Abstract
Evolutionary systems biology aims to uncover the general trends and principles governing the evolution of biological networks. An essential part of this process is the reconstruction and analysis of the evolutionary histories of these complex, dynamic networks. Unfortunately, the methodologies for representing and exploiting such complex evolutionary histories in large scale studies are currently limited. Here, we propose a new formalism, called EvoluCode (Evolutionary barCode), which allows the integration of different evolutionary parameters (eg, sequence conservation, orthology, synteny …) in a unifying format and facilitates the multilevel analysis and visualization of complex evolutionary histories at the genome scale. The advantages of the approach are demonstrated by constructing barcodes representing the evolution of the complete human proteome. Two large-scale studies are then described: (i) the mapping and visualization of the barcodes on the human chromosomes and (ii) automatic clustering of the barcodes to highlight protein subsets sharing similar evolutionary histories and their functional analysis. The methodologies developed here open the way to the efficient application of other data mining and knowledge extraction techniques in evolutionary systems biology studies. A database containing all EvoluCode data is available at: http://lbgi.igbmc.fr/barcodes.
Keywords: systems biology, evolutionary history, multilevel data analysis, data representation, data visualization, data mining
Introduction
Systems biology aims to understand the structure and dynamic behavior of complex biological systems by modeling the components and their interactions at different functional levels.1,2 Such a comprehensive understanding requires the integration of large-scale experimental data with computational analyses and mathematical modeling approaches.3 In particular, successful systems biology will rely on our ability to integrate different types of multi-scale data across various levels of complexity,4 from individual molecules such as proteins, metabolites, etc. to cells, tissues, organisms or even ecosystems. These different levels are now being described by the large volumes of experimental data resulting from genomics technologies such as next-generation sequencing, transcriptomics, interactomics, etc. This high throughput data is characterized by a low signal-to- noise ratio and data mining and extraction of significant, pertinent knowledge are major challenges. In this context, the field of evolutionary systems biology aims to combine the modeling aspects of current systems biology with the long-standing quantitative experience in evolutionary genetics in order to uncover the general trends and principles underlying the evolution and function of complex biological networks.5,6
Evolutionary based inference provides an incredibly powerful tool for comparing multiple sources of data, since features that are maintained in several organisms tend to be functionally important while variations or differences may indicate key innovations. Comparative studies of individual components, such as proteins, have been widely used and are generally based on multiple sequence alignments and the subsequent reconstruction of a phylogenetic tree. Evolutionary histories are then typically represented by mapping major events (duplications, speciations, gene loss, domain reorganization, etc.) onto the tree. Some recent work has applied these methodologies at the genome scale, for example to build the complete collections of gene phylogenies (phylomes) in the PhylomeDB database,7 or in the construction of the Chordate Proteome History Database (ioda.univ-provence.fr). At the level of protein networks or pathways, the reconstruction of the evolutionary histories is more complex, since the interactions between the different molecular components have to be taken into account and changes at one biological level often have consequences on the evolution of other levels.8–11 Therefore, additional information concerning genome context, gene expression, molecular interactions, etc. is needed to successfully model the dynamic behavior of the system.
A number of groups have performed genome-scale studies aimed at investigating the potential correlations between variables characterizing different aspects of protein network functions and evolution.12–14 For example, positive correlations were observed between gene essentiality, duplicability and protein connectivity, estimated by the number of interaction partners in the networks.15,16 Other recent studies have shown negative correlation between expression breadth, ie, the number of tissue types in which genes are expressed, and protein evolutionary rates.17 While these studies were limited to the correlations observed between two variables, others have attempted to compile more diverse sets of evolutionary variables. Thus, principal component analysis was used to investigate the relationships between seven genome-related variables, identifying three main axes reflecting a gene’s “importance”, “plasticity” and “adaptability”.18 Waterhouse et al also examined the links between evolutionary and functional traits, by classifying metazoan orthologs as “essential” or “nonessential” and confronting these classes with various evolutionary variables.19 Although these studies have revealed several interesting trends, new standardized methodologies and tools are now needed that allow the integration of larger, more diverse sets of multilevel data and efficient, quantitative analyses at the genome scale. Similarly, despite some attempts to develop tools providing global overviews of complex evolutionary scenarios,20 original visualization tools will be required to facilitate rapid identification of specific behaviors.
Here we describe a novel formalism, called EvoluCode, or the Evolutionary barCode, which allows the integration of different data types in a unifying framework. Thus, a barcode is assigned to each component in a biological system and diverse evolutionary parameters from different biological levels can be incorporated, facilitating multi-scale evolutionary analyses. Visualization tools have also been developed to allow the human expert to view the barcodes and to identify interesting patterns in both low and high throughput studies. In order to evaluate the pertinence of the evolutionary barcodes and to test their ability to represent complex evolutionary histories, we constructed evolutionary barcodes for the complete proteomes of 17 vertebrate species. In this context, we incorporated a number of different evolutionary variables, including primary sequence data, genome neighborhood and evolutionary conservation, but the barcode formalism can be easily extended to incorporate other variables representing different biological features. At this stage, the values of the barcode parameters are normalized to allow quantitative analyses and automatic comparisons, using standard data mining techniques such as clustering or classification. We show that, in addition to highlighting general evolutionary trends, the barcodes facilitate the identification of specific evolutionary histories, such as strict conservations or significant gene family expansions. Two genome-scale analyses were then performed. First, by mapping the protein barcodes onto the human genome and visualizing the results in our barcode visualization tool, we were able to identify a number of previously described chromosome gene clusters. Second, automatic barcode clustering and functional enrichment analysis allowed us to identify specific sets of proteins that have experienced similar evolutionary histories. In a more detailed study, automatic clustering of multi-pass membrane proteins highlighted a number of particular evolutionary trends that are inherent to these protein families. Finally, as a proof of concept we demonstrate the potential of our evolutionary barcodes for biological pathway analysis. All data described in this publication are available online at: http://lbgi.igbmc.fr/barcodes.
Material and Methods
Protein test set
A reference set of human proteins was retrieved from the Human Protein Initiative (HPI) project.21 This project defined a master human proteome set, according to the quality standards set by the UniprotKB/Swiss- Prot22 databases, resulting in a total of 19778 human reference protein sequences (with 1 protein reference per coding gene). We created our own database of vertebrate proteomes, by selecting an additional 16 vertebrate species that best represent major vertebrate phyla, ie, fish, batracia, sauropsida and mammals (species list in supplementary Table 1). The complete proteomes for these organisms were downloaded from Ensembl (version 51),23 to create a local database with more than 500,000 sequences. Each human protein was then used as a query for a BlastP24 search in this local protein sequence database.
Multiple sequence alignment construction
For each human reference sequence, a modified version of the PipeAlign25 protein analysis pipeline was used to construct a MACS (Multiple Alignment of Complete Sequences) for all sequences detected by the BlastP search with E <10–3 (maximum sequences = 500). PipeAlign integrates several steps, including post-processing of the BlastP results, construction of a MACS with DbClustal,26 verification of the MACS with RASCAL27 and removal of unrelated sequences with LEON.28 In this modified version, DbClustal was replaced by the MAFFT program,29 since the computational speed of MAFFT is better suited to high throughput projects. The MACS obtained from this pipeline were then annotated with structural and functional information thanks to MACSIMS,30 an information management system that combines knowledge-based methods with complementary ab initio sequence-based predictions. MACSIMS integrates several types of data in the alignment, in particular Gene Ontology annotations,31 functional annotations and keywords from Swissprot, and functional/structural domains from the Pfam database.32
Local genome neighborhood conservation
The chromosomal localization of all genes coding for the protein sequences was obtained from Ensembl. Locally developed software was used to identify conserved local synteny between the human genome and each of the 16 other vertebrate genomes. To achieve this, the chromosomes in each genome are represented as a linear sequence of genes. For each human reference sequence, the local syntenic homolog HREF was defined at position i on the human genome and its upstream and downstream neighbors (HREF−1 and HREF+1 respectively) were identified. For each of the 16 vertebrate genomes, the sequences with the highest similarity to HREF−1 and HREF+1 were selected from the MSA, and denoted Vn_Sim−1 and Vn_Sim+1 respectively, where Vn refers to one of the 16 vertebrate genomes. A local synteny homolog, exists for HREF and genome Vn if:
homologs were found in Vn for HREF−1 and HREF+1,
the separation between the highest similarity homologs, denoted Vn_Sim−1 and Vn_Sim+1, on the genome was less than 5 genes,
a homolog of HREF was found on the genome between Vn_Sim−1 and Vn_Sim+1.
The homolog of HREF localized between Vn_Sim−1 and Vn_Sim+1 with the highest similarity to the human reference sequence was then defined as the syntenic homolog. Genes with ambiguous genomic locations, such as scaffolds etc, were discarded since the synteny relationship could not be reliably established. In addition, local or tandem duplications were excluded since the genome contexts of the two gene copies were similar.
Orthology data
Orthologs are homologous genes that diverged from a single ancestral gene in their most recent common ancestor via a speciation event, whereas paralogs are homologs resulting from gene duplications.33 Paralogs are considered as “inparalogs” when they are produced by duplication(s) subsequent to a given speciation event. In this context, several inparalogs of a given species (recently duplicated genes) are “co-orthologs” relative to the non-duplicated ortholog of a second species.
Orthologous relationships were generated with the OrthoInspector software.34 Orthology inference is based on a blast all- vs. -all generated with a 10-9 Expect value threshold. Each human reference sequence was used as a query to retrieve human inparalogs and coorthologs in each of the 16 vertebrate organisms.
Barcode construction for the human proteome
Evolutionary barcodes were constructed for all human reference proteins. Each barcode includes a number of different evolutionary parameters that were extracted from the annotated multiple alignments, synteny analysis and orthology data described above (Fig. 1A). For each of the vertebrate organisms included in this work, the most closely related homolog (based on percent residue identity) was identified in the MACS and seven parameters were extracted:
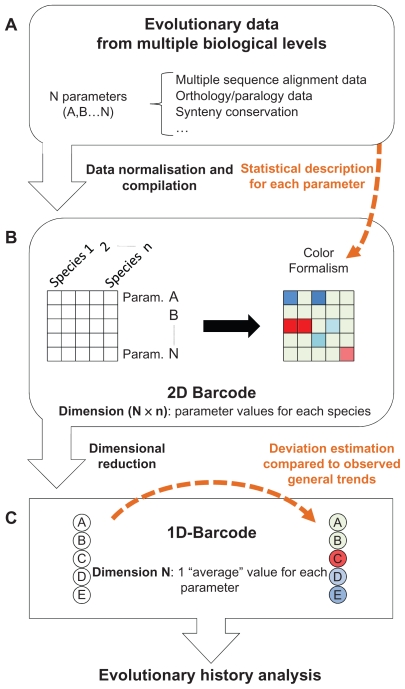
Figure 1.
Schematic view of the methodology used to produce the barcodes representing the evolutionary histories of the human proteome. Three main steps are shown. (A) Multiple evolutionary parameters are selected and described statistically. (B) The values of these parameters for different species are compiled in a 2D barcode. The statistical description of these parameters is used to define a colour code for the barcode. (C) For each barcode, a lower dimensional barcode (1D- barcode) is generated.
length: the length of the vertebrate sequence.
length_difference: the difference in length between the human reference protein and the vertebrate sequence. This parameter may indicate potential genetic events, such as exon/domain gains or losses, but may also highlight protein fragments or sequence prediction errors.
no_of_regions: the number of conserved regions defined by MACSIMS and shared between the human reference protein and the vertebrate sequence.
sequence_identity: the percent residue identity shared between the human reference protein and the vertebrate sequence.
no_of_domains: the number of known protein domains in the vertebrate sequence. These domains are based on annotations from the Pfam database.
domain_conservation: a qualitative parameter indicating changes in the domain structure of the vertebrate sequence compared to the human reference protein. This parameter identifies an unchanged domain organization, domain gains, domain losses or domain shuffling.
hydrophilicity: the average hydrophilicity of the vertebrate sequence.
Two parameters, representing orthology/paralogy data were also extracted from the OrthoInspector database:
inparalog: the number of human inparalogs with respect to the specific vertebrate organism. This parameter represents the recent duplicability of a human gene compared to the other species.
co-ortholog: the number of co-orthologs in the specific vertebrate species with respect to human. This parameter indicates the number of gene duplications in the non human lineage.
Finally, a parameter representing the genome neighborhood between the human and each vertebrate species was calculated:
synteny: categorical parameter with 3 values: (i) synteny on both sides of the gene, (ii) synteny either downstream or upstream of the gene (iii) no synteny.
All these evolutionary parameters were then organized in a 2D matrix, which we will refer to as the “2D-barcode” (Fig. 1B). Each row of the 2D-barcode represents one parameter (denoted A, B … to N). Each column of the 2D-barcode represents one species (denoted 1, 2 … n) and the intersection between rows and columns corresponds to the value or the state of one specific parameter, in one particular species.
To facilitate visualization of the 2D-barcode, a color is assigned to each matrix cell representing typical or atypical parameter values (Fig. 1B). To do this, the distribution of each parameter in each organism is first described by the sample percentiles, using the Emerson-Strenio formulas35 implemented in the R software. These nonparametric statistics are used to avoid bias due to non-Gaussian distributions of some of the parameters. The Emerson median, whiskers and hinges are then used to define three intervals that are assigned color gradients. The first interval (IT1) is assigned a blue-to-green gradient and represents values that are lower than what is generally observed for a specific parameter in a specific organism:
The second interval IT2 (green color) represents values that correspond to what is generally observed for a specific parameter in a specific organism.
The third interval (IT3) is assigned a green-to-red gradient and represents values that are higher than what is generally observed for a specific parameter in a specific organism.
Finally, the 2D-barcodes are reduced to a single dimension (Fig. 1C), called the 1D-barcode. The 1D-barcode is a simple vector representing the “average” state of each evolutionary parameter for the complete set of vertebrate species considered and is designed to facilitate inter barcode comparisons and clustering. The 1D-barcode values are produced by calculating phylum-weighted means: (i) for each parameter, a mean is calculated for 4 phyla: mammals, sauropsida, amphibians and teleostei, (ii) these phylum means are used to calculate a new mean that is the final value for a specific parameter of the 1D-barcode. As in the 2D-barcode, a color is assigned to each 1D- barcode parameter value based on the sample percentiles, for visualization purposes. However, in contrast to the 2D-barcodes, these percentiles are not organism related. They are based on the phylum weighted mean parameter values from the complete set of 1D-barcodes.
Barcode clustering and GO enrichment analysis
The complete set of 1D-barcodes representing the human proteome were used for the clustering analysis, although barcodes with missing values were removed from the test set, leaving a total of 19465 barcodes. Each 1D-barcode was represented by a vector of real values, X =(x1,x2,…xn) and the distance, d(X,Y) between two barcodes was defined as:
The distance between each pair of barcodes was calculated and the complete pairwise distance matrix as used as input to a clustering program that implements an improved Potts clustering model.36 The Potts clustering approach, also known as super-paramagnetic clustering, is based on the physical behavior of an inhomogeneous ferromagnet.37 No assumptions are made about the underlying distribution of the data. Briefly, a Potts spin variable is assigned to each data point and short range interactions between neighboring points are introduced. Spin-spin correlations are measured by a Monte Carlo procedure and are used to partition the data points into clusters.
The GoMiner software38 was then used to analyze the GO enrichment of the resulting barcode clusters. The complete set of human reference sequences was used as a background gene list. As stated by the GoMiner authors, the calculated P-values should be considered as heuristic measures, useful as indicators of possible statistical significance, rather than as the results of formal inference. The P-values can be used, for example, to sort categories to identify those of the most potential interest. In this work, a cluster was considered to be enriched in a GO term if the associated P-value was <0.05, the recommended value for high-throughput GoMiner. We then sorted the clusters according to their mean P-values and selected several top ranking clusters for further manual analysis.
Barcode website
All the data presented in this publication are available online at the following address: http://lbgi.igbmc.fr/barcodes. The website interface allows the user to browse all the human barcodes, as well as the annotated multiple alignments corresponding to each barcode. Barcodes can be selected by textual searches with Uniprot and Ensembl identifiers or by uploading a Fasta sequence followed by a BlastP search. The results of two high throughput analyses are also available: the mapping of all the 1D-barcodes on the human chromosomes and the clustering of the 1D-barcodes generated by the Potts model.
Results and Discussion
Design of the barcode
The objective of the EvoluCode evolutionary barcode is to integrate heterogeneous biological data from different biological levels in order to highlight new evolutionary patterns or scenarios that could not be detected using only one kind of data (genomic context data, sequence data, expression data …). In this study, we applied the barcode formalism to the human proteome to study vertebrate evolution. This barcode (described in detail below) includes data from 17 vertebrate species and 10 evolutionary parameters, representing different biological levels, from the genomic level (synteny) to the clade level (number of co-orthologs). Nevertheless, the barcode can theoretically be of any dimension N × n, with a parameter and species composition depending on the objectives or evolutionary scale (eg, primates, vertebrates, eukaryotes…) of the study.
The barcode combines both continuous parameters, such as sequence conservation or hydrophobicity, and discontinuous parameters, such as local synteny conservation or domain organization. Since the different parameters have very heterogeneous distributions (multi-modal, exponential, normal distribution…) they cannot be described using a single statistical model. We therefore developed a methodology to normalize the values of any given parameter using simple percentile statistics, which are suitable for any kind of parameter distribution. For visualization purposes, the normalized parameters are color-coded to highlight values that are inferior or superior to what is generally observed in a given species.
In order to summarize the diverse data inherent to the 2D-barcode approach, each barcode can also be represented in 1D. The 1D-barcode is thus a vector of continuous values representing the phylum-weighted average state of each evolutionary parameter. In the case of the human proteome barcodes, the 1D- barcode represents the average values observed during the vertebrate evolutionary history. As in the 2D- barcode, the parameters are color-coded to highlight the “expectedness” of a particular value.
Representation of complex evolutionary histories: the human proteome
To demonstrate the applicability of the EvoluCode formalism, we constructed barcodes to represent the evolutionary histories of the complete human proteome since the appearance of the vertebrates. Thus, for 19778 human genes, a representative reference protein was selected and homologs were identified in 16 complete genomes of vertebrate organisms (see Material and Methods). We then constructed 19778 multiple sequence alignments that were annotated with known structural and functional information. In addition, we estimated the synteny between the 19778 human genes and the 16 vertebrate genomes. Finally, orthologous relationships between human and the 16 vertebrates were inferred. Based on these data, we extracted various evolutionary parameters, representing primary sequence characteristics, domain organization, phylogenetic distribution and genome neighborhood conservation. These parameters were then integrated to form an evolutionary barcode representing each human reference protein. Some typical examples of barcodes, representing genes with heterogeneous and complex evolutionary histories, are shown in Figure 2 and described in detail below.
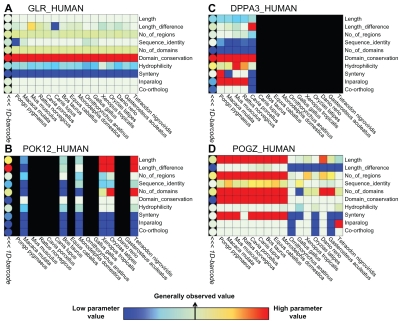
Figure 2.
Four examples of 2D-barcodes (square cells) are shown. Rows represent evolutionary parameters and columns represent vertebrate species. A color gradient highlights parameters having respectively, values lower than what is generally observed (blue), generally observed values (green), values higher than what is generally observed (red). The 1D-barcode is shown on the left hand side (round cells). Each barcode is associated with one human protein: (A) the glucagon receptor (GLR_HUMAN), (B) the HERV-K_1q22 provirus ancestral Pol protein (POK12_HUMAN), (C) the developmental pluripotency-associated protein 3 (DPPA3_HUMAN), and (D) the Pogo transposable element with ZNF domain (POGZ_HUMAN).
The first example (Fig. 2A) corresponds to the glucagon receptor (reference protein GLR_HUMAN). This receptor is essential for blood glucose level regulation, an essential function for all vascular animals.39 For all parameters; the 2D-barcode displays homogenous states over all vertebrates, implying that relatively few genetic events have affected this gene during vertebrate evolution.
The second example (Fig. 2B) corresponds to the barcode of a gene integrated from an endogenous retrovirus (reference protein POK12_HUMAN). In our barcode construction procedure, the human gene was associated with genes from the other vertebrate species that have also integrated endogenous retrovirus genes, characterized by specific sequence motifs. Consequently, the phylogenetic distribution of this barcode is dispersed. Moreover, these genes generally produce polyprotein products, explaining the heterogeneity observed for the number of domains and the fact that these sequences are not detected as orthologs.
The third example (Fig. 2C) represents a gene specific to the rodent and primate lineages (reference protein DPPA3_HUMAN). This gene appeared recently in the mammalian lineage and was previously characterized as playing a role in developmental cell pluripotency and in adult sexual organs.40 The protein product of this gene has several unusual characteristics. Despite its recent evolutionary history, it has very low sequence conservation, with 78% percent identity between human and macaque and only 37% between human and mouse. This is supported by heterogeneous hydrophobicity scores in the different species. Such rapid divergence for reproductive proteins is a well-known phenomena.41
The last example (Fig. 2D) illustrates the ability of our multi-level barcode approach to highlight a potential genetic event. The ‘Pogo transposable element with ZNF domain’ gene (reference protein POGZ_HUMAN) is involved in kinetochore assembly.42 The genetic event highlighted by the 2D- barcode occurred just after the separation of the theria and prototheria lineages. Two different blocks can be distinguished in the 2D-barcode of POGZ_HUMAN. The first block includes all theria and for these species, the gene is characterized by long sequences with conserved synteny and one ortholog in each species. The second block is less homogeneous, characterized by shorter sequences with fewer domains and low percent identities compared to human. The barcode thus suggests a potential domain gain for this gene in the marsupial and placental mammal lineages. This genetic event is particularly interesting because it occurred in a gene implicated in a fundamental process (mitosis) but indicates recent mammalian innovation in this process.42
These examples illustrate the wide range of information that can be extracted using the barcode formalism. By visualizing the evolutionary histories of the different proteins in the form of 2D-barcodes, general evolutionary trends can be observed and specific evolutionary events such as genetic events can be easily identified. The following sections will describe some large-scale analyses of the complete set of barcodes representing the evolutionary histories of the human proteome.
Large scale visualization of evolutionary barcodes
Although the 2D-barcode is a useful tool for visualizing the evolutionary histories of a small number of genes, it is too complex for large-scale visualization. To address this issue, we designed a 1 dimensional version of the evolutionary barcode, called the 1D-barcode. To estimate whether these 1D-barcodes can usefully represent global evolutionary histories, we mapped the human proteome 1D-barcodes to the 24 human chromosomes, resulting in a barcode map of the complete genome.
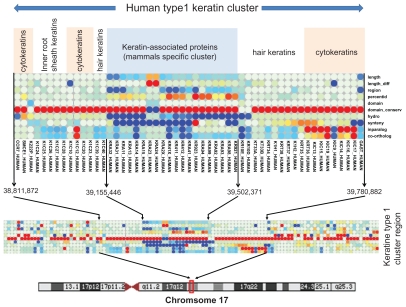
The visual inspection of this map allowed us to distinguish several previously published gene clusters. One example is the case of the keratin I and keratin II gene clusters. Early chordates had one keratin I gene and one keratin II gene.43 During vertebrate evolution, these genes evolved to form gene clusters with evidence of cluster expansion from amphibia and birds to mammals.44 A second gene family appeared during mammalian evolution and separates the type I KR chromosomal cluster in two parts. This family contains keratin associated proteins (KRAP) and represents one of the major components of hair, playing essential roles in the formation of rigid and resistant hair shafts.45 Figure 3 shows the consecutive 1D-barcodes corresponding to the human type I keratin (KR) cluster and highlights different evolutionary histories. The older KRs are the cytokeratins, which are present in the amphibian and bird KR clusters. The number of human inparalogs and the number of co-orthologs in other species have higher values (shown in red) for these cytokeratins compared to the values observed in other human genes. In particular, the number of human inparalogs is relatively high compared to the other vertebrate species, indicating that numerous duplications occurred after the cytokeratin duplications in early vertebrates. Interestingly, the values of these parameters are much lower for hair KR and inner root sheath KR, implying that these genes duplicated more recently. The KRAP cluster splitting the keratin cluster in two parts has very different barcode profiles. The unusual values of the corresponding 1D-barcodes suggest original evolutionary histories. Indeed, the values of the synteny, inparalog, co-ortholog and sequence conservation parameters are low, indicating a gene family that appeared recently with high variability between the species. In fact, these genes are specific to mammals and have evolved and diverged rapidly.45 Thus, this example illustrates the ability of the 1D-barcodes to identify local chromosomal regions that have experienced similar evolutionary histories. Such an approach could be used in the future to identify other chromosomal features, for example evolutionary breakpoints.46
Figure 3.
The 1D-barcodes corresponding to the human type I keratin cluster.
Notes: Each column represents one 1D-barcode of one protein. Several keratin subfamilies are delimited by white vertical lines. The boundaries of the keratin cluster are delimited by black arrows.
Genome-level clustering of evolutionary histories
The goal of this analysis was to identify subsets of genes in the full set of 19778 human genes that share similar barcodes, ie, similar evolutionary histories. To achieve this, we defined a Euclidean distance metric between any two barcodes based on the phylum-weighted mean values of each evolutionary parameter in the 1D-barcode. Since no a priori assumptions can be made about the statistical models underlying the parameter value distributions, we used a clustering algorithm based on nonparametric techniques: the Potts clustering model, also known as super-paramagnetic clustering. The Potts model was first developed for physical systems,47 then recently adapted for clustering purposes in neuroscience and bioinformatics.48–52 The advantage of this technique is that the user does not need to specify the number of clusters required, because this number is estimated in a probabilistic framework. In particular, we used an improved version of this clustering technique called Conditional-Potts Clustering Model.53 This model is based on an improved Potts clustering model37 with an additional prior estimation of the most suitable parameters for an efficient clustering. Using the Potts clustering model, 303 clusters were generated with a maximum cluster size of 380 proteins.
To investigate the potential functional significance of these barcode clusters, we performed a GO enrichment analysis of the 303 generated clusters using the GoMiner software.38 Figure 4 shows the distribution of the mean enrichment P-values obtained by considering all GO terms with a P-value <0.05 (the lower the P-value, the better the enrichment). Most clusters are enriched in at least one GO term, with 75% of the clusters having mean P-values <0.025 and 98% of the clusters having mean P-values <0.03. Several examples of the most enriched clusters are described in Table 1 and some of these clusters are clearly related to specific gene families. One striking example is the cluster 15, which groups numerous olfactory receptors. The family of olfactory receptors experienced a vast expansion during the chordate evolution, with the number of olfactory receptors ranging from a dozen in fishes to over a thousand in rodents.54 Moreover, pseudogenization and decline of olfactory functions has occurred in some lineages and it is thought that half of all primate receptor genes may be pseudogenes.55 The evolutionary history of this family is characterized by barcodes with high hydrophobicity scores, high domain conservation and a variable number of co-orthologs in mammalian species. Interestingly, some keratin-associated proteins, implicated in hair development were clustered together with the olfactory receptors, possibly reflecting their similar, recent expansion during mammalian evolution. Other enriched clusters correspond to highly conserved systems in vertebrates. For example, cluster 46 is enriched in genes linked to the mitochondrial respiratory chain. Similarly, clusters 67 and 153 are enriched in genes linked to translation and mRNA splicing respectively. Interestingly, the barcodes associated with these two clusters are mainly differentiated by the synteny conservation. The synteny tends to be conserved for genes linked to mRNA splicing complexes, but not for the genes involved in translation.
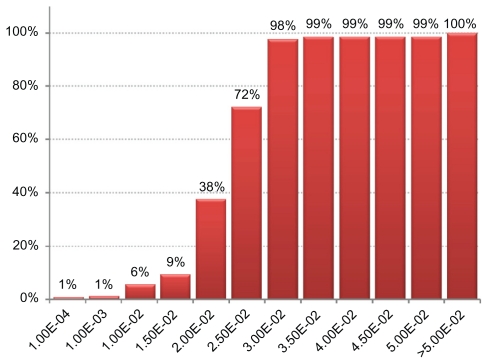
Figure 4.
Percentage of clusters with a mean GO term enrichment P-value below a given threshold.
Note: We calculated the mean P-value of all GO terms having a P-value <0.05. 98% of the clusters have at least one enriched GO term and a mean P-value <3.00E-2, indicating potential biological meaning for most clusters.
Table 1.
Some examples of barcode clusters with high GO enrichment. The most enriched terms for each cluster are shown with their corresponding P-value (10log(p)) and false discovery rate (FDR). The lower the P-value and FDR, the better is the enrichment.
| Cluster id | Representative sequence | Go accession | Go terms | 10log(p) | FDR |
|---|---|---|---|---|---|
| 46 | NDUA7_HUMAN | GO:0022904 | repiratory electron transport chain | −10.894378 | 0 |
| GO:0006796 | phosphate metabolic process | −5.162176 | 0.003 | ||
| 15 | OR2L5_HUMAN | GO:0007608 | sensory perception of smell | −69.573133 | 0 |
| GO:0007606 | sensory perception of chemical stimulus | −66.771345 | 0 | ||
| GO:0007186 | G-protein coupled receptor protein signaling pathway | −55.368505 | 0 | ||
| 95 | D104A_HUMAN | GO:0042742 | defense response to bacterium | −10.156822 | 0 |
| GO:0009607 | response to biotic stimulus | −5.232461 | 0 | ||
| GO:0006950 | response to stress | −4.145167 | 0.018 | ||
| 207 | MYH3_HUMAN | GO:0030029 | actin filament-based process | −8.190798 | 0 |
| GO:0007265 | Ras protein signal transduction | −3.375746 | 0.015 | ||
| GO:0014065 | phosphoinsitide 3-kinase cascade | −2.923239 | 0.031 | ||
| 67 | TF2H2_HUMAN | GO:0006414 | translational elongation | −14.67022 | 0 |
| GO:0042273 | ribosomal large subunit biogenesis | −5.260087 | 0 | ||
| GO:0016072 | rRNA metablic process | −4.21555 | 0 | ||
| 153 | RL15_HUMAN | GO:0044260 | cellular macromolecule metabolic process | −8.336618 | 0 |
| GO:0000398 | nuclear mRNA splicing via spliceosome | −6.719139 | 0 | ||
| GO:0006807 | nitrogen compound metabolic process | −5.889665 | 0 |
In this example analysis, we have studied the functional significance of the barcode clusters, based on GO term enrichment. In the future, we also plan to investigate the correlations between the barcode clusters and other functional data, including gene expression profiles, interactomic data and biological networks.
Multi-dimensional analysis highlights new evolutionary trends
To further illustrate the power of the multi-level barcode analyses, we analyzed the barcodes corresponding to multi-pass membrane proteins. These proteins have strong physico-chemical constraints with a predominant conservation of hydrophobic residues in their alpha helix compared to soluble proteins.56 We extracted from our sequence dataset, the 2674 human proteins that are annotated as “Multi-pass membrane protein” in Uniprot (Uniprot search engine keywords: “location: SL-9909”). In this protein subset study, we wanted to investigate in more detail the contributions of each of the individual parameters to the clustering process. We therefore performed a Multiple Correspondence Analysis (MCA) clustering of the 1D-barcodes, using the FactoMineR R package.57 This package provides visualization tools to display the clustering results. In particular, we can clearly illustrate the correlations between the barcode parameters and the inferred barcode clusters.
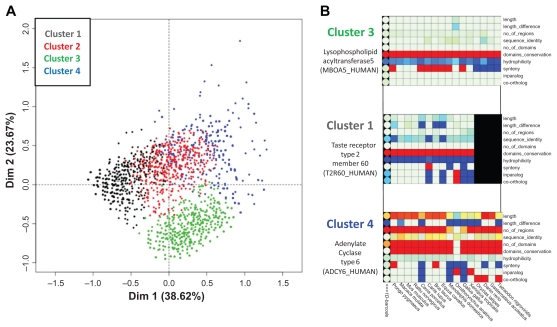
Using the 2674 “multi-pass membrane protein” barcodes, the MCA clustering produced 4 barcode clusters, as shown in Figure 5. The first axis represents parameters linked to the evolutionary history, while the second axis is linked to sequence characteristics. Details of the cluster compositions are provided in supplementary Table 2. All 4 clusters contain similar numbers of barcodes, respectively: 30.3%, 23.7%, 26.6% and 19.4%. Clusters 1, 3 and 4 correspond to three different barcode profiles and are described in detail below. Cluster 2 contains barcodes that are intermediates between clusters 1, 3 and 6.
Figure 5.
(A) MCA clustering of 2674 human membrane multi-pass proteins. (B) Representative barcodes for 3 clusters are shown to illustrate the major differences between the barcodes in each cluster.
Note: Each dot represents one 1D-barcode.
Cluster 1 (black) contains 30% of the 2674 integral membrane proteins and corresponds to proteins with short sequences and low hydrophilicity. From an evolutionary point of view, they are less well conserved, with early mammals, sauropsida and fish often sharing as little as 50% sequence identity. Their phylogenetic distribution is very heterogeneous, with gene gains and losses in many phyla, represented by a wide range of values for the inparalog and co-ortholog parameters. A large proportion (55%) of this cluster is composed of G-protein coupled receptors (GPCRs), mainly olfactory and taste receptors.
Cluster 3 (green) contains 27% of the proteins and is the most homogeneous cluster. It groups barcodes with the number of domains of conserved regions, conserved synteny in most mammals and a single ortholog in most vertebrate species. Thus, the cluster corresponds mainly to genes that are highly conserved in vertebrates with fewer genetic events compared to other multi-pass membrane proteins. To investigate the potential functional significance of this cluster, we mapped the corresponding genes to the KEGG pathway database.58 This analysis linked 41% of the 293 mapped proteins to basal metabolic processes and neural processes (eg, hsa01100-Metabolic systems, hsa04080- Neuroactive ligand-receptor interaction).
Cluster 4 (blue) contains 19% of the proteins and represents a wider distribution of barcodes. It contains average to long sequences, with numerous conserved regions. The associated proteins are not necessarily conserved in vertebrates (heterogeneous sequence identity between barcodes in the cluster), but generally have lower hydrophobicity than the other multi-pass membrane proteins. In fact, the cluster contains many proteins with multiple intra/extracellular regions, which are more conserved and hydrophilic than the hydrophobic α-helix transmembrane regions. Interestingly, 29% of cluster 4 proteins map to KEGG pathways involved in secretion processes (eg, hsa04724- Glutamatergic synapse; hsa04972-Pancreatic secretion; hsa04976-Bile secretion; hsa04970- Salivary secretion; hsa02010-ABC transporters).
This in-depth analysis of the barcodes corresponding to multi-pass membrane proteins identified important evolutionary trends and their correlations with protein function. For example, the proteins in cluster 3 have evolved little during vertebrate evolution and are mostly involved in essential processes, such as metabolic or neural processes. In contrast, cluster 1 highlights a subset of integral membrane protein families, such as GPCRs, that have experienced more genetic events. Interestingly, such behavior seems to be correlated with shorter, more hydrophobic sequences containing few intra/extracellular regions. Thus, membrane proteins that have fewer extramembrane regions are observed to be more divergent. This seems to contradict previous studies indicating that the transmembrane regions of membrane proteins are highly constrained and diverge at slower rates than the extramembrane regions.56
EvoluCode in systems biology: a proof of concept
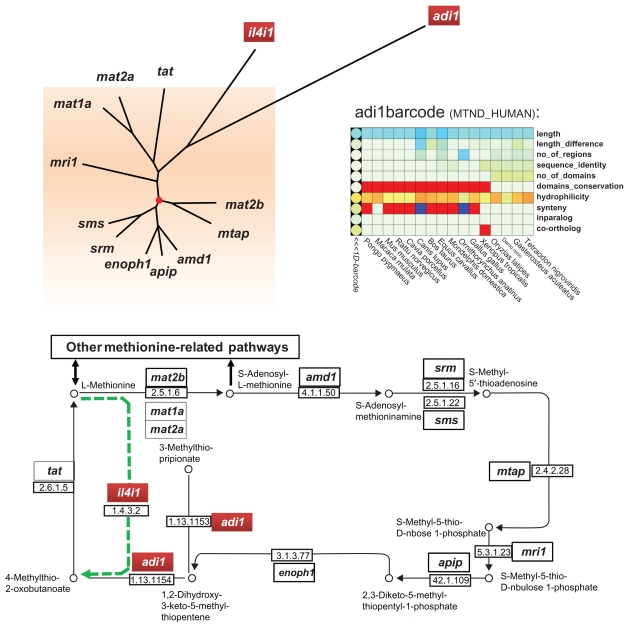
Systems biology aims to analyze genes and proteins in the context of their biological networks. As a proof of concept, we mapped our evolutionary barcodes to the KEGG pathway corresponding to the cysteine and methionine metabolism (hsa00270), in order to identify branches or ‘hot spots’ having particular evolutionary behaviors. Figure 6 shows the human methionine salvage sub-pathway, involving 13 human proteins. This sub-pathway is found in many phyla, such as plants, fungi, mammals, and bacteria (for a review, see Albers, 2009). We then calculated a normalized Euclidean distance between each pair of barcodes and constructed a neighbor-joining tree from the resulting distance matrix (Fig. 6A). This distance between barcodes represents the differences between the corresponding protein evolutionary histories and takes into account, not only sequence similarity, but also other factors, such as domain conservation, gene duplicability and genome context. In the context of the methionine salvage pathway, two barcodes corresponding to the adi1 and il4i1 genes are relatively distant compared to the other barcodes of this metabolic pathway.
Figure 6.
(A) Neighbor-joining tree of barcodes corresponding to genes in the KEGG human methionine salvage sub-pathway (hsa00720). The root of the tree is indicated by a red circle. The most distant barcodes from the root are shown in red boxes. (B) KEGG sub-pathway map, highlighting the positions of the genes corresponding to the most distant barcodes.
First, the ADI1 protein (MTND_HUMAN) is an acireductone dioxygenase. Depending on the ion used as a cofactor, Fe2+ or Ni2+, this enzyme performs different reactions, introducing an “off-pathway” branching.59 Its barcode demonstrates very high hydrophilicity and short sequences for all species, but a variable number of conserved regions and an additional domain in the fish lineage. Interestingly, this enzyme is also implicated in several other processes: the compound produced by this enzyme can cause apoptosis60 and the adi1 gene has been implicated in prostate cancers.61 Thus, it not only generates a new branch in the methionine salvage pathway, but it is also involved in other pathways. These interactions can lead to different evolutionary constraints compared to the other genes implicated in the “canonical” methionine salvage pathway, which might explain its position as an outlier in this analysis.
Second, the IL4I1 protein (OXLA_HUMAN) is an L-amino acid oxidase (LAO). Despite its presence in the KEGG methionine salvage pathway, this protein is mainly expressed in immune defenses of vertebrates and mollusks, in particular in immune system cells and B-cell lymphomas.62 As IL4I1 is not directly implicated in the basal metabolic processes, it is not surprising that the corresponding barcode is seen as an outlier. Moreover, a recent study have shown that the LAO families have undergone repeated duplications and deletions.63 This study supported the hypothesis that IL4I1 and the ancestor of LAO1 and LAO2 arose from an ancient duplication prior to the origin of tetrapods and that IL4I1 was lost in many non-mammalian tetrapods, whereas LAO1 and LAO2 were lost in mouse and human. This evolutionary pattern is in fact characteristic of many families involved in vertebrate immune processes.64
The mapping of the barcodes on the methionine salvage sub-pathway demonstrates their ability to highlight unusual evolutionary patterns, not only related to genomic data, but also to concepts such as centrality in networks or patterns of expression. Interestingly, both outlier barcodes are located in non linear parts of the pathway. Such correlation might indicate different evolutionary constraints for multi-connected pathway nodes. However, this hypothesis will require further investigation. In particular, the identification of such patterns currently requires human expert analysis. Further developments will be needed to automate the process, involving high throughput comparison of the evolutionary barcodes with network and expression data, as well as rigorous mathematical analyses to identify breakpoints and barcode outliers.
Conclusions and Perspectives
The EvoluCode barcode formalism is a powerful tool for the visualization and quantitative analysis of complex evolutionary histories in high throughput studies. Three major advantages are: (i) diverse parameters from different biological levels can be combined in a unifying framework, (ii) the parameter set can be easily modified, facilitating the construction of different barcodes for different purposes, (iii) the parameter values are normalized based on their specific distributions to allow direct comparisons within and between barcodes and to facilitate the rapid identification of typical/atypical values by the user.
We have constructed barcodes representing the evolutionary histories of the complete human proteome. The analysis was restricted to the vertebrate evolutionary scale to ensure the production of high quality multiple alignments, from which several barcode parameters are extracted. Although in principle, the barcode could be applied to higher evolutionary scales (eg, metazoa, eukaryotes …), such an extension would require more robust protocols to evaluate and validate the quality of the alignments.
One critical question that had to be addressed during the design was the selection of pertinent evolutionary parameters. The human proteome barcodes incorporate various multilevel parameters from 17 vertebrate organisms, covering genomic context, primary sequence characteristics, sequence/domain conservation and phylogenetic distributions. However, both the species set and parameter set can be easily adapted to the goals of a specific study. The data mining technique used for the subsequent analysis of the barcodes may also influence the choice of parameters to include. For example, some methods may be sensitive to highly correlated parameters, and a correspondence analysis (CA) may be necessary to select a subset of parameters with low dependency.
The combination of heterogeneous parameters is able to highlight more original and complex evolutionary trends, which could not be detected based on a single parameter such as sequence conservation or orthology. We have demonstrated this in two large scale analyses: chromosome mapping and clustering. However, the EvoluCode formalism opens the way to the application of a wide range of standard data mining or machine learning techniques that have not been possible in evolutionary studies. To illustrate the potential of EvoluCode barcodes in systems biology studies, we described the analysis of a small metabolic pathway. This proof of concept provides the basis for future studies. The automation of such analyses at the scale of all pathways in an organism should provide valuable information for pathway evolution analysis. In particular, the ability to calculate distances between barcodes will allow us to estimate parameters such as pathway “evolutionary rates” and to highlight rapidly evolving sub-pathways.
Future developments will include on the study of other distance metrics, in addition to the Euclidean distance used here. In particular, we will use the Pearson correlation coefficient to estimate the linear dependency between the barcode parameters. This would lead to a barcode clustering based on relative changes in the parameter values, rather than their scale. We will also apply more rigorous mathematical theories to identify outlying parameter values, as well as shifts or breakpoints in the barcode behavior. For example, a formal description of the different blocks in the barcode corresponding to POGZ_HUMAN (Fig. 2) could be a first step towards automatically detecting genetic events. Similarly, the stochastic or heterogeneous nature of a given barcode could be estimated based on the frequency of parameter state changes in the different phyla. This could lead to the development of quantitative indicators of the rate of evolution for a particular gene, facilitating the automatic identification of “original” evolutionary scenarios and signatures of adaptation or innovation. The analysis of the proteome is thus expected to shed more light on the fundamental aspects of the evolutionary processes and the factors that shape contemporary vertebrate genomes.
In the longer term, the methodologies developed here should facilitate, not only the analysis of proteomes from other species, but also the efficient exploitation of evolutionary information in functional genomics (notably, in interactomics and transcriptomics comparisons or in high throughput promoter studies) and large scale systems biology projects.
Supplementary Tables
Supplementary Table 1.
Data collected from Ensembl release 51 (Nov 2008).
| Ensembl identifier | Common name | Scientific name | Number of genes | Number of transcripts |
|---|---|---|---|---|
| ENSP | Human | Homo sapiens | 21971 | 60953 |
| ENSPPY | Orangutan | Pongo pygmaeus | 20068 | 29256 |
| ENSMMU | Macaque | Macaca mulatta | 21905 | 42370 |
| ENSMUS | Mouse | Mus musculus | 23873 | 43630 |
| ENSRNO | Rat | Rattus norvegicus | 22503 | 37672 |
| ENSCPO | Guinea pig | Cavia porcellus | 18673 | 24334 |
| ENSCAF | Dog | Canis familiaris | 19305 | 29804 |
| ENSBTA | Cow | Bos taurus | 21036 | 29517 |
| ENSECA | Horse | Equus caballus | 20322 | 28128 |
| ENSMOD | Opossum | Monodelphis domestica | 19471 | 34132 |
| ENSOAN | Platypus | Ornithorynchus anatinus | 17951 | 29227 |
| ENSGAL | Chicken | Gallus gallus | 16736 | 22945 |
| ENSXET | Xenopus | Xenopus tropicalis | 18023 | 28619 |
| ENSORL | Medaka | Oryzias latipes | 19686 | 25174 |
| ENSDAR | Zebrafish | Danio rerio | 21322 | 35967 |
| ENSGAC | Stickleback | Gasterosteus aculeatus | 20787 | 29096 |
| ENSTNI | Tetraodon | Tetraodon nigroviridis | 19602 | 23909 |
Acknowledgements
We would like to thank Odile Lecompte for stimulating discussions, Raymond Ripp and Laetitia Poidevin for help with database management and Nicolas Wicker and Alejandro Murua for help with the Potts Model clustering. The work was performed within the framework of the Decrypthon program, co-funded by Association Française contre les Myopathies (AFM), IBM and Centre National de la Recherche Scientifique (CNRS). We acknowledge financial support from the ANR (EvolHHuPro: BLAN07-1-198915 and Puzzle-Fit: 09-PIRI-0018-02) and Institute funds from the CNRS, INSERM, and the Université de Strasbourg.
Footnotes
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Kitano H. Systems biology: a brief overview. Science. 2002 Mar 1;295(5560):1662–4. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 2.Snoep JL, Bruggeman F, Olivier BG, Westerhoff HV. Towards building the silicon cell: a modular approach. Biosystems. Feb–Mar;2006;83(2–3):207–16. doi: 10.1016/j.biosystems.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Kohl P, Noble D. Systems biology and the virtual physiological human. Molecular Systems Biology. 2009 Jul;:5. doi: 10.1038/msb.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoehndorf R, Dumontier M, Gennari JH, et al. Integrating systems biology models and biomedical ontologies. Bmc Systems Biology. 2011 Aug 11;:5. doi: 10.1186/1752-0509-5-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loewe L. A framework for evolutionary systems biology. Bmc Systems Biology. 2009;3:27. doi: 10.1186/1752-0509-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medina M. Genomes, phylogeny, and evolutionary systems biology. Proc Natl Acad Sci U S A. 2005 May 3;102(Suppl 1):6630–5. doi: 10.1073/pnas.0501984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huerta-Cepas J, Capella-Gutierrez S, Pryszcz LP, et al. PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acids Res. 2011 Jan;39(Database issue):D556–60. doi: 10.1093/nar/gkq1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cork JM, Purugganan MD. The evolution of molecular genetic pathways and networks. Bioessays. 2004 May;26(5):479–84. doi: 10.1002/bies.20026. [DOI] [PubMed] [Google Scholar]
- 9.Medina M. Genomes, phylogeny, and evolutionary systems biology. Proceedings of the National Academy of Sciences of the United States of America. 2005 May 3;102:6630–5. doi: 10.1073/pnas.0501984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada T, Bork P. Evolution of biomolecular networks—lessons from metabolic and protein interactions. Nature Reviews Molecular Cell Biology. 2009 Nov;10(11):791–803. doi: 10.1038/nrm2787. [DOI] [PubMed] [Google Scholar]
- 11.Lercher MJ, Pal C, Papp B. An integrated view of protein evolution. Nature Reviews Genetics. 2006 May;7(5):337–48. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- 12.Knight CG, Pinney JW. Making the right connections: biological networks in the light of evolution. Bioessays. 2009 Oct;31(10):1080–90. doi: 10.1002/bies.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin EV, Wolf YI. Evolutionary systems biology: links between gene evolution and function. Current Opinion in Biotechnology. 2006 Oct;17(5):481–7. doi: 10.1016/j.copbio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Herbeck JT, Wall DP. Converging on a general model of protein evolution. Trends Biotechnol. 2005 Oct;23(10):485–7. doi: 10.1016/j.tibtech.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001 May 3;411(6833):41–2. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 16.Liang H, Li WH. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends in Genetics. 2007 Aug;23(8):375–8. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Park SG, Choi SS. Expression breadth and expression abundance behave differently in correlations with evolutionary rates. BMC Evol Biol. 2010;10:241. doi: 10.1186/1471-2148-10-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf YI, Carmel L, Koonin EV. Unifying measures of gene function and evolution. Proc Biol Sci. 2006 Jun 22;273(1593):1507–15. doi: 10.1098/rspb.2006.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waterhouse RM, Zdobnov EM, Kriventseva EV. Correlating traits of gene retention, sequence divergence, duplicability and essentiality in vertebrates, arthropods, and fungi. Genome Biol Evol. 2011 Jan;3:75–86. doi: 10.1093/gbe/evq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Procter JB, Thompson J, Letunic I, Creevey C, Jossinet F, Barton GJ. Visualization of multiple alignments, phylogenies and gene family evolution. Nat Methods. 2010 Mar;7(3 Suppl):S16–25. doi: 10.1038/nmeth.1434. [DOI] [PubMed] [Google Scholar]
- 21.O’Donovan C, Apweiler R, Bairoch A. The human proteomics initiative (HPI) Trends Biotechnol. 2001 May;19(5):178–81. doi: 10.1016/s0167-7799(01)01598-0. [DOI] [PubMed] [Google Scholar]
- 22.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 23.Hubbard TJ, Aken BL, Ayling S, et al. Ensembl 2009. Nucleic Acids Research. 2009 Jan;37(Database issue):D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W20–5. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plewniak F, Bianchetti L, Brelivet Y, et al. PipeAlign: A new toolkit for protein family analysis. Nucleic Acids Research. 2003 Jul 1;31(13):3829–32. doi: 10.1093/nar/gkg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Plewniak F, Thierry J, Poch O. DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Research. 2000 Aug 1;28(15):2919–26. doi: 10.1093/nar/28.15.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, Thierry JC, Poch O. RASCAL: rapid scanning and correction of multiple sequence alignments. Bioinformatics. 2003 Jun 12;19(9):1155–61. doi: 10.1093/bioinformatics/btg133. [DOI] [PubMed] [Google Scholar]
- 28.Thompson JD, Prigent V, Poch O. LEON: multiple aLignment Evaluation of Neighbours. Nucleic Acids Research. 2004;32(4):1298–307. doi: 10.1093/nar/gkh294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002 Jul 15;30(14):3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Muller A, Waterhouse A, et al. MACSIMS: multiple alignment of complete sequences information management system. BMC Bioinformatics. 2006;7:318. doi: 10.1186/1471-2105-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000 May;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Research. 2010 Jan;38(Database issue):D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–38. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 34.Linard B, Thompson JD, Poch O, Lecompte O. OrthoInspector: comprehensive orthology analysis and visual exploration. BMC Bioinformatics. 2011;12:11. doi: 10.1186/1471-2105-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emerson JD, Strenio J. Boxplots and batch comparison. In: Hoaglin DC, Mosteller F, Tukey JW, editors. Understanding Robust and Exploratory Data Analysis. Wiley; New York: 1983. pp. 58–96. [Google Scholar]
- 36.Alejandro M, Nicolas W. The Conditional-Potts Clustering Model. [Google Scholar]
- 37.Murua A, Stanberry L, Stuetzle W. On Potts model clustering, kernel K-means, and density estimation. Journal of Computational and Graphical Statistics. 2008 Sep;17(3):629–58. [Google Scholar]
- 38.Zeeberg BR, Feng W, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4(4):R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lok S, Kuijper JL, Jelinek LJ, et al. The human glucagon receptor encoding gene: structure, cDNA sequence and chromosomal localization. Gene. 1994 Mar 25;140(2):203–9. doi: 10.1016/0378-1119(94)90545-2. [DOI] [PubMed] [Google Scholar]
- 40.Clark AT, Rodriguez RT, Bodnar MS, et al. Human STELLAR, NANOG, and GDF3 genes are expressed in pluripotent cells and map to chromosome 12p13, a hotspot for teratocarcinoma. Stem Cells. 2004;22(2):169–79. doi: 10.1634/stemcells.22-2-169. [DOI] [PubMed] [Google Scholar]
- 41.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002 Feb;3(2):137–44. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 42.Nozawa RS, Nagao K, Masuda HT, et al. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol. 2010 Jul;12(7):719–27. doi: 10.1038/ncb2075. [DOI] [PubMed] [Google Scholar]
- 43.Karabinos A, Zimek A, Weber K. The genome of the early chordate Ciona intestinalis encodes only five cytoplasmic intermediate filament proteins including a single type I and type II keratin and a unique IF-annexin fusion protein. Gene. 2004 Feb 4;326:123–9. doi: 10.1016/j.gene.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 44.Zimek A, Weber K. Terrestrial vertebrates have two keratin gene clusters; striking differences in teleost fish. European Journal of Cell Biology. 2005 Jun;84(6):623–35. doi: 10.1016/j.ejcb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Wu DD, Irwin DM, Zhang YP. Molecular evolution of the keratin associated protein gene family in mammals, role in the evolution of mammalian hair. BMC Evol Biol. 2008;8:241. doi: 10.1186/1471-2148-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veron AS, Lemaitre C, Gautier C, Lacroix V, Sagot MF. Close 3D proximity of evolutionary breakpoints argues for the notion of spatial synteny. BMC Genomics. 2011;12:303. doi: 10.1186/1471-2164-12-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blatt M, Wiseman S, Domany E. Superparamagnetic clustering of data. Phys Rev Lett. 1996 Apr 29;76(18):3251–4. doi: 10.1103/PhysRevLett.76.3251. [DOI] [PubMed] [Google Scholar]
- 48.Stanberry L, Murua A, Cordes D. Functional connectivity mapping using the ferromagnetic Potts spin model. Human Brain Mapping. 2008 Apr;29(4):422–40. doi: 10.1002/hbm.20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Getz G, Levine E, Domany E, Zhang MQ. Super-paramagnetic clustering of yeast gene expression profiles. Physica A. 2000 May 1;279(1–4):457–64. [Google Scholar]
- 50.Einav U, Tabach Y, Getz G, et al. Gene expression analysis reveals a strong signature of an interferon-induced pathway in childhood lymphoblastic leukemia as well as in breast and ovarian cancer. Oncogene. 2005 Sep 22;24(42):6367–75. doi: 10.1038/sj.onc.1208797. [DOI] [PubMed] [Google Scholar]
- 51.Radjiman S, Han LY, Wang JS, Chen YZ. Super paramagnetic clustering of DNA sequences. Journal of Biological Physics. 2006 Jan;32(1):11–25. doi: 10.1007/s10867-006-2120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tetko IV, Facius A, Ruepp A, Mewes HW. Super paramagnetic clustering of protein sequences. BMC Bioinformatics. 2005 Apr 1;:6. doi: 10.1186/1471-2105-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murua A, Wicker N. The Conditional-Potts Clustering Model. 2011 submitted. [Google Scholar]
- 54.Zhang X, Firestein S. Genomics of olfactory receptors. Results Probl Cell Differ. 2009;47:25–36. doi: 10.1007/400_2008_28. [DOI] [PubMed] [Google Scholar]
- 55.Kambere MB, Lane RP. Co-regulation of a large and rapidly evolving repertoire of odorant receptor genes. BMC Neurosci. 2007;8(Suppl 3):S2. doi: 10.1186/1471-2202-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oberai A, Joh NH, Pettit FK, Bowie JU. Structural imperatives impose diverse evolutionary constraints on helical membrane proteins. Proc Natl Acad Sci U S A. 2009 Oct 20;106(42):17747–50. doi: 10.1073/pnas.0906390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. Journal of Statistical Software. 2008 Mar;25(1):1–18. [Google Scholar]
- 58.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. discussion 101–103. [PubMed] [Google Scholar]
- 59.Albers E. Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life. 2009 Dec;61(12):1132–42. doi: 10.1002/iub.278. [DOI] [PubMed] [Google Scholar]
- 60.Tang B, Kadariya Y, Murphy ME, Kruger WD. The methionine salvage pathway compound 4-methylthio-2-oxobutanate causes apoptosis independent of down-regulation of ornithine decarboxylase. Biochem Pharmacol. 2006 Sep 28;72(7):806–15. doi: 10.1016/j.bcp.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 61.Oram SW, Ai J, Pagani GM, et al. Expression and function of the human androgen-responsive gene ADI1 in prostate cancer. Neoplasia. 2007 Aug;9(8):643–51. doi: 10.1593/neo.07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carbonnelle-Puscian A, Copie-Bergman C, Baia M, et al. The novel immunosuppressive enzyme IL4I1 is expressed by neoplastic cells of several B-cell lymphomas and by tumor-associated macrophages. Leukemia. 2009 May;23(5):952–60. doi: 10.1038/leu.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hughes AL. Origin and diversification of the L-amino oxidase family in innate immune defenses of animals. Immunogenetics. 2010 Dec;62(11–12):753–9. doi: 10.1007/s00251-010-0482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–52. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Data collected from Ensembl release 51 (Nov 2008).
| Ensembl identifier | Common name | Scientific name | Number of genes | Number of transcripts |
|---|---|---|---|---|
| ENSP | Human | Homo sapiens | 21971 | 60953 |
| ENSPPY | Orangutan | Pongo pygmaeus | 20068 | 29256 |
| ENSMMU | Macaque | Macaca mulatta | 21905 | 42370 |
| ENSMUS | Mouse | Mus musculus | 23873 | 43630 |
| ENSRNO | Rat | Rattus norvegicus | 22503 | 37672 |
| ENSCPO | Guinea pig | Cavia porcellus | 18673 | 24334 |
| ENSCAF | Dog | Canis familiaris | 19305 | 29804 |
| ENSBTA | Cow | Bos taurus | 21036 | 29517 |
| ENSECA | Horse | Equus caballus | 20322 | 28128 |
| ENSMOD | Opossum | Monodelphis domestica | 19471 | 34132 |
| ENSOAN | Platypus | Ornithorynchus anatinus | 17951 | 29227 |
| ENSGAL | Chicken | Gallus gallus | 16736 | 22945 |
| ENSXET | Xenopus | Xenopus tropicalis | 18023 | 28619 |
| ENSORL | Medaka | Oryzias latipes | 19686 | 25174 |
| ENSDAR | Zebrafish | Danio rerio | 21322 | 35967 |
| ENSGAC | Stickleback | Gasterosteus aculeatus | 20787 | 29096 |
| ENSTNI | Tetraodon | Tetraodon nigroviridis | 19602 | 23909 |