Abstract
Animal genes of different lineages, such as vertebrates and arthropods, are well-organized and blended into dynamic chromosomal structures that represent a primary regulatory mechanism for body development and cellular differentiation. The majority of genes in a genome are actually clustered, which are evolutionarily stable to different extents and biologically meaningful when evaluated among genomes within and across lineages. Until now, many questions concerning gene organization, such as what is the minimal number of genes in a cluster and what is the driving force leading to gene co-regulation, remain to be addressed. Here, we provide a user-friendly database—LCGbase (a comprehensive database for lineage-based co-regulated genes)—hosting information on evolutionary dynamics of gene clustering and ordering within animal kingdoms in two different lineages: vertebrates and arthropods. The database is constructed on a web-based Linux-Apache-MySQL-PHP framework and effective interactive user-inquiry service. Compared to other gene annotation databases with similar purposes, our database has three comprehensible advantages. First, our database is inclusive, including all high-quality genome assemblies of vertebrates and representative arthropod species. Second, it is human-centric since we map all gene clusters from other genomes in an order of lineage-ranks (such as primates, mammals, warm-blooded, and reptiles) onto human genome and start the database from well-defined gene pairs (a minimal cluster where the two adjacent genes are oriented as co-directional, convergent, and divergent pairs) to large gene clusters. Furthermore, users can search for any adjacent genes and their detailed annotations. Third, the database provides flexible parameter definitions, such as the distance of transcription start sites between two adjacent genes, which is extendable to genes that flanking the cluster across species. We also provide useful tools for sequence alignment, gene ontology (GO) annotation, promoter identification, gene expression (co-expression), and evolutionary analysis. This database not only provides a way to define lineage-specific and species-specific gene clusters but also facilitates future studies on gene co-regulation, epigenetic control of gene expression (DNA methylation and histone marks), and chromosomal structures in a context of gene clusters and species evolution. LCGbase is freely available at http://lcgbase.big.ac.cn/LCGbase.
Keywords: co-regulated genes, vertebrate, evolution, database
Introduction
Animal genomes harbor several tens of thousands of protein-coding and RNA-coding genes and the rest are regulatory elements adjacent to genes.1 Although there are intergenic sequences, which have been called “gene desert”, it is believed that a majority of them may also be parts of genes that have not yet been discovered.2,3 It is important for the entire genome to be regulated timely and accurately through a battery of processes with distinct mechanisms. In prokaryotes (such as Escherichia coli) and lower eukaryotes (such as Caenorhabditis elegans), operons or clustered genes are major regulatory mechanisms so that genes in a consecutive order share a suite of transcription machinery and its accessories.4 However, gene structures in higher eukaryotes are not only greater in numbers but also more complex than those of prokaryotes.5 For instance, there are large genes in a size range of one million basepairs or more (such as dystrophin) and numerous regulatory elements for transcriptional regulation, including enhancers, insulators, silencers, and repressors.6 In addition, epigenetic regulations, including DNA methylation, hydroxymethylation, various histone marks, and chromatin structure states, may all play essential roles in the construction of a multiple-layer gene expression regulatory network.7,8 In such as a complex gene regulation context, co-regulated genes are first to be scrutinized since they are readily defined based on transcriptomic data and are either adjacent to each other or co-regulated (co-activated, co-suppressed, and antagonism); the minimal co-regulated genes are a pair of genes adjacent to each other and the maxima are several genes that are clustered together over evolutionary time scale, which may be even extendable to large chromosomal regions.4 For instance, some of the clustered genes may perform long-range interaction-based functions or be involved in the same regulatory or metabolic pathways.9 The precise identification of compositional and organizational features for these gene clusters may improve our knowledge on transcriptional controls and RNA processing mechanisms.
Previous studies on minimal gene clustering have been largely focused on genes in three basic categories of paired orientations according to the relative transcription direction between two neighboring genes: divergently-paired (DPGs, positioned head-to-head but transcribed toward opposite directions), co-directionally-paired (CDPGs, positioned head-to-tail and transcribed in the same direction), and convergently-paired genes (CPGs, positioned tail-to-tail and transcribed toward each other).10,11 It has been suggested that tandem duplication may be the major cause leading to these paired genes (especially CDPGs), and promoter sharing is an plausible explanation for the occurrence of DPGs.4,12 It has been reported that the proportion of DPGs is positively correlated with gene densities as DPGs tend to keep their transcription directions throughout relatively larger evolutionary time scale (eg, human to fugu comparison).10 DPGs tend to perform similar biological functions being involved in housekeeping functions, as compared to CDPGs and CPGs, and the expression of DPGs is often positively correlated (albeit minor exceptions) at different developmental stages and under pathologic conditions.10,11 Furthermore, when comparing dynamic structural features of DPGs between vertebrates and insects, we found that all three categories of paired genes in insects are less conserved than their vertebrate counterparts, although DPGs in insects also tend to form functional clusters and to share promoters.13
As to the intergenic distance (longer in metazoa and shorter in fungi), although the distance of transcription starts between two co-regulated DPGs is between a few hundreds and around one thousand basepairs,12 we recognize the possible function of sequences— often tens of kilo-basepairs in length—between the two neighboring DPGs with respect to co-expression and shared regulatory elements.14 Furthermore, the bimodality of intergenic distances observed among mammal gene pairs (but not in other vertebrates) suggests that mammals share certain common features in transcription regulation.11 Until now, how the length of intergenic regions affects the contiguity in regulating multiple genes remains to be illuminated.
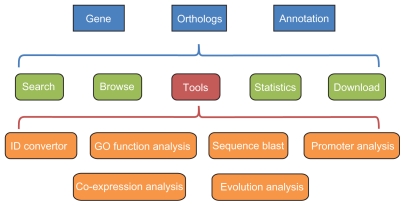
As next-generation sequencing technology matures, both cost and throughput are in favor of more basic data acquisition. In future studies, lineage-based data organization will take over the “one-covers-all” fashion and more tools will be developed for handling both larger and more genomes in addition to those for smaller and single genomes, such as those of mitochondrion,15 plastid,16 and yeast.17,18 A recent study has expanded a gene order browser into 74 species but covers only four mammals.19 In this study we curated 38 mammal and 14 other animal genomes (only use one fungus as out-group) to discover and to display conserved gene clusters across mammals and their sub-groups, such as primates, large mammals, and rodents. In particular, we combine the two concepts that stringently-defined lineage-specific conserved core paired genes (based on both orthology and transcriptional direction) and gene order of ten consecutive genes flanking the core paired genes. We also offer a series of toolkits covering GO functional annotations promoter identification, gene expression, and evolution analysis to help characterizing features of gene clusters (Fig. 1).
Figure 1.
A flowchart to illustrate the content and organization of LCGbase.
Using LCGbase, we would like to address several most imperative questions: (1) Although mammalian gene order or genome organization have been thought to be non-randomly distributed among the chromosomes, what is the precise number of genes that tend to move around or to form clusters? (2) How are clustered genes conserved across various definable lineages? Are the forming-and-breaking events evolutionarily selected and functionally meaningful? What are the mechanisms, including rearrangement, translocation, inversion, recombination, duplication, and transposon-mediated episodes, that alter clustered genes? (3) Are we able to define a “core clustered set” for different lineages or subgroups? Are there identifiable chromosomal regions whose gene clusters are evolutionarily stable? (4) How are gene clusters related to nucleosome positioning and chromosome folding in the nucleus?20,21 The questioning continues but the conclusions will be what we have to know for every single gene and its position on the chromosome, not only physically but also functionally.
Functionality
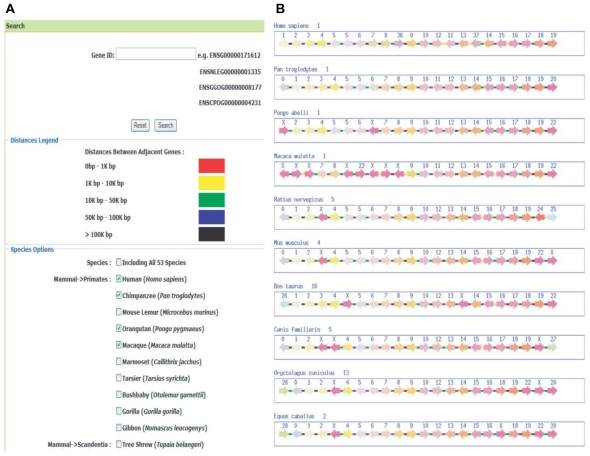
These are several ways to reach available data in LCGbase. First, one can utilize the browse option to direct all annotated genes in the 53 species, and each gene can be found by the link of gene ID. Second, one can take advantage of gene positioning or clustering information to use a gene ID from the neighbouring genes within and across lineages. In particular, the search is strand-sensitive when used to detect strand-specific organizational features of gene clusters and their variations. The database also distinguishes TSS (transcript start site) distances between two adjacent genes in five roughly defined categories: 0–1 kbp, 1–10 kbp, 10–50 kbp, 50–100 kbp, and >100 kbp. It display ten genes left or right of the core gene cluster and high light all the genes on screen in different colours to indicate their orthologous groups. Furthermore, it assigns random group numbers to order all groups (Fig. 2). Genes that are not assigned in groups are labelled with “X”. Users can click on the hyperlink for each gene to check for detailed annotations (eg, location, structure, ontology, and family). Third, the result page also displays gene orders from different species according to taxonomic and lineage definitions, such as mammals (primates, rodents, afrosoricida, carnivora, chiroptera, lagomorpha), birds (galliformes and passeriformes), reptiles (squamata), amphibians (anura), fishes (beloniformes, tetraodontiformes, cypriniformes, gasterosteiformes), insects (diptera), chordata (enterogona), nematoda (rhabditida), and fungi (saccharomycetales). The information helps to reveal lineage-specific dynamic patterns or rules of gene clusters in lineage groups and sub-group. In particular, the database provides three kinds of downloadable files (xls, cvs and html) containing information including species, gene ID, strand category, and group number, which appears on the search result page. Fourth, we also count species number, strand-specificity, and orthologous gene. Fifth, the database also provides blast tools22 (ie, to match cDNA sequence with blastn and protein sequence with blastp or blastx) to help users to study their query sequences and associate them to data in LCGbase as well as other databases.
Figure 2.
An example of the LCGbase browser (A) and a search result (B). The inquired gene is ENSG00000171612.
Due to co-regulation, genes in a cluster may have related functions, share promoters, evolve at a similar rate or in a distinct pattern, and show significantly correlated expressions. LCGbase also provides several easy-to-use tools to facilitate the analysis of these features. Due to the fact that gene ID used in this database is the same as the Ensembl gene ID, an ID Convertor tool takes charge of converting gene IDs of other systems (eg, Entrez Gene ID, Gene Symbol, Refseq mRNA ID and Refseq protein ID) into Ensemble gene ID. GO Function Classification tool is to compare a query gene list with all genes in both species and GO terms (with at least 10 genes)23 and performs gene function enrichment analysis to determine whether gene clusters tend to be functionally related or not. This tool adopts the Fisher Exact Test involved in perl Text-NSP module (http://search.cpan.org/dist/Text-NSP/) combing with four multiple testing correction methods (ie, Bonferroni correction, Bonferroni Step-down [Holm] correction, Benjamini & Hochberg False Discovery Rate, and Not adjusted).24 Four cut-off values are to be chosen: 0.1, 0.05, 0.01, and 0.001. Promoter Analysis tool is to compare a query nucleotide sequence with the upstream and downstream (from −499 bp to 100 bp, or from −9999 bp to 6000 bp) of experimentally-identified transcript start site (TSS) embedded in Eukaryotic Promoter Database (EPD), which is a promoter sequence collection of model organisms.25 To illustrate the co-expressed genes in a cluster, we introduced co-expression data of seven animals including human, mouse, rat, chicken, zebrafish, fly, and nematode from COXPRESdb (Gene Coexpression Database).26 We adopted R package “BioNet” to draw network,27 when a query gene has correlated expression with other query genes. Evolution Analysis tool includes KaKs_Calulator2.0 toolkit28 that adopts multiple algorithms and alternative codon tables to compute nonsynonymous (Ka) and synonymous mutation rates (Ks). The ratio of Ka to Ks is a popular statistical measure for selection between one or multiple pairs of protein-coding genes and one may want to know if several genes in a cluster evolve simultaneously.
In the statistics section, we draw two types of figures to describe TSS distance and minimal distance between three cluster classes: CDPGs, CPGs, and DPGs. Minimal distance is defined as (1) the subtraction of the 5′-end of the downstream transcript and the 3′-end of the upstream transcript for CDPGs, (2) the subtraction of the 3′-end of the downstream transcript and the 3′-end of the upstream transcript for CPGs, and (3) the subtraction of the 5′-end of the downstream transcript and the 5′-end of the upstream transcript for DPGs. In the downloadable page, we also provide the characterized features of gene pairs (“->->”, “-><-” and “<-->” to represent CDPGs, CPGs and DPGs, respectively), including gene pair ID, order class, TSS distance, minimal distance, chromosome, gene ID, transcript ID, protein ID, and strand, as well as transcription start site and transcription end site of both genes.
Case Study
LCA5L (ENSG00000157578, Leber congenital amaurosis 5-like) and SH3BGR (ENSG00000185437, SH3 domain binding glutamic acid-rich protein) are both DPGs on human chromosome 21. Although most of the distances between transcription start sites of paired genes are from 1 Kb to 100 Kb, this gene pair is conserved in all vertebrate lineages across mammals, birds, reptiles, amphibians, and fishes (Fig. S1). When compared the genes among fish and bird lineages, we found that PSMG1, BRWD1, HMGN1, WRB, LCA5L, SH3BGR, and B3GALT5 cluster tightly in three bird species (chicken, turkey, and zebra finch) and that MTMR9, XKR6, CCM2, FAM167A, LCA5, and SH3BGRL2 cluster in medaka and stickleback. The different clustering suggests potential difference in regulated mechanisms between the two vertebrate lineages.
TUB (ENSG00000166402, tubby) and RIC3 (ENSG00000166405, resistance to inhibitors of cholinesterase 3) transcribe in the opposite direction and they are CPGs on human chromosome 11 (Fig. S2). All distances between transcription start sites of the two genes are larger than 10 Kbp. When trying to expand it into distant gene clusters, we found three obviously distinct patterns in the three taxonomic groups: TUB, RIC3, LMO1 and STK33 in mammals (such as human, chimpanzee, mouse, lemur, macaque, marmoset, galago, gibbon, guinea pig, mouse, cow, dog, giant panda, bottle-nose dolphin, rabbit, horse, elephant, and opossum), CYP2R1, PDE3B, COPB1, RRAS2, TUB, and RIC3 in fishes (such as medaka, zebrafish, and stickleback), and INSC, CALCA, CYP2R1, PDE3B, PSMA1, COPB1, RRAS2, TUB, RIC3 and LMO1 in birds (such as chicken, turkey, and zebra finch).
PAPD5 (ENSG00000121274, PAP associated domain containing 5) and ADCY7 (ENSG00000121281, adenylate cyclase 7) are CDPGs on human chromosome 16 (Fig. S3). We found some interesting duplication events happened in long-term evolution of paired genes throughout the vertebrate lineages. There are two patterns in the gene clusters; one contains NSUN2, SRD5A1, PAPD7, ADCY2 and the other have PAPD5, ADCY7, BRD7, and NKD1. We found that the two clusters appear on different chromosomes of several species (eg, chimpanzee, orangutan, macaque, gibbon, turkey, fugu, and zebrafish). This phenomenon suggests that the duplication and rearrangement events forming these clusters happened very early in vertebrate evolution (perhaps at the formation of vertebrates). Moreover, we have observed several species-specific gene insertion or deletion events. For instance, the loss of SRD5A1 gene happened between NSUN2 and PAPD7 on the anole chromosome 4 and the gain of A530095I07Rik gene occurred between SRD5A1 and PAPD7 on the mouse chromosome 13.
Data Collection
We collected positions of genes, transcripts, and proteins as well as other annotation information (eg, Gene Ontology and gene family classification) of 53 species across broad lineages (including vertebrates, insects, nematode, and fungi) from the Ensembl/Biomart Version 62 (www.ensembl.org).29 We only selected transcripts with the longest coding sequence to represent genes or gene loci. Gene orthology relationship was also retrieved from this database, and we defined orthology between human and other 52 species as well as paralogs within human. In details, we assumed that there is a transitive relationship among homologs so that we combine paired homologs into one group until the group number becomes stable or converged. Based on this evolutionary principle and phylogenetic relationship, we classified all genes into homologous groups.
Implementation
This database is built on a GNU/Linux web-database LAMP framework (OS—linux, web server—Apache, database management program—MySQL, and server-side script—PHP language). At the server-side, PHP takes charge of calling Perl scripts and R functions, and uses GD modules across API (application programming interface) to generate 2D graphs. At the browser-side, we use HTML, Javascript, and CSS to allow users to experience better and convenient interfaces. We also chose SQL scripts and appropriate storage engine for MySQL to optimize the database performance, with three heavily-loaded record tables including gene, orthologous group, and gene annotation from the information of the 53 species. To speed up searching process and time-consuming tasks, we created full-text indexes for key fields in the database, and added Enquiry Optimizing of high-performance matching in MySQL database and Structured Query Language Grammar Optimizing.
Future Work
First, we plan to update the database as frequently as when new species are sequenced and new assemblies are released. We will focus on insect or arthropod genomes for comparative analysis with vertebrate genomes. Furthermore, with the I5K initiative (to sequence 5,000 insect genomes in the next five years), a large number of insect genomes may soon be available. Our preliminary analysis on the two dozen or so sequenced plant genomes also revealed clustering features, but due to the lack of contiguity within the genome assemblies, we are not able to include the data into our database at present time. In the future, however, we will bring in plant genomes to the database to study gene clustering/ordering and distinct gene organizational parameters, such as large genes with small intergenic regions in animals and small genes with larger intergenic regions in plants.30 We will also curate new annotations when they are published, including regulatory elements and new genes, such as what from ENCODE (The Encyclopedia of DNA Elements) and similar projects.31,32 Second, we will increase the complexity of our curations. For instance, our current organization of genes and their clusters are basically linear. We should be able to incorporate chromosomal structures and organizational information in a tempo-spatial fashion such as early and later replicated/transcribed genes. We should also be able to map nucleosome positioning and packaging information.33 Third, we can extend the concept “co-expression” or “co-regulation” to genes beyond clusters but neighboring clusters and clusters on chromosomes and chromosome regions (such as subtelomeric and subcentromeric regions). These new additions will lead to a network of genes and their relationships, a path toward systems biology. Finally, we hope to reveal regulatory mechanisms and their related genes that control lineage-specific or species-specific characteristics over evolutionary time scales.
Supplementary Materials
Supplementary figures S1, S2, and S3 are available from 8540 Supplementary Files.zip
Acknowledgements
This work was supported by grants from the National Basic Research Program (973 Program; 2011CB944100 and 2011CB944101), National Natural Science Foundation of China (90919024) awarded to JY.
Footnotes
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Colbourne JK, Pfrender ME, Gilbert D, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011 Feburary;331(6017):555–61. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong GK, Passey DA, Huang Y, Yang Z, Yu J. Is “junk” DNA mostly intron DNA. Genome Res. 2000 Nov;10(11):1672–8. doi: 10.1101/gr.148900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong GK, Passey DA, Yu J. Most of the human genome is transcribed. Genome Res. 2001 Dec;11(12):1975–7. doi: 10.1101/gr.202401. [DOI] [PubMed] [Google Scholar]
- 4.Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004 Apr;5(4):299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 5.Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006 Feb;23(2):450–68. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 6.Narlikar L, Ovcharenko I. Identifying regulatory elements in eukaryotic genomes. Brief Funct Genomic Proteomic. 2009 Jul;8(4):215–30. doi: 10.1093/bfgp/elp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meaney MJ, Ferguson-Smith AC. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat Neurosci. 2010 Nov;13(11):1313–8. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- 8.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010 Oct 29;330(6004):622–7. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemons D, McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006 Sep 29;313(5795):1918–22. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- 10.Li YY, Yu H, Guo ZM, Guo TQ, Tu K, Li YX. Systematic analysis of head-to-head gene organization: evolutionary conservation and potential biological relevance. PLoS Comput Biol. 2006 Jul 7;2(7):e74. doi: 10.1371/journal.pcbi.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davila Lopez M, Martinez Guerra JJ, Samuelsson T. Analysis of gene order conservation in eukaryotes identifies transcriptionally and functionally linked genes. PLoS One. 2010;5(5):e10654. doi: 10.1371/journal.pone.0010654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004 Jan;14(1):62–6. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Yu J. A comparative analysis of divergently-paired genes (DPGs) among Drosophila and vertebrate genomes. BMC Evol Biol. 2009;9:55. doi: 10.1186/1471-2148-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueda R, Iketaki H, Nagata K, et al. A common regulatory region functions bidirectionally in transcriptional activation of the human CYP1A1 and CYP1A2 genes. Mol Pharmacol. 2006 Jun;69(6):1924–30. doi: 10.1124/mol.105.021220. [DOI] [PubMed] [Google Scholar]
- 15.Jameson D, Gibson AP, Hudelot C, Higgs PG. OGRe: a relational database for comparative analysis of mitochondrial genomes. Nucleic Acids Res. 2003 Jan 1;31(1):202–6. doi: 10.1093/nar/gkg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurihara K, Kunisawa T. A gene order database of plastid genomes. Data Science Journal. 2004;3(0):60–79. [Google Scholar]
- 17.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005 Oct;15(10):1456–61. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrne KP, Wolfe KH. Visualizing syntenic relationships among the hemiascomycetes with the Yeast Gene Order Browser. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D452–5. doi: 10.1093/nar/gkj041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez MD, Samuelsson T. eGOB: eukaryotic Gene Order Browser. Bioinformatics. 2011 Apr 15;27(8):1150–1. doi: 10.1093/bioinformatics/btr075. [DOI] [PubMed] [Google Scholar]
- 20.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008 Jan;9(1):15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 21.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009 Mar;10(3):161–72. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000 May;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 25.Schmid CD, Perier R, Praz V, Bucher P. EPD in its twentieth year: towards complete promoter coverage of selected model organisms. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D82–5. doi: 10.1093/nar/gkj146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obayashi T, Kinoshita K. COXPRESdb: a database to compare gene coexpression in seven model animals. Nucleic Acids Res. 2010 Jan;39(Database issue):D1016–22. doi: 10.1093/nar/gkq1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beisser D, Klau GW, Dandekar T, Muller T, Dittrich MT. BioNet: an R- Package for the functional analysis of biological networks. Bioinformatics. 2010 Apr 15;26(8):1129–30. doi: 10.1093/bioinformatics/btq089. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a tool-kit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010 Mar;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flicek P, Amode MR, Barrell D, et al. Ensembl 2011. Nucleic Acids Res. 2011 Jan;39(Database issue):D800–6. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Wong GKS, Wang J, Yang H. Shotgun sequencing (SGS) Encyclopedia of molecular cell biology and molecular medicine. 2005 [Google Scholar]
- 31.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 Jun 14;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Celniker SE, Dillon LA, Gerstein MB, et al. Unlocking the secrets of the genome. Nature. 2009 Jun 18;459(7249):927–30. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosak ST, Groudine M. Gene order and dynamic domains. Science. 2004 Oct 22;306(5696):644–7. doi: 10.1126/science.1103864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1, S2, and S3 are available from 8540 Supplementary Files.zip