TDP-43 interacts with and coactivates NF-κB p65 in the spinal cord of amyotrophic lateral sclerosis (ALS) patients, and an NF-κB inhibitor suppresses ALS disease symptoms and neuromuscular junction denervation in an ALS mouse model.
Abstract
TDP-43 (TAR DNA-binding protein 43) inclusions are a hallmark of amyotrophic lateral sclerosis (ALS). In this study, we report that TDP-43 and nuclear factor κB (NF-κB) p65 messenger RNA and protein expression is higher in spinal cords in ALS patients than healthy individuals. TDP-43 interacts with and colocalizes with p65 in glial and neuronal cells from ALS patients and mice expressing wild-type and mutant TDP-43 transgenes but not in cells from healthy individuals or nontransgenic mice. TDP-43 acted as a co-activator of p65, and glial cells expressing higher amounts of TDP-43 produced more proinflammatory cytokines and neurotoxic mediators after stimulation with lipopolysaccharide or reactive oxygen species. TDP-43 overexpression in neurons also increased their vulnerability to toxic mediators. Treatment of TDP-43 mice with Withaferin A, an inhibitor of NF-κB activity, reduced denervation in the neuromuscular junction and ALS disease symptoms. We propose that TDP-43 deregulation contributes to ALS pathogenesis in part by enhancing NF-κB activation and that NF-κB may constitute a therapeutic target for the disease.
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disorder characterized by the progressive degeneration of motor neurons in the brain and spinal cord. Approximately 10% of ALS cases are familial and 90% are sporadic. Recently, TDP-43 (TAR DNA-binding protein 43) has been implicated in ALS (Neumann et al., 2006). TDP-43 is a DNA/RNA-binding 43-kD protein that contains an N-terminal domain, two RNA recognition motifs and a glycine-rich C-terminal domain, characteristic of the heterogeneous nuclear RNP class of proteins (Dreyfuss et al., 1993). TDP-43, normally observed in the nucleus, is detected in pathological inclusions in the cytoplasm and nucleus of both neurons and glial cells of ALS and frontotemporal lobar degeneration with ubiquitin inclusions (FTLD-U) cases (Arai et al., 2006; Neumann et al., 2006). The inclusions consist prominently of TDP-43 C-terminal fragments of ∼25 kD. The involvement of TDP-43 with ALS cases led to the discovery of TDP-43 mutations found in ALS patients. Dominant mutations in TARDBP, which codes for TDP-43, were reported by several groups as a primary cause of ALS (Gitcho et al., 2008; Kabashi et al., 2008; Sreedharan et al., 2008; Van Deerlin et al., 2008; Corrado et al., 2009; Daoud et al., 2009) and may account for ∼3% of familial ALS cases and ∼1.5% of sporadic cases.
Neuronal overexpression at high levels of WT or mutant TDP-43 in transgenic mice caused a dose-dependent degeneration of cortical and spinal motor neurons but with no cytoplasmic TDP-43 aggregates (Wegorzewska et al., 2009; Stallings et al., 2010; Wils et al., 2010; Xu et al., 2010), raising up the possibility that an up-regulation of TDP-43 in the nucleus rather than TDP-43 cytoplasmic aggregates may contribute to neurodegeneration. The physiological role of TDP-43 and the pathogenic pathways of TDP-43 abnormalities are not well understood. TDP-43 is essential for embryogenesis (Sephton et al., 2010), and postnatal deletion of the TDP-43 gene in mice caused down-regulation of Tbc1d1, a gene which alters body fat metabolism (Chiang et al., 2010). Proteins known to interact with TDP-43 have also been implicated in protein refolding or proteasomal degradation, including ubiquitin, proteasome-β subunits, SUMO-2/3, and Hsp70 (Seyfried et al., 2010).
Because TDP-43 is ubiquitously expressed and several studies have supported the importance of glial cells in mediating motor neuron injury (Clement et al., 2003; Boillée et al., 2006a,b), we have searched for additional proteins that might interact with TDP-43 in LPS-stimulated microglial (BV-2) cells. Our rationale for choosing microglial BV-2 cells was that TDP-43 deregulation may occur not only in neurons but also in microglial cells. Moreover, there are recent reports of increased levels of LPS in the blood of ALS patients (Zhang et al., 2009a) and of an up-regulation of LPS/TLR-4 signaling–associated genes in peripheral blood monocytes from ALS patients (Zhang et al., 2011). Accordingly, we have biased our search for proteins interacting with TDP-43 when microglia are activated by LPS. Surprisingly, coimmunoprecipitation assays and mass spectrometry led us to identify the p65 subunit of NF-κB as a binding partner of TDP-43. Furthermore, we discovered that TDP-43 messenger RNA (mRNA) was abnormally up-regulated in the spinal cord of ALS subjects. The results reported here led us to further explore the physiological significance of the interaction between TDP-43 and p65 NF-κB.
RESULTS
TDP-43 interacts with the p65 subunit of NF-κB
Mass spectrometry analysis and coimmunoprecipitation experiments were performed to identify proteins that interact with TDP-43 in mouse microglia (BV-2) cells after LPS stimulation, as described in Materials and methods. Many proteins were coimmunoprecipitated with TDP-43, including proteins responsible for RNA granule transport (kinesin), molecular chaperones (Hsp70), and cytoskeletal proteins (unpublished data). In addition, our analysis revealed p65 (REL-A) as a novel protein interacting with TDP-43. An interaction between TDP-43 with p65 NF-κB was confirmed by a coimmunoprecipitation assay with a polyclonal antibody against TDP-43 using spinal cord extracts from transgenic mice overexpressing human TDP-43WT and TDP-43G348C mutant (Swarup et al., 2011) by threefold (Fig. 1 B). Additional coimmunoprecipitation experiments performed using BV-2 cells that were transiently transfected with pCMV–TD-P43WT and pCMV-p65 plasmids clearly showed that TDP-43 interacts with p65.
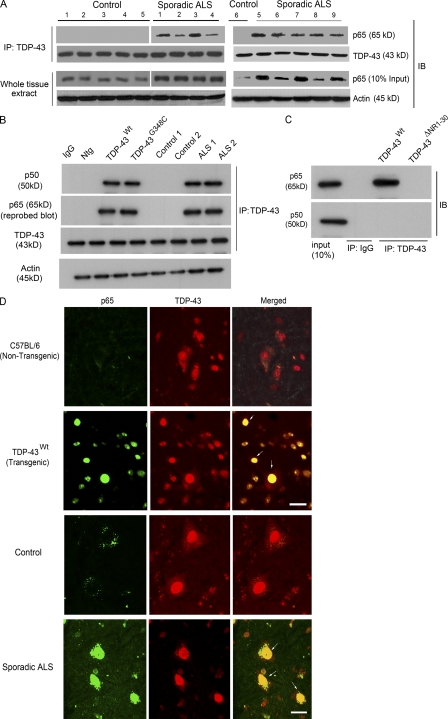
Figure 1.
TDP-43 interacts with NF-κB p65. (A) Protein extracts from the spinal cords of nine sporadic ALS subjects (1–9) and six control individuals (1–6) were used for the immunoprecipitation (IP) with TDP-43–specific polyclonal antibody where indicated. Immunoprecipitates or whole cells extracts were subjected to immunoblot (IB) with the indicated antibodies. Two experiments were performed (one with controls 1–5 and ALS patients 1–4, and the other with control 6 and ALS patients 5–9). (B) Total protein extract from spinal cords of TDP-43WT and TDP-43G348C transgenic mice, B6 nontransgenic mice (Ntg), two control individuals, and two sporadic ALS patients were subjected to immunoprecipitation and immunoblot where indicated. (C) Neuro2a cells were transfected with pCMV-p65 and pCMV-p50 expression vectors along with TDP-43WT or TDP-43ΔNR1-30. Extracts were immunoprecipitated with anti–TDP-43 or control IgG where indicated and immunoblotted with anti-p65 and anti-p50. (B and C) A representative blot from two independent experiments is shown. (D) Spinal cords of B6 nontransgenic or TDP-43WT transgenic mice or control or ALS patients were stained with anti-p65 and anti–TDP-43 and analyzed by immunofluorescence. Brightness and contrast adjustments were made to the whole image to make background intensities equal in control and ALS cases. The images represent at least four sections from two experiments using ALS and control patient material. Arrows indicate colocalization of TDP-43 with p65. Bars, 20 µm.
To further determine the significance of TDP-43 interaction with p65 in the context of human ALS, TDP-43 was pulled down with the polyclonal anti–TDP-43 antibody using spinal cord extracts from nine sporadic ALS cases and six control subjects (Fig. 1 A). In protein extracts from ALS cases, p65 NF-κB was coimmunoprecipitated with TDP-43. In contrast, no p65 was pulled down with TDP-43 using extracts of control spinal cords. To further validate TDP-43–p65 interaction, we performed reverse coimmunoprecipitation using p65 antibody to immunoprecipitate TDP-43 in human spinal cord tissues. Indeed, p65 was able to coimmunoprecipitate TDP-43 in all nine ALS cases but not in six control cases (Fig. S1 A). Along with p65, p50 was also coimmunoprecipitated with TDP-43 from the spinal cord samples of TDP-43WT and TDP-43G348C mice and ALS samples but not from nontransgenic or control spinal cord tissues, suggesting that TDP-43, p50, and p65 are a part of a complex (Fig. 1 B). To determine whether TDP-43 interacts directly with p65 or p50, we have performed overexpression experiments using pCMV expression vectors transfected into mouse neuroblastoma Neuro2a cells (Fig. 1 C). Neuro2a cells were transfected with pCMV-p65 or pCMV-p50 expression vectors along with vectors encoding either hemagglutinin (HA)-tagged TDP-43WT or TDP-43ΔNR1-30, a deletion mutant lacking the region required for binding to p65 as described in the section p65 interacts with the N-terminal and RRM-1 domains of TDP-43. It should be noted that the cells were not stimulated by LPS or any other means. After overexpression of p65 and TDP-43WT in the Neuro2a cells, p65 was coimmunoprecipitated with TDP-43WT but not with TDP-43ΔNR1-30 using anti-HA antibody. In contrast, p50 was not coimmunoprecipitated with TDP-43WT when overexpressed alone with TDP-43. These results suggest that TDP-43 interacts directly with p65 but not directly with p50. Immunofluorescence microscopy corroborated these results. In the spinal cord of sporadic ALS subjects, p65 was detected predominantly in the nucleus of cells in colocalization with TDP-43 (Fig. 1 D). On the contrary, in control spinal cord, there was an absence of p65 in the nucleus, reflecting a lack of p65 activation (Fig. 1 D). It is remarkable that microscopy of the spinal cord from TDP-43WT transgenic mice revealed ALS-like immunofluorescence with active p65 that colocalized perfectly with TDP-43 in the nuclei of cells (Fig. 1 D). To elucidate which cell types in the spinal cord of ALS cases express TDP-43 and p65, we performed three-color immunofluorescence with CD11b as a microglial-specific marker and glial fibrillary acidic protein (GFAP) as an astroglial marker. We found that TDP-43 and p65 colocalized in many microglial and astroglial cells (Fig. 2, D–F, insets). We have quantified our data and found that 20 ± 5% of microglia and 8 ± 3% of astrocytes had TDP-43–p65 colocalization. We also found that many of the TDP-43 p65 colocalization was in neurons, and some also in motor neurons in many ALS cases (Fig. 2, A–C). In many ALS cases in which TDP-43 formed aggregates in the cytoplasm, p65 was still in the nucleus (Fig. 2, A–C, arrowheads). In nontransgenic C57BL/6 mice, the lack of p65 activation resulted in partial colocalization of TDP-43 with p65 mainly in cytoplasm (Fig. 1 D). LPS-stimulated BV-2 cells transfected with pCMV-p65 and pCMV–TDP-43WT had most p65 colocalized with nuclear TDP-43WT, whereas in unstimulated cells, p65 did not colocalize with nuclear TDP-43WT. Although p65 was mainly cytoplasmic in 3-mo-old TDP-43WT spinal cord, there was gradual age-dependent p65 activation in 6- and 10-mo-old TDP-43WT spinal cord (Fig. S1 D).
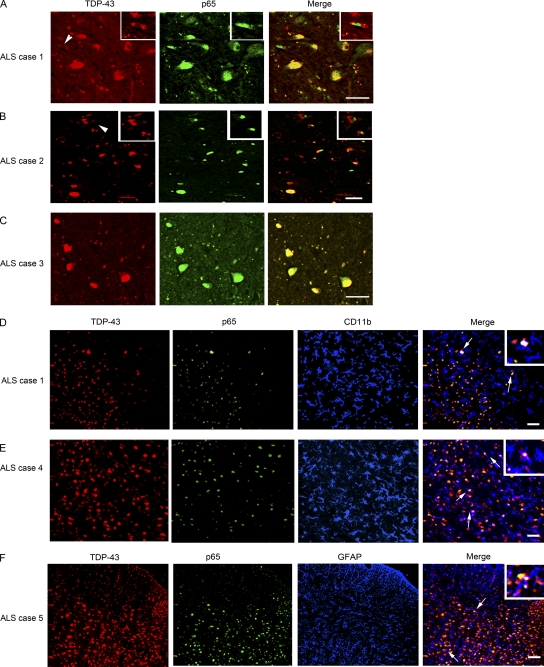
Figure 2.
TDP-43 colocalizes with p65 in neuronal and glial cells. (A–C) TDP-43 and p65 double immunofluorescence was performed in different sporadic ALS cases as indicated. Double immunofluorescence pictures were taken at various magnifications. Arrowheads represent cytoplasmic localization of TDP-43 and nuclear p65 staining. Insets of higher magnification show cytoplasmic localization of TDP-43 and nuclear p65 staining. (D and E) A three-color immunofluorescence was performed using rabbit TDP-43, mouse p65, and rat CD11b (marker for microglia) as primary antibodies and Alexa Fluor 488 (green), 594 (red), and 633 (far-red, pseudo-color blue) as secondary antibody. Insets of higher magnification show triple colocalization (white) of TDP-43–, p65-, and CD11b-positive cells (arrows). (F) A three-color immunofluorescence was performed using rabbit TDP-43, mouse p65, and rat GFAP (marker for astrocytes) as primary antibodies and Alexa Fluor 488 (green), 594 (red), and 633 (far-red, pseudo-color blue) as secondary antibody. An inset of higher magnification shows triple colocalization (white) of TDP-43–, p65-, and GFAP-positive cells (arrows). (A–F) The images shown are representative of at least four sections from two experiments from ALS patients. Bars, 20 µm.
TDP-43 acts as a co-activator of p65
A gene reporter assay was performed to study the effect of TDP-43 on NF-κB–dependent gene expression. The effect of TDP-43 was studied on gene expression of the reporter plasmid 4κBWT-luc by transfecting pCMV–TDP-43WT in BV-2 cells with or without cotransfection of pCMV-p65 (Fig. 3 A). When expressed alone, TDP-43 had no detectable effect on the basal transcription level of plasmid 4κBWT-luc, suggesting that TDP-43 does not alter the basal transcription level of NF-κB. However, in coexpression with p65, TDP-43 augmented the gene expression of plasmid 4κBWT-luc in a dose-dependent manner. 20 ng pCMV-p65 alone activated gene expression of 4κBWT-luc by 10-fold (Fig. 3 A). However, upon cotransfection with 20 ng pCMV–TDP-43WT, the extent of gene activation was elevated to 22-fold (2.2-fold augmentation by the effect of TDP-43). A further increase in NF-κB–dependent gene expression was recorded as the levels of TDP-43WT were elevated to 50 ng (2.8-fold activation) and 100 ng (3.2-fold activation; n = 4; P < 0.05). When using a control luciferase reporter construct, 4κBmut-luc, in which all four κB sites were mutated, neither the activation by pCMV-p65 nor the effect of cotransfection of pCMV–TDP-43WT was detected. The boosting effects of TDP-43 were not caused by increased levels in p65 as shown by immunoblotting (Fig. 3 B). Similarly, pCMV–TDP-43A315T and pCMV–TDP-43G348C augmented p65-mediated gene expression from the reporter plasmid 4κBWT-luc (not depicted).
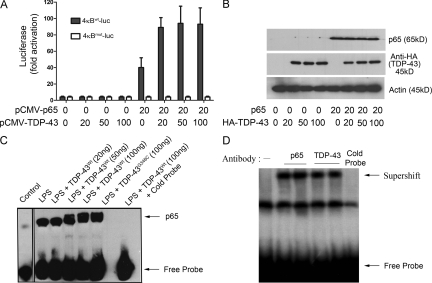
Figure 3.
TDP-43 acts as a co-activator of NF-κB p65. (A) BV-2 cells were transfected with 20 ng 4κBWT-luc (containing WT NF-κB–binding sites) or 4κBmut-luc (containing mutated NF-κB–binding sites) together with the indicated amounts of pCMV–TDP-43WT expression plasmid. Cells were harvested 48 h after transfection, and luciferase activity was measured. Values represent the luciferase activity mean ± SEM of three independent transfections, and statistical analysis was performed by two-way ANOVA with Bonferroni adjustment. TDP-43–transfected BV-2 cells were treated with 100 ng/ml LPS. (B) BV-2 cells were transfected with 20 ng pCMV-p65 and various concentrations of pCMV–TDP-43WT. TDP-43 levels are shown when blotted with anti-HA antibody (Sigma-Aldrich), and actin is shown as a loading control. (C) 48 h after transfection, BV-2 cells were harvested, and nuclear extracts were then incubated with NF-κB p65–binding site–specific oligonucleotides coated with streptavidin. EMSA was then performed using the NF-κB EMSA kit. The specificity of the assay was ascertained by adding cold probe. The control lane was performed on a separate EMSA experiment and added. EMSA shown is a representative image of two independent experiments. (D) Supershift assay was performed by adding anti-HA antibody, which specifically recognizes human TDP-43, during the EMSA assay. p65 antibody was also added in a separate lane as a positive control. Note that all the samples were TDP-43 and p65 transfected and LPS stimulated. Supershift EMSA shown is a representative image of two independent experiments.
To further examine the effect of TDP-43 on the activation of p65, we performed p65 electrophoretic mobility shift assays (EMSAs). Transfection in BV-2 cells of pCMV-p65 with pCMV–TDP-43WT or pCMV–TDP-43G348C and LPS treatment was followed by extraction of nuclear proteins. Subsequently, the interaction between p65 in the protein extract and DNA probe was investigated using the EMSA kit from Panomics according to the manufacturer’s instructions. TDP-43 increased the binding of p65 to the NF-κB DNA probe in a dose-dependent manner. LPS alone induced the binding of p65 to the DNA probe by about twofold as compared with control (Fig. 3 C). The cotransfection of 50 and 100 ng TDP-43WT or of 100 ng TDP-43G348C resulted in a significant dose-dependent increase in the DNA binding of p65. The specificity of the gel shift assay was assessed by adding a cold probe. TDP-43 alone did not bind to p65 EMSA probes (Fig. S1 B). Moreover, adding an anti-HA antibody that recognizes the transfected TDP-43 or an anti-p65 antibody caused supershifts of bands in the p65 EMSA (Fig. 3 D). Along with p65 and TDP-43, p50 is also part of the activated complex as seen by supershifts of bands in p65 EMSA experiments in BV-2 cells using antibodies specific to p65, TDP-43, and p50 (Fig. S1 C).
p65 interacts with the N-terminal and RRM-1 domains of TDP-43
To determine which domains of TDP-43 interact with p65, we constructed a series of deletion mutants of various TDP-43 domains. Various pCMV-HA–tagged deletion mutants like TDP-43ΔN (1–105 aa), TDP-43ΔRRM-1 (106–176 aa), TDP-43ΔRRM-2 (191–262 aa), and TDP-43ΔC (274–414 aa) were transfected in BV-2 cells with pCMV-p65 (Fig. 4 A). TDP-43ΔRRM-1 coimmunoprecipitated p65 partially, whereas TDP-43ΔRRM-2 and TDP-43ΔC interacted well with p65, suggesting that RRM-1 is important but RRM-2 and C-terminal domains are dispensable for interaction with p65. After transfection, we found that TDP-43ΔN had much reduced interaction with p65 (Fig. 4 B), thereby suggesting that the N-terminal domain of TDP-43 is essential for the interaction of TDP-43 with p65. Because the nuclear localization signal is in the N terminus, the reduced interaction of TDP-43ΔN to p65 could have been the result of a mislocalization of TDP-43ΔN. To address this issue and to further define the interacting domain, we constructed a series of N-terminal and RRM-1 deletion mutants, TDP-43ΔNR1-81 (98–176 aa), TDP-43ΔNR1-50 (51–81 and 98–176 aa), and TDP-43ΔNR1-30 (31–81 and 98–176 aa), with the nuclear localization signal attached so that the mutant proteins are able to be directed to the nucleus. Coimmunoprecipitation with these constructs suggested that even though TDP-43ΔNR1-30 is in the nucleus (Fig. 4 C), it cannot effectively interact with p65, TDP-43ΔNR1-81, and TDP-43ΔNR1-50, whereas it can interact with p65 (Fig. 4 B). These results indicate that TDP-43 interacts with p65 through its N-terminal domain (31–81 and 98–106 aa) and RRM-1 (107–176 aa) domain.
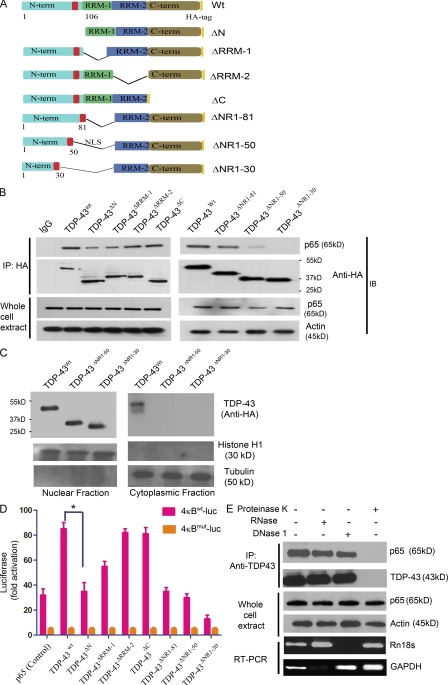
Figure 4.
The N-terminal and RRM-1 domains of TDP-43 are crucial for interaction with p65. (A) Two-dimensional cartoon of TDP-43 protein showing various deletion mutants used in this study. Deletion mutants TDP-43ΔN (1–105 aa), TDP-43ΔRRM-1 (106–176 aa), TDP-43ΔRRM-2 (191–262 aa), and TDP-43ΔC (274–414 aa) and full-length TDP-43 (TDP-43WT) are shown. Serial N-terminal and RRM-1 domain deletion mutants are also shown. TDP-43ΔNR1-81 (98–176 aa), TDP-43ΔNR1-50 (51–81 and 98–176 aa), and TDP-43ΔNR1-30 (31–81 and 98–176 aa) were generated. (B) All constructs (WT and deletion mutants) were cloned in pcDNA3.0 with HA tag at the extreme C terminus of the encoded protein. BV-2 cells were transfected with TDP-43WT or deletion constructs and pCMV-p65. 24 h after transfection, cells were harvested and immunoprecipitated (IP) with anti-HA antibody. Immunoprecipitates or whole cells extracts were subjected to immunoblot (IB) with the indicated antibodies. A representative gel from three independent experiments is shown. (C) BV-2 cells transfected with TDP-43WT, TDP-43ΔNR1-50, or TDP-43ΔNR1-30 were fractionated into nuclear and cytoplasmic fractions using sucrose density gradient centrifugation. These fractions were then probed with anti-HA antibody for the expression of transfected TDP-43 species. Histone H1 is used as a nuclear and tubulin as a cytoplasmic marker. A representative gel from two independent experiments is shown. (D) Various deletion mutants of TDP-43 were cotransfected along with 4κBWT-luc (containing WT NF-κB–binding sites) or 4κBmut-luc (containing mutated NF-κB–binding sites). 48 h after transfection, luciferase activity was measured. Statistical analysis was performed by two-way ANOVA with Bonferroni adjustment (*, P < 0.05). Error bars represent mean ± SEM from three independent experiments. (E) TDP-43 antibody was added to BV-2–transfected cell lysates, and proteins were immunoprecipitated with the indicated antibody. After TDP-43 immunoprecipitation, samples were treated with 1 µg/ml proteinase K, 1 µg/ml RNase, or 1 µg/ml DNase 1. To monitor the effectiveness of RNase and DNase digestion, RNase or DNase was added to cell lysates before immunoprecipitation and subjected to PCR. GAPDH RT-PCR was used to monitor RNase digestion, whereas Rn18s gene (which codes for 18SrRNA) genomic PCR was used to monitor DNase digestion. Representative blots and gels from three different experiments are shown.
To assess the effect of these deletion mutants on the activation of NF-κB gene, we used the gene reporter assay. Various deletion mutants of TDP-43 were cotransfected along with 4κBWT-luc or 4κBmut-luc. When compared with full-length TDP-43WT, TDP-43ΔN had reduced effect (twofold; n = 3; P < 0.05) on the gene activation. TDP-43ΔRRM-1 also exhibited attenuation of gene activation but to a lesser extent than TDP-43ΔN (Fig. 4 D). In contrast, TDP-43ΔRRM-2 and TDP-43ΔC deletion mutants had effects similar to full-length TDP-43WT. As expected, because TDP-43ΔNR1-30 does not effectively interact with p65, the level of NF-κB activation detected by the 4κBWT-luc reporter assay was extremely low, sixfold lower than full-length TDP-43WT (n = 3; P < 0.001; Fig. 4 D). p65 and luciferase vectors were used as controls for the experiment. Note that the amount of pCMV-p65 vector transfected in control was more than in other experiments to keep similar amounts of total transfected DNA. Transfection of a control luciferase reporter construct, 4κBmut-luc, in which all four κB sites were mutated, had no effect on the basal level activation of p65. To determine whether the interaction between TDP-43 and p65 is a protein–protein interaction, we performed immunoprecipitation experiments by adding proteinase K, RNase A, or DNase 1 (Fig. 4 E). The addition of proteinase K abolished the TDP-43–p65 interaction, whereas RNase A or DNase 1 had no effect, suggesting that the interaction is not DNA/RNA dependent.
TDP-43 small interfering RNA (siRNA) inhibits activation of NF-κB
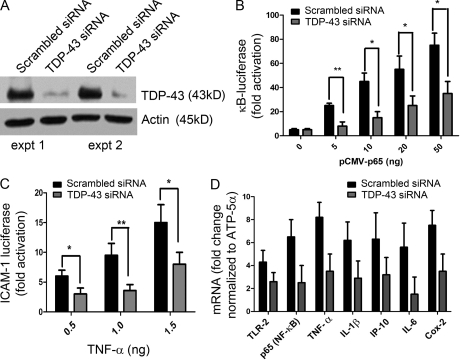
If it is correct that TDP-43 acts as a co-activator of p65, then reducing the levels of TDP-43 should attenuate p65 activation. To reduce the expression levels of TDP-43, microglial BV-2 cells were transfected with either TDP-43 siRNA or scrambled siRNA together with 4κBWT-luc vectors. 72 h after transfection, some of the cells were either stimulated with 100 ng/ml LPS or mock stimulated for 12 h. As shown in Fig. 5 A, TDP-43 siRNA reduced the endogenous mouse TDP-43 levels significantly as compared with scrambled siRNA–transfected cells in two different experiments. To examine the effect of reducing TDP-43 levels on NF-κB activation, BV-2 cells were transfected with pCMV-p65 and 4κBWT-luc vectors. TDP-43 siRNA decreased activation of NF-κB reporter gene in transfected cells. The decrease in NF-κB activation was about threefold for 5 ng pCMV-p65 (n = 4; P < 0.01), ∼2.5-fold for 10 and 20 ng pCMV-p65 (n = 4; P < 0.05), and twofold for 50 ng pCMV-p65 (n = 4; P < 0.05) as compared with scrambled siRNA–transfected cells (Fig. 5 B). To examine the physiological significance of TDP-43 inhibition by siRNA, we transfected BV-2 cells with ICAM-1–luc vector together with TDP-43 siRNA or scrambled siRNA. 72 h after transfection, cells were stimulated with varying concentrations of TNF. When stimulated with 0.5 ng/ml TNF, TDP-43 siRNA–transfected cells exhibited a twofold decrease in ICAM-1 luciferase activity (n = 4; P < 0.05) as compared with cells transfected with scrambled siRNA. Similarly, TDP-43 siRNA–transfected BV-2 cells exhibited at 1.0- and 1.5-ng/ml TNF concentrations a decrease of 2.5-fold (n = 4; P < 0.01) and twofold (n = 4; P < 0.05) in ICAM-1 luciferase activity, respectively (Fig. 5 C). We also tested the effect of TDP-43 siRNA transfected in BM-derived macrophages (BMMs) from normal mice. We compared the level of innate immunity activation when stimulated with LPS. BMMs transfected with TDP-43 siRNA had reduced levels of TLR2 mRNA (1.5-fold; P < 0.05), p65 (threefold; P < 0.01), TNF (threefold; P < 0.01), IL-1β (twofold; P < 0.05), IP-10 (twofold; P < 0.05), IL-6 (2.5-fold; P < 0.01), and Cox-2 (cyclooxygenase-2; twofold; P < 0.05) as compared with scrambled siRNA–transfected BMMs (Fig. 5 D).
Figure 5.
TDP-43 siRNA inhibits activation of NF-κB. BV-2 cells were transfected either with mouse TDP-43 siRNA or scrambled siRNA. 72 h after transfection, some of the cells were either stimulated with 100 ng/ml LPS or mock stimulated for 12 h. (A) Protein extracted from the siRNA experiment was subjected to Western blot analysis. Mouse endogenous TDP-43 levels in TDP-43 siRNA or scrambled siRNA were compared in two different experiments (expt 1 and expt 2) as determined by rabbit polyclonal TDP-43 antibody. (B) Additionally, BV-2 cells were transfected with pCMV-p65 (concentrations as indicated) and 4κBWT-luc vector, and luciferase assay was performed. (C) We transfected BV-2 cells with ICAM-1–luc vector in addition to TDP-43 siRNA or scrambled siRNA in three different experiments. 72 h after transfection, cells were stimulated with varying concentrations (as indicated) of TNF. (D) Real-time quantitative PCR levels of various mRNAs were compared with TDP-43 siRNA–transfected (and LPS stimulated) BMMs and scrambled siRNA–transfected (and LPS stimulated) BMMs. (B-D) Statistical analysis was performed by two-way ANOVA with Bonferroni adjustment (*, P < 0.05; **, P < 0.01). Error bars represent mean ± SEM from three different experiments.
TDP-43 and p65 mRNA levels are up-regulated in the spinal cord of sporadic ALS patients
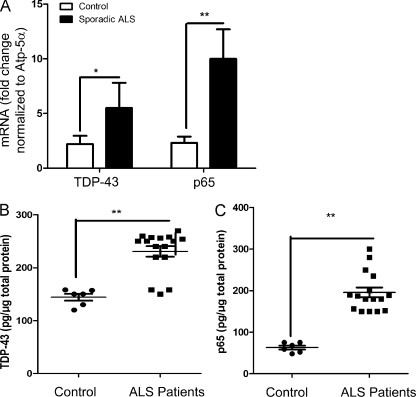
The findings that TDP-43 can interact with p65 and that TDP-43 overexpression in transgenic mice was sufficient to provoke abnormal nuclear colocalization of p65 as observed in sporadic ALS (Fig. 1 D) prompted us to compare the levels of mRNA coding for TDP-43 and p65 NF-κB in spinal cord samples from sporadic ALS cases and control individuals. Real-time RT-PCR data revealed that the levels of TDP-43 mRNA in the spinal cord of sporadic ALS cases (n = 16) were up-regulated by ∼2.5-fold (P < 0.01) compared with controls (n = 6; Fig. 6 A). It is also noteworthy that the levels of p65 NF-κB mRNA were up-regulated by about fourfold (P < 0.001) in ALS cases as compared with controls. Because TDP-43 forms many bands in Western blot analysis, we quantified the total level of TDP-43 protein using sandwich ELISA as described in Materials and methods. The ELISA results suggest that TDP-43 protein levels are in fact up-regulated in total spinal cord protein extracts of ALS cases (n = 16) by 1.82-fold (241.2 ± 8.5 pg/µg of total protein) as compared with control cases (132.8 ± 5.6 pg/µg of total protein; n = 6; Fig. 6 B). For human p65 ELISA, we used an ELISA kit from QIAGEN. The levels of p65 were also up-regulated in total spinal cord extracts of ALS cases (n = 16) by 3.5-fold (222.5 ± 11.5 pg/µg of total protein) as compared with control cases (62.83 ± 3.8 pg/µg of total protein; n = 6; Fig. 6 C).
Figure 6.
Analysis of TDP-43 and NF-κB p65 mRNA expression in sporadic ALS spinal cord. (A) Spinal cord tissue samples from 16 different sporadic ALS patients and 6 controls were subjected to real-time RT-PCR analysis using primers specific for TDP-43 (TARDBP) and p65 (RELA). All real-time RT-PCR values are normalized to Atp-5α levels. (B) Sandwich ELISA was performed for TDP-43 using TDP-43 monoclonal and polyclonal antibodies. After coating the ELISA plates with TDP-43 monoclonal antibody, the plates were incubated with the protein lysates (containing both soluble and insoluble fragments in between) followed by TDP-43 polyclonal antibody and subsequent detection. (C) For p65 ELISA, an ELISA kit from QIAGEN was used. (A–C) Statistical analysis was performed using the unpaired Student’s t test with Welch’s correction (*, P < 0.01; **, P < 0.001). Error bars represent mean ± SEM from three different experiments.
TDP-43 overexpression in glia or macrophages causes hyperactive inflammatory responses to LPS
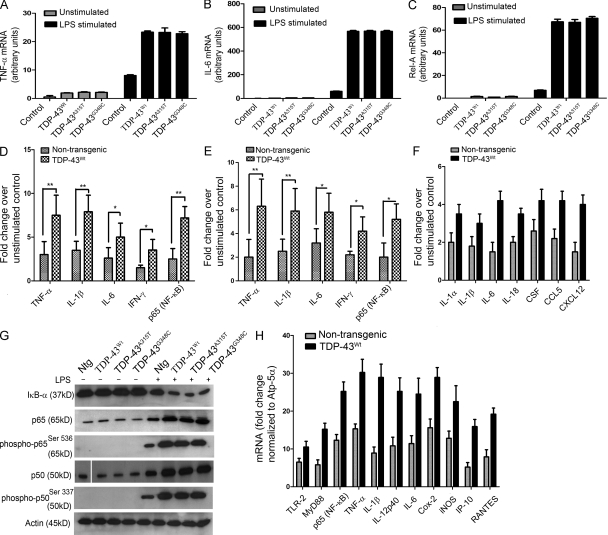
Because NF-κB is involved in proinflammatory and innate immunity response, we tested the effects of increasing TDP-43 mRNA expression in BV-2 cells. Because LPS is a strong proinflammatory stimulator (Horvath et al., 2008), we used it to determine the differences in levels of proinflammatory cytokines produced by TDP-43–transfected or mock-transfected BV-2 cells. BV-2 cells were transiently transfected with pCMV–TDP-43WT, pCMV–TDP-43A315T, pCMV–TDP-43G348C, or empty vector. 48 h after transfection and 12 h after 100-ng/ml LPS challenge, RNA extracted from various samples was subjected to real-time quantitative RT-PCR to determine the mRNA levels of various proinflammatory genes. As expected, there was a fourfold increase in mRNA levels of TNF after LPS stimulation of BV-2 cells compared with controls (Fig. 7 A). However, in LPS-treated cells transfected with WT TDP-43, there was an additional threefold (n = 5; P < 0.05) increase in TNF levels. TDP-43 harboring the A315T and G348C mutations had similar effects on boosting the levels of TNF upon LPS stimulation. Similarly, in response to LPS, the extra levels of TDP-43 species in transfected microglial cells caused a significant fivefold increase (n = 5; P < 0.001) in the mRNA levels of IL-1β (Fig. 8 A) and ninefold increase in mRNA levels of IL-6 (n = 5; P < 0.001; Fig. 7 B) as compared with LPS-treated mock-transfected cells. The levels of NADPH oxidase 2 (Nox-2 gene) was increased by ∼2.8-fold (n = 5; P < 0.05; Fig. 8 B) in LPS-challenged TDP-43–transfected cells as compared with LPS-treated mock-transfected cells. Remarkably, overexpression of TDP-43 species resulted in a 10-fold (n = 5; P < 0.001) increase in levels of p65 (RELA) mRNA in LPS-treated transfected cells as compared with LPS-treated mock-transfected cells (Fig. 7 C). Note that, in the absence of LPS stimulation, microglial cells transfected with TDP-43 species (both WT and mutants) exhibited no significant differences in levels of TNF, IL-1β, Nox-2, and NF-κB when compared with mock-transfected controls.
Figure 7.
Analysis of genes involved in inflammation of mouse microglial and macrophage cells overexpressing human TDP-43. (A–C) Mouse microglial cells BV-2 were either transfected with pCMV–TDP-43WT, pCMV–TDP-43A315T, and pCMV–TDP-43G348C or with empty vectors for 48 h. These cells were then either stimulated with LPS at a concentration of 100 ng/ml or unstimulated (as indicated). 12 h after stimulation, total RNA was extracted with TRIZOL. The total RNA samples were then subjected to real-time quantitative RT-PCR for TNF (A), IL-6 (B), and Rel-A (p65; C). Error bars represent mean ± SEM from five different experiments. (D) Primary microglial cultures from TDP-43WT and B6 nontransgenic mice were stimulated with 100 ng/ml LPS. Proteins from LPS-stimulated microglial cultures were subjected to multianalyte ELISA for inflammatory cytokines and p65. Error bars represent mean ± SEM from four different experiments. (E) Primary microglial cultures from TDP-43WT and B6 nontransgenic mice were treated with 1 mM H2O2 for 1 h and incubated in serum-free media for 12 h to study the effect of ROS. (F) Pure (>90%) primary astrocytes from TDP-43WT and B6 nontransgenic mice were stimulated with LPS and their response studied using real-time PCR for various genes as indicated. (E and F) Error bars represent mean ± SEM from three different experiments. (G) Primary microglial cells from TDP-43WT, TDP-43A315T, TDP-43G348C, and B6 nontransgenic mice (Ntg) were stimulated or unstimulated with LPS. Immunoblots were run to determine the levels of various proteins using specific antibodies as indicated. A representative blot from two independent experiments is shown. (H) BMMs isolated from TDP-43WT and B6 nontransgenic mice were stimulated with 100 ng/ml LPS for 12 h. Total RNA samples were then subjected to real-time quantitative RT-PCR for various genes as indicated. Results are displayed as fold change over unstimulated control. All real-time RT-PCR values are normalized to Atp-5α levels. Error bars represent mean ± SEM from four different experiments. (A–F and H) Statistical analysis was performed by two-way ANOVA with Bonferroni adjustment (*, P < 0.05; **, P < 0.01).
Figure 8.
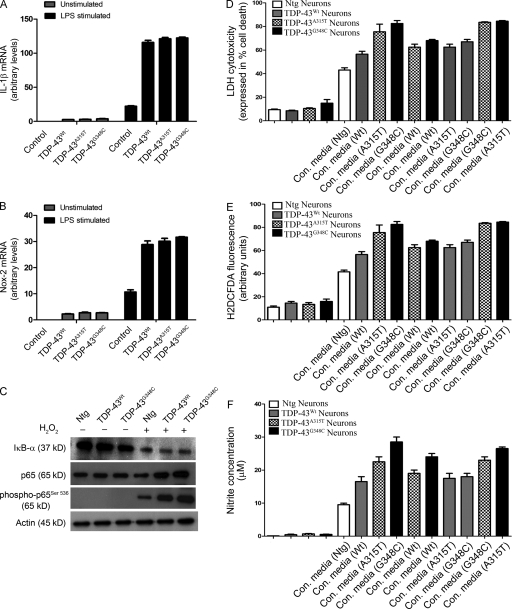
TDP-43 up-regulation enhances neuronal vulnerability to death by microglia-mediated cytotoxicity. (A and B) TDP-43 (WT and mutants)–transfected BV-2 cells were stimulated with LPS. 12 h after stimulation, total RNA was extracted with TRIZOL. The total RNA samples were then subjected to real-time quantitative RT-PCR for IL-1β (A) and Nox-2 (B). Error bars represent mean ± SEM from five different experiments. Statistical analysis was performed by two-way ANOVA with Bonferroni adjustment. (C) Primary microglial cells from TDP-43WT, TDP-43A315T, TDP-43G348C, and B6 nontransgenic mice (Ntg) were stimulated or unstimulated with H2O2. Immunoblots were run to determine the levels of various proteins using specific antibodies as indicated. A representative blot from two independent experiments is shown. (D–F) Primary cortical neurons from TDP-43WT, TDP-43A315T, TDP-43G348C, and control B6 nontransgenic mice were incubated with the conditioned media (con. media) derived from primary microglial cells treated with 100 ng/ml LPS. 12 h after challenging cortical cells, cell culture supernatants were used for LDH assay (D). ROS production was determined by H2DCFDA fluorescence (E), and nitrite production was evaluated by Griess reagent (F). Error bars represent mean ± SEM from four independent experiments.
To further evaluate the effect of LPS stimulation in TDP-43–overexpressing microglia, we prepared primary microglial cultures from C57BL/6 mice and from transgenic mice overexpressing TDP-43WT by threefold. Primary microglial cells were challenged with LPS at a concentration of 100 ng/ml of media. 12 h after LPS challenge, cells were harvested, and total protein was extracted and used for multianalyte ELISA. LPS-treated TDP-43WT transgenic microglia had significantly higher levels of TNF (2.5-fold; P < 0.01), IL-1β (2.3-fold; P < 0.01), IL-6 (twofold; P < 0.05), and IFN-γ (twofold; P < 0.05) as compared with LPS-treated microglia from C57BL/6 nontransgenic mice (Fig. 7 D). However, in the absence of LPS stimulation, no significant differences in cytokines levels were detected between microglia from TDP-43WT transgenic mice and from nontransgenic mice (not depicted). The p65 level was significantly higher (threefold; P < 0.01) in LPS-treated TDP-43WT microglia as compared with nontransgenic microglia (Fig. 7 D). We also treated primary microglial cultures with 1 mM H2O2 for 1 h (and incubated in serum-free media for 12 h) to study the effect of reactive oxygen species (ROS) on primary microglial cultures. H2O2-treated TDP-43WT transgenic microglia had significantly higher levels of TNF (threefold; P < 0.01), IL-1β (2.5-fold; P < 0.01), IL-6 (1.7-fold; P < 0.05), IFN-γ (twofold; P < 0.05), and p65 (RELA) levels (2.2-fold; P < 0.05) when compared with H2O2-treated microglia from C57BL/6 nontransgenic mice (Fig. 7 E) as determined by multianalyte ELISA.
LPS stimulation of primary microglial cells caused degradation of IκB-α as shown in Fig. 7 G. The decrease in IκB-α levels was more pronounced in microglia overexpressing TDP-43 species. After LPS treatment, the increases in levels of p65, phospho-p65Ser536, p50, and phospho-p50Ser337 were also more robust in transgenic microglia overexpressing TDP-43 species (Fig. 7 G). Similarly, H2O2 treatment led to a reduction in IκB-α levels and increase in levels of p65 and phospho-p65Ser536 in TDP-43WT (Fig. 8 C). Again, the effects were more pronounced in transgenic microglia overexpressing TDP-43 species (Fig. 8 C). We then treated primary astrocytes with LPS and studied their response to LPS using real-time RT-PCR. LPS-treated TDP-43WT transgenic astrocytes had significantly higher levels of IL-α (1.75-fold; P < 0.05), IL-1β (1.67-fold; P < 0.05), IL-6 (2.8-fold; P < 0.01), IL-18 (1.8-fold; P < 0.05), and chemokines like CSF (1.6-fold; P < 0.05), CCL5 (1.9-fold; P < 0.05), and CXCL12 (2.67-fold; P < 0.01) as compared with LPS-treated microglia from C57BL/6 nontransgenic mice (Fig. 7 F).
To further evaluate the innate immune response in TDP-43WT transgenic mice, we isolated BMMs from TDP-43WT transgenic mice and from C57BL/6 nontransgenic mice. In LPS-stimulated TDP-43WT macrophages, there was an increase of 1.6-fold (P < 0.05) in TLR2 mRNA levels, 1.8-fold (P < 0.05) in MyD88 levels, and 2.6-fold (P < 0.01) in p65 (RELA; P < 0.01) levels as compared with LPS-stimulated control (nontransgenic) macrophages (Fig. 7 H). We also found in LPS-stimulated TDP-43WT macrophages that there was an increase of 3.2-fold (P < 0.01) in TNF, 3.5-fold in IL-1β (P < 0.01), and 2.6-fold in IL-12p40 levels, 2.5-fold (P < 0.01) in IL-6 levels, twofold (P < 0.05) in Cox-2 and iNOS levels, threefold in IP-10 levels (P < 0.01), and 2.1-fold in RANTES (P < 0.05) mRNA levels as compared with LPS-stimulated control (nontransgenic) macrophages (Fig. 7 H).
TDP-43 up-regulation increases microglia-mediated neurotoxicity
We then examined the effect of TDP-43 overexpression on toxicity of microglia toward neuronal cells. This was done with the use of primary microglia and of cortical neurons derived from transgenic mice overexpressing TDP-43 species (TDP-43WT, TDP-43A315T, or TDP-43G348C) and C57BL/6 nontransgenic mice. Primary cortical neurons were cultured for 12 h in conditioned media from LPS-stimulated microglial cells. All conditioned media from LPS-challenged microglia increased the death of cortical neurons in culture (Fig. 8 D). The media from LPS-stimulated nontransgenic microglial cells increased the neuronal death of nontransgenic mice by 3.5-fold (P < 0.01). However, there were marked increases of neuronal death caused by conditioned media from LPS-challenged microglia (of same genotype) overexpressing TDP-43 species: 5.5-fold (P < 0.001) for TDP-43WT, 6.5-fold (P < 0.001) for TDP-43A315T, and 7.5-fold (P < 0.001) for TDP-43G348C. The increased neurotoxicity of the conditioned media was associated with increased ROS and NO production. The ROS production, as determined by H2DCFDA fluorescence, was significantly higher in conditioned media–challenged neurons from TDP-43WT (1.5-fold; P < 0.05), TDP-43A315T (1.8-fold; P < 0.05), or TDP-43G348C (twofold; P < 0.05) as compared individually with conditioned media–challenged nontransgenic control neurons (Fig. 8 E). Similarly, the nitrite (NO) production was significantly higher in TDP-43WT (1.5-fold; P < 0.05), TDP-43A315T (2.3-fold; P < 0.05), or TDP-43G348C (threefold; P < 0.01) as compared individually with nontransgenic control (Fig. 8 F).
Inhibition of NF-κB activation reduces vulnerability of TDP-43–overexpressing neurons to toxic injury
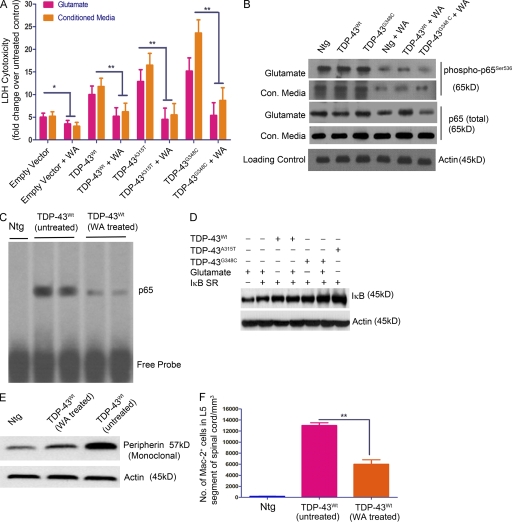
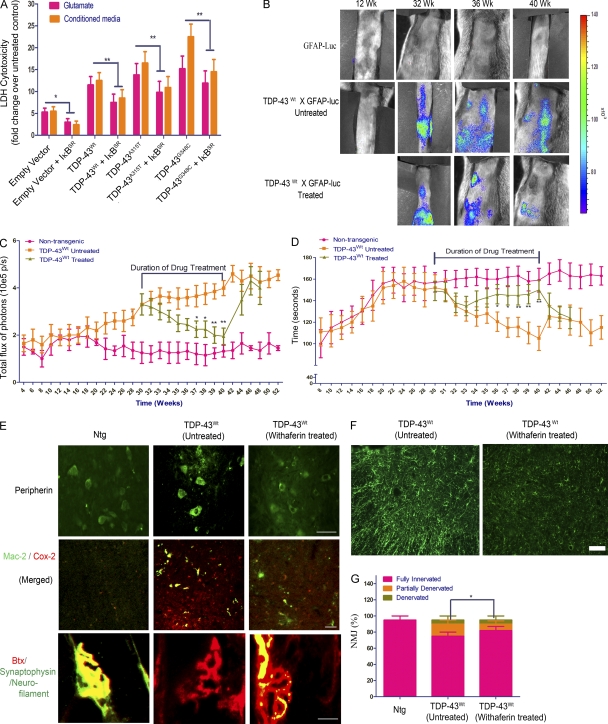
The aforementioned experiments also revealed that the presence of TDP-43 transgenes in cortical neurons increased their vulnerability to microglia-mediated toxicity. NF-κB is known to modulate p53-p38MAPK–dependent apoptosis in neurons when treated with DNA damage–inducing chemicals like camptothecin (Aleyasin et al., 2004), glutamate excitotoxicity (Pizzi et al., 2005), or general bystander-mediated killing of neurons by microglia (Sephton et al., 2010). To assess the potential contribution of NF-κB to the death of TDP-43–overexpressing neurons exposed to toxic injury, we prepared cultures of primary cortical neurons and microglia from transgenic mice overexpressing TDP-43WT or TDP-43 mutants. Cortical neurons were exposed to 10 µM glutamate for 15 min, with or without 1 µM Withaferin A (WA), a known inhibitor of NF-κB (Oh et al., 2008). The lactate dehydrogenase (LDH) cytotoxicity was determined 24 h later (see Fig. 10 A). We found that neurons overexpressing TDP-43 species were more vulnerable than nontransgenic neurons to glutamate cytotoxicity and that inhibition of NF-κB by WA resulted in a marked decrease in cell death: TDP-43WT (twofold; P < 0.01), TDP-43A315T (threefold; P < 0.01), and TDP-43G348C (threefold; P < 0.01). The addition of WA inhibited NF-κB, as detected by reduced levels of phospho-p65Ser536 (see Fig. 10 B). We then incubated cortical neurons with the conditioned media from primary microglial culture, which were challenged with LPS at a concentration of 100 ng/ml of media. Treatment of neuronal cultures with WA resulted in substantial decrease in microglia-mediated death of neurons overexpressing TDP-43WT (twofold; P < 0.01), TDP-43A315T (threefold; P < 0.01), or TDP-43G348C (threefold; P < 0.01). As WA might exert multiple pharmacological actions, we tested a more specific molecular approach for inhibiting NF-κB. Because activation of NF-κB requires its dissociation from the inhibitory molecule, IκB, we expressed a stable mutant super-repressive form of IκB-α (Ser32/Ser36 to alanine mutant; IκBSR) and evaluated its effects on neuronal death. Cultured cortical neurons from TDP-43 transgenic and nontransgenic mice were transfected with a plasmid construct, expressing IκBSR, and exposed to either 10 µM glutamate for 30 min or incubated in conditioned media from LPS-stimulated microglia of the same genotype. Similar to WA treatment, we found that IκBSR inhibited NF-κB activation and it attenuated the glutamate-induced or microglia-mediated death of neurons overexpressing TDP-43WT (1.3-fold; P < 0.01), TDP-43A315T (1.5-fold; P < 0.01), and TDP-43G348C (twofold; P < 0.01; Fig. 9, A and D).
Figure 10.
WA ameliorates TDP-43–mediated toxicity. (A) Primary cortical neurons from TDP-43WT, TDP-43A315T, TDP-43G348C, and B6 nontransgenic mice were exposed to 10 µM glutamate for 15 min or incubated in conditioned media from LPS-stimulated microglia of the same genotype with or without 1 µM WA and were evaluated for LDH cytotoxicity 24 h later. Asterisks represent a statistically significant difference between treated and untreated groups (*, P < 0.05; and **, P < 0.01) using repeated measures two-way ANOVA. Error bars represent mean ± SEM from three independent experiments. (B) Protein samples from cortical neurons (isolated from TDP-43WT, TDP-43A315T, TDP-43G348C, and B6 nontransgenic [Ntg] mice) were subjected to immunoblot against various antibodies as indicated. (C) p65 EMSA was performed on the spinal cord tissue nuclear lysates from WA-treated and untreated GFAP-luc/TDP-43WT mice. A representative EMSA of two independent experiments is shown. (D) IκB levels were measured by Western blot analysis of the cell lysates from cortical neurons of various genotypes as indicated. Actin is shown as loading control. Various conditions are also shown. (E) Western blot analysis of spinal cord sections of nontransgenic (control), TDP-43WT (untreated), and TDP-43WT (WA treated) mice with monoclonal peripherin antibody. (D and E) A representative blot from two different experiments is shown. (F) Quantification of microglial Mac-2–positive cells in the spinal cord sections of nontransgenic (control), TDP-43WT (untreated), and TDP-43WT (WA treated) mice. Mac-2+ cells in TDP-43WT (untreated) L5 spinal cord, 13,000 ± 500/mm3; and TDP-43WT (WA treated) L5 spinal cord, 6,000 ± 300/mm3 (**, P < 0.001). Error bars represent mean ± SEM for four mice of each genotype.
Figure 9.
Inhibition of NF-κB reduces neuronal vulnerability to toxic injury and ameliorates disease phenotypes in TDP-43 transgenic mice. (A) A stable mutant super-repressive form of IκB-α (IκBSR) was expressed, and its effects on neuronal death were evaluated. The phosphorylation-defective IκB-αS32A/S36A acts by sequestering the cytoplasmic NF-κB pool in a manner that is insensitive to extracellular stimuli. Cultured cortical neurons from TDP-43WT, TDP-43A315T, TDP-43G348C, and B6 nontransgenic mice were transfected with a plasmid construct, expressing IκBSR, and exposed to either 10 µM glutamate for 30 min or incubated in conditioned media from LPS-stimulated microglia of the same genotype. Cytotoxicity to the cells was measured by LDH assay using a commercially available kit. Statistical analysis was performed by two-way ANOVA with Bonferroni adjustment (*, P < 0.05; **, P < 0.01). Data represent mean ± SEM from three independent experiments. (B) In vivo bioluminescence imaging of astrocyte activation was analyzed at various time points in the spinal cord of GFAP-luc/TDP-43WT mice. Typical sequence of images of the spinal cord area obtained from of GFAP-luc/TDP-43WT mice at different time points (12, 32, 36, and 40 wk) by in vivo imaging (n = 10 each group). WA was injected in GFAP-luc/TDP-43WT for 10 wk starting at 30 wk of age until 40 wk. Representative images are shown. (C) Longitudinal quantitative analysis of the total photon GFAP signal/ bioluminescence (total flux of photon/s) in WA-treated and untreated GFAP-luc/TDP-43WT mice and control GFAP-luc mice in the spinal cord is displayed. Duration of drug treatment is indicated. (D) Accelerating rotarod analysis was performed in GFAP-luc/TDP-43WT mice at various ages from 8 wk to 52 wk, and time taken by the mice to fall from the rotarod is used as rotarod performance. WA treatment period is marked as drug treatment period. (C and D) Asterisks represent a statistically significant difference between treated and untreated groups (*, P < 0.05; and **, P < 0.01) using repeated measures two-way ANOVA. (C and D) Error bars represent mean ± SEM (n = 10 each group). (E) Immunofluorescence of spinal cord sections of nontransgenic (Ntg; control), TDP-43WT (untreated), and TDP-43WT (WA treated) mice with polyclonal peripherin antibody is shown. Double immunofluorescence of spinal cord sections with activated microglial marker Mac-2 and Cox-2 is shown. Representative images from four different mice per genotype are shown. NMJ staining was performed using anti-synaptophysin/neurofilament antibodies (green) and α-bungarotoxin (BTX; red). Representative images from four different mice per genotype showing fully innervated muscle in 10-mo-old nontransgenic mice, fully denervated muscle in TDP-43WT mice (untreated), and partially denervated muscle in age-matched WA-treated TDP-43WT mice. (F) Immunofluorescence using GFAP antibody was performed in the spinal cord sections of WA-treated and untreated GFAP-luc/TDP-43WT mice. Representative images from five different mice per genotype are shown. (G) 300 NMJs were counted per animal sample. Frequencies of innervation, partial denervation, and denervation were then converted to percentages and plotted as a graph. Statistical analysis was performed by the Student’s t test. The asterisk represents a statistically significant difference between treated and untreated groups (*, P < 0.01) using repeated measures two-way ANOVA. Error bars represent mean ± SEM from three different experiments. Bars, 20 µm.
NF-κB inhibition by WA treatment reduces inflammation and ameliorates motor impairment of TDP-43 transgenic mice
To study the in vivo effect of NF-κB inhibition on disease progression, we injected TDP-43WT;GFAP-luc double transgenic mice with 3 mg/kg body weight of WA twice a week for 10 wk starting at 30 wk. The pharmacokinetic parameters of WA have been published recently (Thaiparambil et al., 2011), and we have determined that this compound passes the blood–brain barrier (unpublished data). We used TDP-43WT;GFAP-luc double transgenic mice because the reporter luciferase allowed the longitudinal and noninvasive biophotonic imaging with charge-coupled device camera of the GFAP promoter activity, which is a target of activated NF-κB. To analyze the spatial and temporal dynamics of astrocyte activation/GFAP induction in the TDP-43 mouse model, we performed a series of live imaging experiments. These live imaging experiments revealed that treatment of TDP-43WT;GFAP-luc mice with WA caused progressive reduction in GFAP-luc expression in the spinal (Fig. 9, B and C) compared with untreated TDP-43WT mice, which continued to exhibit high GFAP-luc expression. The down-regulation of GFAP promoter activity was further confirmed in these mice using GFAP immunofluorescence of spinal cord sections of TDP-43WT mice (both drug treated and untreated; Fig. 9 F).This down-regulation of GFAP in WA-treated mice was actually caused by a reduced amount of active p65 in the nucleus of cells as indicated by p65 EMSA (Fig. 10 C). Down-regulation of GFAP along with reduction in active p65 levels in WA-treated mice prompted us to analyze behavioral changes in these mice. Analysis of motor behavior using accelerating rotarod showed that WA-treated TDP-43WT mice had significantly better motor performance compared with untreated TDP-43WT mice as indicated by improved rotarod testing times (Fig. 9 D). We performed peripherin immunofluorescence and found reduction of peripherin aggregates in WA-treated TDP-43WT mice (Fig. 9 E). Peripherin levels were also reduced in WA-treated TDP-43WT mice as seen by immunoblot (Fig. 10 E). Double immunofluorescence of activated microglial marker Mac-2 and Cox-2 showed a marked reduction in activated microglia in WA-treated TDP-43WT mice (Figs. 9 E and 10 F). The WA-treated mice also had a 40% reduction in the number of partially denervated neuromuscular junctions (NMJs; Fig. 9, E and G).
DISCUSSION
From the data presented in this study, we propose that a TDP-43 deregulation in ALS may contribute to pathogenic pathways through abnormal activation of p65 NF-κB. Several lines of evidence support this scheme: (a) proof of a direct interaction between TDP-43 and p65 NF-κB was provided by immunoprecipitation experiments using protein extracts from cultured cells, from TDP-43 transgenic mice, and from human ALS spinal cord samples; (b) reporter gene transcription assays and gel shift experiments demonstrated that TDP-43 was acting as a co-activator of p65 NF-κB through binding of its N-terminal and RRM-1 domains to p65; (c) the levels of mRNAs for both TDP-43 and p65 NF-κB were substantially elevated in the spinal cord of ALS subjects as compared with non-ALS subjects, whereas immunofluorescence microscopy of ALS spinal cord samples revealed an abnormal nuclear localization p65 NF-κB; (d) cell transfection experiments demonstrated that an overexpression of TDP-43 can provoke hyperactive innate immune responses with ensuing enhanced toxicity on neuronal cells, whereas in neurons TDP-43 overexpression increased their vulnerability to toxic environment; and (e) in vivo treatment of TDP-43 transgenic mice with an inhibitor of NF-κB reduced inflammation and ameliorated motor deficits.
This is the first report of an up-regulation of mRNAs encoding TDP-43 in postmortem frozen spinal cords of sporadic ALS. A recent study has provided evidence of increased TDP-43 immunodetection in the skin of ALS patients (Suzuki et al., 2010), but it failed to demonstrate whether this was caused by up-regulation in TDP-43 mRNA expression. The process that underlies a 2.5-fold increase in TDP-43 mRNA levels in ALS, whether it is transcriptional or mRNA stability, remains to be investigated. It seems unlikely that copy number variants could explain an increase of TDP-43 gene transcription as variations in copy number of TARDBP have not been detected in cohorts of ALS (Guerreiro et al., 2008; Bäumer et al., 2009; Gitcho et al., 2009). Actually, the pathogenic pathways of TDP-43 abnormalities in ALS are not well understood. To date, much attention has been focused on cytoplasmic C-terminal TDP-43 fragments that can elicit toxicity in cell culture systems (Johnson et al., 2008; Dormann et al., 2009; Igaz et al., 2009; Zhang et al., 2009b). However, it is noteworthy that neuronal overexpression at high levels of WT or mutant TDP-43 in transgenic mice caused a dose-dependent degeneration of cortical and spinal motor neurons but without massive cytoplasmic TDP-43 aggregates (Wils et al., 2010). This suggests that an up-regulation of TDP-43 in the nucleus rather than TDP-43 cytoplasmic aggregates may contribute to neurodegeneration in these mouse models. As shown in this study, an overexpression of TDP-43 can trigger pathogenic pathways via NF-κB activation.
The transcription factor NF-κB is a key regulator of hundreds of genes involved in innate immunity, cell survival, and inflammation. Because the nuclear translocation and DNA binding of NF-κB are not sufficient for gene induction (Yoza et al., 1996; Bergmann et al., 1998), it has been suggested that interactions with other protein molecules through the transactivation domain (Schmitz et al., 1995b; Gerritsen et al., 1997; Perkins et al., 1997) as well as its modification by phosphorylation (Schmitz et al., 1995a) might play a critical role. It has been reported that transcriptional activation of NF-κB requires multiple co-activator proteins including CBP (CREB-binding protein)/p300 (Gerritsen et al., 1997; Perkins et al., 1997), CBP-associated factor, and steroid receptor co-activator 1 (Sheppard et al., 1999). These co-activators have histone acetyltransferase activity to modify the chromatin structure and also provide molecular bridges to the basal transcriptional machinery. NF-κB p65 was also found to interact specifically with FUS (fused in sarcoma) protein, another DNA/RNA-binding protein which is involved in ALS (Kwiatkowski et al., 2009; Vance et al., 2009; Deng et al., 2010).
Our results revealed robust effects of TDP-43 on the activation of NF-κB and innate immune responses. After transfection with TDP-43 species, microglial cells challenged with LPS exhibited much higher mRNA levels for proinflammatory cytokines, Nox-2, and NF-κB mRNA when compared with untransfected cells after LPS stimulation. TDP-43 overexpression makes microglia hyperactive to immune stimulation, resulting in enhanced toxicity toward neighboring neuronal cells with involvement of ROS and increased nitrite levels (NO). Moreover, the adverse effects of TDP-43 up-regulation are not limited to microglial cells. TDP-43 overexpression in transgenic astrocytes caused exaggerated responses to LPS (Fig. 7 F), whereas primary cortical neurons overexpressing TDP-43 transgenes by approximately threefold exhibited increased susceptibility to the toxic effects of excess glutamate or LPS-activated microglia (Figs. 8 D and 9 A).
The presence of ALS-linked mutations in TDP-43 (A315T or G348C) did not affect the binding and activation of p65 NF-κB. This is not surprising because our deletion mutant analysis revealed that a region spanning part of the N-terminal domain and RRM-1 of TDP-43 is responsible for interaction with p65, whereas most TDP-43 mutations in ALS occur in the C-terminal domain, which is dispensable for p65 NF-κB activation (Fig. 4). In fact, our cytotoxicity assays with primary cells from TDP-43 transgenic mice revealed that, at similar levels of mRNA expression, the adverse effects of mutant TDP-43 were more pronounced than TDP-43WT. These results could be explained by the observation that ALS-linked mutations in TDP-43 increase its protein stability (Ling et al., 2010). From the data presented here, we propose the involvement in ALS of a pathogenic pathway caused by nuclear increase in TDP-43 levels (Fig. 6). This scheme does not exclude adverse effects caused by cytoplasmic TDP-43 aggregates that might occur concomitantly or later on during the disease process. A recent TDP-43 study with Drosophila melanogaster suggested that the TDP-43 toxicity may occur in the absence of inclusions formation and that neurotoxicity requires the TDP-43 RNA–binding domain (Voigt et al., 2010). These results are consistent with our model of TDP-43 toxicity and with data demonstrating the interaction of TDP-43 with p65 via the RNA recognition motif RMM-1.
Our finding that TDP-43 acts as co-activator of p65 suggests a key role for NF-κB signaling in ALS pathogenesis. This is corroborated by the abnormal fourfold increase of p65 NF-κB mRNA in the spinal cord of human ALS (Fig. 6) and by the nuclear localization of p65 (Fig. 1 D and Fig. 2, insets). Remarkably, an overexpression of TDP-43 species by approximately threefold in transgenic mice (Swarup et al., 2011), at levels similar to the human ALS situation (2.5-fold), was sufficient to cause nuclear translocation of p65 NF-κB in the spinal cord during aging (Fig. 1 D). It should be noted that TDP-43 itself does not cause NF-κB activation (Fig. 7) and that it does not up-regulate p65. It seems that a second hit is required. For example, LPS or other inducers such as pathogen-associated molecular patterns can trigger through TLR signaling p65 NF-κB nuclear localization. Cytokines such as TNF and IL-1β can also trigger p65 activation. In ALS, the second hits triggering innate immune responses remain to be identified. There is recent evidence for involvement of LPS in ALS (Zhang et al., 2009a, 2011) and of endogenous retrovirus (HEVR-K) expression (Douville et al., 2011). In this study, we show that aging is associated with p65 nuclear translocation in the spinal cord of TDP-43 transgenic mice (Fig. S1 D), but the exact factors underlying this phenomenon remain to be defined
There is a recent report of mutations in the gene coding for vasolin-containing protein (VCP) associated with 1–2% of familial ALS cases (Johnson et al., 2010). It is well established that VCP is involved in the control of the NF-κB pathway through regulation of ubiquitin-dependent degradation of IκB-α. For instance, mutant VCP expression in mice resulted in increased TDP-43 levels and hyperactivation of NF-κB signaling (Badadani et al., 2010; Custer et al., 2010). Moreover, some ALS-linked mutations have been discovered in the gene coding for optineurin, a protein which activates the suppressor of NF-κB (Maruyama et al., 2010), further supporting a convergent NF-κB pathogenic pathway. Thus, the data presented in our paper as well as ALS-linked mutations in the VCP and optineurin genes (Badadani et al., 2010; Johnson et al., 2010; Maruyama et al., 2010) are all supporting a convergent NF-κB pathogenic pathway in ALS. Recently, the NF-κB signaling complex was identified as a major contributor of astrocyte mediated toxicity to motor neurons (Haidet-Phillips et al., 2011). In this study, we show that inhibitors of NF-κB activation are able to attenuate the vulnerability of cultured neurons overexpressing TDP-43 species to glutamate-induced or microglia-mediated toxicity. Moreover, pharmacological inhibition of NF-κB by WA treatment attenuated disease phenotypes in TDP-43 transgenic mice. From these results, we propose that NF-κB signaling should be considered as a potential therapeutic target in ALS treatment.
MATERIALS AND METHODS
Human subjects.
The spinal cords of 16 subjects with sporadic ALS and 6 control cases were used in this study. The diagnosis of ALS was made on both clinical and pathological grounds. The ages at death ranged from 42 to 79 yr, and the duration of illness ranged from 21 to 48 mo (Table S3). TDP-43–positive inclusions were found in all ALS cases. We also used spinal cord samples from six neurologically normal individuals (normal controls), aged between 55 and 84 yr. For routine histological examination, the spinal cord of each subject was fixed with 10% buffered formalin for 3 wk and then embedded in paraffin; 4-µm-thick sections were cut and stained with hematoxylin. The use of the human tissue samples described in this article was performed in accordance to the Committee on Research Ethics of Enfant-Jesus Hospital.
Generation of TDP-43 transgenic mice.
TARDBP (GenBank/EMBL/DDBJ accession no. NM_007375) was amplified by PCR from a human BAC clone (clone RPCI-11, clone number 829B14) along with the endogenous promoter (∼4 kB). A315T and G348C mutations in TDP-43 were inserted using site-directed mutagenesis. The full-length genomic TARDBP (TDP-43WT and TDP-43G348C) was linearized by Swa1 restriction enzyme and 18-kb DNA fragment microinjected in 1-d-old mouse embryos (having a background of C3H × C57BL/6). The embryos were implanted in pseudo-pregnant mothers (having ICR CD1 background). Founders were bred with nontransgenic C57BL/6 mice to establish stable transgenic lines (Swarup et al., 2011). Transgene expression was analyzed in brain and spinal cord by real-time PCR and in brain, spinal cord, muscle, and liver by Western blotting using monoclonal human TDP-43 antibody (clone E2-D3; Abnova). All experimental procedures were approved by the Laval University Animal Care Ethics Committee and are in accordance with the Guide to the Care and Use of Experimental Animals of the Canadian Council on Animal Care.
WA treatment.
WA (Enzo Life Sciences) was injected intraperitoneally twice a week for 10 consecutive weeks at 3 mg/kg body weight in 30-wk-old TDP-43WT mice (n = 10). Age-matched control nontransgenic animals (n = 10) and TDP-43WT (n = 10) littermates were injected twice a week with 0.9% saline intraperitoneally. All of the behavioral and imaging experiments were conducted in a double blind manner, and as such the experimenter had no knowledge of the drug treatment or the genotype of animals.
Plasmids.
Mammalian expression vector plasmids pCMV-p65, pCMV-p50, and ICAM1–luc (positions −340 to −25) and luciferase reporter plasmids 4κBWT-luc or 4κBmut-luc, containing four tandem copies of the human immunodeficiency virus-κB sequence upstream of minimal SV40 promoter, and mutant IκB-α (IκBSR), containing Ser32 and Ser36 to alanine mutations, were gifts from the laboratory of M.J. Tremblay (Centre de Recherche du Centre Hospitalier Universitaire de Quebec, Quebec City, Quebec, Canada). To create a human pCMV–TDP-43, the cDNA library from human myeloid cells was amplified by PCR using primers as described in Table S1. These products were subcloned into TOPO vector (Invitrogen) and later digested with Kpn1–BamHI restriction enzymes and subcloned in frame into pcDNA3.0 vector to form pCMV–TDP-43WT. The HA tag was later added by PCR. HA-tagged TDP-43ΔN, TDP-43ΔRRM-1, TDP-43ΔRRM-2, and TDP-43ΔC deletion mutants were constructed by PCR amplification and cloned between Kpn1–BamHI sites using the primers described in Table S1. Point mutations (pCMV–TDP-43A315T and pCMV–TDP-43G348C) were inserted by PCR using site-directed mutagenesis.
Cell culture and transfection.
Mouse microglial BV-2 and mouse neuroblastoma N2a cells were maintained in DME (Invitrogen) with 10% FBS and antibiotics. Cells were transfected using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. At 48 h after transfection, the cells were harvested, and the extracts were prepared for downstream assays.
Primary cell cultures.
Primary microglial culture from brain tissues of neonatal (P0-P1) C57BL/6, TDP-43WT, TDP-43A315T, and TDP-43G348C mice were prepared as described previously (Weydt et al., 2004). In brief, the brain tissues were stripped of their meninges and minced with scissors under a dissecting microscope in DME. After trypsinization (0.5% trypsin, 10 min, 37°C/5% CO2), the tissue was triturated. The cell suspension was washed in culture medium for glial cells (DME supplemented with 10% FBS [Invitrogen], 1 mMl-glutamine, 1 mM Na pyruvate, 100 U/ml penicillin, and 100 mg/ml streptomycin) and cultured at 37°C/5% CO2 in 75-cm2 Falcon tissue culture flasks (BD) coated with 10 mg/ml poly-d-lysine (PDL; Sigma-Aldrich) in borate buffer (2.37 g borax and 1.55 g boric acid dissolved in 500 ml of sterile water, pH 8.4) for 1 h and then rinsed thoroughly with sterile, glass-distilled water. Half of the medium was changed after 6 h in culture and every second day thereafter, starting on day 2, for a total culture time of 10–14 d. Microglia were shaken off the primary mixed brain glial cell cultures (150 rpm, 37°C, 6 h) with maximum yields between days 12 and 16, seeded (105 cells per milliliter) onto PDL-pretreated 24-well plates (1 ml per well), and grown in culture medium for microglia (DME supplemented with 10% FBS, 1 mMl-glutamine, 1 mM Na pyruvate, 50 mM 2-mercaptoethanol, 100 U/ml penicillin, and 100 mg/ml streptomycin). The cells were allowed to adhere to the surface of a PDL-coated culture flask (30 min, 37°C/5% CO2). After removal of primary microglial culture, the remaining cells were mainly astrocytes. Purity of the astrocytes was >90%. Astrocytes were maintained in a medium consisting of DME supplemented with 10% FBS, 1 mMl-glutamine, 1 mM Na pyruvate, 50 mM 2-mercaptoethanol, 100 U/ml penicillin, and 100 mg/ml streptomycin. Primary cortical cultures from brain tissues of gestation day 16 (E16) C57BL/6, TDP-43WT, TDP-43A315T, and TDP-43G348C mice were prepared as described previously (Hilgenberg and Smith, 2007). In brief, dissociated cortical cells (2.5–3.5 hemispheres) were plated onto PDL-coated 24-well plates, containing DME supplemented with 20 mM glucose, 2 mM glutamine, 5% FBS, and 5% horse serum. Cytosine arabinoside was added 4–5 d after the plating to halt the growth of nonneuronal cells. Cultures were maintained at 37°C in a humidified CO2 incubator and used for experiments between 14 and 21 d in vitro. Cells were treated with WA at a final concentration of 1 µM for 24 h. BMMs were isolated and cultured using established protocols as described previously (Davies and Gordon, 2005).
Coimmunoprecipitation and Western blot assays.
After transfection of plasmids, BV-2 cells were cultured for 48 h and then harvested with lysis buffer (25 mM Hepes-NaOH, pH 7.9, 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5% Triton X-100, 1 mM dithiothreitol, and protease inhibitor cocktail). Alternatively, spinal cords from TDP-43 transgenic mice or sporadic ALS subjects along with controls were lysed in the buffer. The lysate was incubated with 50 µl Dynabeads (protein G beads; Invitrogen), anti–TDP-43 polyclonal (ProteinTech), and anti-HA antibody (clone 3F10; Roche). After subsequent washing, the beads were incubated overnight at 4°C with 400 µg of cell lysate. Antibody-bound complexes were eluted by boiling in Laemmli sample buffer. Supernatants were resolved by 10% SDS-PAGE and transferred on nitrocellulose membrane (Bio-Rad Laboratories). The membrane was incubated with anti-p65 antibody, and immunoreactive proteins were visualized by chemiluminescence (PerkinElmer) as described previously (Dequen et al., 2008). In some cases, phospho-p65Ser536 (Cell Signaling Technology) and phospho-p50337 (Santa Cruz Biotechnology, Inc.) were used at a concentration of 1:1,000.
Mass spectrometer analysis.
BV-2 microglial cells were transiently transfected with plasmid vector pCMV–TDP-43WT coding for TDP-43WT tagged with HA and subsequently treated with LPS. 48 h after transfection, the LPS-challenged BV-2 cells were then harvested, and cell extracts were coimmunoprecipitated with anti-HA antibody. Proteins were resolved in 4–20% Tris-glycine gels (Precast gels; Bio-Rad Laboratories) and stained with Sypro-Ruby (Bio-Rad Laboratories). Protein bands from the gel were excised and subjected to mass spectrometer analysis at the Proteomics Platform, Quebec Genomics Centre. The experiments were performed on a Thermo Surveyor MS pump connected to an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Thermo Fisher Scientific). Scaffold (version 1.7; Proteome Software Inc.) was used to validate tandem mass spectrometry–based peptide and protein identifications. Peptide identifications were accepted if they could be established at >90.0% probability as specified by the Peptide Prophet algorithm (Keller et al., 2002).
Immunofluorescence microscopy.
Cells were grown to 70% confluence on glass coverslips and fixed in 2% paraformaldehyde for 30 min. In some cases, BV-2 cells were transiently transfected with the pCMV-TDP-43WT and pCMV-p65 vectors using the Lipofectamine 2000 reagent. After fixation with 4% paraformaldehyde, cells were washed in PBS, and permeabilized with 0.2% Triton X-100 in PBS for 15 min. After blocking coverslips with 5% normal goat serum for 1 h at room temperature, primary antibody incubations were performed in 1% normal goat serum in PBS overnight, followed by an appropriate Alexa Fluor 488 or 594 secondary antibody (Invitrogen) for 1 h at room temperature. Similar procedures were used for staining spinal cord sections from TDP-43 transgenic mice and sections of sporadic ALS cases. Cells were viewed using a 40× or 63× oil immersion objective on a DM5000B microscope (Leica).
Quantitative real-time RT-PCR.
Real-time RT-PCR was performed with a LightCycler 480 (Roche) sequence detection system using LightCycler SYBR green I at the Quebec Genomics Centre. Total RNA was extracted from cell culture experiments using TRIZOL reagent (Invitrogen). Total RNA was treated with DNase (QIAGEN) to get rid of genomic DNA contaminations. Total RNA was the quantified using Nanodrop, and its purity was verified by Bioanalyzer 2100 (Agilent Technologies). Gene-specific primers were constructed using the GeneTools software (Biotools Inc.). Three genes, Atp5, Hprt1, and GAPDH, were used as internal control genes. The primers used for the analysis of genes are given in Table S2.
Cytotoxicity assay.
N2a cells were transfected with pCMV–hTDP-43 (both WT and mutants). 48 h after transfection, cells were treated with the conditioned media derived from BV-2 cells, some of which were treated with LPS (0111:B4 serotype; Sigma-Aldrich). 24 h after challenging N2a cells, culture supernatants were assayed for CytoTox-ONE Homogeneous Membrane Integrity Assay (Promega), a fluorimetric assay which depends on the levels of LDH released as the result of cell death (Swarup et al., 2007a). The assay was performed according to the manufacturer’s protocol. Fluorescence was measured using a SpectraMAX Gemini EM fluorescence plate reader (Molecular Devices) at an excitation wavelength of 560 nm and an emission wavelength of 590 nm. Similar techniques were used for primary cortical neurons derived from TDP-43 transgenic mice.
ELISA.
The levels of TNF, IL-1β, IL-6, and IFN-γ were assayed by multianalyte ELISA and MIX-N-MATCH ELISAarray kits (mouse inflammatory cytokine array; SABiosciences). Mouse p65 ELISA (Stressgen) and human p65 ELISA (SABiosciences) were performed according to manufacturer’s instructions. For TDP-43 ELISA, we used the sandwich-ELISA protocol. In brief, ELISA plates were incubated in mouse monoclonal antibody against TDP-43 (clone E2-D3; Abnova) overnight, and the total protein extracts (both soluble and insoluble fractions) were incubated in precoated plates. A second TDP-43 polyclonal antibody (ProteinTech) was further added, and ELISA was performed as described previously (Kasai et al., 2009; Noto et al., 2011). The standard curve for the ELISA assay was performed with triplicate measurements using 100 µl/well of recombinant TDP-43 protein (molecular mass 54.3 kD, AAH01487, recombinant protein with GST tag; Abnova) solution at different concentrations (0.24, 0.48, 0.97, 1.9, 3.9, 7.8, 15.6, 31.2, 62.5, 125, 250, 500, 1,000, and 1,250 ng/ml) of the protein in PBS. The relative concentration estimates of TDP-43 were calculated according to each standard curve.
Nitrite and ROS assays.
The cell culture supernatants from cortical neurons or N2a cells were assayed for nitrite concentration using Griess reagent (Invitrogen) as described previously (Swarup et al., 2007b). The supernatants were also assayed for ROS using H2DCFDA (Sigma-Aldrich).
EMSA.
48 h after transfection of CMV-p65 with pCMV–TDP-43WT or pCMV–TDP-43G348C and treatment with LPS, BV-2 cells were harvested, and nuclear extracts were prepared. Nuclear proteins were extracted using a protein extraction kit (Panomics) as per the manufacturer’s instructions. Concentrations of nuclear proteins were determined on diluted samples using a Bradford assay (Bio-Rad Laboratories). Interaction between p65 in the protein extract and DNA probe was investigated using the EMSA kit (Panomics) as per the manufacturer’s instructions. These nuclear extracts were incubated with NF-κB–binding site–specific oligonucleotides coated with streptavidin. EMSA was then performed using the NF-κB EMSA kit. For supershift assays, antibodies against p50, p65, or TDP-43 were added during the sample preparation step.
Reporter gene assays.
BV-2 cells were harvested in 120 µl of cell lysis buffer (Promega), and an ensuing 1-min centrifugation step (20,000 g) yielded a luciferase-containing supernatant. In both cases, aliquots of 20-µl supernatant were tested for luciferase activity (luciferase assay kit; Promega) and for β-galactosidase activity (β-galactosidase assay kit; Promega) to adjust for transfection efficiency.
RNA interference.
To selectively prevent TDP-43 expression, we used the RNA interference technology. A double-stranded RNA (siRNA) was used to degrade TDP-43 mRNA and thus to limit the available protein. The siRNA experiments were designed and conducted as described previously (Swarup et al., 2007a). The siRNAs directed against the murine TDP-43 mRNA (GenBank accession no. NM_145556.4) consisted of sequences with symmetrical 3′-UU overhangs using siRNA Target Finder (Invitrogen). The sequence of the most effective TDP-43 siRNAs represented is as follows: 5′-AGGAAUCAGCGUGCAUAUAUU-3′ and 5′-UAUAUGCACGCUGAUUCCUUU-3′. To account for the nonsequence-specific effects, scrambled siRNA was used. The sequence of scrambled siRNA is as follows: 5′-GUGCACAUGAGUGAGAUUU3′ and 5′-CACGUGUACUCACUCUAAA-3′. TDP-43 siRNAs or the scrambled siRNAs were suspended in diethyl pyro-carbonate water to yield the desired concentration. For in vitro transfection, cells were plated in 24-well plates and transfected with 0.6 µmol/l siRNAs with 2 µl Lipofectamine 2000. The cells were then kept for 72 h in OptiMEM medium (Invitrogen).
Accelerating rotarod.
Accelerating rotarod was performed on mice at 4-rpm speed with 0.25-rpm/s acceleration as described previously (Gros-Louis et al., 2008). Mice were subjected to three trials per session and every 2 wk.
In vivo bioluminescence imaging.
As previously described (Maysinger et al., 2007; Cordeau et al., 2008), the images were gathered using the IVIS 200 Imaging System (Caliper Life Sciences). 25 min before imaging session, the mice received intraperitoneal injection of the luciferase substrate d-luciferine (150 mg/kg for mice between 20 and 25 g, 150–187.5 ml of a solution of 20 mg/ml of d-luciferine dissolved in 0.9% saline was injected; Caliper Life Sciences).
Statistical analysis.
For statistical analysis, the data obtained from independent experiments are presented as the mean ± SEM; they were analyzed using a paired Student’s t test with Mann-Whitney test, one-way analysis of variance (ANOVA) with Kruskal-Wallis test, or two-way ANOVA with Bonferroni adjustment for multiple comparisons using Prism software version 5.0 (GraphPad Software). For rotarod and GFAP imaging experiments, repeated measures ANOVA was used. In some experiments, an unpaired Student’s t test followed by a Welch’s test was performed. Differences were considered significant at P < 0.05.
Online supplemental material.
Fig. S1 demonstrates reverse immunoprecipitation of TDP-43 with p65 antibody and EMSA supershift assay and describes how p65 activation is age dependent in TDP-43WT transgenic mice. Table S1 lists the primers used for TDP-43 cloning. Table S2 lists the primers used for quantitative RT-PCR. Table S3 gives details of patients examined during the study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20111313/DC1.
Acknowledgments
We thank Christine Bareil and Genevieve Soucy for technical assistance. We are grateful to the laboratory of Dr. Michel J. Tremblay for p65, IκBSR, and luciferase plasmids.
This work was supported by the Canadian Institutes of Health Research and Neuromuscular Research Partnership, the Robert Packard Center for ALS Research at Johns Hopkins and the Fondation André-Delambre. J.-P. Julien holds a Canada Research Chair Tier 1 in mechanisms of neurodegeneration. V. Swarup is the recipient of the Merit Scholarship for foreign students (Fonds Québécois de la Recherche sur la Nature et les Technologies, Quebec, Canada).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ALS
- amyotrophic lateral sclerosis
- ANOVA
- analysis of variance
- BMM
- BM-derived macrophage
- EMSA
- electrophoretic mobility shift assay
- GFAP
- glial fibrillary acidic protein
- HA
- hemagglutinin
- LDH
- lactate dehydrogenase
- mRNA
- messenger RNA
- NMJ
- neuromuscular junction
- PDL
- poly-d-lysine
- ROS
- reactive oxygen species
- siRNA
- small interfering RNA
- VCP
- vasolin-containing protein
- WA
- Withaferin A
References
- Aleyasin H., Cregan S.P., Iyirhiaro G., O’Hare M.J., Callaghan S.M., Slack R.S., Park D.S. 2004. Nuclear factor-(kappa)B modulates the p53 response in neurons exposed to DNA damage. J. Neurosci. 24:2963–2973 10.1523/JNEUROSCI.0155-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. 2006. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351:602–611 10.1016/j.bbrc.2006.10.093 [DOI] [PubMed] [Google Scholar]
- Badadani M., Nalbandian A., Watts G.D., Vesa J., Kitazawa M., Su H., Tanaja J., Dec E., Wallace D.C., Mukherjee J., et al. 2010. VCP associated inclusion body myopathy and paget disease of bone knock-in mouse model exhibits tissue pathology typical of human disease. PLoS ONE. 5:e13183 10.1371/journal.pone.0013183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumer D., Parkinson N., Talbot K. 2009. TARDBP in amyotrophic lateral sclerosis: identification of a novel variant but absence of copy number variation. J. Neurol. Neurosurg. Psychiatry. 80:1283–1285 10.1136/jnnp.2008.166512 [DOI] [PubMed] [Google Scholar]
- Bergmann M., Hart L., Lindsay M., Barnes P.J., Newton R. 1998. IkappaBalpha degradation and nuclear factor-kappaB DNA binding are insufficient for interleukin-1beta and tumor necrosis factor-alpha-induced kappaB-dependent transcription. Requirement for an additional activation pathway. J. Biol. Chem. 273:6607–6610 10.1074/jbc.273.12.6607 [DOI] [PubMed] [Google Scholar]
- Boillée S., Vande Velde C., Cleveland D.W. 2006a. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 52:39–59 10.1016/j.neuron.2006.09.018 [DOI] [PubMed] [Google Scholar]
- Boillée S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., Cleveland D.W. 2006b. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 312:1389–1392 10.1126/science.1123511 [DOI] [PubMed] [Google Scholar]
- Chiang P.M., Ling J., Jeong Y.H., Price D.L., Aja S.M., Wong P.C. 2010. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc. Natl. Acad. Sci. USA. 107:16320–16324 10.1073/pnas.1002176107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement A.M., Nguyen M.D., Roberts E.A., Garcia M.L., Boillée S., Rule M., McMahon A.P., Doucette W., Siwek D., Ferrante R.J., et al. 2003. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 302:113–117 10.1126/science.1086071 [DOI] [PubMed] [Google Scholar]
- Cordeau P., Jr, Lalancette-Hébert M., Weng Y.C., Kriz J. 2008. Live imaging of neuroinflammation reveals sex and estrogen effects on astrocyte response to ischemic injury. Stroke. 39:935–942 10.1161/STROKEAHA.107.501460 [DOI] [PubMed] [Google Scholar]
- Corrado L., Ratti A., Gellera C., Buratti E., Castellotti B., Carlomagno Y., Ticozzi N., Mazzini L., Testa L., Taroni F., et al. 2009. High frequency of TARDBP gene mutations in Italian patients with amyotrophic lateral sclerosis. Hum. Mutat. 30:688–694 10.1002/humu.20950 [DOI] [PubMed] [Google Scholar]
- Custer S.K., Neumann M., Lu H., Wright A.C., Taylor J.P. 2010. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum. Mol. Genet. 19:1741–1755 10.1093/hmg/ddq050 [DOI] [PubMed] [Google Scholar]
- Daoud H., Valdmanis P.N., Kabashi E., Dion P., Dupré N., Camu W., Meininger V., Rouleau G.A. 2009. Contribution of TARDBP mutations to sporadic amyotrophic lateral sclerosis. J. Med. Genet. 46:112–114 10.1136/jmg.2008.062463 [DOI] [PubMed] [Google Scholar]
- Davies J.Q., Gordon S. 2005. Isolation and culture of murine macrophages. Methods Mol. Biol. 290:91–103 [DOI] [PubMed] [Google Scholar]
- Deng H.X., Zhai H., Bigio E.H., Yan J., Fecto F., Ajroud K., Mishra M., Ajroud-Driss S., Heller S., Sufit R., et al. 2010. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann. Neurol. 67:739–748 10.1002/ana.22051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequen F., Bomont P., Gowing G., Cleveland D.W., Julien J.P. 2008. Modest loss of peripheral axons, muscle atrophy and formation of brain inclusions in mice with targeted deletion of gigaxonin exon 1. J. Neurochem. 107:253–264 10.1111/j.1471-4159.2008.05601.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Capell A., Carlson A.M., Shankaran S.S., Rodde R., Neumann M., Kremmer E., Matsuwaki T., Yamanouchi K., Nishihara M., Haass C. 2009. Proteolytic processing of TAR DNA binding protein-43 by caspases produces C-terminal fragments with disease defining properties independent of progranulin. J. Neurochem. 110:1082–1094 10.1111/j.1471-4159.2009.06211.x [DOI] [PubMed] [Google Scholar]
- Douville R., Liu J., Rothstein J., Nath A. 2011. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 69:141–151 10.1002/ana.22149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M.J., Piñol-Roma S., Burd C.G. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289–321 10.1146/annurev.bi.62.070193.001445 [DOI] [PubMed] [Google Scholar]
- Gerritsen M.E., Williams A.J., Neish A.S., Moore S., Shi Y., Collins T. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA. 94:2927–2932 10.1073/pnas.94.7.2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho M.A., Baloh R.H., Chakraverty S., Mayo K., Norton J.B., Levitch D., Hatanpaa K.J., White C.L., III, Bigio E.H., Caselli R., et al. 2008. TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 63:535–538 10.1002/ana.21344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho M.A., Bigio E.H., Mishra M., Johnson N., Weintraub S., Mesulam M., Rademakers R., Chakraverty S., Cruchaga C., Morris J.C., et al. 2009. TARDBP 3′-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol. 118:633–645 10.1007/s00401-009-0571-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros-Louis F., Kriz J., Kabashi E., McDearmid J., Millecamps S., Urushitani M., Lin L., Dion P., Zhu Q., Drapeau P., et al. 2008. Als2 mRNA splicing variants detected in KO mice rescue severe motor dysfunction phenotype in Als2 knock-down zebrafish. Hum. Mol. Genet. 17:2691–2702 10.1093/hmg/ddn171 [DOI] [PubMed] [Google Scholar]
- Guerreiro R.J., Schymick J.C., Crews C., Singleton A., Hardy J., Traynor B.J. 2008. TDP-43 is not a common cause of sporadic amyotrophic lateral sclerosis. PLoS ONE. 3:e2450 10.1371/journal.pone.0002450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidet-Phillips A.M., Hester M.E., Miranda C.J., Meyer K., Braun L., Frakes A., Song S., Likhite S., Murtha M.J., Foust K.D., et al. 2011. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 29:824–828 10.1038/nbt.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenberg L.G., Smith M.A. 2007. Preparation of dissociated mouse cortical neuron cultures. J. Vis. Exp. 2007:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath R.J., Nutile-McMenemy N., Alkaitis M.S., Deleo J.A. 2008. Differential migration, LPS-induced cytokine, chemokine, and NO expression in immortalized BV-2 and HAPI cell lines and primary microglial cultures. J. Neurochem. 107:557–569 10.1111/j.1471-4159.2008.05633.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz L.M., Kwong L.K., Chen-Plotkin A., Winton M.J., Unger T.L., Xu Y., Neumann M., Trojanowski J.Q., Lee V.M. 2009. Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J. Biol. Chem. 284:8516–8524 10.1074/jbc.M809462200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.S., McCaffery J.M., Lindquist S., Gitler A.D. 2008. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA. 105:6439–6444 10.1073/pnas.0802082105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J., et al. 2010. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 68:857–864 10.1016/j.neuron.2010.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Vande Velde C., Bouchard J.P., Lacomblez L., Pochigaeva K., Salachas F., et al. 2008. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40:572–574 10.1038/ng.132 [DOI] [PubMed] [Google Scholar]
- Kasai T., Tokuda T., Ishigami N., Sasayama H., Foulds P., Mitchell D.J., Mann D.M., Allsop D., Nakagawa M. 2009. Increased TDP-43 protein in cerebrospinal fluid of patients with amyotrophic lateral sclerosis. Acta Neuropathol. 117:55–62 10.1007/s00401-008-0456-1 [DOI] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Kwiatkowski T.J., Jr, Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 323:1205–1208 10.1126/science.1166066 [DOI] [PubMed] [Google Scholar]
- Ling S.C., Albuquerque C.P., Han J.S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D.W. 2010. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. USA. 107:13318–13323 10.1073/pnas.1008227107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H., Morino H., Ito H., Izumi Y., Kato H., Watanabe Y., Kinoshita Y., Kamada M., Nodera H., Suzuki H., et al. 2010. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 465:223–226 10.1038/nature08971 [DOI] [PubMed] [Google Scholar]
- Maysinger D., Behrendt M., Lalancette-Hébert M., Kriz J. 2007. Real-time imaging of astrocyte response to quantum dots: in vivo screening model system for biocompatibility of nanoparticles. Nano Lett. 7:2513–2520 10.1021/nl071611t [DOI] [PubMed] [Google Scholar]
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 314:130–133 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- Noto Y.I., Shibuya K., Sato Y., Kanai K., Misawa S., Sawai S., Mori M., Uchiyama T., Isose S., Nasu S., et al. 2011. Elevated CSF TDP-43 levels in amyotrophic lateral sclerosis: specificity, sensitivity, and a possible prognostic value. Amyotroph. Lateral Scler. 12:140–143 10.3109/17482968.2010.541263 [DOI] [PubMed] [Google Scholar]
- Oh J.H., Lee T.J., Park J.W., Kwon T.K. 2008. Withaferin A inhibits iNOS expression and nitric oxide production by Akt inactivation and down-regulating LPS-induced activity of NF-kappaB in RAW 264.7 cells. Eur. J. Pharmacol. 599:11–17 10.1016/j.ejphar.2008.09.017 [DOI] [PubMed] [Google Scholar]
- Perkins N.D., Felzien L.K., Betts J.C., Leung K., Beach D.H., Nabel G.J. 1997. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 275:523–527 10.1126/science.275.5299.523 [DOI] [PubMed] [Google Scholar]
- Pizzi M., Sarnico I., Boroni F., Benetti A., Benarese M., Spano P.F. 2005. Inhibition of IkappaBalpha phosphorylation prevents glutamate-induced NF-kappaB activation and neuronal cell death. Acta Neurochir. Suppl. (Wien). 93:59–63 10.1007/3-211-27577-0_8 [DOI] [PubMed] [Google Scholar]
- Schmitz M.L., dos Santos Silva M.A., Baeuerle P.A. 1995a. Transactivation domain 2 (TA2) of p65 NF-kappa B. Similarity to TA1 and phorbol ester-stimulated activity and phosphorylation in intact cells. J. Biol. Chem. 270:15576–15584 10.1074/jbc.270.26.15576 [DOI] [PubMed] [Google Scholar]
- Schmitz M.L., Stelzer G., Altmann H., Meisterernst M., Baeuerle P.A. 1995b. Interaction of the COOH-terminal transactivation domain of p65 NF-kappa B with TATA-binding protein, transcription factor IIB, and coactivators. J. Biol. Chem. 270:7219–7226 10.1074/jbc.270.13.7219 [DOI] [PubMed] [Google Scholar]
- Sephton C.F., Good S.K., Atkin S., Dewey C.M., Mayer P., III, Herz J., Yu G. 2010. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 285:6826–6834 10.1074/jbc.M109.061846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried N.T., Gozal Y.M., Dammer E.B., Xia Q., Duong D.M., Cheng D., Lah J.J., Levey A.I., Peng J. 2010. Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyubiquitin chains. Mol. Cell. Proteomics. 9:705–718 10.1074/mcp.M800390-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard K.A., Rose D.W., Haque Z.K., Kurokawa R., McInerney E., Westin S., Thanos D., Rosenfeld M.G., Glass C.K., Collins T. 1999. Transcriptional activation by NF-kappaB requires multiple coactivators. Mol. Cell. Biol. 19:6367–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. 2008. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 319:1668–1672 10.1126/science.1154584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings N.R., Puttaparthi K., Luther C.M., Burns D.K., Elliott J.L. 2010. Progressive motor weakness in transgenic mice expressing human TDP-43. Neurobiol. Dis. 40:404–414 10.1016/j.nbd.2010.06.017 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Mikami H., Watanabe T., Yamano T., Yamazaki T., Nomura M., Yasui K., Ishikawa H., Ono S. 2010. Increased expression of TDP-43 in the skin of amyotrophic lateral sclerosis. Acta Neurol. Scand. 122:367–372 10.1111/j.1600-0404.2010.01321.x [DOI] [PubMed] [Google Scholar]
- Swarup V., Das S., Ghosh S., Basu A. 2007a. Tumor necrosis factor receptor-1-induced neuronal death by TRADD contributes to the pathogenesis of Japanese encephalitis. J. Neurochem. 103:771–783 10.1111/j.1471-4159.2007.04790.x [DOI] [PubMed] [Google Scholar]
- Swarup V., Ghosh J., Duseja R., Ghosh S., Basu A. 2007b. Japanese encephalitis virus infection decrease endogenous IL-10 production: correlation with microglial activation and neuronal death. Neurosci. Lett. 420:144–149 10.1016/j.neulet.2007.04.071 [DOI] [PubMed] [Google Scholar]
- Swarup V., Phaneuf D., Bareil C., Robertson J., Rouleau G.A., Kriz J., Julien J.P. 2011. Pathological hallmarks of amyotrophic lateral sclerosis/frontotemporal lobar degeneration in transgenic mice produced with TDP-43 genomic fragments. Brain. 134:2610–2626 10.1093/brain/awr159 [DOI] [PubMed] [Google Scholar]
- Thaiparambil J.T., Bender L., Ganesh T., Kline E., Patel P., Liu Y., Tighiouart M., Vertino P.M., Harvey R.D., Garcia A., Marcus A.I. 2011. Withaferin A inhibits breast cancer invasion and metastasis at sub-cytotoxic doses by inducing vimentin disassembly and serine 56 phosphorylation. Int. J. Cancer. 129:2744–2755 10.1002/ijc.25938 [DOI] [PubMed] [Google Scholar]
- Van Deerlin V.M., Leverenz J.B., Bekris L.M., Bird T.D., Yuan W., Elman L.B., Clay D., Wood E.M., Chen-Plotkin A.S., Martinez-Lage M., et al. 2008. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 7:409–416 10.1016/S1474-4422(08)70071-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 323:1208–1211 10.1126/science.1165942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A., Herholz D., Fiesel F.C., Kaur K., Müller D., Karsten P., Weber S.S., Kahle P.J., Marquardt T., Schulz J.B. 2010. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS ONE. 5:e12247 10.1371/journal.pone.0012247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegorzewska I., Bell S., Cairns N.J., Miller T.M., Baloh R.H. 2009. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. USA. 106:18809–18814 10.1073/pnas.0908767106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weydt P., Yuen E.C., Ransom B.R., Möller T. 2004. Increased cytotoxic potential of microglia from ALS-transgenic mice. Glia. 48:179–182 10.1002/glia.20062 [DOI] [PubMed] [Google Scholar]
- Wils H., Kleinberger G., Janssens J., Pereson S., Joris G., Cuijt I., Smits V., Ceuterick-de Groote C., Van Broeckhoven C., Kumar-Singh S. 2010. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. USA. 107:3858–3863 10.1073/pnas.0912417107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.F., Gendron T.F., Zhang Y.J., Lin W.L., D’Alton S., Sheng H., Casey M.C., Tong J., Knight J., Yu X., et al. 2010. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J. Neurosci. 30:10851–10859 10.1523/JNEUROSCI.1630-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoza B.K., Hu J.Y., McCall C.E. 1996. Protein-tyrosine kinase activation is required for lipopolysaccharide induction of interleukin 1beta and NFkappaB activation, but not NFkappaB nuclear translocation. J. Biol. Chem. 271:18306–18309 10.1074/jbc.271.31.18306 [DOI] [PubMed] [Google Scholar]
- Zhang R., Miller R.G., Gascon R., Champion S., Katz J., Lancero M., Narvaez A., Honrada R., Ruvalcaba D., McGrath M.S. 2009a. Circulating endotoxin and systemic immune activation in sporadic amyotrophic lateral sclerosis (sALS). J. Neuroimmunol. 206:121–124 10.1016/j.jneuroim.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Hadlock K.G., Do H., Yu S., Honrada R., Champion S., Forshew D., Madison C., Katz J., Miller R.G., McGrath M.S. 2011. Gene expression profiling in peripheral blood mononuclear cells from patients with sporadic amyotrophic lateral sclerosis (sALS). J. Neuroimmunol. 230:114–123 10.1016/j.jneuroim.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.J., Xu Y.F., Cook C., Gendron T.F., Roettges P., Link C.D., Lin W.L., Tong J., Castanedes-Casey M., Ash P., et al. 2009b. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA. 106:7607–7612 10.1073/pnas.0900688106 [DOI] [PMC free article] [PubMed] [Google Scholar]