Abstract
The epidermal growth factor receptor variant III (EGFRvIII) is associated with increased proliferation of glioma cells. However, the impact of EGFRvIII on survival of patients with glioblastoma (GBM) has not been definitively established. In the present study, we prospectively evaluated 73 patients with primary GBM treated with surgical resection and standard radio/chemotherapy. The EGFRvIII was assessed by reverse transcription-polymerase chain reaction (PCR), O6-methylguanine methyltransferase (MGMT) promoter methylation was assessed by methylation-specific PCR, and phosphatase and tension homolog (PTEN) expression was assessed by immunohistochemistry. In 14 patients of this series, who presented with tumor recurrence, EGFRvIII was determined by real-time PCR. Sensitivity to temozolomide (TMZ) was assessed in vitro on GBM neurosphere cell cultures with different patterns of EGFRvIII expression. Age 60 years or younger, preoperative Karnofsky Performance Status score of 70 or higher, recursive partitioning analysis score III and IV, methylated MGMT, and Ki67 index of 20% or less were significantly associated with longer overall survival (OS; P = .0069, P = .0035, P = .0007, P = .0437, and P = .0286, respectively). EGFRvIII identified patients with significantly longer OS (P = .0023) and the association of EGFRvIII/Ki67 of 20% or less, EGFRvIII/normal PTEN, EGFRvIII/methylated MGMT, and EGFRvIII/normal PTEN/methylated MGMT identified subgroups of GBM patients with better prognosis. In recurred GBMs, EGFRvIII expression was approximately two-fold lower than in primary tumors. In vitro, the EGFRvIII-negative GBM neurosphere cells were more resistant to TMZ than the positive ones. In conclusion, in contrast with previous studies, we found that EGFRvIII is associated with prolonged survival of GBM patients treated with surgery and radio/chemotherapy. Depletion of EGFRvIII in recurrent GBMs as well as differential sensitivity to TMZ in vitro indicates that the EGFRvIII-negative cell fraction is involved in resistance to radio/chemotherapy and tumor repopulation.
Introduction
Amplification of the epidermal growth factor receptor (EGFR) gene is the most frequent genetic change associated with glioblastoma (GBM), which results in overexpression of the transmembrane tyrosine kinase receptor, EGFR [1]. GBM showing amplified EGFR frequently overexpresses the receptor variant III (EGFRvIII), which is characterized by a truncated extracellular domain with ligand-independent constitutive activity [2–6]. In vitro and in vivo studies have demonstrated that EGFRvIII confers increased proliferation and invasiveness to glioma cells [5–8]. However, the role of overexpressed EGFRvIII on the prognosis of GBM patients has not definitively been established [6,9–20].
In this report, we prospectively analyzed the relationship between EGFRvIII expression and overall survival (OS) in 73 patients with newly diagnosed GBM treated with gross tumor resection and adjuvant radiotherapy and temozolomide (TMZ). In patients who underwent a second surgery for tumor recurrence, we determined the levels of EGFRvIII both at primary surgery and at recurrence after radio/chemotherapy, and we correlated them with survival after recurrence. Finally, we analyzed the sensitivity to TMZ of GBM stem-like cells with different levels of EGFRvIII expression.
Materials and Methods
Patients
This study includes 73 consecutive adult patients who underwent craniotomy for resection of histologically confirmed GBM (WHO grade 4) [21] and who were treated postoperatively with adjuvant radiotherapy and TMZ at the Università Cattolica del Sacro Cuore, Rome. All patients provided written informed consent according to the research proposals approved by the ethical committee of the Università Cattolica del Sacro Cuore. Patients of pediatric age and patients with secondary GBM were not included. The patients were 20 to 80 years old at the time of primary surgery (median age, 61 years; mean age, 59.9 ± 11.4 years); 45 were men and 28 were women (Table 1 and Table W1). To evaluate the extent of tumor resection, we took into consideration both surgeon's impression at operation and Gd-enhanced axial T1-weighted magnetic resonance image (MRI) obtained 1 month after surgery, that is before radio/chemotherapy. All patients received radiotherapy to limited fields (2 Gy per fraction, once a day, 5 days a week, 60-Gy total dose) and adjuvant TMZ after surgery [22]. Fourteen patients were operated on again for tumor recurrence by 7 to 58 months after primary surgery; nine were men and five were women (Table W2). OS was calculated from the date of surgery to death or end of follow-up.
Table 1.
Patients' Characteristic.
| Characteristic | No. | % | |
| n | 73 | 100 | |
| Median age at diagnosis (years) | 61 | ||
| Range | 20–80 | ||
| Age classes (years) | |||
| 18–39 | 3 | 4 | |
| 40–49 | 8 | 11 | |
| 50–59 | 25 | 33 | |
| 60–69 | 22 | 31 | |
| >70 | 15 | 21 | |
| Sex | |||
| Male | 45 | 61 | |
| Female | 28 | 39 | |
| KPS | |||
| Median (range) | 60 (30–90) | ||
| ≥70 | 35 | 47 | |
| <70 | 38 | 53 | |
| Surgery | |||
| Total resection | 59 | 79 | |
| Partial resection | 14 | 19 | |
| RTOG RPA classes | |||
| RPA III | 4 | 6 | |
| RPA IV | 18 | 24 | |
| RPA V | 35 | 49 | |
| RPA VI | 16 | 22 |
RTOG indicates Radiation Therapy Oncology Group.
Immunohistochemical analysis was performed as previously described [22]. Immunoreactivity for PTEN was performed with mouse monoclonal antibody (1:50; clone 28H6; Novo Castra, Newcastle, United Kingdom). Immunoreactivity was considered as positive when the tumor cells showed a strong nuclear staining similar to control cells and reduced when the nuclear staining was absent or reduced compared with normal. For a semiquantitative immunostaining evaluation, the slides were screened independently by two pathologists (L.M.L. and M.M.) who were unaware of the patient prognosis-related information.
Reverse Transcription-Polymerase Chain Reaction for EGFRvIII
After being deparaffinized, three 10-µm slides were digested overnight at 55°C in 200 µl of TENS 1x (10 mM Tris pH 7.4, 10 mM EDTA, 100 mM NaCl, 1% SDS) with 100 mg/ml proteinase K, and RNA was then extracted by RNAsi mini kit (Qiagen, Milan, Italy), following the manufacturer's protocol. To minimize contamination by normal cells, the tumor areas selected for DNA/RNA extraction contained at least 80% disease-specific cells. We assessed the quantity and quality of the RNA spectrophotometrically (E260, E260/E280 ratio, spectrum 220–320 nm; Biochrom, Cambridge, United Kingdom) and by separation on an Agilent 2100 Bioanalyzer (Palo Alto, CA). RNA was treated with RQ1 RNase-Free DNase (Promega, Milan, Italy), and complementary DNA (cDNA) was synthesized as previously described using random examers [23]. The quality of the reverse transcription synthesis was also tested by amplifying cDNA with primers of housekeeping genes producing fragments with different lengths. The presence of EGFRvIII was assessed using the same primers and following the polymerase chain reaction (PCR) conditions described by Mellinghoff et al. [13] and by Yoshimoto et al. [24]. These two methods showed a 100% concordance in our laboratory. β-Actin amplification was used as an internal control [23]. Specificity of reverse transcription-PCR product was assessed by direct sequencing, using the same primers used for PCR amplification (Figure W2).
Real-time PCR for EGFRvIII
Levels of EGFRvIII messenger RNA (mRNA) were assessed by real-time PCR using SYBR green chemistry. Diluted (1:20) cDNA (4 µl) was added to a PCR mix containing 8.4 µl of sterile water, 12.5 µl of 2x SYBR mix (Qiagen, Milan, Italy), and 0.05 µl each of forward and reverse primers (200 mM) to make up a final volume of 25 µl [23]. Cycling conditions were 95°C for 5 minutes, followed by 40 cycles of 95°C for 10 seconds and 60°C for 30 seconds, and 80 cycles of 55°C ± 0.5°C per cycle for melting curve analysis in a iCycler-iQ multicolor real-time PCR detection system (Bio-Rad, Milan, Italy). Each assay was performed in triplicate, and data were processed by iCycler-iQ optical system software (Bio-Rad). The average obtained for EGFRvIII was normalized to the average amount of β-actin for each sample to determine the relative changes in mRNA expression. A 10-fold serial dilution of 10 ng of plasmid containing the EGFRvIII demonstrated that the sensibility of this assay was less than 10-2 in our laboratory.
DNA Extraction and Methylation-Specific PCR for O6-Methylguanine Methyltransferase
Three 10-µm slides were cut from paraffin-embedded tissues, treated twice with xylene, and then washed with ethanol. DNA was extracted using the QIAamp tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Methylation-specific PCR (MS-PCR) for O6-methylguanine methyltransferase (MGMT) was performed as previously described [23]. Briefly, bisulfite-modified DNA (100–200 ng) was amplified in a mixture containing 1x PCR buffer (20 mM Tris pH 8.3, 50 mM KCl, 1.5 mM MgCl2), dNTPs (200 mM each), primers (20 pM each), and 0.75 U of Taq polymerase platinum (Invitrogen) in a final volume of 25 µl. PCR conditions were an initial denaturation of 95°C for 8 minutes, followed by 35 cycles of 95°C for 60 seconds, 60°C for 60 seconds, and 72°C for 60 seconds. The MS-PCR did not exceed the 35 amplification cycles. PCR products were electrophoresed in a 2.5% agarose gel, stained with ethidium bromide, and visualized under UV illumination. In all samples, MS-PCR analyses were performed in duplicate. Normal lymphocyte DNA supermethylated with SssI methyltransferase (New England Biolabs, Beverly, MA), treated with bisulfite, served as the unmethylated and methylated control, water as a negative control, and untreated DNA as internal PCR control. As control group, we carried out MS-PCR also on granulocyte DNA obtained from 10 healthy individuals.
Establishing GBM Neurosphere Cell Cultures
GBM tissue specimens were collected at surgery from adult patients who had undergone craniotomy at the Institute of Neurosurgery, Catholic University School of Medicine, Rome. Cell suspension obtained by mechanical dissociation of the tumor tissue was passed through a 100-µm mesh to remove aggregates and cultivated in a serum-free medium containing epidermal growth factor and basic fibroblast growth factor as previously described [22,25]. Cell lines actively proliferating required 3 to 4 weeks to be established. Isolated cell lines were expanded and characterized both in vitro and in vivo. In these conditions, cells were able to grow in vitro in clusters called neurospheres and maintain an undifferentiated state, as indicated by morphology and expression of stem cell markers such as CD133, SOX 2, Musashi1, and nestin. The in vivo tumorigenic potential of GBM neurospheres was assayed by intracranial or subcutaneous cell injection in immunocompromised mice. GBM neurospheres were able to generate a tumor identical to the human tumor in antigen expression and histologic tissue organization. Cell lines were used from passage 5 to 10 throughout the study.
In Vitro Sensitivity to TMZ
GBM stem cells were mechanically dissociated and plated at the density of 2000 cells in 96-well plates in triplicate. After 24 hours of incubation at 37°C in a 5% CO2, the cells were treated with TMZ at concentrations of 125, 250, and 500 µM (Schering-Plough, Kenilworth, NJ). Cells' viability was evaluated after 24, 48, 72, and 96 hours of treatment by CellTiter Glo luminescent assay according to the manufacturer's protocol (Promega).
Semiquantitative RT-PCR Analysis of Bcl-XL Expression
After RNA extraction with RNAsi mini kit (Qiagen, Hilden, Germany), first-strand cDNA was synthesized by 1 µg of RNA using Go-Script Reverse Transcription Systemkit according to the manufacturer's protocol (Promega). Three microliters of cDNA was amplified with specific primers for Bcl-XL (5′-TCCTTGTCTACGCTTTCCACG-3′ and 5′-GGTCGCATTGTGGCCTTT-3) and β-actin (5′-TACATGGGTGGGGTGTTGAA-3′ 5′-AAGAGAGGCATCCTCACCCT-5′) in 25 µl of final volume, containing 1 U of GoTaq (Promega), 1 mM of each primer, 200 mM of dNTPs, and 1x reaction buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2). The PCR conditions were as follows: one cycle of 3 minutes at 95°C, followed by 33 cycles at 95°C for 40 seconds, 58°C for 40 seconds, 72°C for 40 seconds. The mixture was separated on a 2% agarose gel, and after staining with ethidium bromide, the PCR product was visualized under ultraviolet light. The Bcl-XL expression were subjected to densitometric analysis by using the Gel-Doc 2000 Quantity One program (Bio-Rad), after normalization with the β-actin intensity.
Statistical Analysis
Statistical analysis was performed using STATA 10 (StataCorp LP, College Station, TX), GraphPad-Prism 5 software (Graph Pad Software, San Diego, CA), and MedCalc version 10.2.0.0 (MedCalc Software, Mariakerke, Belgium). Kaplan-Meier survival curves were plotted, and differences in survival between groups of patients were compared using the log-rank test. Statistical comparison of continuous variables was performed by the Mann-Whitney U test, as appropriate. Comparison of categorical variables was performed by χ2 statistic, using the Fisher exact test. Multivariate analysis was performed using the Cox proportional hazards model, which was adjusted for the major prognostic factors that included age (≤60 vs >60 years), Karnofsky Performance Status (KPS; <70 vs ≥70), extent of surgical resection (total vs partial resection), MGMT methylation status and Ki67 index (≤20% vs >20%). Correlation between EGFRvIII mRNA in recurrent tumors and survival after recurrence was studied using regression analysis and the Spearman correlation coefficient. P < .05 were considered as statistically significant.
Results
Clinical and Molecular Parameters
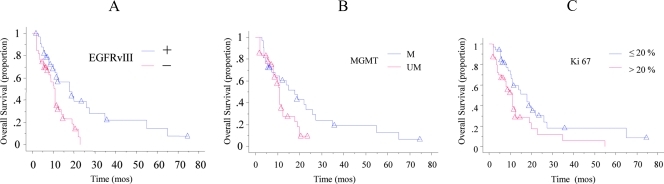
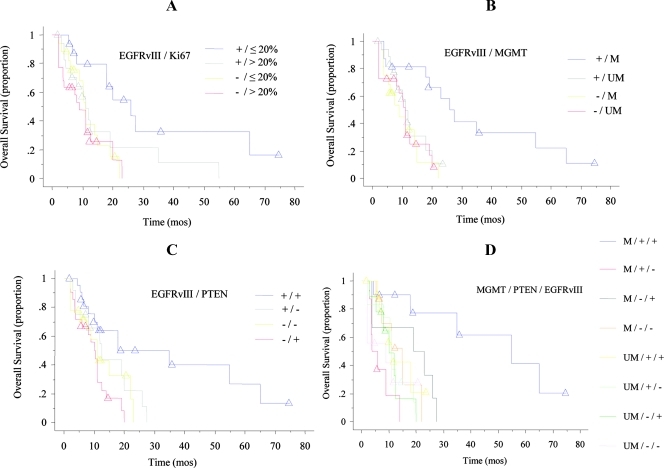
Among the clinical estimates, age 60 years or younger, preoperative KPS score of 70 or higher, and recursive partitioning analysis (RPA) score III and IV were significantly correlated with longer OS (P = .0069, HR = 0.43, 95% confidence interval [CI] = 0.23–0.79 for age; P = .0035, HR = 2.34, 95% CI = 1.32–4.14 for KPS; P = .0007, HR = 0.35, 95% CI = 0.19–0.64 for RPA; Table 2 and Figure W1). Patients with totally resected tumors trended to survive longer than those who had undergone partial tumor resection, although this difference was not significant. Among the biologic features, GBMs with EGFRvIII, with MGMT hypermethylation, and with Ki67 index of 20% or less were associated to a significant longer OS (P = .0023, HR = 2.59, 95% CI = 1.40–4.79; P = .0437, HR = 1.86, 95% CI = 1.02–3.41; P = .0286, HR = 0.53, 95% CI = 0.30–0.93; Figure 1). There was no difference in Ki67 index between EGFRvIII-positive GBMs and those tumors without EGFRvIII (26.6 ± 13.6 and 25.6 ± 12.3, respectively; 26.0 ± 12.8; P = .75). However, GBMs showing EGFRvIII and Ki67 index of 20% or less had significantly longer OS than those with EGFRvIII and Ki67 index greater than 20% (P = .0275, HR= 0.37, 95% CI = 0.15–0.89; Figure 2A). A favorable impact onOS was also demonstrated for the association between the presence of EGFRvIII and MGMT promoter methylation (P = .0108, HR = 3.70, 95% CI = 1.35–10.12; Figure 2B), for the association between the presence of EGFRvIII and normally expressed PTEN (P = .0223, HR = 0.33, 95% CI = 0.13–0.85; Figure 2C), and for the association between the presence of EGFRvIII, normally expressed PTEN, and methylated MGMT (P = .0040, HR = 2.69, 95% CI = 1.37–5.30; Figure 2D).
Table 2.
OS for Clinical and Biologic Parameters.
| Parameter | n | Median OS (Months) | P |
| All patients | 73 | 9 | |
| Age (years) | |||
| ≤60 | 36 | 18 | .0069 |
| >60 | 37 | 9 | |
| KPS | |||
| ≥70 | 35 | 15 | .0035 |
| <70 | 38 | 9 | |
| Surgery | |||
| Total | 59 | 11 | .3822 |
| Partial | 14 | 19 | |
| RTOG RPA score | |||
| III–IV | 22 | 29 | .0007 |
| V | 35 | 11 | |
| VI | 16 | 8 | |
| EGFRvIII | |||
| + | 32 | 19 | .0023 |
| - | 41 | 10.5 | |
| MGMT | |||
| M | 32 | 14 | .0437 |
| UM | 41 | 11 | |
| PTEN | |||
| + | 43 | 11 | .4175 |
| - | 30 | 11 | |
| Ki67 | |||
| ≤20% | 32 | 14 | .0286 |
| >20% | 41 | 9 |
M indicates methylated; UM, unmethylated.
Figure 1.
Kaplan-Meier survival curves of 73 GBM patients stratified by EGFRvIII, MGMT, and Ki67. The presence of EGFRvIII in tumors (A, blue line), methylated MGMT (B, blue line), and Ki67 index of 20%or less (C, blue line), conferred a favorable survival advantage (P =.0023, P = .0437, and P = .0286, respectively).
Figure 2.
Kaplan-Meier survival curves of 73 GBM patients stratified by EGFRvIII/Ki67, EGFRvIII/PTEN, EGFRvIII/MGMT methylation, and MGMT methylation/PTEN/EGFRvIII. (A) Patients stratified by EGFRvIII/Ki67. EGFRvIII-positive GBMs and Ki67 index of 20% or less (blue line) was related with longer OS (P = .0275). There were no differences in survival times between the following three unfavorable groups: EGFRvIII+/Ki67 greater than 20% (gray line), EGFRvIII-/Ki67 20% or less (yellow line), and EGFRvIII-/Ki67 greater than 20% (red line). (B) Patients stratified by EGFRvIII/MGMT methylation. EGFRvIII-positive GBMs, and methylated MGMT (blue line) was related with longer OS (P = .0108). There were no differences in survival times between the following three unfavorable groups: EGFRvIII+/unmethylated MGMT (gray line), EGFRvIII-/methylated MGMT (yellow line), and EGFRvIII-/unmethylated MGMT (red line). (C) Patients stratified by EGFRvIII/PTEN. The presence of EGFRvIII and normal expression of PTEN (blue line) was associated with longer OS (P = .0223). There were no differences in survival times between the following three unfavorable groups: EGFRvIII+/hypoexpression of PTEN (-) (gray line), EGFRvIII-/PTEN- (yellow line), and EGFRvIII-/PTEN+ (red line). (D) Patients stratified by MGMT methylation/PTEN/EGFRvIII. The association of methylated MGMT, normal expression of PTEN, and presence of EGFRvIII (blue line) was associated with longer OS (P = .004).
A multivariate survival model for OS (Cox proportional hazards regression analysis) was established that included age, KPS, extent of resection, Ki67 index, EGFRvIII expression, MGMT promoter methylation, and PTEN expression (Table W3). Age older than 60 years (P = .0182), KPS score of 70 or higher (P = .0055), Ki67 index of 20% or less (P = .0032), and EGFRvIII (P = .0128) emerged as independent prognostic factors for OS.
EGFRvIII Expression in Recurrent GBM
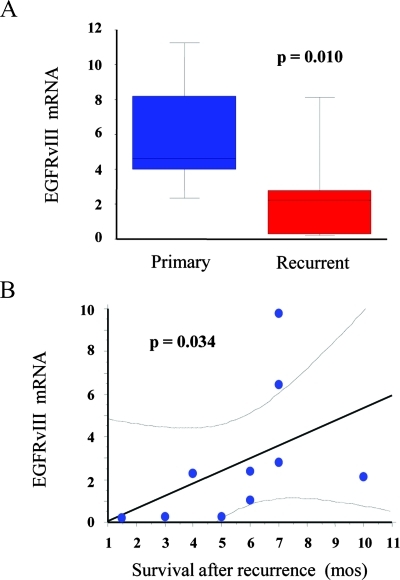
Fourteen patients of this series underwent a second operation for resection of tumors that recurred after radio/chemotherapy (Table W2). The interval for tumor recurrence ranged from 7 to 58 months. Gross total resection was achieved in all cases. Histologic sections both of the primary tumor and of the recurrent one were carefully reviewed and adjacent slices were dissected to eliminate areas of necrosis and hemorrhages. Real-time PCR of such selected regions showed that, relative to GBMs at primary surgery, the level of EGFRvIII in the recurrent tumors decreased by approximately 50%on average (range = 40.4%–77.6%). In four cases, EGFRvIII expression was not detected both in the primary tumor and in the recurrent one (Table W2). In remaining cases, the EGFRvIII levels were reduced in seven cases and unchanged in three cases. Overall, there was a significant depletion of EGFRvIII in recurrent GBMs (P = .01, Wilcoxon signed rank test; Figure 3A). Plotting the EGFRvIII mRNA levels in recurrent tumors against survival after recurrence revealed a linear relationship between the two variables (P = .034, r2 = 0.706, Spearman correlation coefficient; Figure 3B). The same type of relationship was demonstrated when the EGFRvIII values were expressed as the ratio between EGFRvIII mRNA after radio/chemotherapy and EGFRvIII mRNA before radio/chemotherapy, a method that attenuates interindividual variability (P = .029, r2 = 0.724, Spearman correlation coefficient).
Figure 3.
Expression of EGFRvIII in recurrent GBM. (A) Box-and-whisker plots showing significant reduction of EGFRvIIImRNA levels in recurrent GBM in comparison to the primary tumors (Wilcoxon signed rank test). (B)Graph showing the relationship between survival after tumor recurrence and level of EGFRvIII mRNA expression in the recurrent tumor (Spearman correlation coefficient).
Sensitivity to TMZ of GBM Neurosphere Cell Cultures
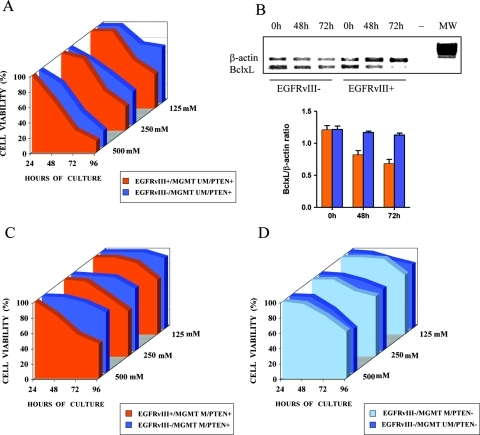
To investigate the relationship between the presence of EGFRvIII and sensitivity to TMZ, we used paired GBM neurosphere cell cultures expressing or not the EGFRvIII and which were obtained from different regions of the same tumor under stem cell culture conditions. These cells provide a unique model that eliminates the confounding variables inherent to cells derived from different patients. We and others have demonstrated that such cells, generally refer red to as GBM stem-like cells, retain many biologic and molecular features of the parental tumor, including resistance to chemotherapeutic agents [26,27]. For this experiment, we used six GBM neurosphere cell cultures that were obtained from three patients and that showed various combinations of EGFRvIII, PTEN expression, and MGMT promoter methylation status (Table W4). All of the GBM neurosphere cell lines had previously been demonstrated to self-renew in vitro and to give rise to tumor xenografts that recapitulate the phenotype and the histologic pattern of the parent tumor on orthotopic transplantation in immunocompromised mice [22,25].
We found that concentrations of TMZ less than 125 µM had no significant effect on the growth of GBM cell lines. At higher concentrations, however, the EGFRvIII-positive GBM cells were less resistant to TMZ compared to their EGFRvIII-negative counterparts (Figure 4). Sensitivity to TMZ correlated with reduction of Bcl-XL mRNA (Figure 4B)—an antiapoptotic member of BCL-2 family that has been demonstrated to increase in EGFRvIII-expressing GBM cells and to be associated with increased chemoresistance [28,29].
Figure 4.
Cell viability percentages of GBM neurosphere cells with different patterns of EGFRvIII expression, MGMT methylation status, and PTEN expression (Table W4) treated for 24, 48, 72, and 96 hours with TMZ at 125, 250, and 500 µM. Each experiment was repeated three times, and mean values were plotted. (A) Cell survival curves comparing sensitivity to TMZ of cell line 1A (blue; EGFRvIII-/MGMT unmethylated/PTEN+) versus cell line 1B (orange; EGFRvIII+/MGMT unmethylated/PTEN+). (B) Semiquantitative RT-PCR analysis of Bcl-XL mRNA expression by the EGFRvIII-positive cell line 1B and by EGFRvIII-negative cell line 1A at 0, 48, and 72 hours exposure to 500 µM TMZ in culture. Upper panel, representative experiment (MW, molecular weight; -, negative control). Lower panel, semi-quantitative analysis of Bcl-xL mRNA expression from three different experiments in EGFR-positive (orange) and in EGFR-negative (blue) neurosphere cell line. (C) Cell survival curves comparing sensitivity to TMZ of cell line 2A (orange; EGFRvIII+/MGMT methylated/PTEN+) versus cell line 2B (blue; EGFRvIII-/MGMT methylated/PTEN+). (D) Cell survival curves comparing sensitivity to TMZ of cell line 3A (light blue; EGFRvIII-/MGMT methylated/PTEN-) versus cell line 3B (dark blue; EGFRvIII-/MGMT unmethylated/PTEN-).
Discussion
In this study, we reevaluated the relationship between EGFRvIII in GBM tumors and survival of patients undergoing gross tumor resection and adjuvant radiotherapy and TMZ. To analyze EGFRvIII presence, we used an RT-PCR assay on selected regions of formalin-fixed paraffin-embedded tumor specimens. This method, which has been proven to be more sensitive than immunohistochemistry, detects EGFRvIII in approximately 27% to 54% of GBMs [12,13,19,24]. Compared with previous studies on the role of EGFRvIII in GBM tumor biology, we used two additional approaches. The first one consisted in measuring the level of EGFRvIII mRNA both in GBMs at primary surgery and in those that recurred after radio/chemotherapy. Enrichment or depletion of EGFRvIII mRNA in the recurrent tumors would inform whether the EGFRvIII status might play any role in radio/chemoresistance and repopulation potential. The second approach used GBM neurosphere cell cultures expressing or not the EGFRvIII to test in vitro sensitivity to TMZ. We and others have recently established multiple stem-like cell lines with distinct expression profiles from different areas of the same GBM tumor [25,30]. These cells probably share a common ancestor but divergent genomic evolution and molecular properties [30,31].
Major results were as follows: 1) the presence of EGFRvIII in GBM tumors correlates with longer OS. The association of EGFRvIII/Ki67 of 20% or less, of EGFRvIII/normal PTEN, and of EGFRvIII/methylated MGMT identified subgroups of GBM patients with better prognosis; 2) EGFRvIII expression is reduced in GBM recurring after adjuvant radiotherapy and TMZ; and 3) EGFRvIII-positive GBM neurosphere cells are less resistant to TMZ than their EGFRvIII-negative counterparts.
Our findings on the prognostic significance of EGFRvIII in GBM diverge from previous studies, where this variant was found either to be unrelated to the patients' outcome [6,11,12,16–20] or to be associated with shorter survival (Table W5) [10,14]. Although in the study by Brown et al. [16] the presence of EGFRvIII was not significantly predictive, patients with high-level (greater than a doubling in EGFR copy number) versus low-level EGFR gene amplification showed a trend to better OS. It is worth noticing that the mechanisms linking EGFRvIII and clinical outcome have not generally been addressed. The few observations that have been made in this regard yielded apparently discrepant results, like that EGFRvIII does not relate with cell proliferation and aggressive tumor features, as expected from in vitro data [11], and that the downstream effectors of Ras are prognostic only in the EGFRvIII-negative patients [14]. In contrast, we found that in the EGFRvIII-positive GBMs, a Ki67 index of 20% or less identified patients with better prognosis. Reportedly, GBMs with high proliferative index are clinically more aggressive and are characterized by deregulation of many different molecular pathways [32–34]. Proliferation rate, however, did not account for the worse prognosis of EGFRvIII-negative GBMs, suggesting different mechanisms of tumor aggressiveness in these neoplasms. The favorable effect of EGFRvIII on prognosis was abrogated either in the absence of the tumor-suppressor protein PTEN or in cases with unmethylated MGMT, which is consistent with the notion that PTEN has a key role in the inhibition of the antiapoptotic signals of the activated PI3K-Akt pathway and that epigenetic silencing of MGMT through promoter methylation is related with improved outcome and response to TMZ in malignant gliomas [13,35,36]. The better survival of patients showing EGFRvIII and PTEN expression seems somewhat contradictory, given that activation of PI3K-Akt pathway is one of the most important transduction signaling of EGFRvIII and that PTEN represents a major inhibitor of PI3K-Akt pathway. We may speculate that loss of PTEN reduced OS in our patients through a PI3K-mediated radioresistance [37,38], whereas the EGFRvIII-positive stem-like fraction of the tumor cells was more sensitive to TMZ therapy.
To resolve the difference in the prognostic value of EGFRvIII between the current study and the previous ones, it may be postulated that very low levels of EGFRvIII would have been detected by RT-PCR as opposed to immunohistochemistry where these specimens would likely have been determined as negative. There may be a threshold of expression necessary that allows for dimerization with possibly differential signaling that results in the negative/neutral prognostic influence of EGFRvIII found in previously published studies. Although EGFRvIII receptor dimerization was not considered initially as factor in the signaling activity [39], this premise is being revisited by investigators who have shown that chemical inducers of EGFRvIII dimerization produce a more oncogenic form of the EGFRvIII [40]. However, the concept of an EGFRvIII expression threshold for dimerization does not seem to be confirmed by our real-time PCR analysis on recurrent tumors, which shows a linear relationship between EGFRvIII levels and survival.
A unique aspect of the present study that may help explaining the discrepancy between our results and previous clinical data is that our patient population is a uniform surgical, radiotherapy, and TMZ cohort. Previous studies included patients treated with surgery and fractionated radiotherapy, where chemotherapy regimens were vastly different. Although some of these studies did contain a substantial number of patients treated with TMZ, the findings about EGFRvIII and prognosis were not placed in the context of TMZ treatment [14,16,17]. For example, in the study by Pelloski et al. [14], patients were not stratified for TMZ treatment. In the studies by Brown et al. [16] and by Van den Bent et al. [17], TMZ-treated patients either concurrently received the EGFR inhibitor erlotinib or were incorporated into the control arm, which included patients treated with carmustine, so that EGFRvIII expression and response to TMZ could not be related to each other.
Our data suggest that sensitivity to TMZ may be related with EGFR expression and that in the tumor context the cell fraction with constitutively activated EGFR may be less resistant to TMZ. In mice grafted with EGFRvIII-positive human GBM cells, TMZ either alone or in combination with inhibitors of the EGFR tyrosine kinase induced significant reduction of the tumor xenografts [41]. Meta-analysis showed that among GBM patients treated with TMZ, EGFRvIII-positive GBMs showed significantly longer survival relative to the EGFRvIII-negative ones (P < .05) [13]. The hypothesis that the EGFRvIII status may affect sensitivity to TMZ of GBM cells has been confirmed by our in vitro experiments, where the absence of EGFRvIII conferred a higher resistance to TMZ. It should be emphasized that these results were obtained using GBM cultures enriched with so-called cancer stem cells, having or not EGFRvIII, and that this model is much closer to the clinical condition than those models based on serum-exposed virus-transfected GBM cell lines [6–8,13]. The higher sensitivity of EGFRvIII-positive GBM cells to TMZ may be ascribed to a mechanism of intrinsic nononcogene addiction [42]. In the EGFRvIII-positive tumor cells, which are chronically dependent on persistent signaling, the rate of spontaneous DNA damage and the degree of replication stress are enhanced. It is likely that in this condition of stress overload, the cancer cells with already elevated levels of DNA damage and replication stress cannot repair the additional damage induced by TMZ.
In the present study, we found a not significant trend to longer survival for patients whose tumors had been totally resected compared with those patients who had undergone partial tumor resection. The role of extent of surgical resection in determining the prognosis of GBM patients is still a matter of debate. Although most authors state that maximal safe resection is associated with better survival, it has been noted that no class I evidence exists [43]. The relatively small number of patients in our series as well as the method we used to evaluate the extent of tumor resection, which was based on surgeon's impression and Gd-enhanced MRI obtained 1 month after surgery and not within 72 hours, may be influential in explaining our result.
Overall, our findings are consistent with a cancer stem cell model for GBM where the mutation of EGFR occurs at the stage of early progenitor cells concurrent with the increase of proliferation from early to late progenitors, where the latter cells are thought to be responsible for tumor bulk but not for tumor recurrence. Mechanistically, a ligand-independent constitutively activated EGFR is hardly plausible in cancer stem cells, which are highly dependent on the exogenous supply of EGF for their in vitro growth.
In summary, this study shows that EGFRvIII is a molecular predictor of longer OS in GBM patients treated with surgery followed by adjuvant radiotherapy and TMZ. This effect may be ascribed to a better response to TMZ and to a lower repopulation potential by the EGFRvIII-positive GBM cells.
Supplementary Material
Footnotes
This work has been partially presented to the 100th annual meeting of the United States and Canadian Academy of Pathology, February 26 to March 4, 2011, San Antonio, TX, and was supported by Ministero della Salute (N.ONC_ORD 15/07 to R.P.), Fondi d'Ateneo Università Cattolica (Linea D1) to R.P. and L.M.L., and Associazione per la Ricerca sul Cancro to R.D.M. The authors thank Fondazione G. Alazio (Via Tasso 22, Palermo; www.fondazionealazio.org) for providing a research grant to Q.G.D. All authors do not present any potential conflicts (financial, professional, or personal) that are relevant to the article.
This article refers to supplementary materials, which are designated by Tables W1 to W5 and Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Huntley BK, Borell TJ, Iturria N, O'Fallon JR, Schaefer PL, Scheithauer BW, James CD, Buckner JC, Jenkins RB. TEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- 2.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci USA. 1990;87:86062–86066. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor receptor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- 4.Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 6.Aldape KD, Ballman K, Furth A, Buckner JC, Giannini C, Burger PC, Scheithauer BW, Jenkins RB, James CD. Immunohistochemical detection of EGFRvIII in high malignancy grade astrocytomas and evaluation of prognostic significance. J Neuropathol Exp Neurol. 2004;63:700–707. doi: 10.1093/jnen/63.7.700. [DOI] [PubMed] [Google Scholar]
- 7.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 8.Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci. 2009;16:748–754. doi: 10.1016/j.jocn.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Feldkamp MM, Lala P, Lau N, Roncari L, Guha A. Expression of activated epidermal growth factor receptors, Ras-guanosine triphosphate, and mitogen-activated protein kinase in human glioblastoma multiforme specimens. Neurosurgery. 1999;45:1442–1453. doi: 10.1097/00006123-199912000-00034. [DOI] [PubMed] [Google Scholar]
- 10.Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 11.Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, Sawaya R, Aldape K. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11:1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Bäcklund LM, Nilsson BR, Grandér D, Ichimura K, Goike HM, Collins VP. Clinical significance of EGFR amplification and the aberrant EGFRvIII transcript in conventionally treated astrocytic gliomas. J Mol Med. 2005;83:917–926. doi: 10.1007/s00109-005-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 14.Pelloski CE, Ballman KV, Furth AF, Zhang L, Lin E, Sulman EP, Bhat K, McDonald JM, Yung WK, Colman H, et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J Clin Oncol. 2007;25:2288–2294. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 15.Viana-Pereira M, Lopes JM, Little S, Milanezi F, Basto D, Pardal F, Jones C, Reis RM. Analysis of EGFR overexpression, EGFR gene amplification and the EGFRvIII mutation in Portuguese high-grade gliomas. Anticancer Res. 2008;28:913–920. [PubMed] [Google Scholar]
- 16.Brown PD, Krishnan S, Sarkaria JN, Wu W, Jaeckle KA, Uhm JH, Geoffroy FJ, Arusell R, Kitange G, Jenkins RB, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol. 2008;26:5603–5609. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MC, Kros JM, Carpentier AF, Clement PM, Frenay M, Campone M, Baurain JF, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC Brain Tumor Group Study 26034. J Clin Oncol. 2009;27:1268–1274. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reardon DA, Vredenburgh JJ, Desjardins A, Peters K, Gururangan S, Sampson JH, Marcello J, Herndon JE, II, McLendon RE, Janney D, et al. Phase 2 trial of erlotinib plus sirolimus in adults with recurrent glioblastoma. J Neurooncol. 2010;96:219–230. doi: 10.1007/s11060-009-9950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiessen B, Stewart C, Tsao M, Kamel-Reid S, Schaiquevich P, ason W, Easaw J, Belanger K, Forsyth P, McIntosh L, et al. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother Pharmacol. 2010;65:353–361. doi: 10.1007/s00280-009-1041-6. [DOI] [PubMed] [Google Scholar]
- 20.Heimberger AB, Suki D, Yang D, Shi W, Aldape K. The natural history of EGFR and EGFRvIII in glioblastoma patients. J Transl Med. 2005;3:38. doi: 10.1186/1479-5876-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louis DN, Ohgaki H, Wiestler OD, Cavenee CW, editors. World Health Organization Classification of the Nervous System. 4th ed. Lyon, France: IARC Press; 2007. pp. 33–49. [Google Scholar]
- 22.Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M, Stassi G, Martini M, Maira G, Larocca LM, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 23.Martini M, Pallini R, Luongo G, Cenci T, Lucantoni C, Larocca LM. Prognostic relevance of SOCS3 hypermethylation in patients with glioblastoma multiforme. Int J Cancer. 2008;123:2955–2960. doi: 10.1002/ijc.23805. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimoto K, Dang J, Zhu S, Nathanson D, Huang T, Dumont R, Seligson DB, Yong WH, Xiong Z, Rao N, et al. Development of a real-time RT-PCR assay for detecting EGFRvIII in glioblastoma samples. Clin Cancer Res. 2008;14:488–493. doi: 10.1158/1078-0432.CCR-07-1966. [DOI] [PubMed] [Google Scholar]
- 25.Ricci-Vitiani L, Pallini R, Larocca LM, Lombardi DG, Signore M, Pierconti F, Petrucci G, Montano N, Maira G, De Maria R. Mesenchymal differentiation of glioblastoma stem cells. Cell Death Diff. 2008;15:1491–1498. doi: 10.1038/cdd.2008.72. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 27.Eramo A, Ricci-Vitiani L, Zeuner A, Pallini R, Lotti F, Sette G, Pilozzi E, Larocca LM, Peschle C, De Maria R. Chemotherapy resistance of glioblastoma stem cells. Cell Death Diff. 2006;13:1238–1241. doi: 10.1038/sj.cdd.4401872. [DOI] [PubMed] [Google Scholar]
- 28.Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3 like proteases. Proc Natl Acad Sci USA. 1998;95:5724–5729. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumor cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 30.Piccirillo SG, Combi R, Cajola L, Patrizi A, Redaelli S, Bentivegna A, Baronchelli S, Maira G, Pollo B, Mangiola A, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28:1807–1811. doi: 10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- 31.Mazzoleni S, Politi LS, Pala M, Cominelli M, Franzin A, Sergi Sergi L, Falini A, De Palma M, Bulfone A, Poliani PL, et al. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70:7500–7513. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- 32.Wakimoto H, Aoyagi M, Nakayama T, Nagashima G, Yamamoto S, Tamaki M, Hirakawa K. Prognostic significance of Ki-67 labeling indices obtained using MIB-1 monoclonal antibody in patients with supratentorial astrocytomas. Cancer. 1996;77:373–380. doi: 10.1002/(SICI)1097-0142(19960115)77:2<373::AID-CNCR21>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 33.Cadieux B, Ching TT, Van den Berg SR, Costello JF. Genomewide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida Y, Nakada M, Harada T, Tanaka S, Furuta T, Hayashi Y, Kita D, Uchiyama N, Hayashi Y, Hamada J. The expression level of sphingosine-1-phosphate receptor type 1 is related to MIB-1 labeling index and predicts survival of glioblastoma patients. J Neurooncol. 2010;98:41–47. doi: 10.1007/s11060-009-0064-5. [DOI] [PubMed] [Google Scholar]
- 35.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 36.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 37.Li HF, Kim JS, Waldman T. Radiation-induced Akt activation modulates radioresistance in human glioblastoma cells. Radiat Oncol. 2009;4:43. doi: 10.1186/1748-717X-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan M, Han ZC. Phosphatidylinositide 3-kinase/AKT in radiation responses. Histol Histopathol. 2004;19:915–923. doi: 10.14670/HH-19.915. [DOI] [PubMed] [Google Scholar]
- 39.Chu CT, Everiss KD, Wilstrand CJ, Batra SK, Kung HJ, Bigner DD. Receptor dimerization is not a factor in the signalling activity of a transforming variant epidermal growth factor receptor (EGFRvIII) Biochem J. 1997;324:855–861. doi: 10.1042/bj3240855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johns TG, Perera RM, Vernes SC, Vitali AA, Cao DX, Cavenee WK, Scott AM, Furnari FB. The efficacy of epidermal growth factor receptor-specific antibodies against glioma xenografts is influenced by receptor levels, activation status, and heterodimerization. Clin Cancer Res. 2007;13:1911–1925. doi: 10.1158/1078-0432.CCR-06-1453. [DOI] [PubMed] [Google Scholar]
- 41.Johns TG, Luwor RB, Murone C, Walker F, Weinstock J, Vitali AA, Perera RM, Jungbluth AA, Stockert E, Old LJ, et al. Antitumor efficacy of cytotoxic drugs and the monoclonal antibody 806 is enhanced by the EGF receptor inhibitor AG1478. Proc Natl Acad Sci USA. 2009;100:15871–15876. doi: 10.1073/pnas.2036503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–827. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhnt D, Becker A, Ganslandt O, Bauer M, Buchfelder M, Nimsky C. Correlation of the extent of tumor volume resection and patient survival in surgery of glioblastoma multiforme with high-field intraoperative MRI guidance. Neuro Oncol. 2011;13:1339–1348. doi: 10.1093/neuonc/nor133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.