A single amino acid shift in TCR recognition of self peptide–MHC determines whether potentially diabetogenic CD4 T cells will be purged in the thymus or have the opportunity to undergo activation in the islets of Langerhans of mice.
Abstract
In nonobese diabetic (NOD) mice, two sets of autoreactive CD4+ T cells recognize the B:9–23 segment of the insulin B chain. One set, type A, recognizes insulin presented by antigen-presenting cells (APCs). These T cells are highly deleted in the thymus. The second set, type B, does not recognize insulin protein but reacts with soluble B chain peptide. This set is not deleted in the thymus but is activated in the islets of Langerhans. In this study, we examine the specificity of these two types of T cells. The protein-reactive set recognizes the stretch of residues 13–21 of the insulin B chain. The set reactive to peptide only recognizes the stretch from residues 12–20. A single amino acid shift of the B chain peptide bound to I-Ag7 determines whether T cells recognize peptides generated by the processing of insulin, and consequently their escape from thymic purging. Biochemical experiments indicate that peptides bound in the 13–21 register interact more favorably with I-Ag7 than peptides that bind in the 12–20 register. Thus, self-reactive T cells can become pathogenic in the target organ where high concentrations of antigen and/or differences in intracellular processing present peptides in registers distinct from those found in the thymus.
A central issue in the study of autoimmunity concerns the nature of the autologous peptides recognized by the autoreactive T cells: do certain self peptides bound to MHC molecules have distinctive features that allow for the activation of autoreactive T cells? Issues of binding affinity, posttranslational modifications, conformational isomers, and register shifting of self peptides have all been the subject of analysis by many laboratories (Liu et al., 1995; Anderton, 2004; Bankovich et al., 2004; Lovitch and Unanue, 2005; Goverman, 2009). Solutions to these various issues could potentially explain why self-reactive T cells escape thymic selection and subsequently become pathogenic in the target tissue. Our own interest centers on whether the peptide–MHC (pMHC) complex presented by tissue APCs may be distinct compared with the pMHC being presented, if at all, by thymic APCs during T cell selection.
In the nonobese diabetic (NOD) mouse, the islet autoimmune response is highly focused against the insulin molecule. The insulin autoreactivity is a major contributing factor and is essential for the development of diabetes (Daniel et al., 1995; French et al., 1997; Jaeckel et al., 2004; Nakayama et al., 2005, 2007). A recent study of ours analyzing the T cell response during diabetogenesis of NOD mice led to the identification of a notable set of autoreactive CD4+ T cells that recognized insulin peptides found in the early islet infiltrates of NOD mice (Mohan et al., 2010). These CD4+ T cells recognized the 9–23 residue stretch of the insulin B chain (B:9–23), a peptide which binds to I-Ag7 at very low affinity with a fast rate of dissociation (Yu et al., 2000; Levisetti et al., 2007). Importantly, most of the anti-insulin T cells did not recognize the B:9–23 segment when the insulin protein was processed by the APCs; these T cells only recognized the B:9–23 segment when it was offered as an exogenous peptide. In brief, we identified subsets of T cells against an insulin epitope that exhibited a pattern of reactivity akin to T cells identified when studying the protein hen egg white lysozyme (HEL), which we had termed type A and B (Lovitch and Unanue, 2005).
These insulin peptide–specific T cells were readily activated by APCs normally inhabiting the islets of Langerhans (Calderon et al., 2008) but not by any other tissue APCs, including those in the thymus. Importantly, the secretory granules from β cells, in addition to containing the high amounts of mature insulin molecules, also contained peptide fragments derived from the insulin protein. These granules were taken up by the intraislet APCs and charged their MHC molecules with insulin-derived peptides, explaining the activation of these peptide-specific T cells. Such peptide-specific T cell lines transferred into nondiabetic NOD mice were specifically recruited to the islets and caused diabetes (Mohan et al., 2010). The fact that these peptide-specific T cells are not clonally deleted in the thymus, even though they are pathogenic, unmistakably underscores their importance.
In this study, we examine and explain the features of the CD4+ type A and B T cells against the B chain of insulin. The explanation for their dissimilar reactivity lies in the peptide segment being recognized: type A T cells recognized the B:13–21 register bound to I-Ag7, whereas type B T cells recognized the B:12–20 register. A single residue shift of the peptide segment bound within the binding groove of the MHC distinguished both sets of autoreactive T cells. An explanation for such distinction lies in the binding properties of each segment.
RESULTS AND DISCUSSION
Insulin-reactive type A and B T cells recognize different registers of the B:9–23 peptide
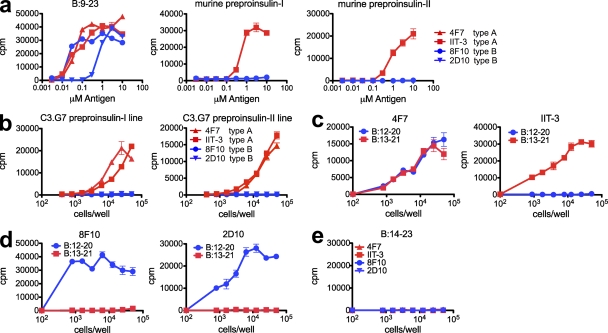
Previously, a cohort of insulin-reactive CD4+ T cell hybridomas was generated from the infiltrated islets of prediabetic NOD mice or by immunization with the insulin B chain peptide encompassing residues 9–23. These B:9–23-reactive T cell hybridomas were divided into two groups based on their ability to recognize the insulin protein processed by APCs in standard T cell assays. Those that recognized both insulin and B:9–23 peptide presented by APCs were called type A T cells, whereas those only recognizing the B:9–23 peptide but not the insulin molecule were called type B (Mohan et al., 2010). In this study, the ability of these T cells to recognize mouse preproinsulin proteins was tested in two experimental systems: in one, the C3.G7 APC line was cultured with recombinant mouse preproinsulin I or II, and in the other manipulation, the same APC was engineered to endogenously express mouse preproinsulin I or II via retroviral transduction of their respective cDNA.
Both type A and B T cells were activated by adding the B:9–23 peptide to the C3.G7 APC line (Fig. 1 a). The type A T cells recognized recombinant mouse preproinsulin I and II when added to the APC (Fig. 1 a) or to either of the proteins expressed in the C3.G7 line (Fig. 1 b). Type B T cells were incapable of recognizing preproinsulin in either situation (Fig. 1, a and b). Collectively, these results confirmed our previous findings using exogenous human insulin (Mohan et al., 2010): the type B T cells did not recognize the B:9–23 epitope generated by the processing of mouse insulin proteins.
Figure 1.
Type A and B reactive T cells recognize distinct registers of the B:9–23 peptide. (a) IL-2 production by T cell hybridomas in response to B:9–23 peptide, mouse preproinsulin I, or mouse preproinsulin II, presented by the C3.G7 APC cell line. (b) IL-2 production in response to C3.G7 cells expressing mouse preproinsulin I and preproinsulin II cDNA constructs. (c and d) Response of two type A hybridomas (4F7 and IIT-3; c) and two type B hybridomas (8F10 and 2D10; d) to the B:12–20 (register 1) and B:13–21 (register 2) peptides covalently linked to I-Ag7 expressed on C3.G7 cells. (e) Response of type A and type B hybridomas to the B:14–23 peptide linked to I-Ag7 (register 3) expressed on M12.C3 cells. Error bars indicate SEM. Data are representative of three independent experiments.
A previous study in the laboratory indicated that T cells reacted only to two partially overlapping nine amino acid registers within the B:9–23 peptide, (SHLVEALYLVCGERG; Levisetti et al., 2007): register 1 encompassing the 12–20 (VEALYLVCG) segment and register 2 using the 13–21 (EALYLVCGE) segment (the P9 residue of each is underlined). To test whether type A or B T cells were restricted to one of the two registers of the peptide bound to the I-Ag7 MHC molecule, APCs were generated containing the minimal registers covalently bound to the I-Ag7 β chain via a flexible glycine/serine linker (Kozono et al., 1994; Landais et al., 2009). These constructs, along with the I-Ag7 α chain were expressed in the M12.C3 B lymphoma cell line. All cell lines used in these experiments expressed comparable levels of I-Ag7 on the cell surface (not depicted).
Constructs containing either register 1 or register 2 segments covalently bound to the I-Ag7 β chain were generated and expressed in the M12.C3 cell line. The cell line expressing the register 2:13–21 segment covalently linked to I-Ag7 (C3.G7B:13–21) was exclusively recognized by the type A T cells but not by the type B T cells (Fig. 1, c and d). We found no examples of a type B T cell activated by the register 2:13–21 complex. In striking contrast, the type B T cells recognized the register 1:12–20 segment covalently linked to I-Ag7 (C3.G7B:12–20; Fig. 1, c and d). Whereas the majority of type A T cells only recognized the C3.G7B:13–21 cell line, interestingly, a subset of them also recognized register 1:12–20 covalently linked to I-Ag7 (Fig. 1 c).
A third potential binding register previously suggested to encompass the B:14–22 segment (register 3:14–22) of the peptide (Stadinski et al., 2010) was evaluated. A cell line expressing the B:14–23 segment covalently linked to I-Ag7 (C3.G7B:14–23) did not stimulate any type A or B hybridomas (Fig. 1 e).
Although no type B T cells recognized register 2, a subset of type A T cells recognized both registers 1 and 2 covalently bound; note in Table S1 their incidence to presentation by C3.G7B:12–20 and to C3.G7B:9–20. Further studies are needed to examine their specificity; such a group may recognize the 13–20 segment plus the adjacent glycine from the linker of the covalent constructs (i.e. EALYLVCGG; putative P9 underlined) or may recognize the 12–20 segment in a cross-reactive way. The recognition of the 12–20 segment by type A T cells in general was considerably weaker than the 13–21 segment, and the best example is that of the T cell shown (Fig. 2 a). Moreover, as summarized in Table S2, many of these dual reactive T cells did not recognize a soluble peptide containing the register 1:12–20, which is another indication of their weak cross-reactivity.
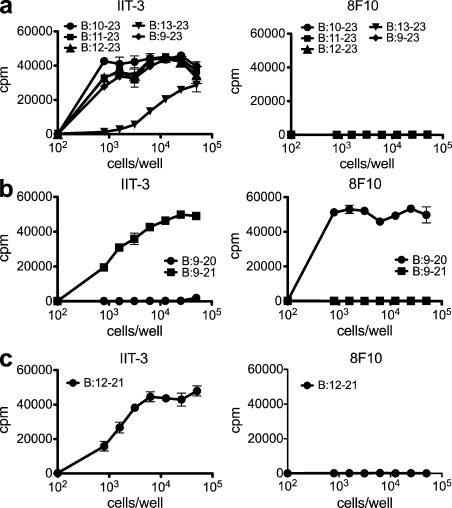
Figure 2.
The glutamic acid at B21 determines register binding of covalently linked insulin pMHC molecules. (a) Response of type A (IIT-3) and type B (8F10) hybridomas to N-terminal truncated variants of the B:9–23 peptide covalently linked to I-Ag7 expressed on M12.C3 cells. (b) Type A (IIT-3) and type B (8F10) T cell response to cells expressing covalently linked B:9–20 and B:9–21 C-terminal truncations of B:9–23. (c) Type A (IIT-3) and type B (8F10) T cell response to the G7B:12–21 cell line. Error bars indicate SEM. Data are representative of three independent experiments.
The B21 glutamic acid influences register selection of covalent pMHC
In our previous study (Mohan et al., 2010), only the type A T cells but not the type B T cells recognized the covalent linkage of the entire B:9–23 peptide to I-Ag7. This indicated that the linked B:9–23 peptide was solely bound in register 2:13–21. To address this issue, a panel of truncated B:9–23 peptides covalently linked to the I-Ag7 molecule was developed to determine which residues influenced register binding. Peptides with various lengths of the N-terminal flank were all recognized by type A T cells but not by type B T cells (Fig. 2 a). Thus, regardless of the N-terminal flank length, the covalently bound peptide was always presented in register 2:13–21.
Next, covalent complexes with truncations of the C-terminal residues were examined. A cell line with truncation of two amino acids at the C terminus of the peptide (C3.G7B:9–21) was only recognized by type A T cells and not by type B T cells (Fig. 2 b). Pointedly, when the C terminus was further truncated by removing the B21 glutamic acid to generate the C3.G7B:9–20 cell line, the opposite outcome was observed. The G7B:9–20 complex was recognized by type B T cells but not by the type A T cells (Fig. 2 b).
These results suggest that the glutamic acid at position 21 in the covalently linked pMHC locks register 2 in the peptide-containing groove of I-Ag7 molecules; register 1 is only presented when this residue is absent in the covalently linked complexes. Therefore, we surmised that the addition of a single amino acid, the B21 glutamic acid, to the B:12–20 peptide should shift the presentation from register 1:12–20 to register 2:13–21. Accordingly, a covalent cell line expressing B:12–21 (VEALYLVCGE), which contains the minimal peptide encompassing the two registers, was only recognized by type A T cells and not by type B T cells (Fig. 2 c).
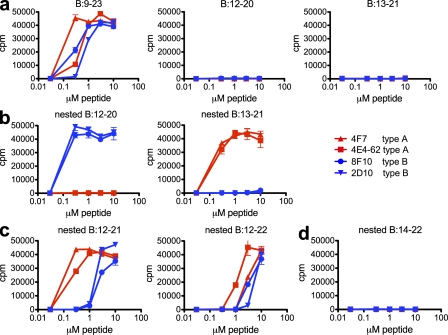
Synthetic peptides confirm register reactivity of A and B T cells
The findings with covalently bound peptides were extended using soluble peptides. Although both sets of T cells recognized the B:9–23 peptide, the type A or B T cells did not recognize the B:12–20 or B:13–21 minimal register peptides when presented by APCs (Fig. 3 a). These findings confirm that the residues flanking the minimal binding core are required for recognition by insulin-reactive T cells (Levisetti et al., 2007). To circumvent this issue, synthetic peptides were designed containing a single register (underlined in the following sequences) nested between artificial flanking residues. The three amino and carboxyl flanking residues present in the covalently linked insulin-pMHC molecules were added to the peptides containing the core registers (Table S2). Akin to the covalently linked cell lines, type B T cells only recognized the soluble peptide containing the nested register 1, with flanking residues (TEGVEALYLVCGGGS; Fig. 3 b). Conversely, a similar peptide containing the nested register 2 (TEGEALYLVCGEGGS) was recognized by type A T cells but not by type B T cells (Fig. 3 b). Similar to the natural B:9–23 peptide, peptides containing both registers as their core, B:12–21 (GTEVEALYLVCGEGGS) and B:12–22 (TEGVEALYLVCGERGGS), were recognized by both type A and B T cells (Fig. 3 c). A peptide containing register 3, B:14–22, nested with the same flanks (TEGALYLVCGERGGS) was not recognized by any of the T cells (Fig. 3 d).
Figure 3.
Type A and B T cells recognize independent registers within the B:9–23 soluble peptide. (a) Response of type A (4F7 and 4E4-62) and type B (8F10 and 2D10) hybridomas to B:9–23, B:12–20, or B:13–21. (b) Response to nested B:12–20 with flanking residues or nested B:13–21 with flanking residues. (c) Response to nested B:12–21 with flanking residues or nested B:12–22 with flanking residues. (d) Response to nested B:14–22 with flanking residues. Error bars indicate SEM. Data are representative of three independent experiments.
In summary, type A T cells specifically recognized register 2:13–21, whereas type B T cells specifically recognized register 1:12–20. The P9 glutamic acid (B21) of register 2 had a seminal role in register selection of covalently bound peptides. All covalently linked peptides containing the B21 glutamic acid were presented on I-Ag7 in register 2, whereas peptides lacking this residue were presented in register 1. Without the constraints imposed by the intracellular handling of insulin by APCs (or by the use of covalent linkages), soluble peptides have the flexibility to be presented in both register 1 and register 2. These findings, summarized in Tables S1 and S2, indicate that register 2 was highly favored under physiological conditions such as when insulin was processed and presented by the APCs.
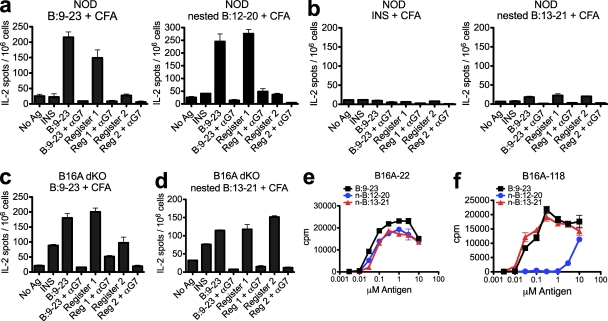
Immunization elicits register 1–reactive type B T cells in vivo
NOD mice were unresponsive to immunization with insulin protein but responded to immunization with the B:9–23 peptide, generating T cells that exhibited type B reactivity, only responding to the peptide but not to insulin (Mohan et al., 2010). Linking these previous findings to the aforementioned results, the expectation would be that the majority of B:9–23 T cells elicited in NOD mice would recognize register 1:12–20. Conversely, immunization with a register 2:13–21 peptide should not elicit a response: mice are highly tolerant to insulin, and insulin processing by APCs mainly generated register 2:13–21. These expectations were fulfilled.
NOD mice immunized with the B:9–23 or nested register 1 peptide emulsified in CFA elicited a strong ELISPOT response to the B:9–23 peptide and also to the nested register 1 peptide but not to the nested register 2 peptide or insulin (Fig. 4 a). The response was inhibited with the addition of anti–I-Ag7 antibody in the assay. Mice that were immunized with insulin protein or the nested register 2 peptide did not elicit a response to any of the peptides (Fig. 4 b).
Figure 4.
Immunization of NOD mice gives rise to T cells that recognize register 1 but not register 2 of B:9–23. (a and b) ELISPOT assay of IL-2 secretion by NOD mice immunized with 10 nmol of B:9–23 or register 1:12–20 nested peptide (TEGVEALYLVCGGGS; a) or insulin or register 2:13–21 nested peptide (TEGEALYLVCGEGGS; b) in CFA. (c and d) ELISPOT assay of IL-2 secretion by B16A-dKO mice immunized with 10 nmol of B:9–23 (c) or register 2:13–21 nested peptide (d) in CFA. (a–d) 106 cells from the draining lymph node were restimulated without antigen (No Ag) or with B:9–23 peptide, insulin (INS), register 1, or register 2 nested peptides (all at a dose of 10 µM) in the presence or absence of a specific blocking antibody to I-Ag7 (αG7). (e and f) Response of insulin-reactive hybridomas B16A-22 (e) and B16A-118 (f) from B16A-dKO mice to B:9–23, nested 12–20 peptide (n-B:12–20), and nested 13–21 peptide (n-B:13–21). Error bars indicate SEM. Data are representative of two or more independent experiments (n = 3 mice per group).
In our previous study, we showed that type A T cells escaped thymic selection and could be elicited after immunization of the insulin mutant strain B16:A-dKO (Mohan et al., 2010). These mice have targeted deletions of the native INS1 and INS2 genes but express a mutant insulin molecule that has a single amino acid substitution (tyrosine to alanine) at the 16th residue of the B chain (Nakayama et al., 2005). When B16A-dKO mice were immunized with the B:9–23 peptide, we detected a response to register 1 similar to that found in NOD mice, but importantly, we detected a response to the insulin protein and to the nested register 2 peptide (Fig. 4 c), both of which are absent in NOD mice (Fig. 4 a). Similarly, when we immunized B16A-dKO mice with the nested register 2 peptide, we detected a specific T cell response to the B:9–23 peptide, insulin protein, and both register 1 and 2 nested peptides (Fig. 4 d). The response to register 1:12–20 was unexpected and required further examination.
Therefore, we tested a panel of type A hybridomas generated from B16A-dKO mice. These hybridomas robustly recognized register 2 and also recognized register 1:12–20 at similar levels (Fig. 4 e) or to a weaker extent (Fig. 4 f). We therefore conclude that the dual reactive nature of these T cells explains the unexpected response to register 1 seen after immunization of these mice with the nested register 2 peptide (Fig. 4 d).
All together, these results confirm that the majority of T cells that escape thymic negative selection in NOD mice are peptide specific and recognize register 1 of the B:9–23 peptide. The presence of register 2–reactive T cells in the periphery of B16A-dKO mice confirms that the B:13–21 register plays a critical role in mediating negative selection of insulin T cells. It is currently unclear whether many of the T cells found in them recognize both registers or whether they recognize a putative hybrid register of the register 1 peptide.
Biochemical features of the two main binding segments
The same nested peptides containing the flanking residues and the core segments (Table S2) were tested in binding assays to purified I-Ag7 molecules. Peptides containing register 2:13–21 bound better than those containing register 1:12–20, 0.7 µM versus 1.4 µM, respectively. The nested 12–20 register peptide required about double the amount of free peptide to inhibit the binding of a standard radioactive peptide in contrast to that of the nested 13–21 register (Table S2; in seven different experiments with P < 0.03). The peptide having the 12–21 segment, i.e., both registers, bound at about the same strength, 0.5 µM, to that of register 2:13–21. Changing the P9 residue to a lysine of register 2:13–21 peptide greatly decreased the binding capacity and completely inhibited T cell reactivity (Fig. 5 a and Table S2), confirming the binding register. The effects on binding of lysine substitution on P9 are less evident with peptides having nonacidic residues. Indeed this was the case with the register 1:12–20 peptide when having a lysine substitution at P9 but nonetheless completely abrogated T cell recognition (Fig. 5 b and Table S2). A peptide with the nested register 3:14–22 bound very weakly at 5.5 µM.
Figure 5.
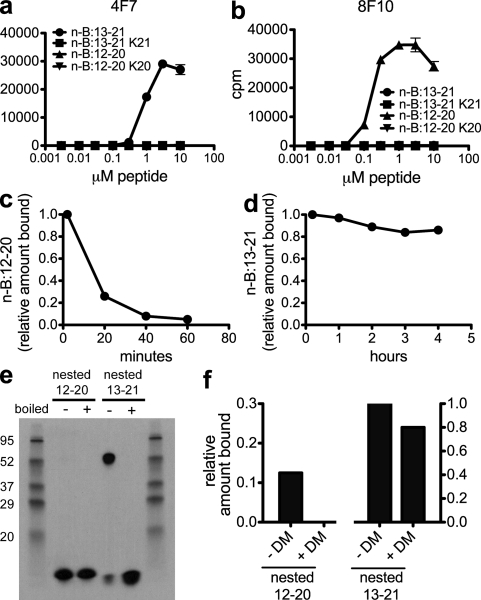
Binding of register 1:12–20 peptide to I-Ag7 molecules is unstable and dissociates rapidly compared with binding of register 2:13–21 peptides. (a and b) Response of type A (4F7; a) and type B (8F10; b) hybridomas to the nested 12–20 peptide (n-B:12–20), nested 12–20 K20 peptide (n-B:12–20 K20), nested 13–21 peptide (n-B:13–21), and nested 13–21 K21 peptide (n-B:13–21 K21). (c and d) Dissociation rate of the 12–20 nested peptide (c) and 13–21 nested peptide (d) from soluble I-Ag7 protein. (e) SDS stability of 12–20 nested peptide I-Ag7 complexes and 13–21 nested peptide I-Ag7 complexes. To denature stable complexes, samples were boiled before loading as indicated. Molecular mass is indicated in kilodaltons. (f) H2-DM editing of soluble peptide I-Ag7 complexes. Bars indicate the percentage of remaining peptide bound to I-Ag7 after 30 min in the presence of 10-fold molar excess of H2-DM. Error bars indicate SEM. Data are representative of 3–10 independent experiments.
The nested peptide with register 1 had a considerably faster dissociation rate from I-Ag7, with >50% of the bound peptide being lost in the first 20 min (Fig. 5 c). In striking contrast, the nested 13–21 register 2 showed a very slow rate of dissociation, and ∼70% was SDS stable (Fig. 5, d and e). In accordance with the weak binding of the nested register 1, most of the bound peptide dissociated when the pMHC complex was run in SDS-PAGE under nonboiling conditions (Fig. 5 e). Addition of H2-DM to the complex of peptide and I-Ag7 increased the fast rate of dissociation of nested 12–20 to a complete loss after 30 min but had little effect on nested 13–21 (Fig. 5 f). In sum, the binding features indicate the register 2:13–21 segment with the B21 glutamic acid at the P9 MHC anchor residue is much more favorable than register 1:12–20 in its interaction with I-Ag7.
The present findings extend the previous report of two major sets of insulin-reactive CD4+ T cells (Mohan et al., 2010), identify their peptide binding segment to MHC, correlate the recognition in in vivo experiments, and present a biochemical explanation for their selection. Both sets of T cells were reactive with the B chain of insulin centered on a core segment represented by residues 12–21. We show here that the 12–21 segment contained two adjacent binding registers, giving rise to T cells that recognized one or the other. More importantly, the register specificity of these T cells corresponded to previously identified subsets of type A and B insulin–reactive T cells (Mohan et al., 2010), which have seminal biological differences.
The minor set of insulin-reactive T cells in vivo, type A, recognizes the register 2:13–21 segment of the B:9–23 peptide. These T cells react with the insulin molecule after its processing by APCs as well as to the B:9–23 peptide. Such T cells are under strong selective pressure in the thymus where insulin is expressed (Mohan et al., 2010). Deletion of register 2–reactive T cells is not complete, and a limited number can be found in the periphery and infiltrated islets of NOD mice (Wegmann et al., 1994; Mohan et al., 2010). In striking contrast, the majority of insulin T cells found in NOD mice, the type B T cells, recognize the 12–20 segment of the B:9–23 peptide. These T cells are not activated by any source of insulin, explaining their ability to escape thymic negative selection. Such T cells are selected by the intraislet APCs, spontaneously infiltrate the islets of prediabetic NOD mice, and cause diabetes upon transfer (Mohan et al., 2010).
The binding properties of each peptide register plus the anatomical features of antigen presentation in the islet of Langerhans explain the presence and features of the two sets of T cells. The binding characteristics of I-Ag7 typically select for the presence of peptides bearing one or more acidic residues at the carboxyl end of the peptide (Corper et al., 2000; Latek et al., 2000; Suri et al., 2002, 2005). A minor set of peptides have small residues bound in the P9 pocket (Stratmann et al., 2000; Suri et al., 2002, 2005). In the case of the two insulin registers found in the 12–21 segment, register 2 has the favorable glutamic acid at P9. This register binds better to I-Ag7, has a longer half-life as a pMHC complex, is more stable in SDS, and is not affected by H2-DM compared with register 1, which has a less favorable glycine at P9. The contrasting findings of each register are compatible with a scenario in which the processing of insulin selects for the better binding segment. The environment of the lysosomal compartment, coupled with the weak binding of register 1 to I-Ag7, leads to the generation of pMHC complexes that are too unstable to compete and survive their sojourn to the cell surface in sufficient quantity to stimulate a T cell response.
The presentation of peptides bound in register 1 was found only in the APCs that normally reside within the islets of Langerhans (Mohan et al., 2010). Such APCs constitutively took up insulin granules, many of which, aside from insulin and C-peptide, also contained peptides derived from the insulin B chain. It stands to reason that the constant exposure to the secretory granule with preformed insulin B chain peptides resulted in a significant number of pMHCs bearing the peptide bound in register 1, a situation akin to the reactivity seen in the culture assays, when APCs are constantly exposed to exogenous synthetic peptides. Whether there are other issues with the islet APCs that allow them to select for peptides that are bound in register 1 needs to be examined. An additional consideration is whether there are unique features of the peptides released from β cells that may favor register 1 binding.
We were unable to find any evidence for insulin T cells recognizing a third register in the B:9–23 segment encompassing 14–22 (Stadinski et al., 2010). T cells against the 14–22 register are unexpected because of the placement of an unfavored arginine within the P9 pocket, which results in very weak binding to I-Ag7, even weaker than that of register 1 and 2 segments. The inability to find T cells that recognize this register indicates that register 3 is unlikely to play an important role in diabetes.
In conclusion, biochemical analyses have given several explanations for the presentation of peptides from self-proteins. Important are the findings emphasized by us, that the pMHC complexes presented by APCs in tissues may differ from those present in the thymic APC system. We called attention to these in the context of the small protein HEL where conformational isomers could be distinguished, which explained the presence of T cells to one set in HEL transgenic mice (Peterson et al., 1999). In the studies of spontaneous diabetes in NOD mice, we find selection for T cells that recognized a segment in which the register shifted to one poorly presented, if at all, in the thymus. Previous studies have shown that more than one binding register can be selected within a relatively short peptide segment, a phenomenon called register shifting, which gave rise to nonoverlapping sets of T cells (Scott et al., 1998; McFarland et al., 1999; Robertson et al., 2000). The issue of register shifting to explain the escape of autoimmune T cells has also been discussed in autoimmune encephalomyelitis (Anderton et al., 2002; Seamons et al., 2003; Bankovich et al., 2004; Goverman, 2009). The conclusion is that self-reactive T cells can potentially become pathogenic in the target organ where high concentrations of antigen and/or differences in intracellular processing requirements lead to the presentation of peptides in alternate conformers or, as shown here, in alternate registers to those found in the thymus.
MATERIALS AND METHODS
Mice.
NOD and NOD.Cg-Tg(Ins2*Y16A)1Ell Ins1tm1Jja Ins2tm1Jja/GseJ (B16:A-dKO) mice were obtained from the Jackson Laboratory and maintained in specific pathogen–free conditions. All animal experiments were approved by the Animal Study Committee of Washington University in St. Louis.
Expression and isolation of mouse preproinsulin proteins.
Mouse preproinsulin I and II cDNAs were generated from RNA isolated from NOD pancreatic islet cells. These cDNAs were cloned into the pCR 2.1 TOPO vector (Invitrogen) using the following primer pairs: preproinsulin I forward primer, 5′-ATGGCCCTGTTGGTGCACTTCCTA-3′; and reverse primer, 5′-TTAGTTGCAGTAGTTCTCCAG-3′; and preproinsulin II forward primer, 5′-ATGGCCCTGTGGATGCGCTTCCTG-3′; and reverse primer, 5′-CTAGTTGCAGTAGTTCTCCAG-3′. The preproinsulin DNA was subsequently amplified to contain a 5′ NcoI site and 3′ BamHI site to facilitate cloning into the pET-32c vector (EMD) using the following primer pairs: NcoI ppINS-1 forward, 5′-TCATTACCATGGCCCTGTTGGTGCACTTCCTA-3′; and BamHI ppINS-1 reverse, 5′-CTTAGAGGATCCTTAGTTGCAGTAGTTCTCCAG-3′; and NcoI ppINS-2 forward, 5′-TCATTACCATGGCCCTGTGGATGCGCTTCCTG-3′; and BamHI ppINS-2 reverse, 5′-CTTAGAGGATCCCTAGTTGCAGTAGTTCTCCAG-3′. pET-32c-preproinsulin constructs were transformed into the origami B (DE3) pLysS Escherichia coli strain (EMD). Bacteria were grown in a shaking incubator (230 rpm) in LB-ampicillin media at 30°C. Protein expression was induced when the culture reached an OD600 ∼0.6 with 0.25 mM IPTG (Roche) for 6 h. Bacteria were isolated by centrifugation, and after gentle lysis, preproinsulin proteins were isolated from the cytoplasmic fraction using Ni-NTA agarose (QIAGEN). Protein was dialyzed against PBS, pH 7.5, overnight at 4°C. The isolated protein was then concentrated with 10,000 MWCO Centricon columns (Millipore) and treated with 1 U/mg thrombin (EMD) to separate expression and solubility tags from the preproinsulin proteins. Proteins were dialyzed twice (24 h each) against PBS and stored at 4°C.
Cell lines.
The T cell hybridomas used in this study were previously described (Mohan et al., 2010). To generate C3.G7 cell lines expressing preproinsulin I and II, DNA was amplified from the pCR 2.1 preproinsulin constructs using the following primer pairs: NotI ppINS-1 forward, 5′-TCATTAGCGGCCGCATGGCCCTGTTGGTGC-3′; and BamHI ppINS-1 reverse (listed in previous section); and NotI ppINS-2 forward, 5′-TCATTAGCGGCCGCATGGCCCTGTGGATGCGC-3′; and BamHI ppINS-2 reverse (listed in previous section). Amplified DNA was digested and cloned into the pRetroX-IRES-dsRedExpress vector (Takara Bio Inc.). C3.G7–preproinsulin I and II stable cell lines were generated by retroviral infection and sorted multiple times based on dsRed expression for high purity. The generation of cell lines with covalently linked pMHC molecules is listed in the next section.
Generation of covalently linked pMHC constructs.
The I-Ad α chain was cloned from NOD splenic cDNA with the following primer pair: NotI G7α forward, 5′-TTATGCGGCCGCATGCCGTGCAGCAGAGCTCT-3′; and BamHI G7α reverse, 5′-TTCAGGATCCTCATAAAGGCCCTGGGTGTCT-3′ and subsequently cloned into the pQCXIP vector (Takara Bio Inc.). The I-Ag7 β chain with the B:9–23 peptide covalently linked was isolated from a previously described cell line (Mohan et al., 2010) and cloned into the pCR 2.1 vector (Invitrogen) with the following primers: NotI G7β forward, 5′-TTATGCGGCCGCATGGCTCTGCAGATCCCCAG-3′; and BamHI G7β reverse, 5′-CATTGGATCCTCACTGCAGGAGCCCTGCTGG-3′. The B:9–23 peptide was removed by XmaI and SpeI restriction digest. Complementary synthetic oligonucleotides of the desired insulin peptide sequences with restriction enzyme sites (XmaI and SpeI) at the 5′ and 3′ ends were annealed, digested, and ligated into the I-Ag7 β chain construct. The G7β chain with the modified linker (GGSLVPRGSGGGGS) and covalently bound insulin peptide constructs were cloned in the pRetroX-IRES-dsRedExpress vector (Takara Bio Inc.) using the NotI and BamHI restriction sites.
Generation of retrovirus.
Phoenix cells (Orbigen) were transfected with the pQCXIP I-Ad α plasmid and the VSV envelope plasmid with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. After 48 h, the supernatant was harvested, centrifuged at 3,000 rpm for 10 min to remove cellular debris, and then applied to M12.C3 cells with 10 µg/ml polybrene (Sigma-Aldrich). Cells were spinfected at 2,500 rpm for 90 min, incubated at 37°C overnight, washed, and expanded for another 48 h. 10 µg/ml puromycin (Sigma-Aldrich) was added for 48 h to select for a stable M12.C3.I-Ad α cell line. This line was subsequently infected with viruses generated in the same manner containing the G7β peptide constructs. After infection with the β chain construct, cells were sorted once for expression of dsRed on a high-speed cell sorter. Expression of I-Ag7 was assessed by flow cytometry using the AG2.42.7 monoclonal antibody.
Antigen presentation assays.
T cell assays were performed in triplicate in 96-well flat-bottom tissue culture plates. T cell hybridomas (5 × 104 per well) were cultured with 5 × 104 C3.G7 cells in the presence of the indicated doses of antigen. All peptides used in this study were synthesized in the laboratory using standard protocols. After incubation for 18 h, the culture supernatant from each well was assayed for IL-2 production by a standard bioassay using the IL-2–dependent cell line CTLL-2.
Immunizations and ELISPOT analysis.
Immunizations and ELISPOT were performed as previously described (Mohan et al., 2010). In brief, mice were immunized in the hind foot pad with either 10 nmol insulin (Sigma-Aldrich) or synthetic peptide emulsified in CFA (Difco). On day 7, the draining popliteal lymph nodes were isolated and dissociated into a single cell suspension and used in an IL-2 ELISPOT assay according to the manufacturer’s protocol (BD). Blocking of I-Ag7 was performed by adding the AG2.42.7 antibody (2 µg/well) 30 min before adding antigen.
Biochemical analysis.
Biochemical analysis was performed using soluble I-Ag7 and H2-DM molecules purified from baculoviruses, the same constructs used in previous studies (Pu et al., 2004; Levisetti et al., 2007). Bindings were performed, as detailed previously (Levisetti et al., 2007), by competition assays of unlabeled peptides to standard 125I-labeled peptide. The labeled peptide (GKKVATTVHAGYG) was used at concentrations that bound ∼25% of input; the concentration of unlabeled peptide that inhibits the binding by 50% was determined. All assays were performed at least three times. For experiments measuring the rate of dissociation, radioactive peptide bearing the nested register 1:12–20 or register 2:13–21 sequence was bound to I-Ag7 overnight at pH 5.5, after which a 1,000-fold excess of cold peptide was added, and the amounts of labeled peptide were then determined in the isolated complex. In some experiments, H2-DM at a 10-fold excess molar ratio was added. Peptides used in the bindings assays shown in Table S2 contained three constant residues at the N terminus (TEG) and three at the C terminus (GGS) and the core sequence indicated in the table. For dissociation assays, the peptides were made with an added tyrosine at the N terminus and changing the tyrosine at B16 to phenylalanine (YEGVEALFLCGGGS and YEGEALFLVGEGGS). The peptides were labeled with 125I by the chloramine T method.
Online supplemental material.
Table S1 lists the sequences of the covalent pMHC molecules and the reactivity of T cells examined. Table S2 lists the sequences of the synthetic peptides, their binding affinity to I-Ag7, and the reactivity of T cells examined. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20111502/DC1.
Acknowledgments
We thank Stephen Horvath and Paul Allen for help with peptide synthesis and members of the Unanue laboratory for comments and advice.
This research was supported by National Institutes of Health grants AI024742, DKo58177, and P60DK20579, by the Juvenile Diabetes Research Foundation grant JDRF 1-2007-731, and by grants from the Kilo Diabetes and Vascular Research Foundation.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- HEL
- hen egg white lysozyme
- NOD
- nonobese diabetic
- pMHC
- peptide–MHC
References
- Anderton S.M. 2004. Post-translational modifications of self antigens: implications for autoimmunity. Curr. Opin. Immunol. 16:753–758 10.1016/j.coi.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Anderton S.M., Viner N.J., Matharu P., Lowrey P.A., Wraith D.C. 2002. Influence of a dominant cryptic epitope on autoimmune T cell tolerance. Nat. Immunol. 3:175–181 10.1038/ni756 [DOI] [PubMed] [Google Scholar]
- Bankovich A.J., Girvin A.T., Moesta A.K., Garcia K.C. 2004. Peptide register shifting within the MHC groove: theory becomes reality. Mol. Immunol. 40:1033–1039 10.1016/j.molimm.2003.11.016 [DOI] [PubMed] [Google Scholar]
- Calderon B., Suri A., Miller M.J., Unanue E.R. 2008. Dendritic cells in islets of Langerhans constitutively present beta cell-derived peptides bound to their class II MHC molecules. Proc. Natl. Acad. Sci. USA. 105:6121–6126 10.1073/pnas.0801973105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corper A.L., Stratmann T., Apostolopoulos V., Scott C.A., Garcia K.C., Kang A.S., Wilson I.A., Teyton L. 2000. A structural framework for deciphering the link between I-Ag7 and autoimmune diabetes. Science. 288:505–511 10.1126/science.288.5465.505 [DOI] [PubMed] [Google Scholar]
- Daniel D., Gill R.G., Schloot N., Wegmann D. 1995. Epitope specificity, cytokine production profile and diabetogenic activity of insulin-specific T cell clones isolated from NOD mice. Eur. J. Immunol. 25:1056–1062 10.1002/eji.1830250430 [DOI] [PubMed] [Google Scholar]
- French M.B., Allison J., Cram D.S., Thomas H.E., Dempsey-Collier M., Silva A., Georgiou H.M., Kay T.W., Harrison L.C., Lew A.M. 1997. Transgenic expression of mouse proinsulin II prevents diabetes in nonobese diabetic mice. Diabetes. 46:34–39 10.2337/diabetes.46.1.34 [DOI] [PubMed] [Google Scholar]
- Goverman J. 2009. Autoimmune T cell responses in the central nervous system. Nat. Rev. Immunol. 9:393–407 10.1038/nri2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeckel E., Lipes M.A., von Boehmer H. 2004. Recessive tolerance to preproinsulin 2 reduces but does not abolish type 1 diabetes. Nat. Immunol. 5:1028–1035 10.1038/ni1120 [DOI] [PubMed] [Google Scholar]
- Kozono H., White J., Clements J., Marrack P., Kappler J. 1994. Production of soluble MHC class II proteins with covalently bound single peptides. Nature. 369:151–154 10.1038/369151a0 [DOI] [PubMed] [Google Scholar]
- Landais E., Romagnoli P.A., Corper A.L., Shires J., Altman J.D., Wilson I.A., Garcia K.C., Teyton L. 2009. New design of MHC class II tetramers to accommodate fundamental principles of antigen presentation. J. Immunol. 183:7949–7957 10.4049/jimmunol.0902493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latek R.R., Suri A., Petzold S.J., Nelson C.A., Kanagawa O., Unanue E.R., Fremont D.H. 2000. Structural basis of peptide binding and presentation by the type I diabetes-associated MHC class II molecule of NOD mice. Immunity. 12:699–710 10.1016/S1074-7613(00)80220-4 [DOI] [PubMed] [Google Scholar]
- Levisetti M.G., Suri A., Petzold S.J., Unanue E.R. 2007. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J. Immunol. 178:6051–6057 [DOI] [PubMed] [Google Scholar]
- Liu G.Y., Fairchild P.J., Smith R.M., Prowle J.R., Kioussis D., Wraith D.C. 1995. Low avidity recognition of self-antigen by T cells permits escape from central tolerance. Immunity. 3:407–415 10.1016/1074-7613(95)90170-1 [DOI] [PubMed] [Google Scholar]
- Lovitch S.B., Unanue E.R. 2005. Conformational isomers of a peptide-class II major histocompatibility complex. Immunol. Rev. 207:293–313 10.1111/j.0105-2896.2005.00298.x [DOI] [PubMed] [Google Scholar]
- McFarland B.J., Sant A.J., Lybrand T.P., Beeson C. 1999. Ovalbumin(323-339) peptide binds to the major histocompatibility complex class II I-A(d) protein using two functionally distinct registers. Biochemistry. 38:16663–16670 10.1021/bi991393l [DOI] [PubMed] [Google Scholar]
- Mohan J.F., Levisetti M.G., Calderon B., Herzog J.W., Petzold S.J., Unanue E.R. 2010. Unique autoreactive T cells recognize insulin peptides generated within the islets of Langerhans in autoimmune diabetes. Nat. Immunol. 11:350–354 10.1038/ni.1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Abiru N., Moriyama H., Babaya N., Liu E., Miao D., Yu L., Wegmann D.R., Hutton J.C., Elliott J.F., Eisenbarth G.S. 2005. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 435:220–223 10.1038/nature03523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Beilke J.N., Jasinski J.M., Kobayashi M., Miao D., Li M., Coulombe M.G., Liu E., Elliott J.F., Gill R.G., Eisenbarth G.S. 2007. Priming and effector dependence on insulin B:9-23 peptide in NOD islet autoimmunity. J. Clin. Invest. 117:1835–1843 10.1172/JCI31368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D.A., DiPaolo R.J., Kanagawa O., Unanue E.R. 1999. Quantitative analysis of the T cell repertoire that escapes negative selection. Immunity. 11:453–462 10.1016/S1074-7613(00)80120-X [DOI] [PubMed] [Google Scholar]
- Pu Z., Lovitch S.B., Bikoff E.K., Unanue E.R. 2004. T cells distinguish MHC-peptide complexes formed in separate vesicles and edited by H2-DM. Immunity. 20:467–476 10.1016/S1074-7613(04)00073-1 [DOI] [PubMed] [Google Scholar]
- Robertson J.M., Jensen P.E., Evavold B.D. 2000. DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323-339 epitope. J. Immunol. 164:4706–4712 [DOI] [PubMed] [Google Scholar]
- Scott C.A., Peterson P.A., Teyton L., Wilson I.A. 1998. Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues. Immunity. 8:319–329 10.1016/S1074-7613(00)80537-3 [DOI] [PubMed] [Google Scholar]
- Seamons A., Sutton J., Bai D., Baird E., Bonn N., Kafsack B.F., Shabanowitz J., Hunt D.F., Beeson C., Goverman J. 2003. Competition between two MHC binding registers in a single peptide processed from myelin basic protein influences tolerance and susceptibility to autoimmunity. J. Exp. Med. 197:1391–1397 10.1084/jem.20022226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadinski B.D., Zhang L., Crawford F., Marrack P., Eisenbarth G.S., Kappler J.W. 2010. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc. Natl. Acad. Sci. USA. 107:10978–10983 10.1073/pnas.1006545107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann T., Apostolopoulos V., Mallet-Designe V., Corper A.L., Scott C.A., Wilson I.A., Kang A.S., Teyton L. 2000. The I-Ag7 MHC class II molecule linked to murine diabetes is a promiscuous peptide binder. J. Immunol. 165:3214–3225 [DOI] [PubMed] [Google Scholar]
- Suri A., Vidavsky I., van der Drift K., Kanagawa O., Gross M.L., Unanue E.R. 2002. In APCs, the autologous peptides selected by the diabetogenic I-Ag7 molecule are unique and determined by the amino acid changes in the P9 pocket. J. Immunol. 168:1235–1243 [DOI] [PubMed] [Google Scholar]
- Suri A., Walters J.J., Gross M.L., Unanue E.R. 2005. Natural peptides selected by diabetogenic DQ8 and murine I-A(g7) molecules show common sequence specificity. J. Clin. Invest. 115:2268–2276 10.1172/JCI25350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D.R., Gill R.G., Norbury-Glaser M., Schloot N., Daniel D. 1994. Analysis of the spontaneous T cell response to insulin in NOD mice. J. Autoimmun. 7:833–843 10.1006/jaut.1994.1066 [DOI] [PubMed] [Google Scholar]
- Yu B., Gauthier L., Hausmann D.H., Wucherpfennig K.W. 2000. Binding of conserved islet peptides by human and murine MHC class II molecules associated with susceptibility to type I diabetes. Eur. J. Immunol. 30:2497–2506 [DOI] [PubMed] [Google Scholar]