Abstract
Evolution can follow predictable genetic trajectories1, indicating that discrete environmental shifts can select for reproducible genetic changes2-4. Conspecific individuals are an important feature of an animal's environment, and a potential source of selective pressures. We show here that adaptation of two Caenorhabditis species to growth at high density, a feature common to domestic environments, occurs by reproducible genetic changes to pheromone receptor genes. Chemical communication through pheromones that accumulate during high-density growth causes young nematode larvae to enter the long-lived but non-reproductive dauer stage. Two strains of Caenorhabditis elegans grown at high density have independently acquired multigenic resistance to pheromone-induced dauer formation. In each strain, resistance to the pheromone ascaroside C3 results from a deletion that disrupts the adjacent chemoreceptor genes serpentine receptor class g (srg)-36 and -37. Through misexpression experiments, we show that these genes encode redundant G protein-coupled receptors for ascaroside C3. Multigenic resistance to dauer formation has also arisen in high-density cultures of a different nematode species, Caenorhabditis briggsae, resulting in part from deletion of an srg gene paralogous to srg-36 and srg-37. These results demonstrate rapid remodeling of the chemoreceptor repertoire as an adaptation to specific environments, and indicate that parallel changes to a common genetic substrate can affect life history traits across species.
Caenorhabditis elegans and many other nematode species evaluate environmental conditions to choose between two alternative developmental trajectories, one leading to rapid reproduction and one leading to arrest in the long-lived, stress-resistant dauer larva stage. High population density, limiting food, and high temperature promote dauer larva formation5 (Figure 1a), a stage that corresponds to the infectious juvenile stage of parasitic nematodes. Dauer larvae do not feed or reproduce, but can survive under conditions that kill other stages, and respond to environmental improvements by exiting the dauer stage and resuming reproductive development. Although the pheromone cues that signal nematode density are normally integrated with food availability, pheromone accumulation in high-density liquid cultures causes animals to form dauer larvae despite the presence of ample food6. Nonreproducing dauer animals would appear to be at a disadvantage relative to those that continue to grow in these conditions. To examine adaptation to high density culture conditions, we measured dauer formation in two laboratory strains of C. elegans, LSJ2 and CC1, that were grown in liquid axenic media for approximately and fifty and four years, respectively, before permanent cultures were frozen down7, 8 (Figure 1b and Methods). Unlike wild-caught strains9 and the standard laboratory strain N210, which readily form dauers in response to partially purified N2 dauer pheromone, CC1 and LSJ2 strains formed virtually no dauer larvae (Figure 1c).
Figure 1.
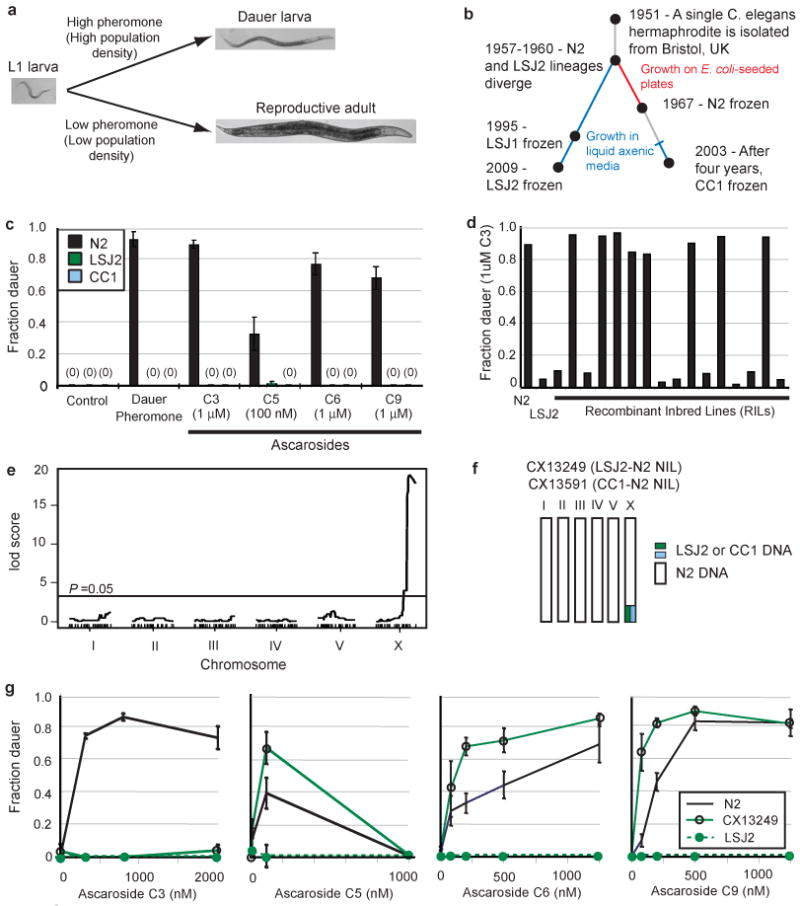
C. elegans cultivated in liquid are resistant to dauer pheromones. a, The developmental decision between reproductive growth and dauer larva formation is regulated by temperature, food, and population density. Population density is assessed by the release and sensation of ascarosides including C3, C5, C6, and C9. b, History of the C. elegans strains N2, LSJ1, LSJ2, and CC1 (see Methods). c, Dauer formation of N2, LSJ2, and CC1 in response to crude dauer pheromone or synthetic ascarosides. d, Dauer formation in response to synthetic C3 ascaroside. e, QTL mapping of C3 resistance. f, Schematic of near isogenic lines (NILs) with a small region from LSJ2 or CC1 introgressed into N2. g, Dauer formation in N2, LSJ2, and CX13249 strains. Error bars in all figures represent s.e.m.
N2, LSJ2, and CC1 arose from a common, inbred C. elegans ancestor after isolation from the wild (Figure 1b), so the pheromone resistance of LSJ2 and CC1 strains must result from new mutations that occurred in the laboratory. The genetic basis of dauer pheromone resistance was characterized by generating 94 recombinant inbred lines (RILs) between LSJ2 and N2 (Figure S1) that were genotyped at 176 informative SNPs (Table S1) identified by whole-genome sequencing of LSJ2 and N2 strains (Tables S2 and S3). Initial genetic mapping of dauer formation using N2-derived dauer pheromone preparations and the N2-LSJ2 RILs indicated that the trait was multigenic (data not shown). The active components of dauer pheromone are ascarosides, a group of small molecules with a common sugar scaffold and variable side chains11-13. Four individual ascarosides that effectively induced N2 dauer formation (C3, C5, C6, and C9) did not induce dauer formation in LSJ2 or CC1 (Figure 1c). To simplify trait mapping, we examined dauer formation in response to individual ascarosides, focusing on the C3 ascaroside, whose receptors and cellular sites of actions are unknown. Among sixteen RILs exposed to 1 μM C3, eight formed dauers at a rate comparable to N2 and eight formed dauers at a rate comparable to LSJ2 (Figure 1d). This bimodal distribution suggests the existence of a single locus that confers C3 resistance.
Quantitative trait locus (QTL) mapping using these sixteen RILs identified a single region on the X chromosome that correlated with the C3 response (Figure 1e). Mapping of the X-linked C3 resistance locus was verified by creating a near isogenic line (NIL) with the candidate region from LSJ2 introgressed into an N2 background by ten generations of backcrossing (Figure 1f). The LSJ2-N2 NIL was resistant to dauer formation induced by C3 ascaroside across a broad concentration range (Figure 1g). Unlike the parental LSJ2 strain, the LSJ2-N2 NIL formed dauer larvae in the presence of three other ascarosides (Figure 1g). These results identify an X-linked C3-resistance locus as one of several loci that confer pheromone resistance on LSJ2.
To identify the genetic changes in LSJ2 associated with C3 resistance, we sequenced the LSJ2 and N2 strains and identified all fixed polymorphisms between the two strains (Table S2 and Table S3). The region of X associated with C3 resistance included four SNPs in intronic or intergenic regions and a 4,906 bp deletion in the LSJ2 strain that disrupts two predicted G protein-coupled receptor genes (srg-36 and srg-37) (Figure 2a). A genomic clone from the N2 strain that contains both srg-36 and srg-37 fully rescued C3 resistance when introduced into the LSJ2-N2 NIL strain, indicating that this deletion causes the C3 resistance associated with the X-linked QTL (Figure 2b). Strikingly, srg-36 and srg-37 were also disrupted by a 6,795 bp deletion in the CC1 strain (Figure 2a). The deletions in CC1 and LSJ2 have different breakpoints, indicating that they occurred independently. To ask whether deletion of srg-36 and srg-37 also caused resistance to C3 in CC1, the region surrounding the srg-36 and srg-37 deletion was introgressed from CC1 into N2 to make a CC1-N2 NIL strain (Figure 1f). The CC1-N2 NIL was resistant to dauer formation induced by C3 ascaroside (Figure 2b) and its C3-resistance phenotype was rescued by a transgene covering the srg-36 and srg-37 genomic regions (Figure 2b). The CC1-N2 NIL readily formed dauers in response to other ascarosides (Figure S2), indicating that additional genetic mutations contribute to pheromone resistance in CC1. The existence of independent C3-resistance mutations affecting srg-36 and srg-37 in LSJ2 and CC1 provides strong genetic evidence linking these two chemoreceptors to dauer formation.
Figure 2.
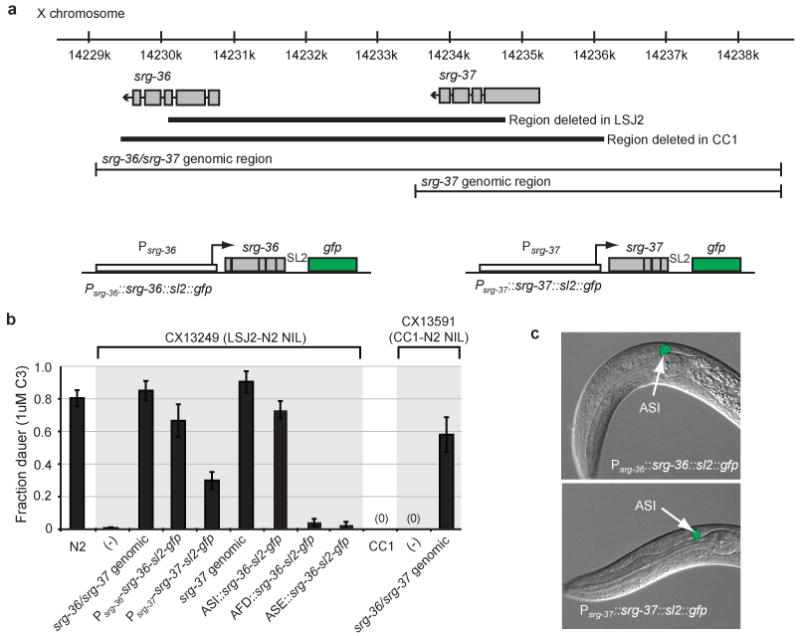
Resistance to C3 ascaroside is caused by deletion of two srg genes. a, Genomic region surrounding srg-36 and srg-37 on X, deletion breakpoints in LSJ2 and CC1 strains, fragments used for transgenic rescue, and design of bicistronic fusion genes. b, Transgenic rescue of dauer formation in response to C3 ascaroside. NIL strains used as recipients for rescue are shown in Figure 1f. ASI promoter was srg-47 (Figure S3), AFD promoter was gcy-8, ASE promoter was flp-6. c, Expression of GFP from bicistronic fusion genes for srg-36 and srg-37 in L1 larvae, showing predominant expression in ASI sensory neurons.
To determine which of the two predicted genes is associated with C3 sensitivity, we introduced srg-36 and srg-37 cDNAs with their respective upstream regions into the C3-resistant LSJ2-N2 NIL strain (Figure 2a). Transgenic strains expressing either cDNA formed dauer larvae in response to C3, although the srg-37 transgene was less active than the srg-36 transgene (Figure 2b). An srg-37 genomic fragment also rescued dauer formation (Figure 2b). These results suggest that the SRG-36 and SRG-37 genes are at least partially redundant; either can support dauer formation in response to C3 ascaroside.
The expression patterns of srg-36 or srg-37 were inferred from bicistronic transcripts expressing green fluorescent protein downstream of the srg-36 or srg-37 promoter and cDNA. These srg-36 and srg-37 reporter transgenes rescued C3-induced dauer formation (Figure 2b), and were most strongly and consistently expressed in the ASI chemosensory neurons, with weak or inconsistent expression in a few other neurons (Figure 2c). srg-36 and srg-37 reporters were robustly expressed during the L1 stage when the dauer decision is made (Figure 2c). The ASI neurons are primary regulators of dauer formation14, and are therefore plausible sites of srg-36 and srg-37 action. An srg-36 cDNA driven by the ASI-selective srg-47 promoter rescued C3-induced dauer formation in the LSJ2-N2 NIL, but expression of srg-36 in AFD or ASE sensory neurons did not (Figure 2b, Figure S3). These results are consistent with the hypothesis that srg-36 acts in ASI to sense ascaroside C3 (Figure S4).
The subcellular localization of SRG-36 was examined by fusing GFP to the srg-36 cDNA and expressing the hybrid gene from an ASI-specific promoter. This fusion protein was primarily localized in the sensory cilia of ASI (Figure 3a), suggesting a sensory function for SRG-36. The selective association of srg-36 and srg-37 with C3 responsiveness, and not with responsiveness to other ascarosides, suggested that they might encode C3 receptors. We tested this hypothesis by a gain of function experiment in which the srg-36 cDNA or the srg-37 cDNA was expressed in ASH neurons, a pair of polymodal nociceptive neurons that direct rapid avoidance behavior15. Unlike control animals, animals expressing the ASH∷srg-36 or ASH∷srg-37 transgene reversed rapidly in response to 1 μM C3 in an acute avoidance assay (Figure 3b). Neither ASH∷srg-36, ASH∷srg-37, nor control animals responded strongly to 1 μM C6 (Figure 3b). These results demonstrate that expression of SRG-36 or SRG-37 in ASH is sufficient for C3-specific behavioral responses.
Figure 3.
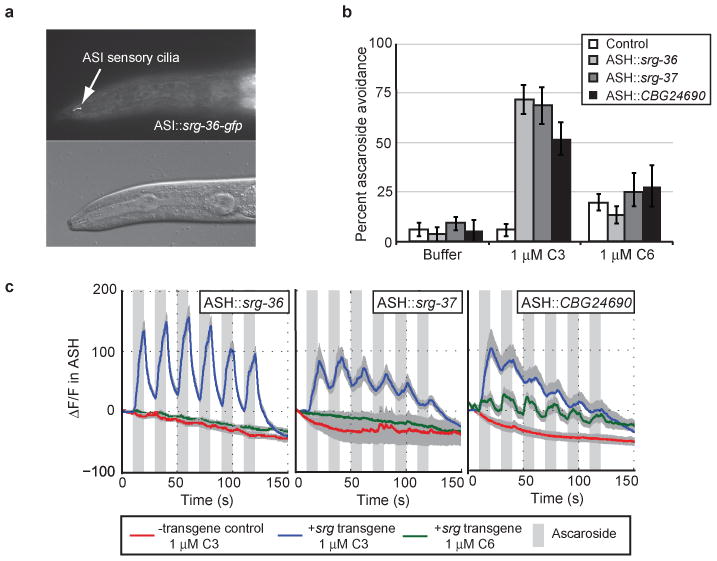
The srg genes encode ascaroside receptors. a, Localization of SRG-36∷GFP to ASI cilia (L4 animal). b, Ascaroside avoidance behaviours of animals with ectopic expression of srg-36, srg-37, or CBG24690 (shown in Figure 4) in the ASH nociceptive neurons. c, Ascaroside-induced Ca++ transients in ASH neurons that ectopically express C. elegans srg-36 or srg-37 or C. briggsae CBG24690 in ASH. Grey bars indicate the presence of C3 or C6 ascaroside, shading indicates s.e.m., n≥10 animals/condition. Ca++ was monitored using the genetically-encoded calcium sensor GCaMP3.030. Δ/F, percentage fluorescence change (baseline fluorescence = 100%).
The ASH neurons respond to repulsive stimuli with increases in intracellular calcium that can be monitored using genetically-encoded calcium indicators16. Animals expressing srg-36 or srg-37 in ASH showed rapid, reliable Ca++ increases in response to 1 μM C3 ascaroside, but not to 1 μM C6 ascaroside (Figure 3c); control animals did not respond to either C3 or C6. These results suggest that SRG-36 and SRG-37 are chemoreceptors (or subunits of chemoreceptors) that sense the C3 ascaroside. Although srg-36 and srg-37 are normally expressed in ASI, ASI neurons did not respond to C3 with calcium transients (data not shown). Little is known about pheromone signaling pathways in ASI, so the reason for this negative result is unclear.
LSJ2 was originally propagated in the Dougherty laboratory in the 1950s and 1960s to study nutrient requirements for nematode growth. A strain of C. briggsae, DR1690, that was grown in the Dougherty laboratory under the same conditions as LSJ2 also acquired resistance to dauer pheromone17 (Figure 4a). C. briggsae and C. elegans are estimated to have diverged 20-30 million years ago18. The C. briggsae genome encodes several genes closely related to srg-36 and srg-37, but does not have one-to-one orthologs to these genes (Figure 4b). Comparing genomic DNA sequences from DR1690 with the reference C. briggsae strain AF16, we discovered a 33 kb deletion in DR1690 that disrupts one of the srg paralogs, CBG24690, and six other genes (Figure 4c). To determine whether this deletion affects pheromone responses, we created a near isogenic line with the CBG24690 deletion introgressed into the AF16 reference background (DR1690-AF16 NIL). As previously reported17, AF16 readily formed dauers in response to C. elegans dauer pheromone, whereas DR1690 animals were resistant to dauer pheromone (Figure 4a). The C. briggsae DR1690-AF16 NIL formed dauers at an intermediate level compared to these two strains (Figure 4a), indicating that this region contains one of several mutations that contribute to pheromone resistance in DR1690. The DR1690-AF16 NIL was also resistant to purified ascaroside C3 compared to the parental AF16 strain (data not shown). A transgene covering the CBG24690 genomic clone rescued dauer formation in the pheromone-resistant DR1690-AF16 NIL strain (Figure 4a). These results demonstrate that the CBG24690 srg gene contributes to pheromone-induced dauer formation in C. briggsae.
Figure 4.
Evolutionary conservation of srg function. a, Rescue of C. briggsae dauer formation in response to partially purified dauer pheromone by genomic fragments containing the CBG24690 gene. CX13431 is a near isogenic line containing the CBG24690 deletion from DR1690 introgressed into the AF16 background. b, Schematic of genes closely related to srg-36 and srg-37 from C. elegans, C. briggsae, and C. remanei (adapted from19). c, CBG24690 genomic region from the AF16 C. briggsae reference strain, and location of a large deletion in the DR1690 C. briggsae strain that was cultivated for an extended period in liquid axenic media.
To ask whether CBG24690 also encodes an ascaroside receptor, we expressed a CBG24690 cDNA in the C. elegans ASH neurons. Animals expressing the ASH∷CBG24690 transgene reversed rapidly when presented with 1 μM C3, but not 1 μM C6 (Figure 3b). Animals expressing the CBG24690 srg gene in ASH also showed rapid Ca++ increases in response to 1 μM C3 (Figure 3c). Unlike animals expressing srg-36 or srg-37 in ASH, animals expressing CBG24690 in ASH showed weaker but reliable Ca++ increases in response to the related ascaroside C6 (Figure 3c).
Our results suggest that srg-36 and srg-37, two members of a large nematode-specific family of G protein-coupled receptors19, encode redundant receptors for the ascaroside C3. The srg gene family is distinct from the srbc gene family previously implicated in sensing ascarosides C6 and C920, suggesting that at least two of the seven chemoreceptor superfamilies of C. elegans can detect ascaroside pheromones. Chemoreceptors are among the fastest-evolving genes in metazoan genomes. They come from entirely different protein families in vertebrates, insects, and nematodes, and change rapidly between species21; only half of the chemoreceptors in C. elegans and C. briggsae are one-to-one ortholog pairs19. Despite the rapid evolution of these genes, the function of srg-like genes in pheromone detection has been conserved since C. briggsae and C. elegans diverged, as C. elegans srg-36 and srg-37, and C. briggsae CBG24690, each sense C3 ascaroside to induce dauer formation. Some differences between species exist, however, as CBG24690 also senses C6 ascaroside at concentrations that are not sensed by srg-36 and srg-37. Differences in pheromone production and sensation by different Caenorhabditis species may allow both species-specific discrimination and general detection of Caenorhabditis species in the vicinity, as is observed with quorum-sensing systems in bacteria22.
These results directly demonstrate a reproducible change in the chemoreceptor repertoire in response to a discrete environmental shift. During high-density growth in the laboratory, resistance to ascaroside pheromones arose independently in C. elegans LSJ2, CC1, and C. briggsae DR1690 through changes in related srg genes. Although numerous single-gene mutations convey resistance to dauer formation in C. elegans23, deletion of srg genes appears to be a favored route contributing to pheromone resistance. It is possible that specific features of the chromosomal region surrounding srg-36 and srg-37 predispose this region to deletion mutations, but these features would also need to be found near the CBG24690 gene in C. briggsae. Alternatively, the spectrum of potential dauer-defective mutants may be constrained because the known single-gene mutations that are resistant to dauer formation have pleiotropic effects on sensory biology, stress resistance, and starvation responses that would reduce their fitness in mixed cultures23. Global analysis of functional genetic variants suggests that evolutionarily relevant mutations are not randomly distributed, but rather cluster in specific genetic loci, or hotspot genes1. These observations suggest that genetic trajectories during evolution are constrained and that adaptation can, at least to some extent, be predictable. One class of known “adaptive” genes are input-output genes, developmental regulators whose complex cis-regulatory motifs provide a molecular substrate that allows sculpting of developmental patterns2, 24. srg-36 and srg-37 appear to fall into a second class of “adaptive” genes, including opsin genes and taste receptors25, 26: sensory receptors whose diversity allows circumscribed adaptation to environmental changes without pleiotropic effects.
Methods Summary
The LSJ2 strain and the N2 laboratory strain are descended from one ancestral hermaphrodite isolated by W. Nicholas. LSJ2 was grown continuously in liquid axenic media starting in ∼1957 at the Kaiser Foundation Research Institute, UC Berkeley and San Jose State University until a sample was frozen in 2009.
Dauer formation assays were performed with crude or synthesized ascarosides as described11. Values report the average fraction of dauer animals 72 hours after eggs were laid on assay plates. With the exception of the mapping experiments, each strain and condition was tested in a minimum of five independent assays.
RILs were generated from reciprocal crosses between LSJ2 and an N2-derived strain and inbred for ten generations. Two laboratory-derived polymorphisms that modify the npr-1 and glb-5 genes in N2 affect many C. elegans behaviors7, 27; to eliminate their effects, the cross was initiated with a strain containing 99% N2 DNA but the ancestral alleles of npr-1 and glb-5 from the CB4856 strain (Figure S1). These RILs were genotyped at 192 SNPs between LSJ2 and N2. The fraction of animals forming dauers in response to 1 μM C3 ascaroside was used as a phenotype for nonparametric QTL mapping.
srg-36, srg-37, and CBG24690 were ectopically expressed in ASH using the sra-6 promoter. Vehicle control, 1 μM of C3, or 1 μM of C6 was dissolved in M13 buffer and presented to animals using the drop test28. Each animal was scored three times for the ability to reverse in response to the stimulus. At least fifty animals were scored blindly for each strain and condition.
ASH imaging was performed in a custom-designed microfluidic device29. The genetically-encoded calcium indicator GCaMP3.030 was expressed in ASH using the sra-6 promoter.
Supplementary Material
Acknowledgments
We thank Nancy Lu for the LSJ2 strain, Matt Rockman for a qgIR1(X,CB4856>N2) introgression strain, Justin Ragains for ascaroside synthesis, Howard Hang for assistance in purifying pheromones, and Scott Dewell, Kevin Foster, Niels Ringstad, Andrés Bendesky, Yasunori Saheki, Manuel Zimmer, Sean Crosson, Evan Feinberg, Esteban Toro, Matt Rockman, and Leonid Kruglyak for comments and advice. P.T.M. was funded by a Damon Runyon Fellowship. Y.X. was supported by MSTP grant GM07739 and a Paul and Daisy Soros Fellowship. J.L.G. was a HHMI fellow of the Helen Hay Whitney Foundation and is funded by National Institutes of Health (NIH) K99 GM092859. R.A.B. is supported by R00GM87533. C.I.B. is an investigator of the Howard Hughes Medical Institute. This work was supported by the HHMI.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: P.T.M. and C.I.B. designed and interpreted experiments and wrote the paper. P.T.M. performed all genetic, molecular, and behavioural experiments, Y.X. conducted calcium imaging experiments, M.A. identified the dauer formation defect in the LSJ2 lineage, R.A.B. characterized and synthesized ascarosides, and J.L.G. contributed reagents.
Competing Financial Interests: The authors declare no competing financial interests.
Reprints and permissions information are available at www.nature.com/reprints.
References
- 1.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–51. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–5. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Protas ME, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–11. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- 4.Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:9107–12. doi: 10.1073/pnas.0602917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102:368–78. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- 6.Stiernagle, T. Maintenance of C. elegans. WormBook (ed. The C. elegans Research Community) doi:10.1895/wormbook. 1.101.1 (11 February 2006); available at http://www.wormbook.org
- 7.McGrath PT, et al. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–9. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szewczyk NJ, Kozak E, Conley CA. Chemically defined medium and Caenorhabditis elegans. BMC Biotechnol. 2003;3:19. doi: 10.1186/1472-6750-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viney ME, Gardner MP, Jackson JA. Variation in Caenorhabditis elegans dauer larva formation. Dev Growth Differ. 2003;45:389–96. doi: 10.1046/j.1440-169x.2003.00703.x. [DOI] [PubMed] [Google Scholar]
- 10.Golden JW, Riddle DL. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science. 1982;218:578–80. doi: 10.1126/science.6896933. [DOI] [PubMed] [Google Scholar]
- 11.Butcher RA, Ragains JR, Kim E, Clardy J. A potent dauer pheromone component in Caenorhabditis elegans that acts synergistically with other components. Proc Natl Acad Sci U S A. 2008;105:14288–92. doi: 10.1073/pnas.0806676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong PY, et al. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature. 2005;433:541–5. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]
- 13.Srinivasan J, et al. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454:1115–8. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–6. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan JM, Horvitz HR. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1993;90:2227–31. doi: 10.1073/pnas.90.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilliard MA, et al. In vivo imaging of C. elegans ASH neurons: cellular response and adaptation to chemical repellents. EMBO J. 2005;24:63–72. doi: 10.1038/sj.emboj.7600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fodor A, Riddle DL, Nelson FK, Golden JW. Comparison of a new wild-type Caenorhabditis briggsae with laboratory strains of C. briggsae and C. elegans. Nemotologica. 1983;29:203–217. [Google Scholar]
- 18.Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 2008;25:778–86. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- 19.Thomas JH, Robertson HM. The Caenorhabditis chemoreceptor gene families. BMC Biol. 2008;6:42. doi: 10.1186/1741-7007-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K, et al. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science. 2009;326:994–8. doi: 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–63. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 22.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–46. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 23.Hu, P.J. Dauer. Wormbook (ed. The C. elegans Research Community) doi:10.1895/wormbook.1.144.1 (8 August 2007); available at http://www.wormbook.org
- 24.Sucena E, Delon I, Jones I, Payre F, Stern DL. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature. 2003;424:935–8. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- 25.Kim UK, et al. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–5. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- 27.de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–89. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 28.Hilliard MA, Bargmann CI, Bazzicalupo P. C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr Biol. 2002;12:730–4. doi: 10.1016/s0960-9822(02)00813-8. [DOI] [PubMed] [Google Scholar]
- 29.Chronis N, Zimmer M, Bargmann CI. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods. 2007;4:727–31. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- 30.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



