Abstract
Nodular sclerosing Hodgkin lymphoma (NSHL) is a distinct, highly heritable Hodgkin lymphoma subtype. We undertook a genome-wide meta-analysis of 393 European-origin adolescent/young adult NSHL patients and 3315 controls using the Illumina Human610-Quad Beadchip and Affymetrix Genome-Wide Human SNP Array 6.0. We identified 3 single nucleotide polymorphisms (SNPs) on chromosome 6p21.32 that were significantly associated with NSHL risk: rs9268542 (P = 5.35 × 10−10), rs204999 (P = 1.44 × 10−9), and rs2858870 (P = 1.69 × 10−8). We also confirmed a previously reported association in the same region, rs6903608 (P = 3.52 × 10−10). rs204999 and rs2858870 were weakly correlated (r2 = 0.257), and the remaining pairs of SNPs were not correlated (r2 < 0.1). In an independent set of 113 NSHL cases and 214 controls, 2 SNPs were significantly associated with NSHL and a third showed a comparable odds ratio (OR). These SNPs are found on 2 haplotypes associated with NSHL risk (rs204999-rs9268528-rs9268542-rs6903608-rs2858870; AGGCT, OR = 1.7, P = 1.71 × 10−6; GAATC, OR = 0.4, P = 1.16 × 10−4). All individuals with the GAATC haplotype also carried the HLA class II DRB1*0701 allele. In a separate analysis, the DRB1*0701 allele was associated with a decreased risk of NSHL (OR = 0.5, 95% confidence interval = 0.4, 0.7). These data support the importance of the HLA class II region in NSHL etiology.
Introduction
Hodgkin lymphoma (HL) is a B-cell lymphoid malignancy defined by the presence of the malignant Hodgkin/Reed-Sternberg cell. It is composed of diverse etiologic and pathologic subtypes distinguished by histology, age at diagnosis, and EBV tumor status. Since the World Health Organization Revised European-American Lymphoma (REAL) classification was introduced in 2000, nodular sclerosing Hodgkin lymphoma (NSHL) has often been combined with mixed-cellularity Hodgkin lymphoma (MCHL) and other subtypes as classic Hodgkin lymphoma (cHL).1 However, abundant evidence suggests that NSHL is an etiologic entity distinct from other subtypes. NSHL is the most common histologic subtype among adolescents and young adults in industrialized countries.2 The risk of NSHL increases according to the level of economic development and is associated with childhood isolation.2–4 This suggests a strong childhood environmental influence, a pattern not seen for MCHL.2–4 NSHL is not associated with a history of infectious mononucleosis, whereas MCHL is strongly associated.5–6 Histologically, most NSHL tumors are EBV− and contain wide bands of sclerotic tissue; accordingly, the mRNA gene-expression pattern is reminiscent of wound healing and collagen synthesis.7–8 In contrast, the majority of MCHL tumors are EBV+ and the gene-expression pattern of MCHL suggests inflammation.7–8 Therefore, NSHL has a morphologic and risk pattern that differs from that of MCHL and should be considered a distinct etiologic entity.1
NSHL is also among the most heritable of neoplasms, with a 100-fold increased risk to identical twins.9–10 Specific HLA types have consistently been associated with NSHL risk,10–12 but polymorphisms from candidate genes have shown inconsistent results.13–19 A recent genome-wide association study (GWAS) confirmed the previously observed association with the HLA class II region and identified additional associated SNPs in proximity to the REL, PVT1, and GATA3 genes.20 However, the GWAS discovery set consisted of cHL, and therefore combined several distinct subtypes of HL. To specifically address genetic susceptibilities unique to the most heritable HL subtype, we undertook a GWAS to identify risk loci for NSHL.
Methods
Subjects
We performed a meta-analysis on 2 discovery sets: 1 from the University of Southern California (USC) and 1 from the University of Chicago (UC). Replication was performed on samples from the Mayo Clinic. This study was approved by the institutional review boards of the Keck School of Medicine of USC, UC, and the Mayo Clinic in accordance with the Declaration of Helsinki. Signed informed consent was obtained from all participants in this study.
USC set.
Cases were 380 European-origin HL patients diagnosed between the ages of 7 and 58; 99% (377) were diagnosed between ages 13 and 46. A total of 233 patients diagnosed between 2000 and 2008 were recruited from the USC Cancer Surveillance Program and the Cancer Prevention Institute of California (the Los Angeles County and Greater San Francisco Bay Area Survey of Epidemiology and End Results registries, respectively), and 147 patients diagnosed with HL from 1975 through 2006 were recruited from the population-based California Twin Program21 and volunteer International Twin Study.22 If the HL-affected twin was unable to provide a sample, the unaffected identical monozygotic twin's DNA sample was used; 255 (67%) were diagnosed as NSHL; 37 (10%) as MCHL; 12 (3%) as cHL; 50 (13%) as HL not otherwise specified; 11 (3%) as lymphocyte-predominant HL; and 15 (4%) as other. Of the 157 specimens tested for EBV, 90% of the NSHL specimens and 50% of the MCHL specimens were EBV−. When an HL-affected twin was unable to provide a sample, the unaffected monozygotic twin's DNA sample was used.
Controls were 2299 European-origin individuals genotyped as part of the Cancer Genetic Markers and Susceptibility Project (CGEMS).23–24 Of these, 1142 female controls were from the CGEMS Breast Cancer GWAS Stage 1 (with samples originally from the Nurse's Health Study, ages 25-42 at enrollment) and 1157 male controls were from the CGEMS Prostate Cancer GWAS Stage 1 (with samples originally from the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial, PLCO, ages 55-74 at enrollment).
UC set.
Cases consisted of 214 European-origin HL patients participating in the Childhood Cancer Survivor Study (CCSS), a retrospective study of 14 358 survivors of childhood cancer diagnosed before 21 years of age and surviving at least 5 years.25 Of these, 144 (67%) were diagnosed as NSHL; 21 (10%) as MCHL; 38 (18%) as HL not otherwise specified; 8 (4%) as lymphocyte predominant; 3 (1%) as lymphocyte depleted or other. Tumor EBV status was not available.
Controls were 1016 cancer-free individuals of European ancestry (466 males and 550 females) from the Genetic Association Informative Network schizophrenia study cohort (phs000021.v1.p1).26 This dataset consists of 6 separate case-control studies of attention deficit hyperactivity disorder, diabetic nephropathy, psoriasis, major depression, schizophrenia, and bipolar disorder (the GAIN collaborative research group), with ages ranging from 18-77 years at enrollment. Permission was obtained for use of CGEMS and GAIN GWAS results from dbGAP (http://dbgap.ncbi.nlm.nih.gov/aa/dbgap).
Mayo Clinic set.
Cases were 113 adolescent/young adult (18-46 years of age at diagnosis) European-origin patients seen at the Mayo clinic with pathologically confirmed NSHL. Controls were 214 cancer-free patients seen in the general internal medicine clinic at the Mayo Clinic (19-91 years of age).
Genotyping
USC set.
DNA was isolated from whole blood using QIAamp 96 DNA Blood Mini kits (QIAGEN/USC Genomics Core) or from saliva using Oragene saliva self-collection kits (DNA Genotek). The Illumina Human610-Quad Beadchip was used to obtain genotypes for all cases, resulting in 599 011 successfully genotyped SNPs. Blinded replicate samples (1%-2%) were genotyped to assess both reproducibility and genotype concordance across stages. The Illumina HumanHap550 (v.1.1) SNP Beadchip was used to obtain genotypes for the CGEMS breast cancer controls, and the Illumina HumanHap250S (v1.0) and HumanHap300 (v1.1) Beadchips were used to obtain genotypes for the CGEMS prostate cancer controls.
The PLINK software package (http://pngu.mgh.harvard.edu/∼purcell/plink) was used to calculate missingness, allele frequencies, and deviations from Hardy-Weinberg Equilibrium for all analyses. Of the 255 NSHL cases, 4 failed quality control metrics, in which we required a genotyping call rate of > 95%, an inbreeding coefficient of < 0.05, and a lack of cryptic relatedness. Analysis of population substructure with Eigenstrat identified 2 outlier subjects, which were subsequently removed. SNPs with a call rate of < 0.95, those with a minor allele frequency (MAF) of < 0.01, those that strongly deviated from Hardy-Weinberg equilibrium (P < 1 × 10−5), and those with genotypes that resulted from plate artifacts were removed. After applying quality control, 423 144 SNPs were successfully genotyped in 249 NSHL cases (call rate = 99.87%). In addition, SNPs were using IMPUTE2 (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html) with the HapMap phase 3 CEU population release 2 (www.hapmap.org) serving as the reference. After imputation, SNPs with a certainty score < 0.8 and an MAF < 0.05 were removed, leaving 923 203 SNPs available for analysis.
UC set.
DNA was isolated from EBV-immortalized LCLs established from nonmalignant peripheral blood lymphocytes using the PureGene DNA extraction kit (Gentra Systems), from whole blood using the PureGene kit (QIAGEN), or from saliva using the Oragene kit (DNA Genotek). We used the Affymetrix Genome-Wide Human SNP Array 6.0 to obtain genotypes for the NSHL cases and GAIN controls. SNPs with a call rate of < 0.95, those with an MAF of < 0.01, those that strongly deviated from Hardy-Weinberg equilibrium (P < 1 × 10−5), and those with genotypes that resulted from plate artifacts were removed, leaving 741 279 SNPs (call rate = 99.6%) for analysis.
To obtain genotypes for SNPs found on the Illumina Human610_Quad array not present on the Affymetrix Genome-Wide Human SNP Array 6.0, we imputed genotypes using the MACH software package (www.sph.umich.edu/csg/abecasis/MACH) with genotypes from the HapMap phase 3 CEU population serving as the reference. After imputation, we retained only imputed SNPs with an MAF of > 0.05 and with imputation quality of > 0.3, leaving 1 065 076 SNPs for analysis.
Two different software packages were used for imputation as a consequence of the separate GWAS conducted at each institution, but this is very unlikely to have affected the results.
Mayo Clinic set.
DNA was extracted using an automated platform (AutoGen FlexStar; QIAGEN). Genotyping of SNPs that surpassed the threshold for genome-wide significance in the discovery phase was performed using the Illumina Veracode Platform. Two SNPs in almost perfect linkage disequilibrium (LD; rs9268542 and rs9268528, r2 = 0.981) failed Illumina scoring and were replaced with a highly correlated tag SNP (rs9268544, r2 = 1.0 for both SNPs).
Statistical analysis
A total of 705 591 SNPs were directly genotyped in at least 1 discovery set and directly genotyped or confidently imputed in the other. The association of each SNP with risk of NSHL for each set was calculated separately using multivariable unconditional logistic regression after adjusting for sex and the top 10 eigenvectors identified in a principal component analysis using Eigenstrat27 to control for cryptic stratification. We performed a meta-analysis to obtain combined estimates using an inverse variance weighting of study-specific estimates. An association was considered significant if the P value from the meta-analysis was < 5 × 10−8.
For replication in the Mayo Clinic set, logistic regression, adjusting for sex, was performed to estimate the effect of the SNPs on risk of NSHL.
We estimated extended haplotypes using SNPs surpassing the threshold for genome-wide significance in the discovery analysis for all cases and controls and determined their association with NSHL, ORs, and 95% confidence intervals (CIs) by logistic regression adjusted by sex and the top 10 eigenvectors.27
A 3-way meta-analysis combining the USC, UC, and Mayo datasets was conducted to assess the significance of replicated SNPs and haplotypes.
We used data from the Hapmap CEU individuals to determine the link between our SNP haplotypes and HLA-DRB1-HLA-DQB1 alleles. HLA-DRB1-HLA-DQB1 haplotype frequencies were estimated using Estihaplo28 (Table 5). HLA alleles in USC and UC cases and controls were then imputed from the GWAS data and the association between the putatively associated HLA allele and NSHL risk was determined unconditional logistic regression to obtain ORs, 95% CIs, and P values, combining the estimates in a meta-analysis.
Table 5.
SNP-HLA haplotype frequencies in the HapMap CEU dataset
| rs204999-rs9268528-rs9268542-rs6903608-rs2858870 | HLA-DRB1-HLA-DQB1 | HLA haplotype frequency | Distribution of HLA haplotypes by SNP haplotype |
|---|---|---|---|
| A-G-G-C-T | 1101-0301 | 2.99% | 31% |
| A-G-G-C-T | 1401-0503 | 2.99% | 31% |
| A-G-G-C-T | 1301-0603 | 1.49% | 14% |
| A-G-G-C-T | 0301-0201 | 0.77% | 8% |
| A-G-G-C-T | 1404-0503 | 0.75% | 8% |
| A-G-G-C-T | 1305-0301 | 0.75% | 8% |
| G-A-A-T-C | 0701-0303 | 5.22% | 50% |
| G-A-A-T-C | 0701-0201 | 5.21% | 50% |
Results
Characteristics of the USC and UC discovery sets are shown in Table 1. The median age at diagnosis was older in the USC set compared with the UC set (29 vs 16 years, respectively); however, 88% of the patients in each group were in the adolescent/young adult range typical of NSHL. The proportion of female patients was higher in the UC set (74%) compared with the USC set (47%) as a consequence of selection criteria for another study. Principal component analysis using Eigenstrat27 revealed no evidence for population stratification (supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). A quantile-quantile plot for the combined set revealed no overdispersion of significant P values (genomic control λ = 1.071; supplemental Figure 2).29 When limited to only genotyped SNPs, the overdispersion parameter for the USC set (λUSC) was 1.02 and for the UC set (λUC), it was 1.03.
Table 1.
Demographic and clinical information for adolescent/young adult NSHL patients analyzed in each discovery set
| USC | UC | Combined | |
|---|---|---|---|
| Total patients, n | 249 | 144 | 393 |
| Sex, n (%) | |||
| Male | 133 (53) | 37 (26) | 170 (43) |
| Female | 116 (47) | 107 (74) | 223 (57) |
| Median age at diagnosis, n | 29 | 16 | 22 |
| Age at diagnosis, range | 7-46 | 4-21 | 4-46 |
| 5-9 | 1 (0) | 6 (4) | 7 (3) |
| 10-19 | 38 (15) | 125 (87) | 163 (41) |
| 20-29 | 93 (37) | 13 (9) | 106 (27) |
| 30-39 | 88 (35) | 0(0) | 88 (22) |
| 40-46 | 29 (12) | 0(0) | 29 (7) |
Five SNPs on chromosome 6p21.32 achieved genome-wide significance for association with NSHL in the meta-analysis: rs6903608 (OR = 1.6, P = 3.52 × 10−10), which had been identified in an earlier GWAS of cHL,20 rs9268542 (OR = 1.6, P = 5.35 × 10−10), rs204999 (OR = 0.5, P = 1.44 × 10−9), rs9268528 (OR = 1.6, P = 1.19 × 10−9), and rs2858870 (OR = 0.4, P = 1.69 × 10−8; Figure 1 and Table 2). Genotyping accuracy was confirmed in the USC set by genotyping the 5 variants surpassing the threshold for genome-wide significance in all 249 cases using TaqMan. In the UC set, 15 samples were sequenced for the 3 imputed SNPs to confirm the imputed genotypes for rs9268528, rs6903608, and rs2858870 (5 samples carrying each genotype). Because there were differences in the age distribution between the 2 case samples, even within the adolescent/young adult age range, we performed a sensitivity analysis limiting the cases from each sample to < 21 years of age at diagnosis. The effect measures for each SNP were nearly identical to the results obtained when the entire patient sample was used, although the P values were slightly larger because of the loss in sample size (supplemental Table 1). We did not observe an interaction between sex and any of the genome-wide significant SNPs on NHL risk (data not shown).
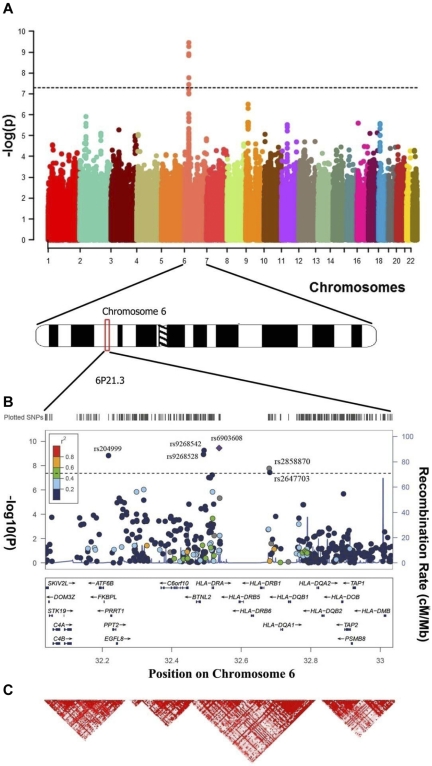
Figure 1.
Results of a meta-analysis of 2 GWAS on 393 cases and 3315 controls. (A) Manhattan plot of the genome-wide results. P values were determined for each SNP based on the meta-analysis of the UC and USC samples. Five SNPs surpassed the genome-wide significance threshold (P value = 5 × 10−8): rs9268542, rs9268528, rs204999, rs2858870, and rs6903608 6p21.3. (B) Regional plot of the 6p21.3 for the combined genome-wide association results. The blue lines represent recombination rates. The previously reported SNP rs6903608 is designated by a diamond; all other SNPs are depicted by circles. (C) Linkage disequilibrium map of the 6p21.3 region (red represents r2 > 0.9).
Table 2.
Chromosome 6 SNPs associated with adolescent/young adult NSHL
| SNP | BP* | USC |
P | UC |
Combined |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor allele | MAF (Ca) | MAF (Co) | OR (95% CI)† | MAF (Ca) | MAF (Co) | OR (95% CI)† | P | OR (95% CI)† | P | |||
| rs6903608‡ | 32536263 | C | 0.48 | 0.33 | 1.7 (1.4-2.1) | 4.50 × 10−8 | 0.40 | 0.32 | 1.5 (1.2-1.9) | 1.43 × 10−3 | 1.6 (1.4-1.9) | 3.52 × 10−10 |
| rs9268542§ | 32492699 | G | 0.51 | 0.38 | 1.6 (1.3-1.9) | 1.37 × 10−5 | 0.53 | 0.39 | 1.8 (1.4-2.4) | 5.76 × 10−6 | 1.6 (1.4-1.9) | 5.35 × 10−10 |
| rs9268528‡ | 32491086 | G | 0.51 | 0.38 | 1.5 (1.3-1.9) | 1.78 × 10−5 | 0.50 | 0.37 | 1.8 (1.4-2.3) | 1.07 × 10−6 | 1.6 (1.4-1.9) | 1.19 × 10−9 |
| rs204999§ | 32217957 | G | 0.16 | 0.27 | 0.5 (0.4-0.7) | 1.78 × 10−7 | 0.21 | 0.28 | 0.6 (0.4-0.8) | 1.60 × 10−3 | 0.5 (0.4-0.7) | 1.44 × 10−9 |
| rs2858870‡ | 32680229 | G | 0.06 | 0.13 | 0.4 (0.3-0.6) | 1.64 × 10−6 | 0.07 | 0.13 | 0.5 (0.3-0.8) | 2.35 × 10−3 | 0.4 (0.3-0.6) | 1.69 × 10−8 |
Chromosome location based on National Center for Biotechnology Information Human Genome Build 36 coordinates.
OR (95% CI) adjusted for gender and top 10 eigenvectors.
Directly genotyped in cases and controls analyzed at USC; imputed in cases and controls analyzed at UC using the MACH program.
Directly genotyped in all case and control samples.
rs9268542 and rs9268528 were in almost perfect LD (r2 = 0.981), rs204999 and rs2858870 were in weak LD (r2 = 0.257), and there was no notable LD between the remaining pairs of SNPs (r2 < 0.10; supplemental Table 2). When adjusted for rs6903608, rs9268542 and rs9268528 retained genome-wide significance (rs9268542: P = 4.51 × 10−10 and rs9268528: P = 2.81 × 10−9) and rs204999 and rs2858870 remained nominally significant (rs204999: P = 2.57 × 10−6 and rs2858870: P = 7.22 × 10−6; Table 3).
Table 3.
Association of SNPs surpassing the threshold for genome-wide significance in the adolescent/young adult NSHL meta-analysis conditioned on rs6903608
| USC |
UC |
Combined |
||||
|---|---|---|---|---|---|---|
| OR (95% CI)* | P | OR (95% CI)* | P | OR (95% CI)* | P | |
| rs204999 | 0.6 (0.5-0.8) | 1.12 × 10−4 | 0.6 (0.5-0.9) | 7.08 × 10−3 | 0.6 (0.5-0.8) | 2.57 × 10−6 |
| rs9268542 | 1.6 (1.3-1.9) | 5.75 × 10−6 | 1.7 (1.4-2.2) | 1.51 × 10−5 | 1.6 (1.4-1.9) | 4.51 × 10−10 |
| rs9268528 | 1.5 (1.3-1.9) | 1.31 × 10−5 | 1.7 (1.3-2.2) | 4.49 × 10−5 | 1.6 (1.4-1.9) | 2.81 × 10−9 |
| rs2858870 | 0.5 (0.3-0.7) | 2.59 × 10−4 | 0.5 (0.3-0.8) | 8.94 × 10−3 | 0.5 (0.4-0.7) | 7.22 × 10−6 |
OR and 95% CI adjusted for rs6903608, sex, and top 10 eigenvectors.
To replicate our findings, we genotyped rs6903608, rs204999, rs2858870,and rs9268544 in an independent set of 113 young adult NSHL cases and 214 controls (supplemental Tables 3-4). rs6903608 (OR = 1.9, P = .000 24) and rs2858870 (OR = 0.6, P = .04077) were significantly associated with NSHL, whereas rs204999 was comparably, but not significantly, associated (OR = 0.7, P = .1426). No association between NSHL and rs9268544 was seen in the replication sample (OR = 1.1, P = .7155).
A meta-analysis combining the USC, UC, and Mayo Clinic datasets yielded associations with increased statistical significance for the replicated SNPs rs6903608 (OR = 1.6, P = 1.19 × 10−12) and rs2858870 (OR = 0.4, P = 5.61 × 10−9), and an association with slightly weaker significance for rs204999 (OR = 0.6, P = 2.34 × 10−8). When conditioned on the previously reported SNP rs6903608, replicated SNPs rs2858870 (P = 5.82 × 10−6) and rs204999 (P = .001) remained significant in the 3-way meta-analysis.
Our results suggest the presence of risk loci located in a region of high LD that contains HLA class II genes as well as other immune-response genes. A characteristic feature of the HLA class II region is extensive LD. To determine whether the extended haplotype containing these 5 SNP variants captured more information about risk than individual SNPs, we estimated haplotypes and determined their association with NSHL. We found that the 5-variant haplotype model of risk resulted in the strongest overall predictor of NSHL risk (P = 1.19 × 10−17). Two distinct haplotypes were significantly associated with NSHL risk (Table 4): one haplotype contained the risk alleles for all 5 SNPs (Hap3: AGGCT) and was associated with a 70% increased risk of NSHL (OR = 1.7, P = 1.71 × 10−6); the other haplotype (Hap6: GAATC) contained the protective alleles for all 5 SNPs and was associated with a 60% decreased risk (OR = 0.4, P = 1.16 × 10−4). Similar associations (ORs) between NSHL risk and these 2 haplotypes were observed in the replication set, although only the association with haplotype 3 was statistically significant (Table 4). When data from the 3 centers were combined in a meta-analysis, statistical significance of the associations with haplotypes 3 (OR = 1.7, P = 2.13 × 10−7) and 6 (OR = 0.4, P = 4.75 × 10−5) increased and the global P decreased (P = 7.62 × 10−18).
Table 4.
The association of 6p21.32 haplotypes with young adult NSHL risk
| Structure (SNP)* | USC† |
UC‡ |
Combined§ |
Replication¶ |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq | OR (95% CI)6 | P | Freq | OR (95% CI)# | P | OR (95% CI)# | P | Freq | OR (95% CI)# | P | ||
| Hap1 | A-A-A-T-T | 0.23 | 1 | 0.21 | 1 | 1 | 0.22 | 1 | ||||
| Hap2 | A-G-G-T-T | 0.17 | 1.0 (0.7-1.4) | 0.851 | 0.17 | 1.8 (1.2-2.8) | 0.004 | 1.3 (1.0-1.7) | 0.066 | 0.19 | 0.9 (0.5-1.7) | 0.855 |
| Hap3 | A-G-G-C-T | 0.17 | 1.7 (1.2-2.2) | 6.37 × 10−4 | 0.16 | 1.9 (1.3-2.67) | 0.001 | 1.7 (1.4-2.4) | 1.71 × 10−6 | 0.16 | 1.7 (1.0-3.0) | 0.045 |
| Hap4 | A-A-A-C-T | 0.16 | 1.0 (0.7-1.5) | 0.881 | 0.15 | 1.4 (0.9-2.1) | 0.161 | 1.2 (0.8-1.6) | 0.294 | 0.18 | 1.8 (1.0-3.3) | 0.063 |
| Hap5 | G-A-A-T-T | 0.12 | 0.6 (0.4-1.0) | 0.028 | 0.13 | 1.0 (0.6-1.6) | 0.949 | 0.8 (0.6-1.1) | 0.119 | 0.11 | 1.1 (0.6-2.3) | 0.71 |
| Hap6 | G-A-A-T-C | 0.08 | 0.3 (0.1-0.5) | 5.72 × 10−5 | 0.08 | 0.6 (0.4-1.1) | 0.121 | 0.4 (0.3-0.7) | 1.16 × 10−4 | 0.08 | 0.5 (0.2-1.3) | 0.162 |
| Others | 0.08 | 0.09 (0.5-1.3) | 0.476 | 0.09 | 1.3 (0.8-2.1) | 0.241 | 1.1 (0.8-1.5) | 0.733 | 0.06 | 1.3 (0.5-3.5) | 0.53 | |
rs204999-rs9268528-rs9268542-rs6903608-rs2858870.
Global P = 4.21 × 10−13.
Global P = 6.44 × 10−7.
Combined global P = 1.19 × 10−17.
Replication with 113 European origin adolescent/young adult NSHL patients and 214 controls: global P = 0.055; rs968544 substituted for rs9268528 and rs9268542.
OR (95% CI) adjusted for gender and top 10 eigenvectors.
Because HLA class II alleles have been previously associated with NSHL,10–12 we used data from the HapMap CEU individuals to determine whether our SNP haplotypes tagged specific HLA-DRB1-HLA-DQB1 alleles28 (Table 5). Whereas individuals with haplotype 3 (AGGCT) had multiple HLA-DRB1 alleles, all individuals with haplotype 6 (GAATC) carried the DRB1*07:01 allele. In our combined USC and UC datasets, the HLA class II allele DRB1*0701 was associated with a significant 50% decreased risk of NSHL (OR = 0.5, 95% CI = 0.4-0.7).
Discussion
We performed a GWAS of NSHL and found significant associations between NSHL risk and SNPs at chromosome 6p21.32. While this paper was in review, another study reported an association between cHL and one SNP identified in our GWAS, rs6903608.20 We replicated this finding and also identified additional risk loci, rs204999 and rs2858870, in the same region. When accounting for rs6903608, we found that rs204999 and rs2858870 remained nominally significant at the genome-wide level. Protective alleles for these SNPs were also contained in haplotypes significantly associated with NSHL risk, one of which appears to tag a protective HLA-DRB1 allele. The highly correlated SNPs rs9268542 and rs9268528 did retain genome-wide significance with rs6903608 in the model, but could not be replicated in the small sample from Mayo clinic.
The 6p21.32 region contains more than 200 genes with SNPs in strong LD, the majority of which are expressed and involved in immune function,30 including HLA-DRB1 and HLA-DQB1, which code the corresponding HLA class II alleles. The HLA class II region has been most strongly associated with autoimmune disease30–31 and generally not with solid tumors. However, there is a long-known association between HL, particularly NSHL, and specific HLA class II alleles of DRB1, DQA1, DQB1, and DPB1.10–12 A recent study reported associations between cHL and multiple HLA-DR alleles, including a significant protective association with HLA-DRB1*070132 similar in magnitude to the 50% decreased risk we observed associated with this allele. Follicular lymphoma risk has also been linked to a genetic signal in the HLA class II region,33 and HLA alleles, including DRB1*0701, have been associated with multiple myeloma risk.34 The importance of the region suggests a role for immune response to antigen in the etiology of B-cell diseases, including lymphoma and autoimmune disease.35
Substantial evidence supports the hypothesis that NSHL results from an atypical immune response to a virus4 or another biologic trigger in the setting of a Th2-skewed immune response.6,13,15,16 Genetic variation in HLA class II genes may underlie this aberrant response, because such variation results in structural alterations in the HLA molecule-binding pockets, and therefore potentially in binding capacity difference for specific antigens.36 In addition, HLA class II alleles can influence CD4+ T-cell polarization to either the Th1 or Th2 subtype with subsequent alterations in cytokine responses.37–38 NSHL tumors produce large amounts of Th2 and inflammatory cytokines,39 and susceptibility is associated with increased Th2 and decreased Th1 cytokine production.13,15–16 Therefore, our SNPs could code for HLA allele variation, which in turn could affect antigen-binding capacity and CD4+ cell polarization, thereby contributing to a protective or risk immunophenotype.
Some posit that EBV tumor status is a more important etiologic marker than histology.40 Enciso-Moral et al examined GWAS differences by EBV tumor status, but not histology, and found that the rs6903608 SNP was associated more strongly with EBV− than with EBV+ disease.20 Age and EBV tumor status are highly correlated7 and the majority of young adult HL patients in economically developed countries have the NSHL subtype with EBV− tumors. Because our study was restricted to adolescent/young adult NSHL, the majority of which is EBV− (90% in our study), we did not have sufficient power to examine effect modification by EBV tumor status.
A possible limitation of our study was the difference in length of follow-up for the USC and UC subsets if survival is differentially associated with etiological HL subtype. All of the patients analyzed at UC (CCSS) and 88% of the patients analyzed at USC had survived at least 5 years before participation. The remaining 12% of the patients analyzed at USC were recruited from the Los Angeles USC Cancer Surveillance Program (SEER registry) via rapid case ascertainment within 6 months of diagnosis; all are still living, although the follow-up period is less than 5 years. Given the very high survival rate of young adult NSHL patients (> 92%), a survival bias is unlikely. The age range of the control sets was broad but skewed toward older ages, which is unlikely to bias our results, because even the younger controls have an extremely low probability of developing NSHL (peak age-specific incidence = 4-6/100 000/y; see http://seer.cancer.gov/).
In conclusion, in the present study, we identified an association between SNPs in the 6p21.32 region and NSHL risk, including a previously reported SNP.20 The SNPs occur on 2 mirror haplotypes that are significantly associated with NSHL risk, and at least 1 may be linked to a reported protective HLA allele.32 Because of the limitations of the association study design and the strong LD in the region, we cannot determine whether the newly identified SNPs (rs204999 and rs2858870) are independent of the previously reported SNP or whether they simply provide additional information by extending the known region. Larger studies will be required to determine whether these loci are actually a single associated region, if they are indeed independent, or if they show evidence for epistasis. This study supports a possible role for HLA-DRB1 polymorphisms in NSHL susceptibility.
Acknowledgments
The authors thank Jorge Oksenberg and Christopher Haiman for thoughtful discussion of the manuscript, Xiang Hua for assistance with population stratification analysis, and the participating patients and their family members, without whom this work would not have been possible.
This work was supported by grants from the National Institutes of Health (CA110836 to W.C.; HD0433871, CA129045, and CA40046 to K.O.; CA55727 to L.L.R.; CA58839 to T.M.M.); the United States Army Medical Research and Materiel Command (Department of Defense PR054600 to W.C.); the American Cancer Society Illinois Division (to K.O.); the American Lebanese Syrian Associated Charities (to L.L.R.); the Leukemia & Lymphoma Society (TR6137-07 to W.C.); and the Cancer Research Foundation (to K.O.). This project was funded in whole or in part with federal funds from the National Cancer Institute Surveillance Epidemiology and End Results Population-based Registry Program, National Institutes of Health, Department of Health and Human Services, under contracts N01-PC-35139 (to W.C.) and N01-PC-35136 (to the Cancer Prevention Institute of California), and from the National Cancer Institute contract 263-MQ-417755 (to S.L.G.). The collection of incident HL patients used in this publication was supported by the California Department of Health Services as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885.
The ideas and opinions expressed herein are those of the authors, and no endorsement by the State of California, Department of Health Services, is intended or should be inferred. This publication was made possible by grant number 1U58DP000807-01 from the Centers for Disease Control and Prevention. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the federal government.
Footnotes
The online version of this article contains a data supplement.
Presented as an abstract and poster at the 52nd ASH Annual Meeting and Exposition, December 5, 2010, Orlando, FL, and at the 10th InterLymph Meeting, June 10, 2011, Cagliari, Sardinia, Italy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: W.C., D.L., D.V.C., and K.O. designed the research and the data analysis; W.C., D.V.C., and K.O. supervised the overall project; W.C., T.M.M., S.L.G., S.B., L.C.S., B.K.L., and L.L.R. collected the patient samples and data; T.M.M., V.K.C., F.R.S., and A.D.S. provided study design input; B.N.N. and L.M.W. validated the histopathology; D.J.V.D.B. planned and supervised the genotyping; D.L and T.B. conducted the statistical analysis; A.E.H. constructed and maintained the database; C.K.E. provided statistical and database support; P-A.G. performed the HLA allele analysis; J.R.C., T.M.H., and B.K.L. collected the samples from patients and controls used in the replication; D.L., S.L.S., and Z.S.F. performed the replication analysis; and W.C. wrote the manuscript with input from D.L., D.V.C., and K.O.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wendy Cozen, DO, MPH, Department of Preventive Medicine, USC Norris Comprehensive Cancer Center, 1441 Eastlake Ave, MC 9175, Los Angeles, CA 90089-9175; e-mail: wcozen@usc.edu.
References
- 1.Mani H, Jaffe ES. Hodgkin lymphoma: an update on its biology with newer insights into classification. Clin Lymphoma Myeloma. 2009;9(3):206–216. doi: 10.3816/CLM.2009.n.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curado MP, Edwards B, Shin HR, et al. Cancer incidence in five continents. Volume IX. Lyon, France: World Health Organization Publications; 2009. IARC scientific publication number 160. [Google Scholar]
- 3.Cozen W, Katz J, Mack TM. Hodgkin's disease varies by cell type in Los Angeles. Cancer Epidemiol Biomarkers Prev. 1992;1(4):261–268. [PubMed] [Google Scholar]
- 4.Mueller NE, Grufferman S. Hodgkin lymphoma. In: Schottenfeld D, Fraumeni JF Jr, editors. Cancer Epidemiology and Prevention. New York, NY: Oxford University Press; 2006. pp. 872–897. [Google Scholar]
- 5.Hjalgrim H, Askling J, Rostgaard K, et al. Characteristics of Hodgkin's lymphoma after infectious mononucleosis. N Engl J Med. 2003;349(14):1324–1332. doi: 10.1056/NEJMoa023141. [DOI] [PubMed] [Google Scholar]
- 6.Cozen W, Hamilton AS, Zhao P, et al. A protective role for early childhood exposures and young adult Hodgkin lymphoma. Blood. 2009;114(19):4014–4020. doi: 10.1182/blood-2009-03-209601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glaser SL, Gulley ML, Clarke CA, et al. Racial/ethnic variation in EBV-positive classical Hodgkin lymphoma in California populations. Int J Cancer. 2008;123(7):1499–1507. doi: 10.1002/ijc.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birgersdotter A, Baumforth KRN, Porwit A, et al. Inflammation and tissue repair markers distinguish the nodular sclerosis and mixed cellularity subtypes of classical Hodgkin's lymphoma. Br J Cancer. 2009;101(8):1393–1401. doi: 10.1038/sj.bjc.6605238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mack TM, Cozen W, Shibata DK, et al. Concordance for Hodgkin's disease in identical twins suggests genetic susceptibility to the young-adult form of the disease. N Engl J Med. 1995;332(7):413–418. doi: 10.1056/NEJM199502163320701. [DOI] [PubMed] [Google Scholar]
- 10.Harty LC, Lin AY, Goldstein AM, et al. HLA-DR, HLA-DQ, and TAP genes in familial Hodgkin disease. Blood. 2002;99(2):690–693. doi: 10.1182/blood.v99.2.690. [DOI] [PubMed] [Google Scholar]
- 11.Staratschek-Jox A, Shugart YY, Strom SS, Nagler A, Taylor GM. Genetic susceptibility to Hodgkin's lymphoma and to secondary cancer: workshop report. Ann Oncol. 2002;13(suppl 1):30–33. doi: 10.1093/annonc/13.s1.30. [DOI] [PubMed] [Google Scholar]
- 12.Klitz W, Aldrich C, Fildes N, Horning S, Begovich A. Localization of predisposition to Hodgkin's disease in the HLA class II region. Am J Hum Genet. 1994;54(3):497–505. [PMC free article] [PubMed] [Google Scholar]
- 13.Cozen W, Gill PS, Ingles SA, et al. IL-6 levels and genotype are associated with risk of young adult hodgkin lymphoma. Blood. 2004;103(8):3216–3221. doi: 10.1182/blood-2003-08-2860. [DOI] [PubMed] [Google Scholar]
- 14.Cordano P, Lake A, Shield L, et al. Effect of IL-6 promoter polymorphism on incidence and outcome in Hodgkin's lymphoma. Br J Haematol. 2005;128(4):493–495. doi: 10.1111/j.1365-2141.2004.05353.x. [DOI] [PubMed] [Google Scholar]
- 15.Nieters A, Beckmann L, Deeg E, Becker N. Gene polymorphisms in Toll-like receptors, interleukin-10, and interleukin-10 receptor alpha and lymphoma risk. Genes Immun. 2006;7(8):615–624. doi: 10.1038/sj.gene.6364337. [DOI] [PubMed] [Google Scholar]
- 16.Cozen W, Gill PS, Salam MT, et al. Interleukin-2, interleukin-12 and interferon-gamma levels and risk of young adult Hodgkin lymphoma. Blood. 2008;111(7):3377–3382. doi: 10.1182/blood-2007-08-106872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broderick P, Cunningham D, Vijayakrishnan J, et al. IRF4 polymorphism rs872071 and risk of Hodgkin lymphoma. Br J Haematol. 2010;148(3):413–415. doi: 10.1111/j.1365-2141.2009.07946.x. [DOI] [PubMed] [Google Scholar]
- 18.Mollaki V, Georgiadis T, Tassidou A, et al. Polymorphisms and haplotypes in TLR9 and MYD88 are associated with the development of Hodgkin's lymphoma: a candidate-gene association study. J Hum Genet. 2009;54(11):655–659. doi: 10.1038/jhg.2009.90. [DOI] [PubMed] [Google Scholar]
- 19.Salipante SJ, Mealiffe ME, Wechsler J, et al. Mutations in a gene encoding a midbody kelch protein in familial and sporadic classical Hodgkin lymphoma lead to binucleated cells. Proc Natl Acad Sci U S A. 2009;106(35):14920–14925. doi: 10.1073/pnas.0904231106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enciso-Mora V, Broderick P, Ma Y, et al. A genome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p15.1 (REL), 8q24.21 and 10p14 (GATA3). Nat Genet. 2010;42(12):1126–1230. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cockburn MG, Hamilton AS, Zadnick J, Cozen W, Mack TM. Development and representativeness of a large population-based cohort of native California twins. Twin Res. 2001;4(4):242–250. doi: 10.1375/1369052012461. [DOI] [PubMed] [Google Scholar]
- 22.Mack TM, Deapen D, Hamilton AS. Representativeness of a roster of volunteer North American twins with chronic disease. Twin Res. 2000;3(1):33–42. doi: 10.1375/136905200320565670. [DOI] [PubMed] [Google Scholar]
- 23.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP Database of Genotypes and Phenotypes. Nat Genet. 2007;39(7):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robison LL, Armstrong GT, Boice JD, et al. The Childhood Cancer Survivor Study: A National Cancer Institute-supported resource for outcome and intervention research. J Clin Oncol. 2009;27(14):2308–2318. doi: 10.1200/JCO.2009.22.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GAIN Collaborative Research Group. Manolio TA, Rodriguez LL, Brooks L, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39(9):1045–1451. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Gourraud PA, Génin E, Cambon-Thomsen A. Handling missing values in population data: consequences for maximum likelihood estimation of haplotype frequencies. Eur J Hum Genet. 2004;12(10):805–812. doi: 10.1038/sj.ejhg.5201233. [DOI] [PubMed] [Google Scholar]
- 29.Devlin B, Roeder K. Genomic control for associations. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 30.The MHC Sequencing Consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401(6756):921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 31.de Bakker PIW, McVean G, Sabeti PC, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38(10):1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, Kushekhar K, Nolte I, et al. Multiple HLA class I and II associations in classical Hodgkin lymphoma and EBV status defined subgroups. Blood. 2011;118(19):5211–5217. doi: 10.1182/blood-2011-04-342998. [DOI] [PubMed] [Google Scholar]
- 33.Conde L, Halperin E, Akers NK, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet. 2010;42(8):661–664. doi: 10.1038/ng.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcoceba M, Marin L, Balanzategui A, et al. The presence of DRB1*01 allele in multiple myeloma patients is associated with an indolent disease. Tissue Antigens. 2008;71(6):548–551. doi: 10.1111/j.1399-0039.2008.01048.x. [DOI] [PubMed] [Google Scholar]
- 35.Conde L, Bracci PM, Halperin E, Skibola CF. A search for overlapping genetic susceptibility loci between non-Hodgkin lymphoma and autoimmune diseases. Genomics. 2011;98(1):9–14. doi: 10.1016/j.ygeno.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and disease: a structural perspective. Nat Rev Immunol. 2006;6(4):271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 37.Ovsyannikova IG, Jacobson RM, Ryan JE, et al. HLA class II alleles and measles virus-specific cytokine immune response following two doses of measles vaccine. Immunogen. 2005;56(11):798–807. doi: 10.1007/s00251-004-0756-0. [DOI] [PubMed] [Google Scholar]
- 38.Ovsyannikova IG, Ryan JE, Jacobson RM, Vierkant RA, Pankratz VS, Poland GA. Human leukocyte antigen and interleukin 2, 10 an d12p40 cytokine responses to measles: Is there evidence of the HLA effect? Cytokine. 2006;36(3-4):173–179. doi: 10.1016/j.cyto.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skinnider BF, Mak TW. The role of cytokines in classical Hodgkin lymphoma. Blood. 2002;99(12):4283–4297. doi: 10.1182/blood-2002-01-0099. [DOI] [PubMed] [Google Scholar]
- 40.Jarrett RF. Risk factors for Hodgkin's lymphoma by EBV status and significance of detection of EBV genomes in serum of patients with EBV-associated Hodgkin's lymphoma. Leuk Lymphoma. 2003;44(suppl 3):S27–S32. doi: 10.1080/10428190310001623801. [DOI] [PubMed] [Google Scholar]