involved in the recognition of danger-associated molecular patterns (Lamkanfi and Dixit, 2009) and in the establishment of adaptive immunity to influenza virus infection (Ichinohe et al., 2009) and protective responses to mycobacterial species and C. albicans (Harris et al., 2010). Poly ICLC induced the expression of TNFSF13B (BAFF), which triggers Ig class switching in human mucosal B cells (Xu et al., 2008) and binds TNFRSF17, a B cell growth factor which predicts antibody responses after YF17D vaccination (Querec et al., 2009). Poly ICLC also increased the expression of components of the complement system (C1QB, C3AR1, and SERPING1; Fig. 3 A and Table S2), as well as the expression of EIF2AK2 (an EIF2AK4 homologue also known as protein kinase RNA-activated, PKR), which regulates protein synthesis by phosphorylating eIF2α during viral infections and is also part of the integrated stress response (García et al., 2006). C1QB and EIF2AK4 have been shown to be predictors of CD8+ T cell responses to the YF17D vaccine (Querec et al., 2009). Thus, poly ICLC not only induced the IFN signaling pathway but also up-regulated gene expression of other components of major effector pathways of innate immunity (inflammasomes, complement, and IFNs).
Validation of the transcriptional profiling
To validate the gene expression findings, we performed real-time PCR using the OpenArray Real-Time PCR Platform on 20 selected IRGs from Fig. 2 and samples collected at baseline from volunteers receiving poly ICLC or placebo and from days 1 and 7 after poly ICLC. The OpenArray data confirmed the highly integrated up-regulation of IRGs by poly ICLC at day 1 and high correlation (mean r = 0.81) with the corresponding microarray gene expression (Fig. S3).
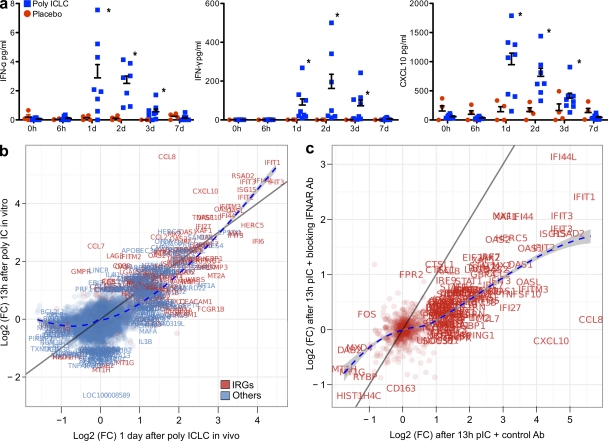
In addition, we measured the plasma concentrations of IFNs and the IFN-γ–induced chemokine CXCL10. Significant differences in concentrations (P < 0.001) between poly ICLC and placebo groups were observed at days 1–3 and levels returned to baseline at day 7 (Fig. 4 A).
Figure 4.
Validation of the transcriptional profiling on PBMCs. (a) Poly ICLC induced systemic secretion of IFNs. Plasma concentrations of IFN-γ, IFN-α, and CXCL10 at time points after poly ICLC (blue, n = 8) or placebo (red, n = 4) were analyzed by two-way ANOVA. Error bars represent the standard error of the mean concentrations. Poly ICLC effects were significant at days 1–3 (*, P < 0.001). (b and c) PBMCs from four donors were stimulated with 50 µg/ml poly IC for 13 h plus 10 µg/ml IFNAR antibody or isotype control antibody. RNA was extracted for transcriptional analysis. (b) Differential gene expression in blood 1 d after in vivo poly ICLC (x axis, n = 8) was compared with differential gene expression in PBMCs after 13 h of poly IC stimulation in vitro (y axis, n = 4). Genes falling on the identity line (dark gray) are equally regulated in both systems. A loss curve (dashed blue) is used to infer the local trend between the two systems. IRGs are in red. Text labels are added to points for genes with FC > 2 at 5% FDR in at least one comparison. (c) Log2(FC) of IRGs after in vitro poly IC with blocking anti-IFNAR mAb (y axis) and after in vitro poly IC with control antibody (x axis).
To obtain further validation for type-I IFN signaling induced by poly ICLC in vivo, freshly isolated PBMCs from healthy volunteers were cultured with 10 µg/ml IFN-α receptor (IFNAR) blocking mAb or control mAb for 1 h, and then poly IC (parent dsRNA) at 50 µg/ml was added to the cell culture. We first determined the kinetics of the in vitro effects of poly IC on PBMCs. Cells were stimulated with 50 µg/ml poly IC and collected at 1, 3, 6, 12, 18, and 24 h. STAT1 phosphorylation was detected by flow cytometry at 6 h and at later time points (unpublished data). We estimated that the peak gene regulation would occur earlier after direct in vitro stimulation of PBMCs with poly IC (∼12 h) than the peak regulation in vivo (24 h). Gene expression analysis revealed that 104 genes were regulated after 6 h of poly IC in vitro and 238 genes after 13 h in comparison with unstimulated cells. Gene regulation was defined by FC > 2 at 5% FDR. We therefore chose to focus our analyses on gene expression changes after 13 h of poly IC in the presence of IFNAR blocking Ab or control Ab.
We then compared the transcriptional profiles of PBMCs stimulated with poly IC in vitro (13 h), in the absence of IFNAR blockade, to those induced in whole blood after in vivo poly ICLC (Fig. 4 B). We focused our comparison on the top regulated genes (FC > 2 at 5% FDR) by dsRNA in vivo or in vitro and found that 106 were commonly regulated by both in vivo poly ICLC and in vitro poly IC. As expected, 66 of these commonly regulated genes are known IRGs (Table S5). The discordance in gene regulation for the other top regulated genes (106 unique to in vivo poly ICLC and 101 to in vitro poly IC) may be accounted for by the fact that in vivo poly ICLC acts not only on hematopoietic but also on nonhematopoietic cells, the main source of type I IFN secretion after poly IC in a mouse model (Longhi et al., 2009), whereas the in vitro analysis was performed on purified PBMCs. In addition, the kinetics of gene regulation between the in vitro and in vivo systems might differ.
We then analyzed the effects of IFNAR blocking on gene modulation induced by poly IC in vitro. This blockade led to an obvious dampening of genes that responded to poly IC. Among the 207 genes most affected by poly IC (FC > 2, FDR 5%), 97/207 were described as IRGs (Fig. 4 C), and 83/207 (or 48/83 IRGs) had their response to poly IC strongly affected by IFNAR blockade. These results demonstrated that IRGs are among the top regulated genes induced by poly ICLC both in vitro and in vivo.
A defined innate agonist mimics an attenuated live viral vaccine
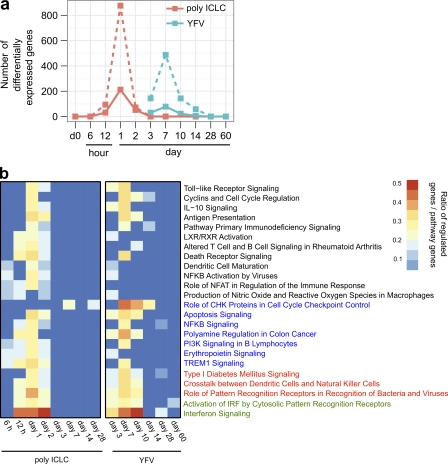
To compare poly ICLC, which is considered to be a mimic of viral double-stranded RNA and is known to interact with TLR-3 and MDA-5, with a live viral replication-competent vaccine, we analyzed the gene expression data in this study to prior datasets from volunteers immunized with YF17D vaccine (Gaucher et al., 2008). The comparison was performed at both the individual gene and the gene set levels. Both poly ICLC and YF17D induced a large transcriptional response in blood, but the response to poly ICLC was faster and involved more genes (212 genes 1 d after poly ICLC and 78 genes 7 d after YF17D with FC > 2 at 5% FDR). Using Ingenuity Pathway Analysis, we found that a large number of similar transcriptional and signal transduction canonical pathways were up-regulated at the peak time points, day 1 after poly ICLC administration and day 7 after the viral vaccine (Fig. 5 B). Pathways induced by poly ICLC at day 1, depicted in Fig. 3 A, were similar and shared many identical genes with the pathways induced by YF-17D (Table S6). As expected, some immune-related pathways were uniquely regulated by YF-17D vaccine, such as IL-1 signaling and protein ubiquitination among others. This difference in regulation is likely a result of the fact that YFV can signal through TLRs 2, 7, and 9 (Querec et al., 2006), and it has been demonstrated that signaling through different TLRs leads to regulation of a common core of genes, but genes that are more specific to the pathogen-sensing pathway are also triggered (Amit et al., 2009).
Figure 5.
Poly ICLC activated similar innate immune pathways as YF17D. (a) Differentially expressed genes (DEGs) after poly ICLC (n = 8, versus placebos; n = 4, at same time points) or YF17D (versus day 0, n = 15; Gaucher et al., 2008) at several time points after administration. The threshold of differential expression is set at FDR 5%, with either FC > 2 (solid line) or FC > 1.3 (dashed line). Peak transcriptional changes are shown after poly ICLC (212 DEGs, day 1) and YF17D (78 DEGs, day 7). (b) Heat map showing statistically significant canonical pathways (Ingenuity Pathway Analysis Software) commonly regulated by poly ICLC and YF17D at least at four different time points. Columns represent time points after either poly ICLC or YF17D. Rows represent significantly regulated canonical pathways. Heat map colors represent the ratio of regulated genes/pathway genes after poly ICLC or YF17D (pathways not overrepresented are dark blue). Maximum pathway modulation similarity was obtained between poly ICLC at day 1 and YF17D at day 7. Pathways were significantly regulated at 7 of 14 different time points analyzed (green) and at 6, 5, and 4 time points in red, blue, and black, respectively. The overrepresentation test was performed using Fisher’s Exact Test. Statistical significance was achieved at P < 0.05.
Only two vaccine adjuvants (alum and MPL-A) are approved in the United States for prophylactic vaccination in humans, and currently approved adjuvants do not always induce the desired immune response against specific pathogens, especially T cell immune responses. We had reported that double-stranded RNAs—poly IC and its analogue poly ICLC—are superior adjuvants for T cell immunity in animal models (Longhi et al., 2009; Stahl-Hennig et al., 2009). To help identify and develop adjuvants that are safe for humans, it is important to define the innate response to synthetic ligands for microbial innate receptors, like poly ICLC for TLR3 and MDA-5, in healthy subjects and to compare this response to complex microbial vaccines that contain intrinsic adjuvants.
Our results highlight the feasibility of performing such research in humans. They also demonstrate the value of using systems biology, as it is the only approach that allows the simultaneous identification of integrated immune pathways that can lead to protection and a thorough comparison of the innate response to different adjuvants.
Our study was designed to compare activation of innate immune pathways after YF17D (Gaucher et al., 2008; Querec et al., 2009) to the immune modulation after poly ICLC. In addition to the reliability of the response of human subjects to this adjuvant, we found extensive overlap in transcriptional profiles, as the top regulated innate immune pathways were shared between the attenuated viral vaccine YF17D and the synthetic double-stranded RNA poly ICLC. These results were unexpected because it has been reported in vitro that YF17D signals a large number of TLRs (Querec et al., 2006). Our findings underscore the fact that a synthetic dsRNA to just MDA-5 and TLR3 is able to mimic the early effects of a replication-competent and protective vaccine in vivo. Poly ICLC is therefore a true viral mimic, and these features of innate immunity need to be evaluated with other new adjuvants that are ready for investigational study in humans.
MATERIALS AND METHODS
Study design and objectives.
The study was conducted at the Rockefeller University Hospital in New York City. This study was approved by the Institutional Review Board of The Rockefeller University Hospital. Individual participants in this study provided written informed consent after appropriate review, discussion, and counseling by the clinical study team. The study was conducted in compliance with the International Conference on Harmonization Good Clinical Practice (ICH-GCP) guidelines and registered on www.clinicaltrials.gov (NCT01012700). Healthy men and women aged 18–60 yr were eligible for participation. Exclusion criteria included chronic medical conditions, clinically significant abnormal laboratory parameters, infection with HIV or Hepatitis B or C virus, and recent receipt of a live-attenuated vaccine or blood transfusion.
The study design is summarized in Fig. S1. This study was randomized, double blinded, and placebo controlled. The randomization schedule was prepared by the Rockefeller University Hospital Pharmacy using a web-based program (http://www.randomization.com/). Subjects were randomized to either 1.6 mg poly ICLC or placebo (sterile saline) in a 2:1 ratio, administered s.c. Dose levels were based on safety data from previous trials (Rosenfeld et al., 2010). 12 subjects were enrolled. Safety and tolerability of poly ICLC or placebo in each cohort were monitored. Adverse events were graded according to the Toxicity Grading Scale for Healthy Adult and Adolescent Volunteers Enrolled in Preventive Vaccine Clinical Trials. After receipt of the study drug or placebo, subjects were followed for 28 d.
Microarray analysis.
Blood was collected from each volunteer in PAXgene Blood RNA tubes (BD) for whole-blood RNA isolation at baseline (day 0) and at 6 and 12 h and 1, 2, 3, 7, 14, and 28 d after poly ICLC or placebo administration. In brief, RNA was extracted using PAXgene Blood RNA kits (QIAGEN) according to the manufacturer’s protocol. Total RNA was checked for quantity and quality using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) and Experion automated electrophoresis system (Bio-Rad Laboratories). Only RNA samples with a 28S/18S ratio >1.5 were processed for array analysis. 500 ng of total RNA was amplified using the Illumina TotalPrep RNA amplification kits (Applied Biosystems) as recommended by manufacturer. 750 ng of the biotinylated cRNA was hybridized onto HumanRef-8 Expression BeadChips v3 (Illumina) at 58°C for 20 h and quantified using an iScan System (Illumina) and GenomeStudio software (Illumina).
Analysis of the GenomeStudio output data was conducted using the R statistical language (R Development Core Team) and various software packages from Bioconductor, an open source project for the analysis and comprehension of high-throughput genomic data (Gentleman et al., 2004). First, arrays displaying unusually low median intensity, low variability, or low correlation relative to the bulk of the arrays were discarded from the rest of the analysis. Quantile normalization was applied, followed by a log2 transformation and batch effect subtraction using the ComBat procedure (Johnson et al., 2007). The LIMMA package (Bioconductor; Smyth, 2004) was used to fit a linear model to each probe and to perform a (moderated) Student’s t test on various differences of interest. For the poly ICLC in vivo experiment, the differences calculated were between the poly ICLC and placebo groups, separately at each time point. For the in vitro IFNAR-blockade experiment, the differences calculated were between PBMCs stimulated with poly IC and unstimulated PBMCs, with or without IFNAR blockade separately, whereas the blocking effect was evaluated using the interaction term (defined as the difference between the last two differences). For the comparison with YFV, the differences calculated were between each time point and day 0. The expected proportions of false positives (FDR) were estimated from the unadjusted p-value using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). The microarray data are available through the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession no. GSE32862.
OpenArray real-time PCR.
1 µg of total RNA was reverse transcribed using the SuperScript VILO cDNA synthesis kit (Invitrogen) with random primers. The cDNA was analyzed in duplicate on an OpenArray NT Cycler (BioTrove) using TaqMan Real-Time PCR plates (Applied Biosystems) preloaded with primer pairs (see Fig. S3 for TaqMan gene expression assay identifiers). cDNA samples were loaded into the plates using an OpenArray AccuFill instrument (Applied Biosystems) according to the manufacturer’s protocols. Each sub-array was loaded with 2.5 µl 2× GeneAmp Fast PCR Master Mix (Applied Biosystems), 1 µl 5× TaqMan OpenArray Remix (Applied Biosystems), 0.3 µl RNase-free water, and 1.2 µl cDNA. The thermal cycling protocol was to manufacturer defaults. PCR data were analyzed by DataAssist v3.0 (Applied Biosystems) using the −ΔCT method. Values were normalized by the mean of three housekeeping genes.
Pathway analysis.
Ingenuity Pathway Analysis software (Ingenuity Systems) was used to identify canonical signaling pathways regulated by poly ICLC or YF17D (published microarray dataset in Gaucher et al., 2008; GEO accession no. GSE13699). Canonical pathway analysis identified the pathways from the Ingenuity Pathway Analysis library of canonical pathways that were most significant to the dataset. Illumina Probe IDs were imported into the Ingenuity software and mapped to the Gene Symbol from Ingenuity database. Genes that had a nonadjusted p-value <0.05 at each time point after poly ICLC administration (6 and 12 h and 1, 2, 3, 7, 14, and 28 d) or YF17D immunization (3, 7, 10, 14, 28, and 60 d) and were associated with a canonical pathway in Ingenuity’s Knowledge Base were used for pathway analysis. The significance of the association between the dataset and the canonical pathway was measured in two ways: (1) A ratio of the number of genes from the dataset that map to the pathway divided by the total number of genes that map to the canonical pathway was displayed; (2) over-representation Fisher’s exact test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone. The pathways were ranked by −log p-value. This score was used as the cutoff for identifying canonical pathways significantly (P < 0.05) affected by poly ICLC or YF17D.
IFN gene analysis.
IRGs were selected using the IFN-α–, IFN-β–, IFN-γ–, and IFN-ω–induced gene lists from human peripheral blood cells published by Waddell et al. (2010). We supplemented this list with a query consisting of 46 IRGs (Table S3) chosen from our own transcriptional database of innate immune IFN responses with additional annotation supplied by the Genecards V3 public database (http://www.genecards.org) and the National Center for Biotechnology Information Biosystems Database (http://www.ncbi.nlm.nih.gov/biosystems).
Phenotyping of blood cell populations.
Freshly isolated PBMCs were isolated by density gradient centrifugation. Absolute PBMC counts were calculated by an automated cell counter (Vi-Cell XR), and PBMCs were then cryopreserved in FBS plus 10% DMSO. Frozen PBMCs collected at baseline and 1 and 2 d after poly ICLC or placebo administration were rapidly thawed at 37°C. 2 × 106 cells were stained in 20 µl PBS 2% FBS according to standard protocols. The following antibodies were used to identify peripheral blood DCs, monocytes, and lymphoid cells in whole PBMCs: CD3 (clone S4.1), CD19 (clone SJ25-C1), CD56 (clone MEM-188), CD14 (clone Tük4), CD16 (clone 3G8), CD11c (clone 3.9), and CD123 (clone AC145). Dead cells were excluded using LIVE/DEAD stain kit (Invitrogen). Flow cytometry was performed on an LSRII cytometer (BD), and data were analyzed with FlowJo (Tree Star). Total PBMC counts and relative frequencies of blood cell subsets between poly ICLC and placebo groups were compared using likelihood ratio tests. Statistical significance was achieved with p-values <0.05.
Cytokine secretion assays.
Plasma samples collected at baseline, 6 h, and 1, 2, 3, and 7 d after poly ICLC or placebo injection were stored frozen until analysis for cytokine production. Plasma concentrations of IFN-γ, IL-1β, IL-12p70, IL-6, IL-8, IL-2, IL-10, GM-CSF, and TNF were measured with a proinflammatory multiplex kit by Meso Scale Discovery (MSD), according to manufacturer’s instructions. Cytokine concentrations were calculated using Sector Image Reader (SI2400). Concentration of IFN-α 2a was also measured using MSD technology. CXCL10 and IFN-β were measured by ELISA (R&D Systems). Concentration levels were analyzed by two-way ANOVA.
In vitro stimulation of PBMCs with poly IC.
PBMCs were isolated from blood of healthy donors via density gradient centrifugation. 106 freshly isolated PBMCs were pretreated with 10 µg/ml anti-IFNAR mAb (Fitzgerald Industries) or control-IgG (mouse IgG2a; BioLegend) for 1 h in 96-well round bottom plates and subsequently stimulated with poly IC at 50 µg/ml (InvivoGen) or left unstimulated. Cells were harvested 13 h later and preserved in RLT buffer (QIAGEN) for future RNA extraction and microarray analyses.
Online supplemental material.
Fig. S1 shows a flow diagram of clinical trial design. Fig. S2 shows plots of 2D classical MDS and the first two PCAs to demonstrate that gene regulation was homogenous and peaked early on after s.c. poly ICLC. Fig. S3 shows the correlation of gene expression changes after poly ICLC by microarray and OpenArray PCR for selected genes. Table S1 shows demographic information of study volunteers. Table S2 lists DEGs 1 d after s.c. administration of poly ICLC in comparison to placebo. Table S3 lists IRGs. Table S4 shows the canonical pathways significantly regulated at 1 d after poly ICLC s.c. Table S5 lists the top commonly regulated genes in whole blood 1 d after poly ICLC s.c. and after in vitro stimulation of PBMCs with poly IC. Table S6 shows selected pathways and their genes commonly modulated after poly ICLC and YF17D. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20111171/DC1.
Acknowledgments
We thank the Clinical and Translational Science Award at the Rockefeller University Hospital and their associated staff for assistance in conducting this study (J. Gonzalez for ELISA assays and S. Tian for statistical assistance), A. Smith and P. Wilkinson at the Vaccine and Gene Therapy Institute of Florida Collaborative Genomic Center for their genomics and bioinformatics assistance, and J. Adams for graphics.
Funding was provided by National Institutes of Health Grants AI081677 (R.M. Steinman), K23AI084855 (M. Caskey), and UL1RR024143 (Rockefeller University Hospital Center Clinical and Translational Science Award), by the Bill and Melinda Gates Foundation Grant #38650 of The Collaboration for AIDS Vaccine Discovery (R.P. Sékaly), by the Foundation for the National Institutes of Health through the Grand Challenges in Global Health initiative of Bill and Melinda Gates Foundation Grant GC#334 (R.M. Steinman and S.J. Schlesinger), and by Génome Québec (R.P. Sékaly).
A.M. Salazar heads Oncovir Inc., which provided poly ICLC. The authors have no other financial interest.
Footnotes
Abbreviations used:
- DEG
- differentially expressed gene
- FC
- fold change
- FDR
- false discovery rate
- IFNAR
- IFN-α receptor
- IRG
- IFN-regulated gene
- MDS
- multidimensional scaling
- PCA
- principal component analysis
- poly IC
- polyinosinic:polycytidylic acid
- poly ICLC
- poly IC stabilized with poly-l-lysine
- PRR
- pattern recognition receptor
- TLR
- Toll-like receptor
- YFV
- yellow fever vaccine
References
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 413:732–738 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- Amit I., Garber M., Chevrier N., Leite A.P., Donner Y., Eisenhaure T., Guttman M., Grenier J.K., Li W., Zuk O., et al. 2009. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 326:257–263 10.1126/science.1179050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300 [Google Scholar]
- Beutler B. 2004. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 430:257–263 10.1038/nature02761 [DOI] [PubMed] [Google Scholar]
- Coffman R.L., Sher A., Seder R.A. 2010. Vaccine adjuvants: putting innate immunity to work. Immunity. 33:492–503 10.1016/j.immuni.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M.A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060 10.1128/MMBR.00027-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher D., Therrien R., Kettaf N., Angermann B.R., Boucher G., Filali-Mouhim A., Moser J.M., Mehta R.S., Drake D.R., III, Castro E., et al. 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 205:3119–3131 10.1084/jem.20082292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain R.N. 2010. Vaccines and the future of human immunology. Immunity. 33:441–450 10.1016/j.immuni.2010.09.014 [DOI] [PubMed] [Google Scholar]
- Guarda G., So A. 2010. Regulation of inflammasome activity. Immunology. 130:329–336 10.1111/j.1365-2567.2010.03283.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J., Sharp F.A., Lavelle E.C. 2010. The role of inflammasomes in the immunostimulatory effects of particulate vaccine adjuvants. Eur. J. Immunol. 40:634–638 10.1002/eji.200940172 [DOI] [PubMed] [Google Scholar]
- Huang C.C., Duffy K.E., San Mateo L.R., Amegadzie B.Y., Sarisky R.T., Mbow M.L. 2006. A pathway analysis of poly(I:C)-induced global gene expression change in human peripheral blood mononuclear cells. Physiol. Genomics. 26:125–133 10.1152/physiolgenomics.00002.2006 [DOI] [PubMed] [Google Scholar]
- Ichinohe T., Lee H.K., Ogura Y., Flavell R., Iwasaki A. 2009. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 206:79–87 10.1084/jem.20081667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E., Li C., Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 8:118–127 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- Kolumam G.A., Thomas S., Thompson L.J., Sprent J., Murali-Krishna K. 2005. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 202:637–650 10.1084/jem.20050821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M., Dixit V.M. 2009. Inflammasomes: guardians of cytosolic sanctity. Immunol. Rev. 227:95–105 10.1111/j.1600-065X.2008.00730.x [DOI] [PubMed] [Google Scholar]
- Le Bon A., Schiavoni G., D’Agostino G., Gresser I., Belardelli F., Tough D.F. 2001. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 14:461–470 10.1016/S1074-7613(01)00126-1 [DOI] [PubMed] [Google Scholar]
- Le Bon A., Durand V., Kamphuis E., Thompson C., Bulfone-Paus S., Rossmann C., Kalinke U., Tough D.F. 2006. Direct stimulation of T cells by type I IFN enhances the CD8+ T cell response during cross-priming. J. Immunol. 176:4682–4689 [DOI] [PubMed] [Google Scholar]
- Longhi M.P., Trumpfheller C., Idoyaga J., Caskey M., Matos I., Kluger C., Salazar A.M., Colonna M., Steinman R.M. 2009. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 206:1589–1602 10.1084/jem.20090247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E., Tschopp J. 2006. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol. Cell. 22:561–569 10.1016/j.molcel.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Morse M.A., Chapman R., Powderly J., Blackwell K.L., Keler T., Green J., Riggs R., He L.Z., Ramakrishna V., Vitale L., et al. 2011. Phase I study utilizing a novel antigen-presenting cell-targeted vaccine with Toll-like receptor stimulation to induce immunity to self-antigens in cancer patients. Clin. Cancer Res. 17:4844–4853 10.1158/1078-0432.CCR-11-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Kalinski P., Ueda R., Hoji A., Kohanbash G., Donegan T.E., Mintz A.H., Engh J.A., Bartlett D.L., Brown C.K., et al. 2011. Induction of CD8+ T-cell responses against novel glioma-associated antigen peptides and clinical activity by vaccinations with alpha-type 1 polarized dendritic cells and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in patients with recurrent malignant glioma. J. Clin. Oncol. 29:330–336 10.1200/JCO.2010.30.7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S.A. 2008. Vaccines: correlates of vaccine-induced immunity. Clin. Infect. Dis. 47:401–409 10.1086/589862 [DOI] [PubMed] [Google Scholar]
- Pulendran B., Ahmed R. 2006. Translating innate immunity into immunological memory: implications for vaccine development. Cell. 124:849–863 10.1016/j.cell.2006.02.019 [DOI] [PubMed] [Google Scholar]
- Querec T., Bennouna S., Alkan S., Laouar Y., Gorden K., Flavell R., Akira S., Ahmed R., Pulendran B. 2006. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J. Exp. Med. 203:413–424 10.1084/jem.20051720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B., et al. 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10:116–125 10.1038/ni.1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S.G., Bertholet S., Coler R.N., Friede M. 2009. New horizons in adjuvants for vaccine development. Trends Immunol. 30:23–32 10.1016/j.it.2008.09.006 [DOI] [PubMed] [Google Scholar]
- Rosenfeld M.R., Chamberlain M.C., Grossman S.A., Peereboom D.M., Lesser G.J., Batchelor T.T., Desideri S., Salazar A.M., Ye X. 2010. A multi-institution phase II study of poly-ICLC and radiotherapy with concurrent and adjuvant temozolomide in adults with newly diagnosed glioblastoma. Neuro-oncol. 12:1071–1077 10.1093/neuonc/noq071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 472:481–485 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:e3. [DOI] [PubMed] [Google Scholar]
- Stahl-Hennig C., Eisenblätter M., Jasny E., Rzehak T., Tenner-Racz K., Trumpfheller C., Salazar A.M., Uberla K., Nieto K., Kleinschmidt J., et al. 2009. Synthetic double-stranded RNAs are adjuvants for the induction of T helper 1 and humoral immune responses to human papillomavirus in rhesus macaques. PLoS Pathog. 5:e1000373 10.1371/journal.ppat.1000373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S.J., Popper S.J., Rubins K.H., Griffiths M.J., Brown P.O., Levin M., Relman D.A. 2010. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS ONE. 5:e9753 10.1371/journal.pone.0009753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Santini P.A., Matthews A.J., Chiu A., Plebani A., He B., Chen K., Cerutti A. 2008. Viral double-stranded RNA triggers Ig class switching by activating upper respiratory mucosa B cells through an innate TLR3 pathway involving BAFF. J. Immunol. 181:276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]