Abstract
MCF-10F a spontaneously immortalized ERα negative human breast epithelial cell line derived from breast tissues containing Lobules type 1, is able to form normal ductal structures in a tridimensional collagen matrix system. MCF-10F cells that are Estrogen-transformed [trMCF cells] progressively express phenotypes of in vitro cell transformation, including colony formation in agar methocel, and loss of the ductulogenic capacity. Further selection of these trMCF cells for invasiveness in a Matrigel invasion system identified cells [bcMCF] that formed tumors in severe combined immunodeficient [SCID] mice. The cell lines derived from those tumors [caMCF] were poorly differentiated ERα, PR and ERBB2 negative adenocarcinomas. These characteristics are similar to the human basal cell-like carcinomas. This in vitro in vivo model demonstrates the importance of the basal cell type as a stem cell that reconstitute the branching pattern of the breast and that is also target of a carcinogenic insult leading to transformation and cancer.
Keywords: Basal stem cell, epithelial mesenchymal transition, triple negative breast cancer
I. INTRODUCTION
Breast cancer is an invasive and ultimately fatal disease whose incidence in postmenopausal women has gradually increased in most Western societies over the last few decades, but has sharply increased in younger women [1]. Invasive breast cancer is a heterogeneous disease that encompasses a variety of pathological features that are associated with specific clinical behavior [2]. The classification and grading of the tumors, which are essential for selecting therapeutic approaches and the prediction of their biological behavior and patient prognosis, are currently based on the Nottingham modification of the Scarff-Bloom-Richardson system (NSBR) grading scheme. This system, however, is hindered by the subjectivity of the morphological assessment of nuclear grade, mitosis, and tubular formation [3]. The discovery that the morphological heterogeneity of breast cancer is also reflected at the transcriptome level has allowed the classification of breast cancer into five main groups: luminal A and B, normal breast-like, ERBB2 [HER2] and basal-like breast carcinomas [4–6]. The luminal-like subtypes display moderate to high expression of ERα and luminal cytokeratins. The basal-like carcinomas, which have been reported to have a more aggressive clinical behavior, are the focus of this work. They are composed of cells that consistently express genes usually found in normal basal/myoepithelial cells of the breast, including basal cytokeratins (5/6, 14 and 17), E-cadherin, caveolin 1 and p53 [7, 8]. Molecular analyses of basal-like carcinomas have confirmed the frequent lack of expression of estrogen (ER) and progesterone (PR) receptors and HER2, high levels of expression of proliferation-related genes, and frequent mutation of the TP53 gene. Morphologically, basal-like breast carcinomas present with high histological grade, high mitotic indices, central necrotic zones, pushing borders and a conspicuous lymphocytic infiltrate. In addition, metaplastic elements and medullary/atypical medullary features have been reported. Similarities have been found between basal-like tumors and breast carcinomas occurring in BRCA1 mutation carriers, in premenopausal African American women, Hispanic women, and in general, in the younger breast cancer patient population. Breast cancer is more aggressive in African American women [9]; this phenomenon has been partly explained by a later stage at diagnosis [10], larger size of the tumors that are more commonly high grade, and both ER and HER2 negative [11–14].
Basal-like breast cancer has been strongly associated with African American race and Hispanic ancestry [15–18]. Surveillance, Epidemiology, and End Results (SEER) data [19] also show a shift toward more aggressive subtypes in African-American and Hispanic women that is consistent across several studies that include basal cell type with a 5-year relative survival of only 14% [20, 21]. Basal-like breast cancers are measured with specificity by adding positive markers such as cytokeratin 5/6 or epidermal growth factor receptor [22]. Nonetheless, specific markers for basal-like breast cancer are not presently available in most studies, even though distinguishing true basal-like from triple negative breast cancers has important implications for clinical prognosis [22,23].
II. BIOLOGICAL AND MOLECULAR UNDERSTANDING OF BASAL BREAST CANCER AND EPITHELIAL MESENCHYMAL TRANSITION (EMT)
II.i. The in vitro model of basal-like breast cancer
Primary mammary epithelial cells grown in collagen matrix are able to form tree-like structures resembling in vivo ductulogenesis [24]. The human breast epithelial cells MCF-10F form tubules when grown in type I collagen. The advantage of an in vitro model of three-dimensional (3-D) growth is that it reproduces the epithelial architecture of the breast (Figure 1) [24–30]. Normal epithelial cells form duct-like structures, having apical-basal polarity and well-organized tubular structures with stable adherent junctions and cell-basement communications. The cell’s neoplastic transformation is associated with the loss of apical-basal polarity and monolayer morphology and significant deviations from normal epithelial behavior in 3-D cultures [24–30].
Figure 1.
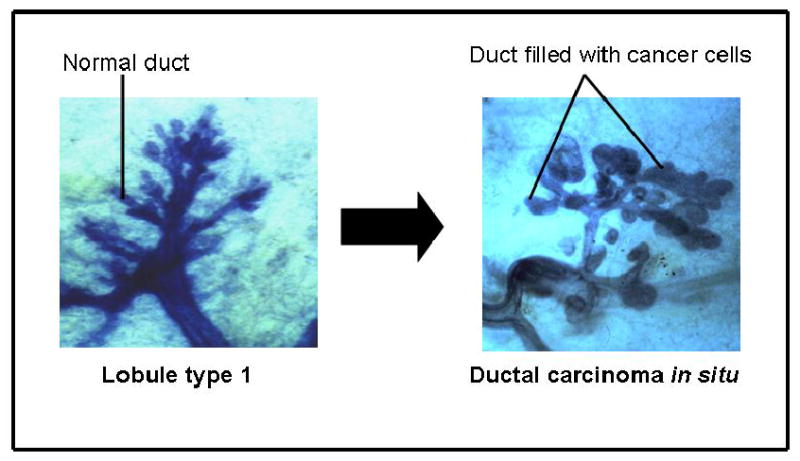
Lobule type 1 or the terminal ductal lobular unit is the site of origin of ductal carcinoma in situ Adapted from: Russo, J, Hu, Y.F., Yang, X. and Russo, I.H. Developmental, cellular and molecular basis of human breast cancer: J. Natl. Cancer Inst. Monograph 27, 2000, pp 17–38.
Our observations that ductal carcinomas originate in the Lob.1 f the immature breast (Figure 1) [31], which are the structures with the highest proliferative activity, and the fact that the cells that do proliferate in culture are ERα negative suggest that the stem cells that originate normal ductal structures and cancer are the ERα negative proliferating cells. This idea is supported by our observations that MCF-10F, a spontaneously immortalized ERα negative human breast epithelial cell line derived from breast tissues containing Lob.1 and Lob.2, is able to form normal ductal structures in a tridimensional collagen matrix system (Figure 2). The ductal structures are lined by a monolayer of well polarized epithelial cells that become malignant after exposure to either the chemical carcinogen benz[a]pyrene [32] or the natural estrogen 17β-estradiol (E2) [24, 29].
Figure 2.
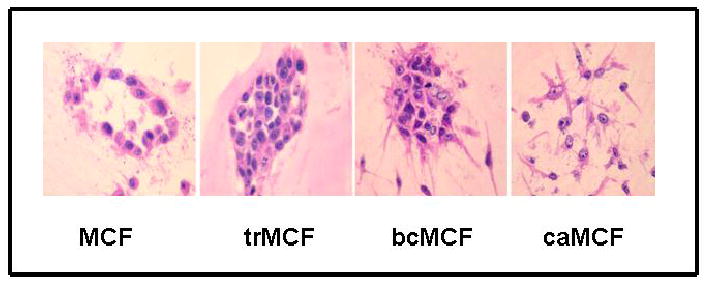
In vitro-in vivo model of cell transformation. A) Different stages in the in vitro-in vivo model. B) The MCF basal cell type from normal ductal structures that loss the ductulogenic capacity forming the trMCF cells that evolves to bcMCF and caMCF cells. Adapted from: Russo, J., Fernandez, S.V., Russo, P.A., Fernbaugh, R., Sheriff, F.S., Lareef, H.M., Garber, J., and Russo, I.H. 17 beta estradiol induces transformation and tumorigenesis in human breast epithelial cells. FASEB J 20:1622–1634, 2006 and Tiezzi, D. G., Fernandez, S.V., Russo, J. Epithelial to Mesenchymal transition during breast cancer progression. Int J Oncol 31: 823–827, 2007
We have developed an in vitro-in vivo model of human breast epithelial cells transformation induced by estradiol (Figure 3) [29, 30]. In this model, the human breast epithelial cell line MCF-10F that is estrogen receptor α (ESRα) negative, was transformed by estradiol and different cell lines that represent different stages of breast cancer progression were isolated [29, 30]. The MCF-10F progression model consists of four derived cell lines: a) the spontaneously immortalized cell line MCF-10F, which does not show any characteristics of invasiveness or tumor formation and therefore is considered to be a normal-like breast epithelial cell line; b) the transformed trMCF cells; c) the invasive bsMCF cell line and, d) the tumor cell lines, caMCFs, which shown all characteristics of a fully malignant breast cancer cell types [29, 30] (Figure 2). The bsMCF cells induced tumors in SCID mice that were poorly differentiated adenocarcinomas that were ESRα, progesterone receptor [PR] and ERBB2 negatives and are also metastatic to the lung. (Figure 3).
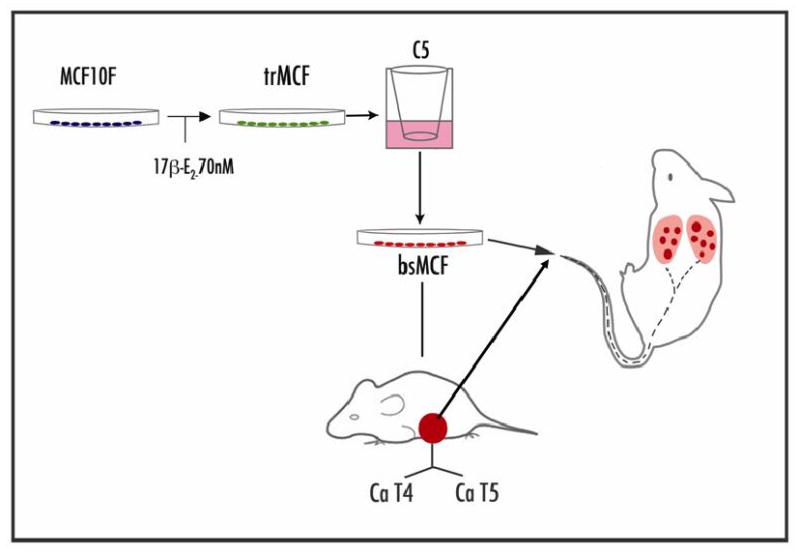
Figure 3.
MCF-10F and derived cell lines (trMCF, bcMCF and caMCF) classify as Basal cell lines. The expression data for these cell lines (26) was combined with the published expression values of the 364 gene set (14) and used to classify the breast cell lines by hierarchical clustering as described (14). The MCF-10F and derived cell lines occupy a distinct branch of the basal subtype (left branch) because the expression values were calculated independently from different batches of normalized genechips.
Loss of the ductulogenic capacity is the earliest phenotype observed accompanied by increasing cell proliferation and the activation of genes related to DNA cell replication, inhibition of apoptosis and the expression of genes related to cell polarity, cell positioning and cellular architecture. Further selection of these trMCF cells for invasiveness in a Matrigel invasion system identified cells (bcMCF) that formed tumors in severe combined immunodeficient (SCID) mice. The cell lines derived from those tumors (caMCF) were poorly differentiated ERα, PR and ERBB2 negative adenocarcinomas [29, 30]. These characteristics are similar to the human basal cell-like carcinomas previously described [6]. To better understand the molecular events associated with the progressive phenotypic changes that were observed during estrogen-mediated malignant cell transformation, we analyzed chromosomal copy number (CN), loss of heterozygosity (LOH), and gene expression changes that occurred at different stages of cell transformation. By integrating these data we were able to identify associations between CN changes, LOH, transcript expression and phenotypes of invasion and tumorigenicity, including a strong gene signature of epithelial to mesenchymal transition (EMT) that was confirmed by immunohistochemistry [30]. The bcMCF (invasive) and caMCF (tumor-derived) cells showed dramatic changes in morphology, losing epithelial characteristics of polarity and acquiring mesenchymal characteristics of a fibroblast-like spindle shape and increased migratory behavior, invasiveness and metastatic capabilities. Changes in gene and protein expression were characteristic of epithelial-mesenchymal transition (EMT), namely loss of intercellular adhesion (E-cadherin and occludins), down-regulation of epithelial makers (cytokeratins), and up-regulation of mesenchymal markers (vimentin and smooth muscleactins) [29, 30].
II. ii. The Molecular Pathway
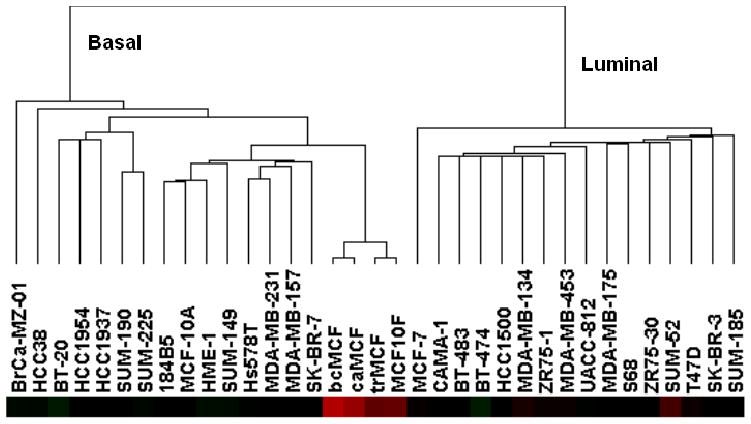
Immortal MCF-10F cells are non-transformed, non-tumorigenic and ER negative. Because malignant cell transformation of these cells produced poorly differentiated tumors characteristic of basal-like carcinomas, we chose to classify these cells relative to the breast cell lines described by Charafe-Jauffret et al. [33]. As shown in Figure 4, these MCF-10F and derived cell lines clustered in the branch containing the basal breast cell lines. In our molecular characterization of malignant cell transformation [30] we identified the “intermediate filament” component enriched in Gene Ontology [GO] analysis, separating the non-tumorigenic MCF-10F and trMCF cells from the tumorigenic bcMCF and caMCF cells. Numerous cytokeratins were suppressed or absent, whereas vimentin was strongly induced in bcMCF (7.0-fold) and caMCF (8.1-fold). Because of these findings, we generated a gene list from published literature for EMT markers and their regulators [30]. The 52 genes in this list were filtered by low stringency criteria of combined coefficient of variation > 0.3 and ‘Present calls’ in more than 30% of the samples. The 27 genes passing these criteria were used for sample and gene clustering (Figure 5A). Two sample groups and two gene groups were identified. The non-tumorigenic MCF-10F and trMCF cells were grouped into sample cluster κ, while the tumorigenic bcMCF and caMCF cells were grouped into cluster λ. On the other side, the genes were grouped into cluster α and β based on their expression pattern. The epithelial markers E-cadherin, occludin, desmoplakin and cytokertins were decreased, while the mesenchymal markers fibronectin, vimentin and N-cadherin were increased in bcMCF and caMCF cells (Figure 5A).
Figure 4.
Expression profile of EMT markers and their regulators during malignant cell transformation.
A. A list of EMT markers and promoting genes was generated a priory by literature search (26). Hierarchical clustering of cell lines and genes was performed using dChip software. Two sample clusters (κ and λ) and two gene clusters (α and β) were identified. The red, white, and blue colors represent level above, at, and below mean expression, respectively. B. Detection of epithelial and mesenchymal markers by immunochytochemistry (100x). a: Histological sections of MCF-10F cells, reacted with pre-immune mouse serum, were used as the negative control; b, c, d: MCF-10F reacted for EMA, E-Cadherin, vimentin, respectively; e: trMCF cells reacted with pre-immune mouse serum used as negative control; f, g, h: trMCF cells reacted for EMA, E-cadherin and vimentin, respectively; i: bsMCF cells reacted with pre-immune mouse serum as a negative control; j, k, l: bsMCF cells reacted for EMA, E-cadherin and vimentin, respectively; m: caMCF tumor cell line cells reacted with pre-immune mouse serum used as negative control; n, o, p: caMCF tumor cell lines reacted for EMA, E-cadherin and vimentin, respectively; q and r: invasive ductal carcinoma of the breast as positive control and immunoreacted for EMA and E-cadherin, respectively; s: histological section of an invasive adenocarcinoma immunoreacted for vimentin. From: Huang, Y., Fernandez, S., Goodwin, S., Russo, P.A., Russo, I. H., Sutter, T., and Russo, J. Epithelial to Mesenchymal Transition in Human Breast Epithelial Cells Transformed by 17- beta- Estradiol. Cancer Res 67 11147–11157, 2007.
Figure 5.
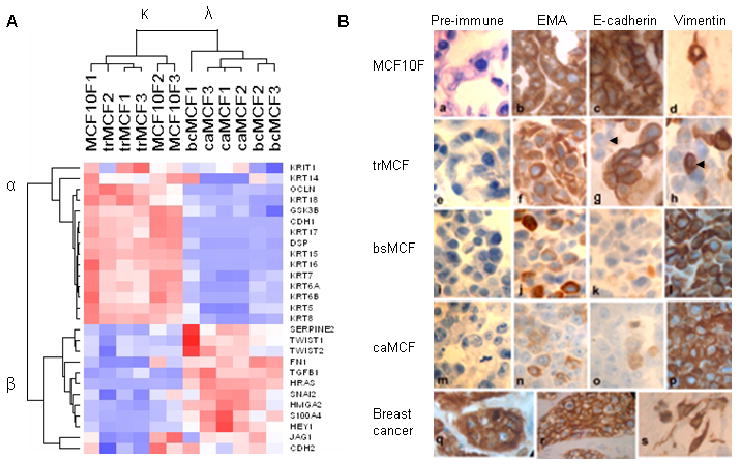
Classification of breast cell lines using an EMT gene signature identifies subtype and invasiveness. Gene expression values were extracted from the CEL files of the 30 cell line subset of breast cancer cell lines that were previously characterized for invasive potential (See Ref. 15, Figure 4). The invasive Basal B cell lines are indicated by the red bar; non-invasive by a green bar.
By real time RT-PCR, it was confirmed that the expression of FN1, S100A4, SNAI2, HRAS and TGFβ1 was increased, while CDH1 (E-cadherin) was decreased in bcMCF and caMCF cells (see 30). Immunocytochemical analysis using antibodies against epithelial membrane antigen EMA (also called MUC1) and E-cadherin displayed significant loss of these epithelial markers and increased expression of the mesenchymal marker vimentin in tumorigenic cells (Figure 5B). These findings confirmed the EMT phenotype revealed by gene expression profile in Figure 5A.
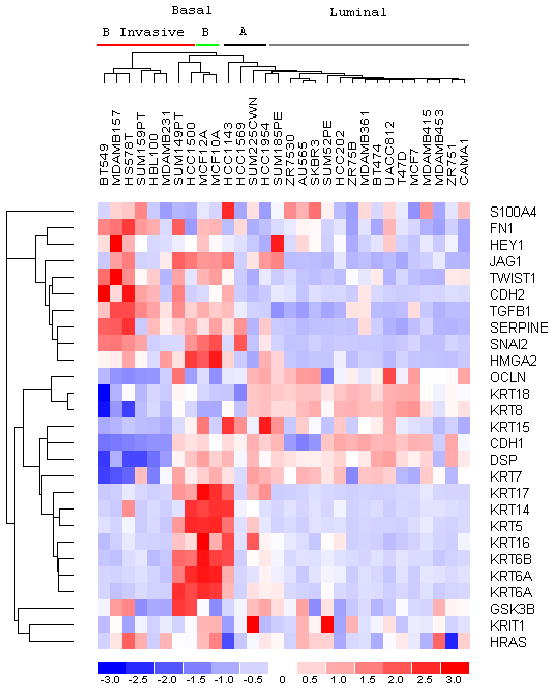
In order to determine whether there is a relationship between our EMT gene signature (Figure 5A) and the classification of the Basal A and Basal B breast cell lines reported in Neve et al. [34], we identified nine genes present in both our EMT gene signature and the 396 gene classifier set of Neve et al. [34]. By Prediction Analysis of Microarray (PAM), we have shown that the parent MCF-10F cells and trMCF cells can be classified as Basal A, whereas the bcMCF (invasive) and caMCF (tumor-derived) cells are classified as Basal B (Table 1). Based on these data, we hypothesize that both Basal subtypes A and B can arise from the same cell of origin, and may reflect differing degrees of EMT and invasive potential. To explore this hypothesis further, we extracted our EMT gene signature from the Genechip expression files of the 30 cell lines that were characterized for invasiveness in a modified Boyden chamber assay. As shown in Figure 6, this EMT gene signature classified, with complete concordance to [34], the cell lines into luminal, Basal A and Basal B sub-types. Of importance to this proposal, the Basal B cell lines that scored as invasive grouped to the far left (Figure 6). This result along with the recent report showing that EMT occurs more frequently in basal-like tumors [35] indicates the relevance of these breast cell lines for molecular analysis of the networks regulating EMT.
Table 1.
Genes differentially expressed genes in bcMCF compared to MCF10F
| Suymbol | Gene name | Fold change |
|---|---|---|
| AZGP1 | Alpha-2-glycoprotein 1, zinc-binding | −31.8 |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | −497.8 |
| EPB41L5 | Erythrocyte membrane protein band 4.1 like 5 | −17.7 |
| FRMD3 | FERM domain containing 3 | −3.3 |
| GRB14 | growth factor receptor-bound protein 14 | −4.1 |
| GPR56 | G protein-coupled receptor 56 | −27.8 |
| KLF5 | Kruppel-like factor 5 (intestinal) | −2.9 |
| MEST | mesoderm specific transcript homolog (mouse) | −4.3 |
| MTUS1 | mitochondrial tumor suppressor 1 | −2.4 |
| RAB25 | RAB25, member RAS oncogene family | −46.2 |
| SFRP1 | secreted frizzled-related protein 1 | −5.8 |
| SFRP1 | secreted frizzled-related protein 1 | −5.5 |
| SFRP1 | secreted frizzled-related protein 1 | −4.8 |
| SRPX | sushi-repeat-containing protein, X-linked | −6.4 |
| ST14 | suppression of tumorigenicity 14 (colon carcinoma) | −5.8 |
| ST14 | suppression of tumorigenicity 14 (colon carcinoma) | −4.2 |
| S100A9 | S100 calcium binding protein A9 | −18.2 |
| SIK1 | salt-inducible kinase 1 | −6.9 |
Figure 6.
Transformation of MCF-10F cells by 17β-estradiol treatment. Experimental protocol: MCF-10F cells treated with 70nM 17β-estradiol (E2) that expressed high colony efficiency (CE) and loss of ductulogenic capacity in collagen-matrix were classified as transformed (trMCF). Transformed cells that were invasive in a Matrigel Boyden type invasion chambers were selected (bsMCF) and plated at low density for cloning (bcMCF). MCF-10F, trMCF, bsMCF, and bcMCF were tested for carcinogenicity by injecting them into the mammary fat pad of 45-day-old female SCID mice. MCF-10F and trMCF cells did not induce tumors; bsMCF, formed solid tumors from which four cell lines, identified as caMCF, were derived and cells proven to be tumorigenic in SCID mice. bsMCF or caMCF when injected in the tail of SCID mice develop metastatic foci in the lung.
While the regulation of EMT is not fully understood, a network of several signaling pathways affecting the expression and/or function of a complex hierarchical network of transcription factors (TFs) has been partially elaborated [36, 37]. Known signaling pathways include multiple tyrosine kinase receptors leading to Ras-mediated activation of MAPK and PI3K pathways, TGF-β, Notch and Wnt.From our studies we have evidence that enhanced TGF-β and Wnt signaling pathways are found in the EMT expressing bcMCF and caMCF cells [30]. TGF-β acting through Smad transcriptional complexes can repress expression of the Id TFs (Id1, Id2, Id3) and activate HMGA2, a DNA binding protein important for chromatin architecture [38]. Expression of HMGA2 is known to regulate several EMT controlling TFs including TWIST1, SNAI1, and SNAI2 (Slug) [30, 36, 37]; (Figures 5 and 6). TGF-β and Wnt signaling also affect the expression of several additional EMT-regulating TFs including ZEB1 (TCF8), TCF3 (E2A encoding E12 and E47), and LEF1 [37].
Analysis of the EMT expressing bcMCF cell line revealed the absence of expression of the secreted frizzled-related protein 1 (SFRP1), a repressor of Wnt signaling [30]. One allele of SFRP1 was deleted in these cells, with the remaining apparently silenced by methylation, accounting for the 28-fold reduction of this transcript. Loss and epigenetic inactivation of SFRP1 occurs often in invasive breast cancer and is associated with poor prognosis [39]. Inspection of the SFRP1 expression levels in Basal B cell lines [30] showed absent calls for 4 of the 8 invasive cell lines; and 8-fold decreases in another 3 invasive cell lines relative to the non-invasive MCF-10A cells. Inspection of the expression files for bcMCF cells and the 8 invasive Basal B cell lines [30] revealed that LEF1 was always absent, while TCF 3 and TCF 8 were expressed.
III. THE METASTATIC PHENOTYPE
As indicated above the bsMCF cells induced tumors in SCID mice that were poorly differentiated adenocarcinomas that were ESRα, progesterone receptor (PR) and ERBB2 negatives. When bsMCF or caMCF (T4 or T5) cells are inoculated in the tail of SCID mice induced metastatic foci in the lung (Figure 3). This unique model allow us: i- to study the genomic and epigenomic changes present in the cell with metastatic capabilities, ii-to use metastatic and non metastatic cells from the same genetic background, and iii-to elucidate the functional role of each of the genes thus identified. We have found that more than 74 genes are either down or upregulated in the invasive and metastatic phenotype of the bsMCF (Figure 7 and Tables 2 and 3). Several genes controlling invasion and metastasis are significantly down regulated in bsMCF (C5) cells (Figure 7 and Table 2). Many of these genes are silenced by methylation in specific GpC islands and the role in the process of invasion and metastasis have been already determined or suspected. For example AZGP1 is known to be downregulated in malignant prostate epithelium [40, 41] but its precise role in metastasis is not clear. CLDN7 or Claudin 7 is significantly downregulated in bsMCF cells as well as in breast [42, 43] and other types of cancer [44]. Hypermethylation at the CLDN7 promoter was detected in 20% of colon cancer cells with low CLDN7 expression. EPB41L5 erythrocyte membrane protein band 4.1 like 5 is involved in cell polarity and in maintaining by separation of the apical and basolateral domains through specialized cell-cell junctions [45–46] and could be an early marker of metastasis. GPR56 or G protein-coupled receptor 56 is downregulated in bsMCF cells and is markedly down-regulated in the metastatic variants of melanoma. Functional studies have shown that over expression of GPR56 suppresses tumor growth and metastasis, whereas reduced expression of GPR56 enhances tumor progression [47, 48]. KLF5 Kruppel-like factor 5 is downregulated also in bsMCF cells and reduced in expression in many types of human tumor [49].
Figure 7.
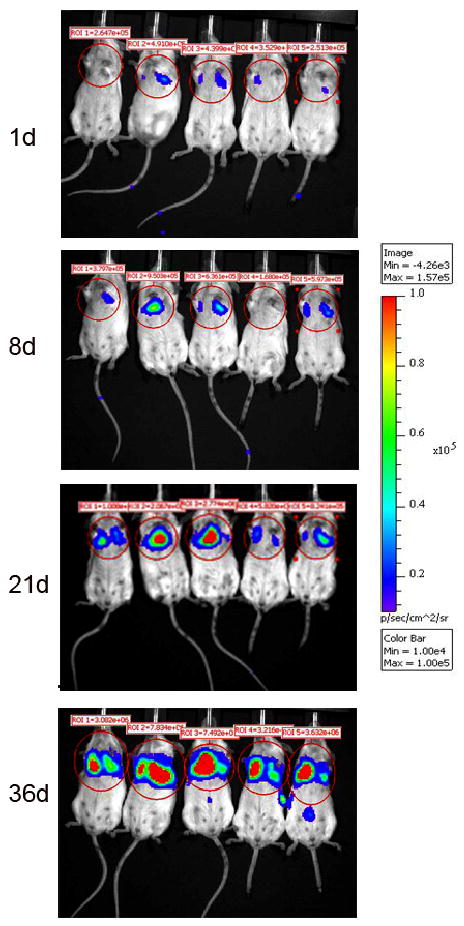
bcMCF cells were transfected by Lipofectamine/Plus Reagent from Life Technologies. The plasmids that were used for the cotransfection were pGL3 control red (SV40-luc) from Promega/C. Contag, Standford University, and pSV40/Zeo from Invitrogen. After the cells were transfected, selection occurred over a period of 10–12 days using 1ug/ml of Zeocin (Invitrogen). After selection, the cells were allowed a period of time to recover after which they were expanded. To ensure the presence of luciferase in the cells, a Luciferase Assay (Promega) was performed using the EnVision Workstation plate reader. Mice were placed in a plastic restraining device equipped with a hole from which we could access the tail. The tails of the mice were placed in warm water in order to dilate the lateral tail veins and to allow easy visualization. Two million (2 × 106) cells suspended in PBS were injected into the lateral tail vein using a 26 gauge needle. The animals were followed over a period of 5 weeks by Bioluminescence Imaging using the Caliper LS/Xenogen IVIS Spectrum System to determine the location of the bcMCF cells.
Table 2.
Upregulated genes differentially expressed genes in bcMCF compared to MCF10F
| Symbol | Gene name | Fold change |
|---|---|---|
| FHL1 | Four and a half domains 1 | 6.5 |
| HEY1 | hairy/enhancer-of-split related with YRPW motif 1 | 5.4 |
| ZEB1 | zinc finger E-box binding homeobox 1 | 12.0 |
| ZEB2 | zinc finger E-box binding homeobox 2 | 11.5 |
| COL6A3 | Collagen, type VI, alpha 3 | 77.1 |
| FN1 | Fibronectin 1 | 4.9 |
| FOSL1 | FOS-like antigen 1 | 3.8 |
| GNG11 | Guanine nucleotide binding protein (G protein), gamma 11 | 10.5 |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | 6.0 |
| NRP1 | Neuropilin 1 | 6.9 |
| RHOB | Ras homolog gene family, member B | 5.7 |
| S100A4 | S100 calcium binding protein A4 | 18.8 |
| TGFB1 | Transforming growth factor, beta 1 | 3.1 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | 2.9 |
| TNS1 | Tensin 1 | 14.9 |
FRMD3 is a member of the protein 4.1 superfamily and is a putative tumor suppressor [50] significantly downregulated in bsMCF cells. Grbl4 a growth factor receptor-bound protein 14 member of the Grb7 family of adapters, is an inhibitor of FGFR signaling. Grbl4 induces an arrest of the signaling transduction cascades in the MDA-MB-231cells by blocking PLCy, ERK2, JNK1 and AKT [51, 52]. Another role of GRB14 is a binding partner of tankyrase 2. Tankyrase is an ankyrin repeat-containing poly[ADP-ribose] polymerase originally isolated as a binding partner for the telomeric protein TRF1. MEST or mesoderm specific transcript homolog is an hypermethylated gene is that is highly enriched for targets of the PRC2 (Polycomb repressive complex 2) in embryonic stem cells. MTUS1 or microtubule associated tumor suppressor 1 is downregulated in bsMCF cells and is significantly down-regulated in colon cancer and in a breast cancer [53–55] an in the triple negative (ER- PR- HER2-) breast carcinomas, a subgroup of highly proliferative tumors with poor outcome and no available targeted therapy. Functional studies indicate that silencing MTUS1 expression by siRNA increases cellular proliferation. Conversely, restoring endogenous levels of MTUS1 expression leads to reduced cancer cell proliferation, clonogenicity, anchorage-independent growth, and reduces the incidence and size of xenografts grown in vivo [56]. Loss of SFRP1, secreted frizzled-related protein 1, expression is observed in breast, along with several other cancers [57–61], and is associated with poor patient prognosis. SFRP1 is significantly downregulated in bsMCF cells and it has been shown to be methylated in many pre and neoplastic breast cancer cell lines. SFRP1 antagonizes the Wnt/beta-catenin signaling pathway by competing with the Frizzled receptor for Wnt ligands resulting in an attenuation of the signal transduction cascade leading to the development of several human cancers, including breast cancer. SRPX sushi-repeat-containing protein, X-linked is downregulated in bsMCF cells and is also markedly reduced in carcinomas of colon, bladder, and ovary and closely linked to the progression of T-cell leukemia/lymphoma. The SRPX gene was originally isolated as a novel suppressor gene of v-src transformation and was shown to induce apoptosis in human cancer cells. It has been observed by us and others that TWIST that is an important transcription factor during embryonic development and has recently been found to promote the EMT phenomenon seen during the initial steps of tumor metastasis, is upregulated in bsMCF cells whereas SRPX is downregulated. Small interfering RNA (siRNA)-mediated depletion of TWIST there is upregulation of SRPX. Indicating an important role of the SRPX gene in invasion and metastasis. Another gene downregulated in bsMCF cells is SNF1LK or the serine-threonine kinase SIK1 (salt-inducible kinase 1) as a regulator of p53-dependent anoikis. Inactivation of SIK1 compromised p53 function in anoikis and allowed cells to grow in an anchorage-independent manner. In vivo, SIK1 loss facilitated metastatic spread and survival of disseminated cells as micrometastases in lungs. The presence of functional SIK1 was required for the activity of the kinase LKB1 in promoting p53-dependent anoikis and suppressing anchorage-independent growth, matrigel invasion, and metastatic potential. Decreased expression of the gene encoding SIK1 closely correlated with development of distal metastases in breast cancers from three independent cohorts. Together, these findings indicate that SIK1 links LKB1 to p53-dependent anoikis and suppresses metastasis [62, 63]. SIK is an inducible gene target of TGFbeta/Smad signaling. Loss of endogenous SIK results in enhanced gene responses of the fibrotic and cytostatic programs of TGFbeta [64].
Among the upregulated genes in the bsMCF cells are the one listed in table 3 are FHL1, HEY1, ZEB1, ZEB2, FOSL1 and S100A4. FHL1 four and a half LIM domains 1 and may play an important role in ER signaling as well as breast cancer cell growth regulation [65, 66]. HEY1 hairy/enhancer-of-split related with YRPW motif 1 and NOTCH3 are upregulated also in bsMCF cells and may be involved in the epithelial mesenchymal transition process [66–68]. Transforming growth factor-beta (TGF-beta) is upregulated in bsMCF cells and is involved in the epithelial-mesenchymal transition (EMT) through activation of Smad and non-Smad signaling pathways. EMT is the differentiation switch by which polarized epithelial cells differentiate into contractile and motile mesenchymal cells. Cell motility and invasive capacity are activated upon EMT. Multiple transcription factors, including deltaEF1/ZEB1, SIP1/ZEB2, and Snail/SNAI1, are induced by TGF-beta-Smad signaling and play critical roles in TGF-beta-induced EMT. In addition, both non-Smad signaling activated by TGF-beta and cross-talk with other signaling pathways play important roles in induction of EMT. Of these, Ras signaling synergizes with TGF-beta-Smad signaling, and plays an important role in the induction of EMT. FOSL1 is upregulated in bsMCF cells and has been shown to be over expressed in MCF 7 cells after development of antiestrogen resistance. Fos is a component of the dimeric transcription factor activator protein-1 (Ap-1), which is composed mainly of Fos (c-Fos, FosB, Fra-1 and Fra-2) and Jun proteins (c-Jun, JunB and JunD). Unlike Fra-1 (encoded by Fosl1), c-Fos contains transactivation domains required for oncogenesis and cellular transformation [69]. The Fos related antigen-1 (Fra-1) is activated in multiple cancers and gene ablation can suppress the invasive phenotypes of many tumor cell lines [69–71]. S100A4 calcium binding protein A4 is upregulated in bsMCF cells and many other cancers [72, 73]. It promotes metastasis in several experimental animal models, and S100A4 protein expression is associated with patient outcome in a number of tumor types and possesses a wide range of biological functions, such as regulation of angiogenesis, cell survival, motility, and invasion [74–75].
SUMMARY AND CONCLUSIONS
From these preliminary data we concluded that the epithelial mesenchymal transition occurring in the breast basal cells depends predominately on TGF-β and Wnt signaling pathways, which increase the expression and function of transcription and chromatin organization factors that repress the epithelial and enhance the mesenchymal phenotype, thus favoring increased invasion and metastatic activity.
Figure 8.

Heat map of the bcMCF cells compared with the MCF10F cells
Acknowledgments
Work supported by grant U01 ES/CA 12771 from the National Institute of Environmental Health Sciences (NIEHS) and P30 CA06927 a CORE grant from the National Cancer Institute (NCI). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS or NCI, NIH.
References
- 1.Bouchardy C, Fioretta G, Verkooijen HM, Vlastos G, Schaefer P, Delaloye J-F, Neyroud-Caspar I, Balmer Majno S, Wespi Y, Forni M, Chappuis P, Sappino A-P, Rapiti E. Recent increase of breast cancer incidence among women under the age of forty. British Journal of Cancer. 2007;96:1743–1746. doi: 10.1038/sj.bjc.6603783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25:5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 3.Tawfik O, Kimler BF, Davis M, Stasik C, Lai SM, Mayo MS, Fan F, Donahue JK, Damjanov I, Thomas P, Connor C, Jewell WR, Smith H, Fabian CJ. Grading invasive ductal carcinoma of the breast: advantages of using automated proliferation index instead of mitotic count. Virchows Arch. 2007;450(6):627–636. doi: 10.1007/s00428-007-0400-0. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Rijn M, Perou CM, Tibshirani R, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991–6. doi: 10.1016/S0002-9440(10)64476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 9.Chlebowski RT, Chen Z, Anderson GL, Rohan T, Aragaki A, Lane D, Dolan NC, Paskett ED, McTiernan A, Hubbell FA, Adams-Campbell LL, Prentice R. Ethnicity and breast cancer: factors influencing differences in incidence and outcome. J Natl Cancer Inst. 2005;97:439–48. doi: 10.1093/jnci/dji064. [DOI] [PubMed] [Google Scholar]
- 10.Newman LA, Mason J, Cote D, Vin Y, Carolin K, Bouwman D, Colditz GA. African-American ethnicity, socioeconomic status, and breast cancer survival: a meta-analysis of 14 studies involving over 10,000 African-American and 40,000 White American patients with carcinoma of the breast. Cancer. 2002;94:2844–54. doi: 10.1002/cncr.10575. [DOI] [PubMed] [Google Scholar]
- 11.Li CI, Malone KE, Daling JR. Differences in breast cancer hormone receptor status and histology by race and ethnicity among women 50 years of age and older. Cancer Epidemiol Biomarkers Prev. 2002;11:601–7. [PubMed] [Google Scholar]
- 12.Joslyn SA. Hormone receptors in breast cancer: racial differences in distribution and survival. Breast Cancer Res Treat. 2002;73:45–59. doi: 10.1023/a:1015220420400. [DOI] [PubMed] [Google Scholar]
- 13.Lippman ME. The development of biological therapies for breast cancer. Science. 1993;259:631–2. doi: 10.1126/science.8430312. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham JE, Butler WM. Racial disparities in female breast cancer in South Carolina: clinical evidence for a biological basis. Breast Cancer Res Treat. 2004;88:161–76. doi: 10.1007/s10549-004-0592-9. [DOI] [PubMed] [Google Scholar]
- 15.Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, Smith LV, Labbok MH, Geradts J, Bensen JT, Jackson S, Nyante S, Livasy C, Carey L, Earp HS, Perou CM. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109:123–139. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vona-Davis L, Rose DP. The influence of socioeconomic disparities on breast cancer tumor biology and prognosis: a review. J Womens Health (Larchmt) 2009;18(6):883–93. doi: 10.1089/jwh.2008.1127. [DOI] [PubMed] [Google Scholar]
- 17.Troester MA, Swift-Scanlan T. Challenges in studying the etiology of breast cancer subtypes. Breast Cancer Res. 2009;11(3):104. doi: 10.1186/bcr2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin NU, Claus E, Sohl J, Razzak AR, Arnaout A, Winer EP. Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: high incidence of central nervous system metastases. Cancer. 2008 Nov 15;113(10):2638–45. doi: 10.1002/cncr.23930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson WF, Jatoi I, Devesa SS. Distinct breast cancer incidence and prognostic patterns in the NCI’s SEER program: suggesting a possible link between etiology and outcome. Breast Cancer Res Treat. 2005;90:127–137. doi: 10.1007/s10549-004-3777-3. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Chao L, Li X, Ma G, Chen L, Zang Y, Zhou G. Elevated expression of phosphorylated c-Jun NH2-terminal kinase in basal-like and “triple-negative” breast cancers. Hum Pathol. 2009 Nov 12; doi: 10.1016/j.humpath.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)- negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–8. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 22.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by fivebiomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 23.Phipps AI, Malone KE, Porter PL, Daling JR, Li CI. Body size and risk of luminal, HER2-overexpressing, and triple-negativebreast cancer in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2008;17:2078–2086. doi: 10.1158/1055-9965.EPI-08-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo IH, Russo J. Molecular basis of breast cancer prevention and treatment. Springer-Verlag; Heidelberg: 2004. In vitro models for human breast cancer; pp. 227–80. [Google Scholar]
- 25.Russo J, Hasan Lareef M, Balogh G, Guo S, Russo IH. Estrogen and its metabolites are carcinogenic agents in human breast epithelial cells. J Steroid Biochem Mol Biol. 2003;87:1–25. doi: 10.1016/s0960-0760(03)00390-x. [DOI] [PubMed] [Google Scholar]
- 26.Lareef MH, Garber J, Russo PA, et al. The estrogen antagonist ICI-182–780 does not inhibit the transformation phenotypes induced by 17-beta-estradiol and 4-OH estradiol in human breast epithelial cells. Int J Oncol. 2005;26:423–9. [PubMed] [Google Scholar]
- 27.Fernandez SV, Russo IH, Russo J. Estradiol and its metabolites 4-hydroxyestradiol and 2-hydroxyestradiol induce mutations in human breast epithelial cells. Int J Cancer. 2006;118:1862–8. doi: 10.1002/ijc.21590. [DOI] [PubMed] [Google Scholar]
- 28.Russo J, Lareef MH, Tahin Q, et al. 17Beta-estradiol is carcinogenic in human breast epithelial cells. J Steroid Biochem Mol Biol. 2002;80:149–62. doi: 10.1016/s0960-0760(01)00183-2. [DOI] [PubMed] [Google Scholar]
- 29.Russo J, Fernandez SV, Russo PA, et al. 17-Beta-estradiol induces transformation and tumorigenesis in human breast epithelial cells. FASEB J. 2006;20:1622–34. doi: 10.1096/fj.05-5399com. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y, Fernandez SV, Goodwin S, Russo PA, Russo IH, Sutter TR, Russo J. Epithelial to mesenchymal transition in human breast epithelial cells transformed by 17β-estradiol. Cancer Res. 2007;67:11147–57. doi: 10.1158/0008-5472.CAN-07-1371. [DOI] [PubMed] [Google Scholar]
- 31.Russo J, Gusterson BA, Rogers AE, et al. Comparative study of human and rat mammary tumorigenesis. Lab Invest. 1990;62:244–78. [PubMed] [Google Scholar]
- 32.Calaf G, Zhang PL, Alvarado MV, Estrada S, Russo J. C-Ha-ras enhances the neoplastic transformation of human breast epithelial cells treated with chemical carcinogens. Int J Oncol. 1995;6:5–11. doi: 10.3892/ijo.6.1.5. [DOI] [PubMed] [Google Scholar]
- 33.Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, Fekairi S, Xerri L, Jacquemier J, Brinbaum D, Bertucci F. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–84. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 34.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, et al. A collection of breast cancer cell lines for the study of funtionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymanl transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–97. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 36.Moustakas A, Heldin C-H. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peinado H, Olmeda D, Cano A. Snail, ZEB and bHLH factors in tumor progression: an alliance against the epithelial phenotype? Nature Rev. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 38.Han H-J, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–95. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 39.Klopocki E, Kristiansen G, Wild PJ, Klaman I, Castanos-Velez E, Singer G, Stohr R, Sauter G, Leibiger H, Essers L, Weber B, Hermann K, Rosenthal A, Hartmann A, Dahl E. Loss of SFRP1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int J Oncol. 2004;25:641–9. [PubMed] [Google Scholar]
- 40.Henshall SM, Horvath LG, Quinn DI, Eggleton SA, Grygiel JJ, Stricker PD, Biankin AV, Kench JG, Sutherland RL. Zinc-alpha2-glycoprotein expression as a predictor of metastatic prostate cancer following radical prostatectomy. J Natl Cancer Inst. 2006;98(19):1420–4. doi: 10.1093/jnci/djj378. [DOI] [PubMed] [Google Scholar]
- 41.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park D, Kåresen R, Axcrona U, Noren T, Sauer T. Expression pattern of adhesion molecules (E-cadherin, alpha-, beta-, gamma-catenin and claudin-7), their influence on survival in primary breast carcinoma, and their corresponding axillary lymph node metastasis. APMIS. 2007;115:52–65. doi: 10.1111/j.1600-0463.2007.apm_524.x. [DOI] [PubMed] [Google Scholar]
- 43.Sauer T, Pedersen MK, Ebeltoft K, Naess O. Reduced expression of Claudin-7 in fine needle aspirates from breast carcinomas correlate with grading and metastatic disease. Cytopathology. 2005;16:193–8. doi: 10.1111/j.1365-2303.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- 44.Usami Y, Chiba H, Nakayama F, Ueda J, Matsuda Y, Sawada N, Komori T, Ito A, Yokozaki H. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol. 2006;37:569–77. doi: 10.1016/j.humpath.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 45.Usami Y, Satake S, Nakayama F, Matsumoto M, Ohnuma K, Komori T, Semba S, Ito A, Yokozaki H. Snail-associated epithelial-mesenchymal transition promotes oesophageal squamous cell carcinoma motility and progression. J Pathol. 2008;215:330–9. doi: 10.1002/path.2365. [DOI] [PubMed] [Google Scholar]
- 46.Gosens I, Sessa A, den Hollander AI, Letteboer SJ, Belloni V, Arends ML, Le Bivic A, Cremers FP, Broccoli V, Roepman R. FERM protein EPB4iL5 is a novel member of the mammalian CRB-MPP5 polarity complex. Exp Cell Res. 2007;313:3959–70. doi: 10.1016/j.yexcr.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 47.Xu L, Begum S, Hearn JD, Hynes RO. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc Natl Acad Sci U S A. 2006;103:9023–8. doi: 10.1073/pnas.0602681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu L, Hynes RO. GRP56 and TG2: possible roles in suppression of tumor growth by the microenvironment. Cell Cycle. 2007;6:160–5. doi: 10.4161/cc.6.2.3760. [DOI] [PubMed] [Google Scholar]
- 49.Kwak MK, Lee HJ, Hur K, Park do J, Lee HS, Kim WH, Lee KU, Choe KJ, Guilford P, Yang HK. Expression of Krüppel-like factor 5 in human gastric carcinomas. J Cancer Res Clin Oncol. 2008;134:163–7. doi: 10.1007/s00432-007-0265-2. [DOI] [PubMed] [Google Scholar]
- 50.Haase D, Meister M, Muley T, Hess J, Teurich S, Schnabel P, Hartenstein B, Angel P. FMRD3, a novel putative tumour suppressor in NSCLC. Oncogene. 2007;26:4464–8. doi: 10.1038/sj.onc.1210225. [DOI] [PubMed] [Google Scholar]
- 51.Kairouz R, Parmar J, Lyons RJ, Swarbrick A, Musgrove EA, Daly RJ. Hormonal regulation of the Grb14 signal modulator and its role in cell cycle progression of MCF-7 human breast cancer cells. J Cell Physiol. 2005;203:85–93. doi: 10.1002/jcp.20199. [DOI] [PubMed] [Google Scholar]
- 52.Lyons RJ, Deane R, Lynch DK, Ye ZS, Sanderson GM, Eyre HJ, Sutherland GR, Daly RJ. Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J Biol Chem. 2001;276:17172–80. doi: 10.1074/jbc.M009756200. [DOI] [PubMed] [Google Scholar]
- 53.Rodrigues-Ferreira S, Di Tommaso A, Dimitrov A, Cazaubon S, Gruel N, Colasson H, Nicolas A, Chaverot N, Molinié V, Reyal F, Sigal-Zafrani B, Terris B, Delattre O, Radvanyi F, Perez F, Vincent-Salomon A, Nahmias C. 8p22 MTUS1 gene product ATIP3 is a novel anti-mitotic protein underexpressed in invasive breast carcinoma of poor prognosis. PLoS One. 2009;4:e7239. doi: 10.1371/journal.pone.0007239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frank B, Bermejo JL, Hemminki K, Sutter C, Wappenschmidt B, Meindl A, Kiechle-Bahat M, Bugert P, Schmutzler RK, Bartram CR, Burwinkel B. Copy number variant in the candidate tumor suppressor gene MTUS1 and familial breast cancer risk. Carcinogenesis. 2007;28:1442–5. doi: 10.1093/carcin/bgm033. [DOI] [PubMed] [Google Scholar]
- 55.Di Benedetto M, Bièche I, Deshayes F, Vacher S, Nouet S, Collura V, Seitz I, Louis S, Pineau P, Amsellem-Ouazana D, Couraud PO, Strosberg AD, Stoppa-Lyonnet D, Lidereau R, Nahmias C. Structural organization and expression of human MTUS1, a candidate 8p22 tumor suppressor gene encoding a family of angiotensin II AT2 receptor-interacting proteins, ATIP. Gene. 2006;380:127–36. doi: 10.1016/j.gene.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 56.Zuern C, Heimrich J, Kaufmann R, Richter KK, Settmacher U, Wanner C, Galle J, Seibold S. Down-regulation of MTUS1 in human colon tumors. Oncol Rep. 2010;23:183–9. [PubMed] [Google Scholar]
- 57.Huang D, Yu B, Deng Y, Sheng W, Peng Z, Qin W, Du X. SFRP4 was overexpressed in colorectal carcinoma. J Cancer Res Clin Oncol. 2009 Sep 3; doi: 10.1007/s00432-009-0669-2. [DOI] [PubMed] [Google Scholar]
- 58.Saini S, Liu J, Yamamura S, Majid S, Kawakami K, Hirata H, Dahiya R. Functional significance of secreted Frizzled-related protein 1 in metastatic renal cell carcinomas. Cancer Res. 2009;1(69):6815–22. doi: 10.1158/0008-5472.CAN-09-1254. [DOI] [PubMed] [Google Scholar]
- 59.Hu J, Dong A, Fernandez-Ruiz V, Shan J, Kawa M, Martínez-Ansó E, Prieto J, Qian C. Blockade of Wnt signaling inhibits angiogenesis and tumor growth in hepatocellular carcinoma. Cancer Res. 2009;69:6951–9. doi: 10.1158/0008-5472.CAN-09-0541. [DOI] [PubMed] [Google Scholar]
- 60.Lin YW, Chung MT, Lai HC, De Yan M, Shih YL, Chang CC, Yu MH. Methylation analysis of SFRP genes family in cervical adenocarcinoma. J Cancer Res Clin Oncol. 2009;135:1665–74. doi: 10.1007/s00432-009-0613-5. [DOI] [PubMed] [Google Scholar]
- 61.Gauger KJ, Hugh JM, Troester MA, Schneider SS. Down-regulation of sfrp1 in a mammary epithelial cell line promotes the development of a cd44high/cd24low population which is invasive and resistant to anoikis. Cancer Cell Int. 2009;9:11. doi: 10.1186/1475-2867-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng H, Liu P, Wang ZC, Zou L, Santiago S, Garbitt V, Gjoerup OV, Iglehart JD, Miron A, Richardson AL, Hahn WC, Zhao JJ. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Sci Signal. 2009;2:ra35. doi: 10.1126/scisignal.2000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takemori H, Katoh Hashimoto Y, Nakae J, Olson EN, Okamoto M. Inactivation of HDAC5 by SIK1 in AICAR-treated C2C12 myoblasts. Endocr J. 2009;56:121–30. doi: 10.1507/endocrj.k08e-173. [DOI] [PubMed] [Google Scholar]
- 64.Kowanetz M, Lönn P, Vanlandewijck M, Kowanetz K, Heldin CH, Moustakas A. TGFbeta induces SIK to negatively regulate type I receptor kinase signaling. J Cell Biol. 2008;182:655–62. doi: 10.1083/jcb.200804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ding L, Niu C, Zheng Y, Xiong Z, Liu Y, Lin J, Sun H, Huang K, Yang W, Li X, Ye Q. FHL1 interacts with estrogen receptors and regulates breast cancer cell growth. J Cell Mol Med. 2009 Oct 16; doi: 10.1111/j.1582-4934.2009.00938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin J, Ding L, Jin R, Zhang H, Cheng L, Qin X, Chai J, Ye Q. Four and a half LIM domains 1 and receptor interacting protein of 140kDa (RIP140) interact and cooperate in estrogen signaling. Int J Biochem Cell Biol. 2009;41:1613–8. doi: 10.1016/j.biocel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 67.Engin F, Bertin T, Ma O, Jiang MM, Wang L, Sutton RE, Donehower LA, Lee B. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum Mol Genet. 2009;18:1464–70. doi: 10.1093/hmg/ddp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyazono K. Transforming growth factor-beta signaling in epithelial-mesenchymal transition and progression of cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:314–23. doi: 10.2183/pjab.85.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pennanen PT, Sarvilinna NS, Ylikomi TJ. Gene expression changes during the development of estrogen-independent and antiestrogen-resistant growth in breast cancer cell culture models. Anticancer Drugs. 2009;20:51–8. doi: 10.1097/CAD.0b013e32831845e1. [DOI] [PubMed] [Google Scholar]
- 70.Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, Wagner EF. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat Genet. 2000;24:184–7. doi: 10.1038/72855. [DOI] [PubMed] [Google Scholar]
- 71.Young MR, Colburn NH. Fra-1 a target for cancer prevention or intervention. Gene. 2006;379:1–11. doi: 10.1016/j.gene.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 72.Wang HY, Zhang JY, Cui JT, Tan XH, Li WM, Gu J, Lu YY. Expression status of S100A14 and S100A4 correlates with metastatic potential and clinical outcome in colorectal cancer after surgery. Oncol Rep. 2010;23:45–52. [PubMed] [Google Scholar]
- 73.Hua J, Chen D, Fu H, Zhang R, Shen W, Liu S, Sun K, Sun X. Short hairpin RNA-mediated inhibition of S100A4 promotes apoptosis and suppresses proliferation of BGC823 gastric cancer cells in vitro and in vivo. Cancer Lett. 2009 Nov 27; doi: 10.1016/j.canlet.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 74.Boye K, Mælandsmo GM. S100A4 and Metastasis: A Small Actor Playing Many Roles. Am J Pathol. 2009 Dec 17; doi: 10.2353/ajpath.2010.090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ismail TM, Zhang S, Fernig DG, Gross S, Martin-Fernandez ML, See V, Tozawa K, Tynan CJ, Wang G, Wilkinson MC, Rudland PS, Barraclough R. Self-association of calcium-binding protein S100A4 and metastasis. J Biol Chem. 2010;285:914–22. doi: 10.1074/jbc.M109.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]


