Abstract
The evolution of cancer and RNA viruses share many similarities. Both exploit high levels of genotypic diversity to enable extensive phenotypic plasticity and thereby facilitate rapid adaptation. In order to accumulate large numbers of mutations, we have proposed that cancers express a mutator phenotype. Similar to cancer cells, many viral populations, by replicating their genomes with low fidelity, carry a substantial mutational load. As high levels of mutation are potentially deleterious, the viral mutation frequency is thresholded at a level below which viral populations equilibrate in a traditional mutation-selection balance, and above which the population is no longer viable, i.e., the population undergoes an error catastrophe. Consequently viral populations are susceptible to further increases in mutation load and, recently this phenomenon has been exploited therapeutically by a concept that has been termed lethal mutagenesis. Here we review the application of lethal mutagenesis to the treatment of HIV and discuss how lethal mutagenesis may represent a novel therapeutic approach for the treatment of solid cancers.
Keywords: Lethal mutagenesis, mutator phenotype, mutation rate, cancer genome
1. Introduction
Evolutionary changes are driven by selection of stochastically generated pre-existing variants. The key processes of spontaneous mutation, competition and selection, which underlie adaptation and drive evolution, are evident throughout biology. Human cancers, for example, represent a microcosm of Darwinian evolution: tumor progression is a mutation-driven process that results from the adaptation of a heterogeneous cell population to different microenvironments through the preferential replication of the most suitable variants [1, 2]. Similarly, viruses, by mutating at exceptionally high rates, extensively explore phenotypic space and maximize adaptability to their environment [3]. Thus a high mutation rate offers a powerful mechanism to provide a spectrum of mutants for rapid adaptation to changes in the environment, including, for example, evading the host's immunological defenses. In addition, the high frequency of mutations in viral and tumor populations facilitates the rapid emergence of resistance to therapies. In both cases the underlying evolutionary principle is the same: adaptation occurs through phenotypic selection from a large number of randomly generated mutants.
Spontaneous mutations, which underlie selection, recombination, gene flow, and genetic drift, alter fitness and ultimately facilitate adaptation [4]. In all organisms these processes combine as the predominant mechanism for adaptive response to changing environments. Spontaneous mutations in-of-themselves, however, are more likely to be deleterious than beneficial [5, 6], and in the absence of the need for adaptation to an environmental pressure, random mutation leads to a reduction in overall population fitness. In a changing environment, however, survival depends on the production of new mutations [7]. This balancing between fitness reduction and the need for variation to facilitate adaptation results in an optimized mutation rate that is characteristic for each species and organism (see Table 1). The key to adaptation is therefore genetic variation or, more precisely, productive variation – namely sequence variations that do not compromise organismal fitness under the current state but maintain the potential to adapt to new states [8].
Table 1.
Relationship between species, genome size and mutation rate.
| Genome Size | Mutation | Rate1 | |
|---|---|---|---|
| (bp) | (mutations/ basepair/ replication) |
(mutations/ genome/ replication) |
|
| Riboviruses | |||
| Bacteriophage Qβ | ~3.5 × 103 | 1.9 × 10−3 | 6.5 |
| Poliovirus | ~7.5 × 103 | 1.1 × 10−4 | 0.8 |
| Vesicular stomatitis | ~1.1 × 104 | 3.2 × 10−4 | 3.5 |
| Influenza A | 1.36 × 104 | 7.4 × 10−5 | ~1.0 |
| Retroviruses | |||
| Murine leukemia virus | ~8 × 103 | 3.3 × 10−5 | 0.2 |
| Human immunodeficiency virus type 1 | 9.75 × 103 | 2.1 × 10−5 | 0.2 |
| DNA-based | |||
| Escherichia coli | 4.6 × 106 | 5.4 × 10−10 | 0.0025 |
| Mus musculus | 2.7 × 109 | 1.8 × 10−10 | 0.49 |
| Homo sapiens | 3.2 × 109 | 5.0 × 10−11 | 0.16 |
Data are from reference [13]
Because of their high mutation rates, certain viruses are susceptible to further increases in mutational load [9, 10]. This can be exploited therapeutically by what has been termed lethal mutagenesis [11]. An error catastrophe occurs once the level of mutation induced is sufficient to reduce the overall population fitness and so prevent further propagation of the virus [3]. Here we extend this concept to cancer and propose that lethal mutagenesis of cancer may offer a new therapeutic avenue in selected solid tumors.
2. The high mutation rates of viral genomes
The mutation rates of RNA viruses range from 10−3 to 10−5 substitutions per nucleotide copied [12]; more than one million fold greater than the mutation rate exhibited by human cells [13, 14]. As a result, a significant proportion of viral progeny are non-viable [15, 16]. This high mutation rate coupled with rapid replication, however, allows the virus to extensively explore sequence space and, for example, to evade the host’s immune system [17]. The mutation rate of retroviruses is nearly as high as riboviruses; the mutation rate of the retrovirus HIV-1 is ~8.5 × 10! 5 mutations per base pair per replication cycle (reviewed in [18]). Viral mutations are, for the most part, caused by infidelity during replication of the viral genome, with studies on purified reverse transcriptase documenting a frequency of single base mis-incorporation as great as 10! 4 to 10! 5 [19]. In order to overcome the detrimental effects of this level of self-mutagenesis, many viruses maintain their population density by rapidly replicating their genomes. For example, during the acute stage of HIV-infection as many a 1010 to 1011 new virions are produced daily [15, 17]. Recombination provides another mechanism to counterbalance the negative consequences of high mutation rates [20], and allows the virus to make large leaps in sequence space that would otherwise be difficult to bridge by sequentially accumulated mutations. Recombination can also facilitate the rescue of viral genomes from nonviable parental strains.
2.1. Viral quasispecies
Based initially on mathematical considerations, Manfred Eigen hypothesized that RNA viruses within an infected individual exist not as a single unique variant but rather are a complex, self-perpetuating population of diversely related entities acting as a whole [21]. While the initial infection may only require a few viable virions, viral diversity is generated by the progressive accumulation of mutations during subsequent viral replication, producing a “cloud” of genetically distinct yet related genotypes, termed a quasispecies [9, 22, 23]. In a viral quasispecies, it is the fitness of the entire population, not the fitness of individual members, that determines infectivity and the wildtype of a species refers, not to a particularly fit individual, but to an average for all members [9]. However, as the proportion of a single mutant in the quasispecies depends on its individual fitness, well-adapted mutants have a better chance of producing offspring while deleterious mutants fail to do so. As the chances of finding a well-adapted or advantageous mutant is greatest in a region of sequence space associated with high fitness, there is a large bias towards the accumulation of these mutants [21]. In addition, the viral quasispecies is not simply a collection of diverse mutants but rather a group of interactive variants, which cooperate to contribute to maintaain the population. Direct complementation between members of a viral quasispecies indicates that selection indeed occurs at the population level rather than on individual variants [24]. Thus, this collection of genotypes exists at a mutation-selection balance and modeling show that this, in effect, speeds up the “evolutionary opportunization” of viruses by many orders of magnitude, as compared to random accumulation of mutations within a population [3].
2.2. Error catastrophe
A direct prediction of the quasispecies model is the existence of an error threshold [9, 25], a frequency of mutation above which population extinction occurs due to loss of a significant fraction of genotypes through deleterious mutation, i.e., the virus undergoes an error catastrophe. Indeed Eigen and Schuster showed that there are states in which an apparently trivial elevation in the mutation rate could lead to a fundamental change in the composition of genotypes within a population [21]. The mutation rate of a quasispecies is consequently fine-tuned below this error threshold such that the viral population equilibrates in a traditional mutation-selection balance [3, 26]. This error-catastrophe model has inspired treatments aimed at extinguishing viral populations by elevating their mutation frequencies [11]. The concept, termed “lethal mutagenesis,” predicts that even a modest increase in mutation rate can result in the extinction of the viral population and has been experimentally verified for several viruses [11, 27–38].
2.3. Lethal mutagenesis of viruses
Experiments with RNA viruses provide proof for the concept of lethal mutagenesis: a small increase in the frequency of mutations in the viral genome can ablate the viral population [11]. Chemical and X-irradiation of poliovirus- or vesicular stomatitis virus-infected cells, for example, results in a two-fold increase in viral mutation frequency and is associated with a much larger decrease in viral replicative capacity [39]. These observations argue that the mutation rates of retroviruses and other RNA viruses do approach the maximal value that is compatible with sustained production of infectious progeny and increases exceeding this threshold results in lethal mutagenesis. Similarly, lethal mutagenesis may be one of the mechanism underlying ablation of hepatitis C infection by ribavirin, which in combination with interferon alpha is the most frequently used treatment for chronic liver inflammation caused by hepatitis C and other RNA viral infections [28]. Specifically, ribavirin, once it has entered the cell, is phosphorylated to ribavirin triphosphate, is incorporated into viral RNA by the virally encoded RNA polymerase, and during subsequent RNA amplification mis-pairs at a high frequency [40]. Ribavirin induces multiple changes in cells; it also limits viral replication directly by inhibiting HCV RNA-dependent RNA-polymerase and inosine monophosphate dehydrogenase, reduce the immune response by affecting the secretion of interleukins and modify the activity of cytotoxic lymphocytes [41, 42].
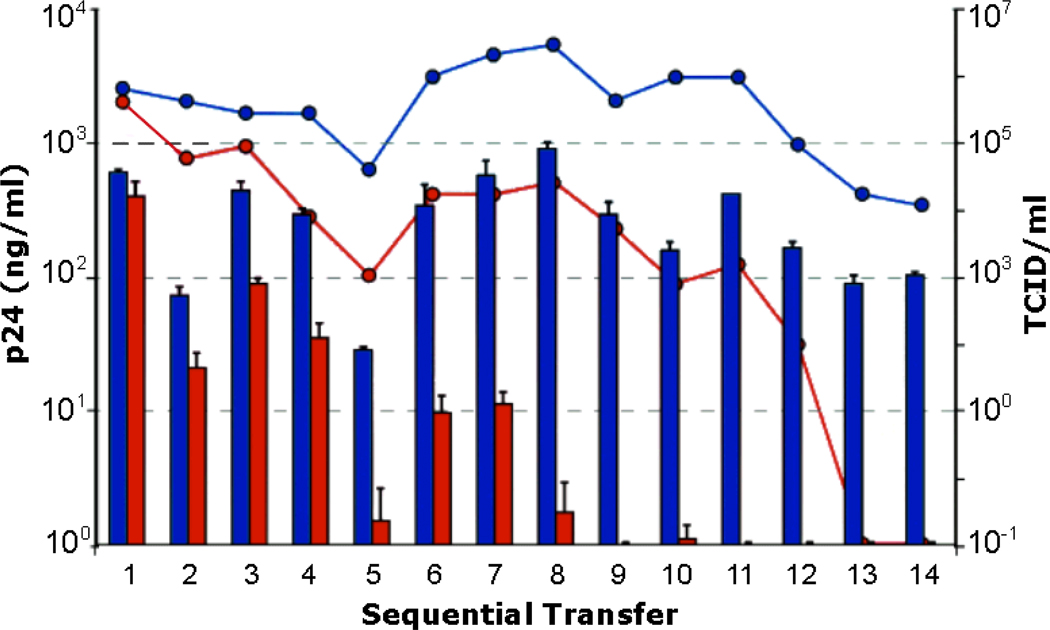
While riboviruses require only a modest ~1.1- to 2.8-fold increase in their mutation frequency in order to reach error catastrophe [39], the retroviral genome may be more tolerant to further increases in mutation frequency [43]. Nonetheless, studies with 5-hydroxy-2'-deoxycytidne (5-OH-dC) and 5-aza-5,6,-dihydro-2'-deoxycytidine (KP-1212) unequivocally demonstrate the potential of lethal mutagenesis for the treatment of HIV. Serial transfer of culture supernatants from HIV infected cells grown in the presence of 0.5 mM 5-OH-dC or a little as 10 nM KP-1212 resulted in ablation of HIV infection after 19–48 and 9–13 transfers, respectively (Fig. 1) [11, 33]. In the case of treatment with KP-1212, HIV reverse transcriptase incorporates KP-1212 triphosphate in place of dCTP (Anderson and Loeb, unpublished results), resulting in a two-fold increase in mutation frequency [33]. The analog-induced mutations are predominantly A>G and G>A transitions, consistent with the predicted base-pairing properties of KP-1212 as determined by NMR (Li, Essigmann and Loeb, unpublished results). In a subsequent phase II clinical trial with KP-1212, newly introduced mutations progressively increased in treated individuals after 56 and 125 days of treatment (Mullins et al., unpublished results). While there was no significant reduction in viral titer over the short course of the trial, the types of mutations observed in the treated group again showed an excess of A>G and G>A single nucleotide substitutions. Depending on the turnover of HIV in sequestered locations, lethal mutagenesis may therefore have the potential to completely eradicate active HIV-infection.
Figure 1. Lethal mutagenesis by 0.1 µM 5-aza-5,6,-dihydro-2'-deoxycytidine (KP-1212) ablates HIV infection.
Supernatants from HIV infected human lymphoblastoid CEM cells, cultured in the absence (blue) or presence (red) of 0.1µM of the mutagenic nucleoside analog KP-1212, were serially transferred to uninfected CEM cells. Viral production was quantified by the detection of p24 antigen (histogram; left scale) and by viral infectivity, measured by TCID50 (lines; right scale). In cells incubated with 0.1µM KP-1212, the amount of p24 was permanently reduced to less than the limit of detection (4 ng/ml) by passage 8, and no infectious HIV was recovered after passage 12.
3. Mutation frequencies in normal and malignant cells
Unlike viruses, eukaryotes replicate their DNA with remarkable accuracy [13]. This accuracy is achieved through a network of conserved and frequently redundant pathways that correct replication errors and repair DNA damage [44]. The multiple mechanisms for the repair of DNA damage in human cells are adequate to guarantee the genetic integrity of cells despite the large number of DNA damaging events that occur each day [45]. Roach et al., for example, have recently shown that as few as 70 mutations accumulate between successive human generations [14]. In contrast to the rarity of mutations in normal human cells, cancer cells contain multiple mutations. We have argued that normal mutation rates cannot account for the number of mutations found in human cancers, and thus we proposed that cancers must exhibit a mutator phenotype, i.e., the mutation rate of cancer cells must be much greater than that of normal cells [1, 46]. The mutator phenotype results from disruption of the function of genes that maintain genetic stability in normal cells and is therefore the driving force for the accumulation of large numbers of mutations in tumors. The resulting genetic diversity, by enabling the selection of tumor promoting events, provides the basis for the emergence of adaptive phenotypes that allow incipient cancer cells to evolve, invade, and metastasize.
3.1. Early evidence for the Mutator Phenotype
Until recently, evidence for the involvement of large numbers of mutations in tumor progression was based mainly on chromosomal aberrations and molecular features of certain hereditary cancers. Early indications of a central role of genome alterations in cancer development emerged in the late nineteenth and early twentieth centuries from studies by David von Hansemann and Theodor Boveri [47, 48]. Techniques such as array comparative genomic hybridization and spectral karyotyping have enabled higher resolution than early cytological observations [49, 50], and have been used to demonstrate that individual metastatic cancer cells harbor a diverse spectrum of unique chromosomal aberrations [51]. Using complementary techniques Stoler et al., estimated that the mean number of genomic events per carcinoma cell is greater than 10,000 [52]. Additional evidence for thousands of mutations in cancer cells came from the observation of changes in the length of microsatellites in tumor DNA from patients with Lynch syndrome (also known as hereditary nonpolyposis colorectal cancer or HNPCC). These patients harbor mutations in mismatch repair (MMR) genes [53, 54], and as a result accumulate thousands of point mutations as well as mutations in as many as 100,000 repetitive sequences per cancer genome [55]. Microsatellite instability has also been detected in tumors without mutations in MMR genes, and in premalignant conditions associated with chronic inflammation [56]. These findings suggest that changes in cellular environments, such as hypoxia [57], may result in a transient deficiency in MMR and give rise to a mutator phenotype. Alterations in the length of poly(dG) repeats in otherwise normal appearing colonic epithelium have even been shown to identify colon cancers at distant sites (see Salk and Horwitz in this issue) [58].
The importance of maintaining genome integrity in preventing tumorigenesis is highlighted by a number of inherited diseases which are associated with elevated risks of specific cancers, and are caused by germline mutations in genes involved in DNA repair,. This association between DNA repair and suppression of carcinogenesis was established, for example, by the seminal findings of the UV-induced DNA damage repair defects in patients with xeroderma pigmentosum [59]. Inherited defects in components of several other DNA-repair pathways also underlie a variety of cancer predisposing syndromes including: mismatch repair (Lynch syndrome) [53, 54], base excision repair (MYH-associated polyposis) [60], homologous recombination (early onset breast cancer) [61], non-homologous DNA end joining (Lig4 syndrome) [62], and translesion synthesis (xeroderma pigmentosum variant) [63]. Hereditary mutations in other genes that are believed to be required for DNA maintenance are also associated with cancer. For instance, mutations in TP53 are found in Li-Fraumeni syndrome [64, 65], a highly cancer-prone condition most frequently associated with sarcomas and breast adenocarcinomas. Additionally, polymorphisms in a large number of genetic stability genes, including OGG1 and XRCC1, are emerging as risk alleles for many cancers (reviewed in [66]).
3.2. Recent evidence for the Mutator Phenotype
Recent evidence strongly supporting the mutator phenotype hypothesis comes primarily from three sources (reviewed in [67, 68]): first, mathematical models that quantitatively predict the efficiency of carcinogenesis with and without a mutator phenotype, indicate that mutator mutations are required for the multiple steps involved in tumor progression (reviewed in this issue by R.A. Beckman); second, human tumors have been shown to have a high frequency of random single base substitutions [69, 70]; and third, DNA sequencing projects have now catalogued large numbers of clonal mutations in individual tumors. These sequencing studies, in particular, have shown that the mutational load in cancer is substantial and highly heterogeneous [71–82].
The International Cancer Genome Consortium, formed in 2008, is currently coordinating efforts to sequence 500 tumors from each of 50 cancer types [83]. It includes two older large-scale projects: the Cancer Genome Atlas and the Cancer Genome project. Both of these projects were initially undertaken with the expectation that exhaustive sequencing of tumor DNA would reveal a small number of key mutations in each cancer type, which would then serve as targets for novel, molecularly directed anticancer therapies [82]. The opposite, however, was found: very few genes are commonly mutated in human cancers. While early cancer genome studies focused primarily on protein coding regions of the genome, the most recent phase of these studies has seen the whole genome characterization of a number of specimens (Table 2). These later studies have unequivocally established that tens of thousands of clonal mutations are present in each cancer genome. As predicted by the mutator phenotype hypothesis, mutations were found to be distributed throughout the nuclear genome of these tumors, with on average one to ten mutations per million basepairs [72, 76–80].
Table 2.
Number of clonal mutations per cancer identified by whole genome sequencing.
| Genome | Clonal Mutations | Non-silent Mutations | Reference |
|---|---|---|---|
| Acute Myeloid Leukemia (n=1) | 500–1,000 | 10 | [75] |
| Acute Myeloid Leukemia (n=1) | 750 | 12 | [76] |
| Small cell lung cancer (n=1) | 22,910 | 101 | [78] |
| Melanoma (n=1) | 33,345 | 182 | [77] |
| Breast cancer (n=3) | 27,173 | ~200 | [70] |
| Non-small cell lung cancer (n=1) | 50,675 | 302 | [74] |
| Normal Human (between generations) | 70 | <<1 | [14] |
Most of the mutations identified by these studies do not appear to be causally involved in the pathogenesis of cancer and only a small subset of the nonsynonymous substitutions are even believed to have been affected by selection [84]. What then do these “passenger” mutations represent? While the substitution trends may partially reflect underlying mutational processes, their distribution may also correspond to hotspots for mutagenesis. If so, one would anticipate that many of these mutations are found in regions of DNA that can assume secondary structures such as hairpins, triple-stranded or quadruplex DNA [85]. Also, these studies fail to characterize the subclonal mutational load of individual cancers [86]. A logical predication of the mutator phenotype hypothesis is that subclonal mutations would be present in large numbers. Many of these mutations are lilely to be in the clonal “driver” genes identified by current methods of DNA sequencing. In addition to the extensive clonal heterogeneity being uncovered, additional mutational diversity exists within individual tumors themselves. A large number of subclonal and random mutations are also present, conferring a unique mutational signature on each cell [51, 69]. This deeper mutational complexity provides a genetic basis for the wide variations observed in tumor behavior and responsiveness to therapy [67].
3.3. Mutation rate as a therapeutic target in cancer
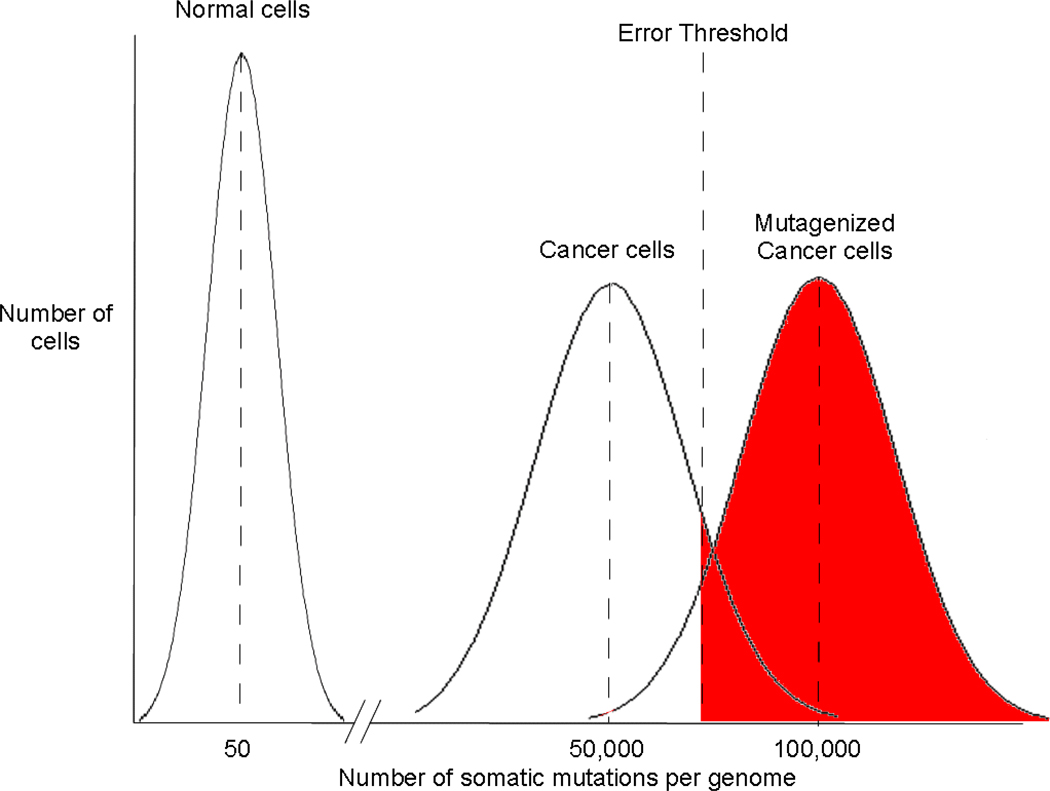
While the extensive genetic variation within a cancer cell population represents the clinically most important consequence of the mutator phenotype, it may also provide unique therapeutic options. Genetic instability in cancer, similar to that of a viral quasispecies, is likely thresholded such that appropriate levels of instability exist to allow selection barriers to be overcome, but excessive instability, which would lead to extinction of the unstable clone, is limited (Fig. 2). The mutation burden of cancer may itself present an unexplored therapeutic target [68]. Conceivably, modulating the mutation frequency of the cancer genome to decrease the overall fitness of the tumor cell population could be achieved either (1) by reducing the mutation rate and thus delaying tumor progression or (2), similar to lethal mutagenesis of viruses, by increasing the mutation burden beyond an error threshold for tumor cell viability.
Figure 2.
Lethal mutagenesis of cancer. Due to the high fidelity of DNA replication and multiple mechanisms for the repair of DNA damage, normal human cells accumulate few mutations (on average approximately 70 per sexual generation). Cancer cells, however, accumulate large numbers of mutations (see Table 2). We propose that genetic instability in cancer cells is limited and, analogous to the situation for RNA viruses, a threshold of mutations exists above which cancer cells are no longer viable. Given their pre-existing mutational load, cancers can therefore be selectively ablated by the incorporation of mutagenic nucleosides.
3.3.1.Treatment and Prevention by delay
Vignuzzi et al., have shown that increasing the fidelity of poliovirus replication markedly limits viral adaptation and pathogenicity [24]. Since mutation accumulation is likely rate limiting for tumor progression, a reduction in mutation rate would also decrease the overall fitness of the tumor cell population and so lengthen the interval between the initiation of cancer and its clinical sequelae. For example, an individual diagnosed with prostatic hypertrophy at age fifty will usually not develop overt malignancy until his eighties [87]. If one could double the number of years it takes for tumor cells to accumulate the requisite number of mutations required for invasiveness and/or metastasis, one would significantly reduce the life-threatening manifestations of cancer.
For most solid tumors, there is more than a 20-year interval between exposure of an individual to a carcinogenic insult and detection of malignancy. Epidemiologically, the link between chronic infection with hepatitis B virus (HBV) and hepatocellular carcinoma (HCC) is, for example, well established. In a prospective population study of 22,707 men, the incidence of HCC in adulthood was found to be greater than 200-fold higher among individuals infected in infancy with HBV as compared to age-matched non-carriers [88]. In the developing world, while hepatitis B infection usually occurs before adolescence, the median age at presentation for HCC is 45 years [89]. Therefore even a two-fold reduction in the rate of mutation accumulation would result in a substantial reduction in associated morbidity and mortality. However, while preventing exposure to carcinogenic insults is a well-established means for reducing cancer incidence, strategies for directly attenuating mutation accumulation in incipient cancer cells have not yet been developed.
3.3.2. Lethal mutagenesis of human cancers
As discussed in section 2, the evolutionary success of many RNA viruses is attributable to the persistent generation of high levels of diversity within the viral population. To maximize adaptive potential, the mutation rate of the viral quasispecies is fine-tuned to establish a mutation-selection balance beyond which the population undergoes an error catastrophe, i.e. no Darwinian selection operates [9, 90]. Consequently, it has been demonstrated experimentally, both in cell culture and in vivo, that increasing the mutation frequency with mutagenic agents can results in extinction of certain viral populations [11, 27–38]. The fitness of a tumor cell population results from a similar balance between the beneficial effects of mutational variation, which can facilitate adaptation under changing environmental pressures, and the detrimental effects of mutation. A limit to the amount of genetic instability that can be tolerated by cancer cells also must exist [68, 91–94], and we propose that human cancers can be selectively ablated by the incorporation of mutagenic nucleosides. Given the pre-existing mutational load of cancer cells, their capacity to tolerate further mutagenesis is most probably thresholded in a manner analogous to the error threshold displayed by RNA viruses (Figure 2). Indeed many commonly used chemotherapeutic agents, such as 5-fluorouracil and temozolomide, are mutagenic and the resultant mutations may, in part, be responsible for their anticancer effects [95, 96].
Studies with RNA viruses demonstrate the possibility of inducing an error catastrophe using mutagenic nucleoside analogs. While the use of mutagenic deoxynucleoside analogs, as opposed to agents that induce DNA adducts, will minimize damage to non-nucleic acid cellular macromolecules [97, 98], several factors have to be considered in selecting compounds for the induction of lethal mutagenesis [27]. First, the compounds must readily enter human cells, be converted to nucleoside triphosphates by normal cellular nucleosides/nucleotide kinases or phosphotransferases, and thereafter be efficiently incorporated into nuclear DNA [99, 100]. Second, the analogs must not be subject to significant DNA repair once incorporated or subject to sanitization while in the nucleotide pool [101]. Lastly, the analogs must mispair at high frequency during replication, leading to the progressive accumulation of mutations. The accumulation of these analogs may be augmented in certain cancers where the mutator phenotype is due to mutations in replicative DNA polymerases that decrease base selection [1], or to dysregulation of low-fidelity specialized DNA polymerases (see Hoffmann and Cazaux in this issue) [102].
A major limitation to molecularly targeted therapies, both antiviral and anticancer, has been the emergence of resistance [103, 104]. One potentially attractive feature of lethal mutagenesis is that the mechanism of killing is uncoupled from exposure. Molecularly targeted therapies, such as azidothymidine (AZT) for HIV [105], create a direct selective pressure for resistant sub-populations. Even in the instance of the important BCR-Abl kinase inhibitors used in the treatment of chronic myelogenous leukemia, such as iminitab (Gleevec), nilotinib and dasatnib, resistance to third line inhibitors has emerged [106, 107]. However, as the deleterious consequences of lethal mutagenesis will not manifest for several generations after incorporation of the mutagenic analog, the possibility of directly selecting for resistance to these agents is minimized. Resistance of viral populations to certain lethal mutagens has been demonstrated experimentally, however only following exposure to very high concentrations of these agents [108].
Enhanced mutagenesis is a major causative factor in the induction of human cancers and the use of mutagenic nucleoside analogs for the treatment of human tumors may therefore have certain limitations. For example, base analogues can be toxic to cells by mechanisms other than lethal mutation induction; thus this strategy will only be useful with analogs that are effective at doses which do not produce acute toxic effects. We appreciate that, irrespective of acute toxicity, the frequency of mutations in non-malignant cells may also increase, potentially resulting in secondary tumors. However, because tumor cells are inherently more error-prone than normal cells, they should preferentially accumulate mutagenic nucleosides, and we envision that it may be possible to calibrate the exposure of normal cells such that it is largely within levels tolerable by their repair capacities but saturates the repair mechanisms of cells possessing a mutator phenotype. The emergence of secondary tumors could be carefully monitored for and would be predicted not to be an immediate event [109, 110]. Lastly, we propose that lethal mutagenesis of cancer would, at least initially, be restricted to patients who have failed extensive prior conventional chemotherapy and as such, have predictably limited life expectancies. Concerns regarding the induction of secondary malignancies would thus be reduced, and as tumors in these individuals will likely have accumulated additional mutations due to prior chemotherapy, they may be more susceptible to lethal mutagenesis.
4. Concluding remarks
The discovery that the mutation rate of viral quasispecies is fine-tuned below an error threshold lead to the prediction that even modest increases in mutation rate could result in the extinction of a viral population. This has been experimentally verified, both in cell culture and in vivo, for several viruses [11, 27–38]. Cancers express a mutator phenotype, and their mutational burden may be limited in a manner analogous to the error threshold displayed by RNA viruses. We envision that treatment of cancer cells with mutagenic nucleoside analogs will result in the accumulation of mutations until a critical level is obtained that results in an error catastrophe-like ablation of the tumor.
Acknowledgments
We thank Drs. Scott Kennedy, Ray Monnat, James Mullins and Marc Prindle for critical comments on the manuscript, and many other colleagues at the University of Washington for critical discussions and unique insights. National Cancer Institute Grants CA102029 and CA115802 supported this work.
Abbreviations
- 5-OH-dC
5-hydroxy-2'-deoxycytidine
- AZT
azidothymidine
- HBV
Hepatitis B virus
- HCV
Hepatitis C virus
- HCC
hepatocellular carcinoma
- KP-1212
5-aza-5, 6,-dihydro-2'-deoxycytidine
- MMR
mismatch repair
- TCID50
50% Tissue Culture Infective Dose
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest statement
The authors declare that there are no conflicts of interest. LAL is a member of the SAB of Koronis Pharmaceuticals.
References
- 1.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 2.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Eigen M. Viral quasispecies. Sci Am. 1993;269:42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 4.Drake JW. Spontaneous mutation. Annu Rev Genet. 1991;25:125–146. doi: 10.1146/annurev.ge.25.120191.001013. [DOI] [PubMed] [Google Scholar]
- 5.Fay JC, Wyckoff GJ, Wu CI. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao F, Chen Y, Levy DN, Conway JA, Kepler TB, Hui H. Unselected mutations in the human immunodeficiency virus type 1 genome are mostly nonsynonymous and often deleterious. J Virol. 2004;78:2426–2433. doi: 10.1128/JVI.78.5.2426-2433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjedov I, Tenaillon O, Gerard B, Souza V, Denamur E, Radman M, et al. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 8.Gould SJ. The evolution of life on the earth. Sci Am. 1994;271:84–91. doi: 10.1038/scientificamerican1094-84. [DOI] [PubMed] [Google Scholar]
- 9.Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 10.Eigen M. Error catastrophe and antiviral strategy. Proc Natl Acad Sci U S A. 2002;99:13374–13376. doi: 10.1073/pnas.212514799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci U S A. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake JW. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann N Y Acad Sci. 1999;870:100–107. doi: 10.1111/j.1749-6632.1999.tb08870.x. [DOI] [PubMed] [Google Scholar]
- 14.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 16.Hughes SH, Shank PR, Spector DH, Kung HJ, Bishop JM, Varmus HE, et al. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978;15:1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- 17.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 18.Smith RA, Loeb LA, Preston BD. Lethal mutagenesis of HIV. Virus Res. 2005;107:215–228. doi: 10.1016/j.virusres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 20.Mikkelsen JG, Pedersen FS. Genetic reassortment and patch repair by recombination in retroviruses. J Biomed Sci. 2000;7:77–99. doi: 10.1007/BF02256615. [DOI] [PubMed] [Google Scholar]
- 21.Eigen M, Schuster P. The hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften. 1977;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 22.Domingo E, Martinez-Salas E, Sobrino F, de la Torre JC, Portela A, Ortin J, et al. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance--a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 23.Steinhauer DA, Holland JJ. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- 24.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eigen M, Schuster P, Sigmund K, Wolff R. Elementary step dynamics of catalytic hypercycles. Biosystems. 1980;13:1–22. doi: 10.1016/0303-2647(80)90002-7. [DOI] [PubMed] [Google Scholar]
- 26.Bull JJ, Meyers LA, Lachmann M. Quasispecies made simple. PLoS Comput Biol. 2005;1 doi: 10.1371/journal.pcbi.0010061. e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson JP, Daifuku R, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu Rev Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 28.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 29.Dapp MJ, Clouser CL, Patterson S, Mansky LM. 5-Azacytidine can induce lethal mutagenesis in human immunodeficiency virus type 1. J Virol. 2009;83:11950–11958. doi: 10.1128/JVI.01406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domingo E, Pariente N, Airaksinen A, Gonzalez-Lopez C, Sierra S, Herrera M, et al. Foot-and-mouth disease virus evolution: exploring pathways towards virus extinction. Curr Top Microbiol Immunol. 2005;288:149–173. doi: 10.1007/3-540-27109-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graci JD, Too K, Smidansky ED, Edathil JP, Barr EW, Harki DA, et al. Lethal mutagenesis of picornaviruses with N-6-modified purine nucleoside analogues. Antimicrob Agents Chemother. 2008;52:971–979. doi: 10.1128/AAC.01056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harki DA, Graci JD, Edathil JP, Castro C, Cameron CE, Peterson BR. Synthesis of a universal 5-nitroindole ribonucleotide and incorporation into RNA by a viral RNA-dependent RNA polymerase. Chembiochem. 2007;8:1359–1362. doi: 10.1002/cbic.200700160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris KS, Brabant W, Styrchak S, Gall A, Daifuku R. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antiviral Res. 2005;67:1–9. doi: 10.1016/j.antiviral.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann WP, Polta A, Herrmann E, Mihm U, Kronenberger B, Sonntag T, et al. Mutagenic effect of ribavirin on hepatitis C nonstructural 5B quasispecies in vitro and during antiviral therapy. Gastroenterology. 2007;132:921–930. doi: 10.1053/j.gastro.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Loeb LA, Mullins JI. Lethal mutagenesis of HIV by mutagenic ribonucleoside analogs. AIDS Res Hum Retroviruses. 2000;16:1–3. doi: 10.1089/088922200309539. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz-Jarabo CM, Ly C, Domingo E, de la Torre JC. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) Virology. 2003;308:37–47. doi: 10.1016/s0042-6822(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 37.Sierra M, Airaksinen A, Gonzalez-Lopez C, Agudo R, Arias A, Domingo E. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J Virol. 2007;81:2012–2024. doi: 10.1128/JVI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tapia N, Fernandez G, Parera M, Gomez-Mariano G, Clotet B, Quinones-Mateu M, et al. Combination of a mutagenic agent with a reverse transcriptase inhibitor results in systematic inhibition of HIV-1 infection. Virology. 2005;338:1–8. doi: 10.1016/j.virol.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Holland JJ, Domingo E, de la Torre JC, Steinhauer DA. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J Virol. 1990;64:3960–3962. doi: 10.1128/jvi.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potter CW, Phair JP, Vodinelich L, Fenton R, Jennings R. Antiviral, immunosuppressive and antitumour effects of ribavirin. Nature. 1976;259:496–497. doi: 10.1038/259496a0. [DOI] [PubMed] [Google Scholar]
- 42.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 43.Pathak VK, Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedberg EC. DNA repair and mutagenesis. 2nd ed. Washington, D.C: ASM Press; 2006. [Google Scholar]
- 45.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 46.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 47.Boveri T. Uber mehrpolige mitosen als mittel zur analyse des zellkerns. Verh D Phys Med Ges Wurzberg. 1902;35:67–90. [Google Scholar]
- 48.von Hansemann D. Ueber asymmetrische Zelltheilung in epithel Krebsen und deren biologische Bedeutung. Virchows Arch A Pathol Anat Histopathol. 1890;119:299. [Google Scholar]
- 49.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 50.Schrock E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 51.Klein CA, Schmidt-Kittler O, Schardt JA, Pantel K, Speicher MR, Riethmuller G. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci U S A. 1999;96:4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoler DL, Chen N, Basik M, Kahlenberg MS, Rodriguez-Bigas MA, Petrelli NJ, et al. The onset and extent of genomic instability in sporadic colorectal tumor progression. Proc Natl Acad Sci U S A. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 54.Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 55.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 56.Brentnall TA, Crispin DA, Bronner MP, Cherian SP, Hueffed M, Rabinovitch PS, et al. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res. 1996;56:1237–1240. [PubMed] [Google Scholar]
- 57.Kondo A, Safaei R, Mishima M, Niedner H, Lin X, Howell SB. Hypoxia-induced enrichment and mutagenesis of cells that have lost DNA mismatch repair. Cancer Res. 2001;61:7603–7607. [PubMed] [Google Scholar]
- 58.Salk JJ, Salipante SJ, Risques RA, Crispin DA, Li L, Bronner MP, et al. Clonal expansions in ulcerative colitis identify patients with neoplasia. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0909428106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 60.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 61.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 62.Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 63.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, et al. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 65.Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348:747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 66.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 67.Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–3557. doi: 10.1158/0008-5472.CAN-07-5835. discussion 7. [DOI] [PubMed] [Google Scholar]
- 68.Prindle MJ, Fox EJ, Loeb LA. The Mutator Phenotype in Cancer: Molecular Mechanisms and Targeting Strategies. Curr Drug Targets. 2010 doi: 10.2174/1389450111007011296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng L, Dai H, Zhou M, Li M, Singh P, Qiu J, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- 71.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 77.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 82.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 83.Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rubin AF, Green P. Mutation patterns in cancer genomes. Proc Natl Acad Sci U S A. 2009;106:21766–21770. doi: 10.1073/pnas.0912499106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McMurray CT. DNA secondary structure: a common and causative factor for expansion in human disease. Proc Natl Acad Sci U S A. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fox EJ, Salk JJ, Loeb LA. Cancer genome sequencing--an interim analysis. Cancer Res. 2009;69:4948–4950. doi: 10.1158/0008-5472.CAN-09-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balducci L, Khansur T, Smith T, Hardy C. Prostate cancer: a model of cancer in the elderly. Arch Gerontol Geriatr. 1989;8:165–187. doi: 10.1016/0167-4943(89)90060-5. [DOI] [PubMed] [Google Scholar]
- 88.Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 89.Kirk GD, Bah E, Montesano R. Molecular epidemiology of human liver cancer: insights into etiology, pathogenesis and prevention from The Gambia, West Africa. Carcinogenesis. 2006;27:2070–2082. doi: 10.1093/carcin/bgl060. [DOI] [PubMed] [Google Scholar]
- 90.Schuster P. RNA based evolutionary optimization. Orig Life Evol Biosph. 1993;23:373–391. doi: 10.1007/BF01582087. [DOI] [PubMed] [Google Scholar]
- 91.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–M60. [PubMed] [Google Scholar]
- 92.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci U S A. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sole RV, Deisboeck TS. An error catastrophe in cancer? J Theor Biol. 2004;228:47–54. doi: 10.1016/j.jtbi.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 94.Bull JJ, Wilke CO. Lethal mutagenesis of bacteria. Genetics. 2008;180:1061–1070. doi: 10.1534/genetics.108.091413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Glaab WE, Mitchell LS, Miller JE, Vlasakova K, Skopek TR. 5-fluorouracil forward mutation assay in Salmonella: determination of mutational target and spontaneous mutational spectra. Mutat Res. 2005;578:238–246. doi: 10.1016/j.mrfmmm.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 96.Skopek TR, Thilly WG. Rate of induced forward mutation at 3 genetic loci in Salmonella typhimurium. Mutat Res. 1983;108:45–56. doi: 10.1016/0027-5107(83)90108-2. [DOI] [PubMed] [Google Scholar]
- 97.Rechkoblit O, Kolbanovskiy A, Malinina L, Geacintov NE, Broyde S, Patel DJ. Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4. Nat Struct Mol Biol. 2010;17:379–388. doi: 10.1038/nsmb.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Upton DC, Wang X, Blans P, Perrino FW, Fishbein JC, Akman SA. Mutagenesis by exocyclic alkylamino purine adducts in Escherichia coli. Mutat Res. 2006;599:1–10. doi: 10.1016/j.mrfmmm.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 99.Arner ES, Eriksson S. Mammalian deoxyribonucleoside kinases. Pharmacol Ther. 1995;67:155–186. doi: 10.1016/0163-7258(95)00015-9. [DOI] [PubMed] [Google Scholar]
- 100.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 101.Mo JY, Maki H, Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc Natl Acad Sci U S A. 1992;89:11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lemee F, Bergoglio V, Fernandez-Vidal A, Machado-Silva A, Pillaire MJ, Bieth A, et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci U S A. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Darby G, Larder BA. The clinical significance of antiviral drug resistance. Res Virol. 1992;143:116–120. doi: 10.1016/s0923-2516(06)80091-9. [DOI] [PubMed] [Google Scholar]
- 104.Hochhaus A, Hughes T. Clinical resistance to imatinib: mechanisms and implications. Hematol Oncol Clin North Am. 2004;18:641–656. doi: 10.1016/j.hoc.2004.03.001. ix. [DOI] [PubMed] [Google Scholar]
- 105.Larder BA, Darby G, Richman DD. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 106.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 107.Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 108.Agudo R, Ferrer-Orta C, Arias A, de la Higuera I, Perales C, Pérez-Luque R, et al. A Multi-Step Process of Viral Adaptation to a Mutagenic Nucleoside Analogue by Modulation of Transition Types Leads to Extinction-Escape. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001072. e1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baker DB, Landrigan PJ. Occupationally related disorders. Med Clin North Am. 1990;74:441–460. doi: 10.1016/s0025-7125(16)30572-7. [DOI] [PubMed] [Google Scholar]
- 110.Wogan GN, Hecht SS, Felton JS, Conney AH, Loeb LA. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14:473–486. doi: 10.1016/j.semcancer.2004.06.010. [DOI] [PubMed] [Google Scholar]