Abstract
Fragments of 16S ribosomal RNA from E. coli and B. stearothermophilus, respectively comprising the 49 and 52 3' terminal nucleotides have been studied thermodynamically using high sensitivity differential scanning calorimetry. The fragments were isolated after cleavage of 16S rRNA in the ribosome by the bacteriocin cloacin DF13. Comparison of the thermal properties of the E. coli fragments with those derived from a kasugamycin resistant mutant, which specifically lacks dimethylation of two adjacent adenosines was employed to study the effect of the methylgroups on the thermal stability. Both E. coli species exhibit similar complex melting patterns with several transitions. Overall molar transition enthalpies are equal and do not depend significantly on buffer conditions (120 kcal/mol at 15 mM Na+ to 136 kcal/mol at 215 mM Na+). However, the transition with the highest Tm, corresponding to unfolding of a nine basepair central helix is lowered by the dimethylation of the adenines in the four-membered loop. This decrease amounts to 4 degrees C at 15 mM Na+ and 2 degrees C at 215 mM Na+. The corresponding nine basepair long hairpin in the Bacillus fragment melts at a temperature of 70 degrees C at 15 mM Na+. This Tmax is much higher than expected on the basis of the sequence in the hairpin.
Full text
PDF
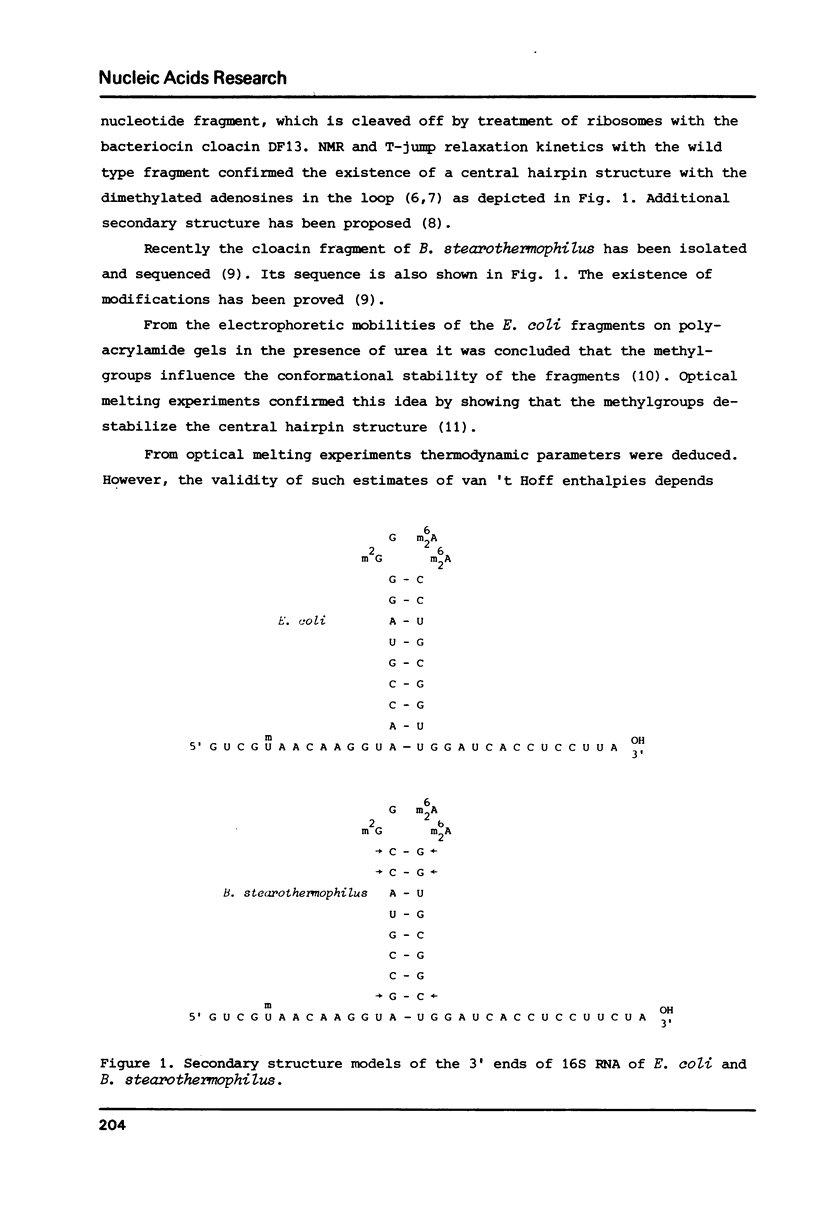



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baan R. A., Hilbers C. W., Van Charldorp R., Van Leerdam E., Van Knippenberg P. H., Bosch L. High-resolution proton magnetic resonance study of the secondary structure of the 3'-terminal 49-nucleotide fragment of 16S rRNA from Escherichia coli. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1028–1031. doi: 10.1073/pnas.74.3.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baan R. A., van Charldorp R., van Leerdam E., van Knippenberg P. H., Bosch L., de Rooij J. F., van Boom J. H. The 3'-terminus of 16 S ribosomal RNA of Escherichia coli. Isolation and purification of the terminal 49-nucleotide fragment at a milligram scale. FEBS Lett. 1976 Dec 1;71(2):351–355. doi: 10.1016/0014-5793(76)80968-4. [DOI] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Cole P. E., Yang S. K., Crothers D. M. Conformational changes of transfer ribonucleic acid. Equilibrium phase diagrams. Biochemistry. 1972 Nov 7;11(23):4358–4368. doi: 10.1021/bi00773a024. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E., Dahlberg J. E. Binding of ribosomal protein S1 of Escherichia coli to the 3' end of 16S rRNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2940–2944. doi: 10.1073/pnas.72.8.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helser T. L., Davies J. E., Dahlberg J. E. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat New Biol. 1971 Sep 1;233(35):12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- Hinz H. J., Filimonov V. V., Privalov P. L. Calorimetric studies on melting of tRNA Phe (yeast). Eur J Biochem. 1977 Jan 3;72(1):79–86. doi: 10.1111/j.1432-1033.1977.tb11226.x. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Itakura K., Breslauer K. J. Extra adenosine stacks into the self-complementary d(CGCAGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):445–451. doi: 10.1021/bi00532a004. [DOI] [PubMed] [Google Scholar]
- Poldermans B., Bakker H., Van Knippenberg P. H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16S ribosomal RNA of Escherichia coli. IV. The effect of the methylgroups on ribosomal subunit interaction. Nucleic Acids Res. 1980 Jan 11;8(1):143–151. doi: 10.1093/nar/8.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldermans B., Van Buul C. P., Van Knippenberg P. H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli. II. The effect of the absence of the methyl groups on initiation of protein biosynthesis. J Biol Chem. 1979 Sep 25;254(18):9090–9093. [PubMed] [Google Scholar]
- Privalov P. L., Khechinashvili N. N. A thermodynamic approach to the problem of stabilization of globular protein structure: a calorimetric study. J Mol Biol. 1974 Jul 5;86(3):665–684. doi: 10.1016/0022-2836(74)90188-0. [DOI] [PubMed] [Google Scholar]
- Van Charldorp R., Heus H. A., Van Knippenberg P. H. Adenosine dimethylation of 16S ribosomal RNA: effect of the methylgroups on local conformational stability as deduced from electrophoretic mobility of RNA fragments in denaturing polyacrylamide gels. Nucleic Acids Res. 1981 Jan 24;9(2):267–275. doi: 10.1093/nar/9.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Heus H. A., Van Knippenberg P. H., Joordens J., De Bruin S. H., Hilbers C. W. Destabilization of secondary structure in 16S ribosomal RNA by dimethylation of two adjacent adenosines. Nucleic Acids Res. 1981 Sep 11;9(17):4413–4422. doi: 10.1093/nar/9.17.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Van Kimmenade A. M., Van Knippenberg P. H. Sequence and secondary structure of the colicin fragment of Bacillus stearothermophilus 16S ribosomal RNA. Nucleic Acids Res. 1981 Oct 10;9(19):4909–4917. doi: 10.1093/nar/9.19.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Van Knippenberg P. H. Sequence, modified nucleotides and secondary structure at the 3'-end of small ribosomal subunit RNA. Nucleic Acids Res. 1982 Feb 25;10(4):1149–1158. doi: 10.1093/nar/10.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R. C., Steitz J. A., Moore P. B., Crothers D. M. The 3' terminus of 16S rRNA: secondary structure and interaction with ribosomal protein S1. Nucleic Acids Res. 1979 Dec 20;7(8):2399–2418. doi: 10.1093/nar/7.8.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]



