Abstract
The tools for predicting clinical outcome after radiotherapy are not yet optimal. To improve on this, we applied the COXEN informatics approach to in vitro radiation sensitivity data of transcriptionally profiled human cells and gene expression data from untreated head and neck squamous cell carcinoma (HNSCC) and bladder tumors to generate a multigene predictive model that is independent of histologic findings and reports on tumor radiosensitivity. The predictive ability of this 41-gene model was evaluated in patients with HNSCC and was found to stratify clinical outcome after radiotherapy. In contrast, this model was not useful in stratifying similar patients not treated with radiation. This led us to hypothesize that expression of some of the 41 genes contributes to tumor radioresistance and clinical recurrence. Hence, we evaluated the expression the 41 genes as a function of in vitro radioresistance in the NCI-60 cancer cell line panel and found cyclophilin B (PPIB), a peptidylprolyl isomerase and target of cyclosporine A (CsA), had the strongest direct correlation. Functional inhibition of PPIB by small interfering RNA depletion or CsA treatment leads to radiosensitization in cancer cells and reduced cellular DNA repair. Immunohistochemical evaluation of PPIB expression in patients with HNSCC was found to be associated with outcome after radiotherapy. This work demonstrates that a novel 41-gene expression model of radiation sensitivity developed in bladder cancer cell lines and human skin fibroblasts predicts clinical outcome after radiotherapy in head and neck cancer patients and identifies PPIB as a potential target for clinical radiosensitization.
Introduction
Radiation therapy is an important treatment modality for head and neck and bladder cancer, either alone or in combination with chemotherapy [1,2]. However, the individual response to radiotherapy can be variable, and hence, any tool that would predict response to this modality would allow enhanced patient stratification among the various treatment options [3]. In addition, once optimally selected, pharmacologic approaches toward radiosensitization promise to further enhance the benefit these patients derive from such treatment [4]. Currently, clinical characteristics of the patient and tumor are primarily used to determine whether treatment with radiotherapy is appropriate [5–7], whereas tumor imaging [8,9] and expression of genes in the tumor tissue [10–12] have been proposed to possibly enhance this. However, even when used together, these are not yet highly predictive of radiation sensitivity of patient tumors before treatment.
Made possible by the development of gene expression microarray or multiplex polymerase chain reaction technologies, mathematical models involving expression measurements of multiple genes have been developed to serve as prognostic indicators of disease aggressiveness or patient survival and to predict response to specific chemotherapeutic agents or regimens [13,14]. Associations of tumor gene expression to radiation response have been developed for cell lines [15–17] and for specific tumors such as cervical cancer [18], breast cancer [19], colorectal adenocarcinoma [20], and cancers of the head and neck [21,22]. In addition, a radiosensitivity signature as an indicator of concurrent chemoradiation therapeutic response has been tested in small sets of rectal, esophageal, and head and neck cancers [17]. Although exciting, the predictive value of these models across different histologic tumor types requires validation on larger and more diverse sample sets. In addition, none have identified genes that are both biomarkers and potential targets for radiosensitization.
Here, we hypothesize that merging three sources of data, namely in vitro radiation sensitivity of cell lines, baseline gene expression of these cell lines, and gene expression of human tumors from multiple cancer histologies would provide a gene profile associated with clinical outcome after radiotherapy as well as potentially identify those genes that may be targets for radiosensitization. This approach would also be less susceptible to identifying tumor histology-specific processes while being more likely to identify broadly relevant targets to enhance the effectiveness of radiotherapy.
Given successful chemosensitivity prediction of human cancer based on in vitro drug sensitivity of cell lines, the Coexpression Extrapolation (COXEN) informatics method [23–25] seemed uniquely suited to test the hypothesis above.We applied this tool on the triumvirate sources of data mentioned above to develop a multigene predictive model. This model was tested for its ability to predict outcome in patients with head and neck squamous cell carcinoma (HNSCC) treated with radiotherapy. In vitro depletion studies of several genes in the model demonstrated that some of these are both clinical response biomarkers and determinants of cellular radiation sensitivity. Furthermore, we demonstrate that pharmacologic inhibition of one such gene, cyclophilin B (PPIB), leads to decreased DNA repair in cancer cells after irradiation. In summary, our approach has provided both biomarkers of clinical outcome after radiotherapy and potential therapeutic targets for radiosensitization.
Materials and Methods
Patient and Cell Line Data Sets
The patient and cell line data sets used in this study and their specific roles are listed in Tables W1A–W1B and in the Supplementary Methods. Use of human tissue samples here was approved by the University of Virginia institutional review board.
Development of the Radiation Response Prediction Gene Expression Model
As Table W1A indicates, gene expression data sets were available on different microarray platforms. To generate a prediction gene expression model (GEM) across all platforms, gene array data processing and calibration were carried out as described in the Supplementary Methods. We used COXEN [23,25] to develop a model predictive of radiation response. Figure W1A shows a schematic depiction of the methodology. We used the BLA-40 bladder cancer cell line data set (Table W1A) to discover genes differentially expressed according to radiosensitivity. To determine the subset of these genes whose expression is relevant to human primary tumors, we determined which of these shared similar patterns of coexpression with the human bladder tumor sample data set (Smith et al. in Table W1A). For each of the remaining probe sets, we measured the significance of differential expression between the 17 most radiosensitive (SF2 range = 0.19–0.51; mean = 0.39) and the 10 most radioresistant (SF2 range = 0.72–0.98; mean = 0.86) cell lines using Student's t test. These numbers were chosen to maximize the number of genes differentially expressed as a function of intrinsic sensitivity to radiation as measured by SF2. We selected the 300 most significantly differentially expressed probe sets as candidate biomarkers for radiation response prediction.
We used a linear discriminant analysis (LDA) approach [26,27] to develop the optimal gene model predictors of radiation response from the 300 probes above as described in Supplementary Methods. To select a GEM that effectively predicts radiation response of several cancer types, we tested the performance of candidate GEMs at predicting radiation sensitivity within the Brock cell panel (Table W1B), as measured by the correlation between predicted sensitivities and actual SF2 values. Candidate GEMs were iteratively constructed from the 300 candidate probe sets above, starting with a model consisting of the top three candidate biomarkers and then successively adding biomarkers until all candidate biomarkers were used. GEMs that resulted in a higher magnitude of correlation between predicted and actual sensitivity were more accurate at response prediction. We selected a GEM that had the highest correlation with the smallest number of probes.
Evaluation of the Radiation Response Prediction GEM in Patient Data Sets
After using the human skin fibroblast (HSFs) cell line data set (Brock) to guide the selection of the optimal GEM, we evaluated the predictive ability of this model on two independent patient tumor expression data sets (Figure W1B). We generated predictions for the HNSCC patient data sets (Table W1A), such that each patient received a GEM score reflective of the relative probability of response to radiation therapy. Importantly, because data sets did not contain information on tumor response (i.e., reduction in tumor size or stage, etc) after radiotherapy, we used overall survival and event-free survival (future distant metastasis) as defined in Rickman et al. (Oncogene. 2008;27:6607–6622) as surrogates of radiocurability. The implicit limitations of this assumption are discussed below.
To assess the accuracy of the GEM prediction, we generated receiver operating characteristic (ROC) curves for the 73 patients treated with radiation for the Rickman HNSCC data set. To determine whether our prediction model correctly classifies true responders as predicted responders without available response information, we made the assumption that patients surviving more than 3 years without evidence of disease were more likely to have responded to treatment than patients who died of disease. Hence, for the purposes of this ROC analysis, we considered patients surviving more than 3 years without evidence of disease to be “true responders” and patients who died of disease as “true nonresponders.” From this ROC curve, we selected a threshold GEM score value that yielded optimal sensitivity and specificity, to dichotomize patients into predicted responders and predicted nonresponders in subsequent Kaplan-Meier analysis. We used the χ2 test to determine the significance of the separation of Kaplan-Meier survival curves between predicted responders and nonresponders.
To determine the utility of the GEM in relation to other clinical variables in predicting patient outcome, we used Cox proportional hazards regression model analysis to assess the significance of these variables and continuous GEM scores. Because the number of patients in the HNSCC test set studies (Table W1A) was relatively small compared with the number of potentially predictive variables, we used a stepwise AIC-based model selection technique to selectively remove insignificant variables [27,28]. This technique leads to a more parsimonious model yet retains explanatory power [27,28].
PPIB and p16 Immunohistochemistry
For the Virginia HNSCC patient data set, 4-µm histologic sections were cut, and PPIB and p16 immunohistochemistry (IHC) were carried out as described in the Supplementary Methods. Immunohistochemical staining for cyclophilin was scored for the intensity of cytoplasmic staining (0 = absent, 1 = weak, 2 =moderate, 3 = strong), and for the purpose of analysis, the scores were dichotomized into two groups: weak to absent (0–1) versus moderate to strong (2–3) expression. p16 IHC expression was scored as positive or negative for use in Kaplan-Meier analysis, using the criteria devised by Reimers et al. [29].
Miscellaneous Procedures
All other procedures used in this article can be found in the Supplementary Methods section.
Results
Development of a GEM Predictive of Cellular Response to Radiation
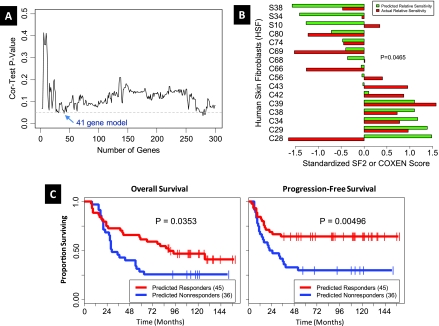
A schematic of model generation and validation process is depicted in Figure W1. Radiosensitivity data for both the bladder cancer (BLA-40, Table W1A) and primary HSFs developed from skin biopsies collected from areas outside the radiation field in patients undergoing radiotherapy (Brock, Table W1A), in the form of SF2 are shown in Tables W2A and W2B, respectively. Of the 8470 Affymetrix HG-U133A probe sets that had matching Illumina probes, we found that 7515 probe sets survived the COXEN coexpression step between bladder cell lines and human tumors. The 300 probe sets most differentially expressed between the 17 most radiosensitive (SF2 range = 0.19–0.51; mean = 0.39) and 10 most radioresistant (SF2 range = 0.72–0.98; mean = 0.86) cell lines were chosen as candidate biomarkers for inclusion in the radiation response GEM. We then assessed how well GEMs, constructed reiteratively from these candidate biomarkers as described in methods, were able to predict radiation sensitivity in the HSF cell line panel and selected a 41-probeset model (Table W3) that was best able to predict radiosensitivity in this panel (Figure 1A). A comparison of the predicted and true radiosensitivity data for the HSF panel using the 41-gene GEM is shown in Figure 1B.
Figure 1.
Selection of the optimal 41-probe model GEM. (A) A plot of the prediction performance of candidate GEMs used to guide selection of optimal GEM. For each candidate GEM, we calculated the correlation between the GEM scores and the actual survival fractions for the human fibroblast data set.Weplot the correlation test P value for each model versus the number of genes in the model. The model with 41 genes balances prediction performance and parsimony. (B) Normalized survival fraction values and GEM scores on the HSF data set are plotted in a bar plot. The Spearman rank-based correlation between normalized GEM score and survival fraction is -0.434, with a one-sided P = .0465. Standardization of SF2 and COXEN GEM score involved reciprocal evaluation because higher SF2 indicated radioresistance and higher COXEN GEM score was indicative of radiosensitivity. (C) Evaluation of the predictive ability of the 41-probe GEM in patients with HNSCC. Kaplan-Meier curves for the HNSCC cancer patients stratified into predicted responders and predicted nonresponders. Kaplan-Meier curves for patients only treated with radiation (n = 72). The left panel shows the overall survival time, whereas the right panel shows the progression-free survival time.
Utility of the 41-Gene GEM in Stratifying Clinical Outcome of Patients Treated with Radiotherapy
We next used the 41-gene GEM to predict the clinical response of patients with HNSCC enrolled in independent clinical studies. The Rickman HNSCC patient data set (Tables W1A and W1B) comprised 81 patients, of whom 73 were treated using radiotherapy alone, whereas 8 patients were treated with an unspecified chemotherapeutic regimen in addition to radiotherapy. Because chemotherapy may have a significant influence on patient response and because the subset of patients treated with radiotherapy alone was sufficiently large, we restricted our prediction assessment analysis to these 73 patients. Each patient was first assigned a GEM score indicating the predicted relative probability of response to radiation. To assess prediction performance, we generated an ROC curve that had a Wilcoxon rank-sum test P = .015, indicating that the predictor was significantly better than random, and an area under the curve (AUC) of 0.61. This ROC curve was used to select a GEM score threshold value that allowed stratification of this group into predicted responders and predicted nonresponders. Kaplan-Meier analysis revealed significant separation in survival time between the predicted responders and nonresponders in overall and progression free survival (Figure 1C).
Because in addition to radiation response, other patient and tumor characteristics influence patient survival, we used Cox proportional hazards regression model to determine the contribution of the GEM response scores in addition to these other characteristics. All patients with complete clinical information (72/73) and all available covariates were included in this analysis with the stepwise AIC model selection discarding the variables least significant to the end point. Results shown in Table W4A (for overall survival) and Table W4B (for progression free survival) indicate the GEM score is a significant variable predicting the overall survival hazard rate (P = .047). Table W4B shows that no variable relates to progression-free survival hazard rate, but the GEM score has the lowest P value (P = .068). We also sought to determine whether two recently published predictive models of radiation response in patients could predict outcome in our data sets [17,19]. Importantly, neither model predicted clinical outcome in the Rickman data set (Supplementary Results).
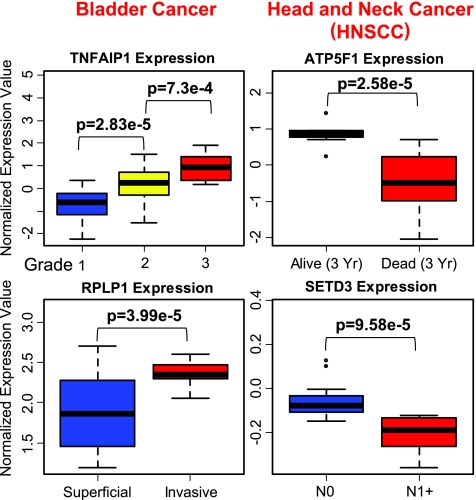
Supporting the notion that the 41-gene GEM informs about clinical outcome after radiation and not general tumor aggressiveness in bladder or HNSCC cancer is the finding that only 4 of 41 genes are associated with stage, grade, invasive ability, or clinical outcome in independently profiled cancer data sets found in Oncomine (Figure 2). Together, these two analyses suggest the 41-gene GEM informs about clinical outcome after radiation rather than a general reflection of tumor aggressiveness.
Figure 2.
Association of gene expression with stage, grade, and outcome in bladder and head and neck cancer (HNSCC). Box plots showing the association of 4 genes of the 41-gene GEM (Table W3) that were found to be significantly associated with tumor stage, grade, or clinical outcome in bladder and head and neck cancer databases found in Oncomine. P values for the specific comparison and tumor parameter used are indicated in the figure. The data sets that contained these significant associations have been previously published [50–56].
Characteristics and Network Analysis of the 41 Genes in the GEM
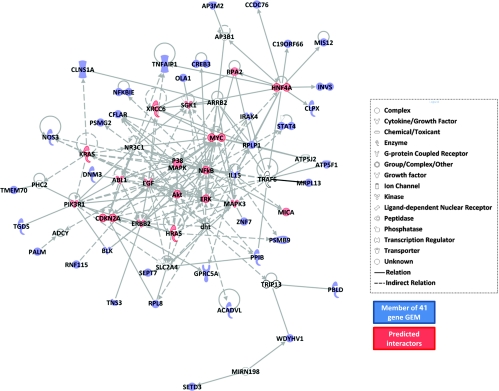
The genes corresponding to the probes in the 41-gene model were identified, and gene ontology (GO) information was obtained from the PANTHER database (Supplementary Results and Figure W2). We then used the Ingenuity Pathways Analysis program to determine whether genes in the model are found in a common network and, if so, discover biomolecules that are known interactors with the latter. Interestingly, unsupervised analysis revealed the top scoring network comprised 39 of the 41 genes of the GEM (Figure 3). In this network, the 39 genes of the radiation response model were found to interact with genes such as KRAS, HRAS, MYC, MYCN, ABL1, ERBB2, PIK3R1, P38MAPK, NFκB, and ERK that are known to be involved in radiation response. These findings suggested that genes in the model might also have causal roles in radioresistance.
Figure 3.
Network analysis of the 41-gene GEM. Ingenuity Pathways Analysis network containing 39 of the 41 genes in the radiation response prediction GEM and functionally related genes. Symbols colored in blue represent the genes in the 41-gene GEM, whereas red and uncolored symbols represent genes with known direct or indirect functional interactions with the GEM genes. Symbols colored in red represent genes known to be associated with radiation.
PPIB and Acidic Ribosomal Phosphoprotein P1 and Response to Radiosensitivity In Vitro
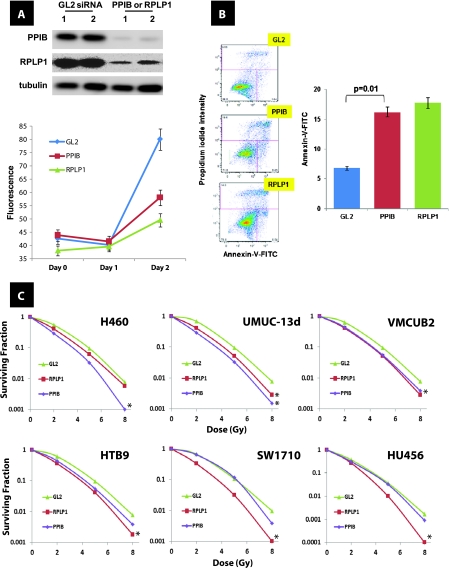
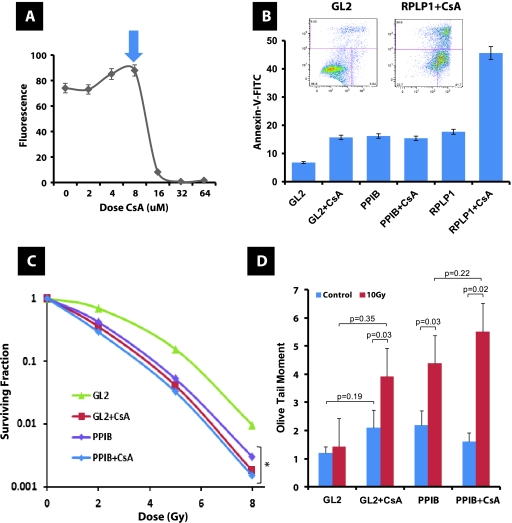
To determine which of the 39 genes in this network have causal roles in radioresistance, we identified those whose expression was most strongly and directly related with this phenotype in a third cell panel, the NCI-60 [16] (Table W1A). The top 2 genes with the strongest correlation of expression to radioresistance were PPIB, and acidic ribosomal phosphoprotein P1 (RPLP1; Figure W3). Given this finding, we sought to evaluate whether the expression of these genes regulated this phenotype. Use of small interfering RNA (siRNA) provided depletion of both proteins in UMUC-13d bladder cancer cells (Figure 4A). Reduced levels of PPIB and RPLP1 were associated with reduction in cell number after depletion (Figure 4A), and this was due to enhanced apoptosis (Figure 4B). When six human cancer cell lines were transiently depleted of either PPIB or RPLP1 and irradiated, we noted that cells with reduced levels of either PPIB or RPLP1 had reduced clonogenicity (Figure 4C). Because cyclophilins are bound and inhibited by cyclosporine A (CsA), we sought to evaluate whether CsA recapitulated the observed reduction in cell number observed with PPIB depletion, we carried out a dose response in UMUC-13d bladder cancer cells. Figure 5A indicates CsA can diminish overall cell numbers during a 48-hour period compared with vehicle-treated cells, yet this effect occurs only with doses greater than 8 µM. Interestingly, transient depletion of either PPIB or RPLP1 with and without CsA in UMUC-13d cells indicated that only depletion of RPLP1 with CsA addition results in enhanced apoptosis compared with all other experimental groups, suggesting functional equivalence of PPIB and CsA about this phenotype (Figure 5B). We next examined the effect of PPIB depletion with and without CsA addition on the in vitro clonogenic ability of UMUC-13d cells after exposure to radiation and found that PPIB depletion or CsA had similar effects, whereas PPIB depletion combined with CsA did not result in further reduction in clonogenic potential (Figure 5C). Finally, our data reveal that both PPIB depletion and CsA inhibit DNA repair, and their combined use does not reduce this further, suggesting that these act on the same pathway(s) regulating DNA repair (Figure 5D). This result mirrors the findings on clonogenicity (Figure 5C).
Figure 4.
Effect of PPIB and RPLP1 depletion on in vitro growth and radiosensitivity in human cancer cell lines. (A) In vitro cell number evaluation using alamarBlue (Invitrogen) assay after depletion of GL2 (control), PPIB or RPLP1 through siRNA in duplicate samples of UMUC-13d bladder cancer cells. Inset: Western blot analysis after siRNA depletion of PPIB and RPLP1 as described in Supplementary Methods. Antibodies against PPIB (clone k2e2; Santa Cruz Biotechnology) and RPLP1 (polyclonal; Sigma) were used. (B) Apoptosis was assessed by the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences) per the manufacturer's instructions 48 hours after transfection with the siRNA duplexes for PPIB and RPLP1. Insets show FACS output. (C) Clonogenic survival in human cancer cell lines after radiation at indicated dose and depletion of GL2 (control), PPIB, or RPLP1 through siRNA. *P < .05 (ANOVA) at the 8-Gy dose.
Figure 5.
Effect of PPIB depletion and CsA on in vitro growth and radiosensitivity and DNA repair of human bladder cancer cells. (A) In vitro cell number evaluation using alamarBlue (Invitrogen) assay after the addition of CsA at indicated doses to duplicate samples of UMUC-13d bladder cancer cells. Arrow indicates dose used in panels B to D. (B) Apoptosis was assessed by the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences) per the manufacturer's instructions 48 hours after transfection with the siRNA duplexes for PPIB and RPLP1 with and without 8 µM CsA. Inset axis labels are similar to those in Figure 4B. (C) Clonogenic survival in UMUC-13d bladder cancer cells after radiation at the indicated dose and depletion of GL2 (control), PPIB through siRNA with and without 8 µM CsA. *P < .05 (ANOVA) at the 8-Gy dose. (D) Comet assay carried out as described in Materials and Methods after transfection with the siRNA duplex for PPIB. Dose of CsA was 8 µM. Each assay was normalized to cloning efficiency given the apoptosis induced by siRNA depletion. Indicated P values generated using Student's t test. The extent of DNA damage was measured 1 hour after a 10-Gy exposure.
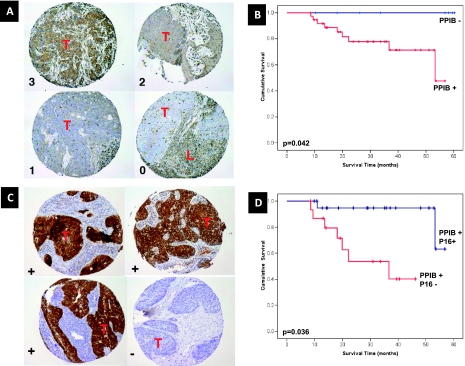
Association of PPIB Protein Expression with Patient Outcome after Radiation
Given that PPIB RNA expression was strongly correlated to radio-resistance (Figure W3), we evaluated the role of PPIB protein expression as a predictor of clinical outcome after radiation treatment of patients with HNSCC at the University of Virginia (Table W1A). PPIB protein levels were found to predict of clinical outcome in these patients (Figure 6, A and B). Interestingly, expression of CDKN2A (p16), a cyclin-dependent kinase inhibitor and surrogate marker of human papillomavirus (HPV) infection, was recently found to predict radiation response in patients with HNSCC [30]. Because this gene was part of the signaling network associated with our 41-gene GEM (Figure 3 and Table W3), we sought to determine whether its level of protein expression provided additional predictive ability when combined with that of PPIB. IHC evaluation revealed that p16 levels provided significant stratification of patients with high PPIB IHC (Figure 6, C and D) levels supporting relevance of the 39-gene network in radiosensitivity of human cancer.
Figure 6.
PPIB and CDKN2A (p16) expression in HNSCC. PPIB and p16 IHC and their relevance to outcome in HNSCC tumors treated at the University of Virginia (n = 72). (A) Examples of the IHC scoring of PPIB. Each panel represents a tissue core in a tissue microarray; the numbers 0 to 3 indicate the expression score given to the tumor in the specimen. Lymphocytes (L) present and infiltrating around the tumor (T) also express this protein at high levels and serve as “internal controls” of staining intensity. (B) Kaplan-Meier curves of overall survival of patients as a function of PPIB IHC score. (C) Examples of the IHC scoring of p16. Each panel represents a tissue core in a tissue microarray; the characters (+) and (-) indicate the expression score (positive or negative, respectively), given to the tumor in the specimen. (D) Kaplan-Meier curves of overall survival of PPIB-positive patients as a function of p16 IHC score.
Discussion
We developed a multigene predictor of clinical outcome after radiation therapy that is applicable across several cancer types. Whereas other predictors have been described to predict response to radiation, they are based on clinical characteristics, single biomarkers, or panels designed specifically for one cancer type. Our model uses genes concordantly regulated between bladder cancer cell lines and human bladder cancer patients as previously successfully done for chemotherapy response prediction [23,24]. Interestingly, this model provided additional predictive ability even in patients who had other concomitant and potentially confounding treatments such as chemotherapy treatment yet, importantly, did not offer any predictive ability in patients who were not treated with radiation. This last observation is similar to a recent breast cancer model [19] and suggests the existence of genomic classifiers that can separately predict prognosis or therapy response.
Providing functional credibility to the model, we note that some genes in the model are implicated in radiation response. The induction of HRAS is associated with radiation resistance [31], and MYC acts synergistically with HRAS to induce resistance [32]. p38 is involved in regulating cellular responses to stress including stress induced by ionizing radiation [33]. It is reassuring that NFκB transcription factor is centrally represented in our network. This factor is involved in multiple cellular processes [34], and its constitutive activation has been described in many cancer types and supports cancer cell survival and to reduce the sensitivity against both radiation and chemotherapeutic drugs [35–38].
We identified PPIB, a peptidylprolyl isomerase (PPIase), as a gene and protein that is strongly related to both in vitro radiation response and is a predictor of clinical outcome in patients with HNSCC. The cyclophilins are members of a larger class of PPIase proteins widely expressed throughout the body, known as the immunophilins—targets for the immunosuppressive agents FK506, CsA, and rapamycin [39,40]. PPIB as a secreted protein is also thought to serve as a ligand for the CD147 receptor, thereby regulating the motility of cells expressing this receptor [41]. A recent study also indicates that PPIB present in the conditioned medium of the MDA-MB-231 breast carcinoma cell line promoted chemotaxis of bone marrow-derived mesenchymal stromal cells [42]. Here we noted that depletion of PPIB protein enhanced cell killing after radiation likely by enhancing the apoptotic process, the latter phenocopied by exposure of cancer cells to CsA which binds PPIB. In addition to an enhanced level of apoptosis as described here for CsA exposure or PPIB knockdown, there may also be a role for DNA repair through PPIB knockdown. CsA binds and inhibits PPIB, thus interfering with DNA repair [43,44] by decreasing calcineurin-mediated expression of DNA polymerase β [45,46]. Using a dominant negative form of polymerase β after ionizing radiation, cell cycle position-dependent radiosensitization, higher numbers of chromatid-type aberrations that result in replication-dependent secondary DNA double-strand breaks, and a higher number of chromosomal deletions were all seen [47–49], and the chromosomal deletions were described as the mechanism of enhanced cell killing. Also supporting the notion of reduced DNA repair capacity are the results from the comet assay that measures both single- and double-strand breaks. The induction of single-strand breaks occurs at a frequency of almost two orders of magnitude over double-strand breaks and is ordinarily rapidly repaired. Within 1 hour, more than 90% of the total strand breaks would be repaired as seen in Figure 5C. Although it is residual DNA lesions that drive cell death, given the extent of apoptosis seen at 24 hours after irradiation, it is conceivable that unrepaired DNA lesions, stalled replication forks, and others, may initiate the apoptotic process in cells compromised by exposure to CsA or reduced PPIB and RPLP1.
Our work has several limitations. Despite the fact that we used all the suitable publicly available data sets of patients treated with radiation to evaluate our model, the number of patients in there is relatively small. In addition, because we did not have data on primary tumor response, we were obliged to make the assumption that progression free and overall survival are radiocurability surrogates. While we realize that lack of progression at the primary tumor site can be associated with distant failure unrelated to the effect of radiation on the primary, we posit that, given that our biomarker model predicted outcome only in patients treated with radiation, whereas not in those who did not have radiation, the model likely captures the elements of radiation response in this albeit limited clinical scenario.
In summary, this work provides a novel approach for the discovery of biomarkers that predict clinical outcome after radiation therapy across multiple tumor histologic types. It also identifies PPIB as both a biomarker of outcome after radiation therapy and a potential druggable target for improving the effects of this modality. Despite limitations because of the limited data sets used, these findings provide an approach and framework that may improve treatment selection by discovering markers that identify patients who can benefit most for functional organ-sparing approaches with radiotherapy and, once selected, offer these individuals even better outcomes by enhancing their response to this treatment.
Supplementary Methods
Patient Data Sets
As part of a study to develop predictors of metastasis among HNSCC patients, 81 tumor samples were transcriptionally profiled on Affymetrix HG-U133 Plus 2.0 GeneChips, and this information is publicly available on ArrayExpress (www.ebi.ac.uk/arrayexpress accession number E-TABM-302) [1,2]. The clinical characteristics of the patients in this data set are summarized in Table W1A. Importantly, 73 of these patients were treated only with radiation therapy.
Additional validation was carried out on a set composed of 118 patients with HNSCC treated between 2002 and 2008 at the University of Virginia with definitive radiation therapy alone or in combination with induction and/or concurrent chemotherapy regimens. Patients with T1–T2 primary tumors and N0-1 nodal disease were treated with radiation alone and patients with T3–4 primary tumors or N2–3 nodal disease received radiation and chemotherapy. Archived pretreatment primary tumor biopsies were available for 72 patients and were used to create a tissue microarray. Survival was calculated from the date of diagnosis to the date of death of any cause or last date of follow-up. The clinical characteristics of the patients in this data set are published [3].
Cell Line Data Sets
The data sets used in this study are listed in Tables W1A–W1B. The bladder cancer cell line data set was used to select biomarkers and train the prediction model. The gene expression profiles of unirradiated samples of these cell lines have previously been measured using Affymetrix HG-U133A GeneChips [4]. We used a human bladder tumor sample data set to find genes concordantly regulated in these bladder cell lines and human tumors. This data set consisted of 60 expression profiles downloaded from the Gene Expression Omnibus (GEO) Web site (GEO Accession Number GSE3167) [5,6], as well as 25 profiles obtained locally [7]. The gene expression profile and radiation sensitivity of a panel of 16 primary HSF cell lines developed from skin biopsies (Brock) from radiotherapy patients (collected from areas outside the radiation field) was used to test prediction results derived from the bladder cell lines to refine model selection. These 16 cell lines were hybridized to Illumina WholeGenome6 v2 and v3 chips. Samples of these cell lines were exposed to a total absorbed dose of 2 Gy of ionizing radiation, after which the survival fraction was measured.
Gene Array Data Processing
Affymetrix data sets were background adjusted, quantile normalized, and summarized using the Robust Multichip Average technique [8–10]. Illumina data were processed according to the manufacturer. As Tables W1A–W1B indicate, gene expression profiles were measured using several different microarray platforms. To generate a consistent prediction model that could be applied to any of these data sets, we limited subsequent analyses to genes that were present on all platforms. First, because all 22215 Affymetrix HG-U133A probe sets are also represented among the 54613 probe sets present on the Affymetrix HG-U133 Plus 2.0 chip, we kept the expression values for this subset in the head and neck human tumor data set and discarded the rest. Second, because the head and neck cell line data set was profiled using two versions of the Illumina WholeGenome6 chip, we used a file downloaded from the Illumina Web site (illumina.com) to map identical or closely matching probes between the two versions. Most of the cell lines in this experiment were profiled using version 2 of the WG6 chip, which has 48,701 probes. The cell lines C42, S34, and S38 were profiled using version 3, which has 48,803 probes. There are 43,071 probes that are identical or closely matching in the two versions. Affymetrix probe sets were mapped to Illumina probes using a file downloaded from the Illumina Web site (illumina.com), which indicated the probe sets and probes that corresponded to the same RefSeq identifier. With these matching steps completed, 8406 unique Illumina probes corresponded to 8470 unique Affymetrix Probe Sets.
GO and Network Analysis
To explore the functional properties of the genes in the radiotherapy response prediction GEM, we found the gene information corresponding to the probe sets from the NetAffx Web site. We queried the PANTHER Classification System database at pantherdb.org for GO information corresponding to the genes in the model [11,12]. We used the Ingenuity Pathways Analysis program (Ingenuity Systems, www.ingenuity.com) to generate networks that include genes in the radiation response GEM. We also searched the Oncomine database [13] to determine which genes from the radiation response GEM were associated with tumor stage, grade, and outcome in bladder and HNSCC.
Linear Discriminant Analysis
We used an LDA approach [14,15] to develop the optimal gene model predictors of radiation response from the 300 probes above. In general, for any given GEM that consists of a list of probe sets, we train a linear discriminant using the expression values of the probe sets in the model for the radiation-sensitive and radiation-resistant bladder cancer cell lines. We apply this discriminant on the expression values of an “optimization” cell line or patient data set to classify the sample as a responder or a nonresponder. Each sample receives a GEM score, which represents the posterior probability that the sample is sensitive to radiation.
PPIB and p16 IHC
For the Virginia HNSCC patient data set, 4-µm histologic sections were cut, placed on charged glass slides (SuperFrost Plus; Fisher Scientific, Pittsburgh, PA), deparaffinized in xylene, and rehydrated in a graded series of ethanol baths. The sections were immersed in target retrieval solution, citrate pH 6.0 (Dako, Glostrup, Denmark), and antigen retrieval was performed in a Pascal pressure chamber (Dako), achieving 22 psi pressure for 30 seconds at 125°C. IHC was performed on a robotic platform (Autostainer; Dako). Endogenous peroxidases were blocked using peroxidase and alkaline phosphatase blocking reagent (Dako). Polyclonal rabbit antibody to PPIB (catalog no. Ab16045; Abcam) was diluted 1:400, and mouse monoclonal antibody to p16 (catalog no. 550834; BD Pharmingen, San Diego, CA) was diluted to 1:100 and applied at ambient temperature for 30 minutes. Antibody binding was visualized by incubation with Envision Dual Link (Dako) followed by incubation with 3,3′-diaminobenzidine tetrahydrochloride. Immunohistochemical staining for PPIB was scored for the intensity of cytoplasmic staining (0 = absent, 1 = weak, 2 = moderate, 3 = strong). p16 status was determined by IHC as specified by Reimers et al. [16]. Tumors were considered positive for p16 when strong nuclear and cytoplasmic staining was present in more than 60% of cells [16].
Cell Line Irradiation, Clonogenic Survival Assay, and Estimation of Radiosensitivity and DNA Repair
Thirty-nine bladder cancer cell lines from a previously described panel of 40 (BLA-40 [4]) were cultured and irradiated with a total dose of 2 Gy, and the fraction surviving was determined (SF2) as described. In a similar fashion, using a panel of 16 primary HSF cell lines developed from skin biopsies (Brock) from radiotherapy patients (collected from areas outside the radiation field), survival curves were generated and SF2 was calculated from those curves using a linear quadratic fit (α/β). Preradiation RNA from these 16 HSF cell lines was hybridized to Illumina WholeGenome6 v2 and v3 chips.
Exponentially growing cells were transfected with siRNA and/or treated with CsA as described in figures legends. After the treatments, cells were irradiated at ambient temperature with 2 and 6 Gy of x-ray (250 keV) and replated into 100-mm-diameter culture dishes at densities calculated to yield 50 to 100 cell colonies per dish. After 10 to 14 days of incubation, cells were fixed and stained with crystal violet in 20% ethanol, and colonies more than 50 cells were counted. The number of surviving colonies divided by the number of plated cells was used to calculate the plating efficiency and survival fraction for each treatment.
Analysis of DNA Damage Repair by the Comet Assay
Analysis of DNA damage repair by the comet assay was carried out as described [17]. We used the standard comet assay (Trevigen, Gaithersburg, MD) to compare the differences in DNA damage repair between wild-type and siRNA knockdown cells. Briefly, exponentially growing cells were irradiated (x-ray, 10 Gy) and allowed to recover for 1 hour. Cells were harvested, mixed with low-melting agarose, and applied to comet slides. After lysis and alkaline unwinding, the electrophoresis was performed under alkaline (pH > 13) denaturating conditions at 1 V/cm for 30 minutes. Slides were stained with SYBR green dye for 10 minutes. One hundred randomly selected cells per sample were captured under a fluorescent microscope and analyzed. The relative length and intensity of SYBR green-stained DNA tails to heads were proportional to the amount of DNA damage present in the individual nuclei and were measured by the Olive tail moment [17]. Cyclosporine was purchased from Sigma (St Louis, MO) and used in vitro as described [18].
In Vitro Cell Growth
Bladder cancer cells were seeded in 96-well cell culture plates at a density of 5000 per well in a volume of 200 µl. Twenty-four hours later, the cells were transfected with the siRNA duplexes described previously (6.25 nM) using Oligofectamine according to the manufacturer's instructions in triplicate. Twenty-four hours later, the cells were treated either with indicated concentrations of CsA or with equal volumes of carrier (100% ethanol) in triplicate. Plates were incubated for 24 to 48 hours with carrier or drug. The cell numbers were assessed by alamarBlue (Invitrogen, Carlsbad, CA) per the manufacturer's instructions.
Apoptosis Assay
Bladder cancer cells were seeded in six-well cell culture plates at a density of 83,000 per well in a volume of 2 ml. Twenty-four hours later, the cells were transfected with the siRNA duplexes described previously (6.25 nM) using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Apoptosis was assessed by the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences, Franklin Lakes, NJ) per the manufacturer's instructions.
Small Interfering RNA
The following siRNA duplexes were chemically synthesized, deprotected, and annealed by Dharmacon (Lafayette, CO): PPIB siRNA duplex: 5′-GGUGGAGAGCACCAAGACA-3′ was described [19]; Red Fluorescent Protein Ctrl siRNA duplex: 5′-(DY547)AAUUCUCCGAACGUGUCACGU-3′ served as a transfection efficiency control and as a negative control. Luciferase (GL2) siRNA duplex: 5′-CGUACGCGGAAUACUUCGA-3′ served as a negative control.
Ribosomal protein, large, P1 (RPLP1) siRNA duplex was chemically synthesized, deprotected and annealed by Sigma-Proligo (catalog no. SASI_Hs01_00160252). Bladder cancer cells were grown at 50% confluence in 100-mm plates, and six-well plates were transfected with the siRNA duplexes (6.25 nM) using Oligofectamine (Invitrogen) according to the manufacturer's instructions.
Western Blot Analysis
Western blot analysis was performed as detailed previously [20]. Antibodies against PPIB (clone k2e2; Santa Cruz Biotechnology, Inc, Santa Cruz, CA) and RPLP1 (polyclonal; Sigma) were used. Immunoblots were developed using Super Signal Femto Chemiluminescence (Pierce, Rockford, IL), and results were visualized and quantified using the Alpha Innotech (San Leandro, CA) imaging system. Monoclonal anti-α-tubulin (clone AA13; Santa Cruz Biotechnology, Inc) was used to detect tubulin expression for normalization of expression of PPIB or RPLP1.
Data Sets Searched for Associations of 41-Gene GEM and Tumor Parameters and Clinical Outcome (Figure 2)
Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P, et al. (2005). Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res 11(11), 4044–4055.
Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, Young J, Dembélé D, Thibault C, Muller D, et al. (2004). Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene 23(14), 2484–2498.
Dyrskjøt L, Kruhøffer M, Thykjaer T, Marcussen N, Jensen JL, Møller K, and Ørntoft TF (2004). Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res 64, 4040–4048.
Dyrskjøt L, Zieger K, Kruhøffer M, Thykjaer T, Jensen JL, Primdahl H, Aziz N, Marcussen N, Møller K, and Orntoft TF (2005). A molecular signature in superficial bladder carcinoma predicts clinical outcome. Clin Cancer Res 11(11), 4029–4036.
Dyrskjøt L, Zieger K, Real FX, Malats N, Carrato A, Hurst C, Kotwal S, Knowles M, Malmström PU, de la Torre M, et al. (2007). Gene expression signatures predict outcome in non-muscle-invasive bladder carcinoma: a multicenter validation study. Clin Cancer Res 13(12), 3545–3551.
Frierson HF Jr, El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA, and Hampton GM (2002). Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol 161(4), 1315–1323.
Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL, and Gaffney PM (2004). Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res 64(1), 55–63.
Hensen EF, De Herdt MJ, Goeman JJ, Oosting J, Smit VT, Cornelisse CJ, and Baatenburg de Jong RJ (2008). Gene-expression of metastasized versus nonmetastasized primary head and neck squamous cell carcinomas: a pathway-based analysis. BMC Cancer 8, 168.
Kuriakose MA, Chen WT, He ZM, Sikora AG, Zhang P, Zhang ZY, Qiu WL, Hsu DF, McMunn-Coffran C, Brown SM, et al. (2004). Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci 61(11), 1372–1383.
Modlich O, Prisack HB, Pitschke G, Ramp U, Ackermann R, Bojar H, Vögeli TA, and Grimm MO (2004). Identifying superficial, muscle-invasive, and metastasizing transitional cell carcinoma of the bladder: use of cDNA array analysis of gene expression profiles. Clin Cancer Res 10(10), 3410–3421.
O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B, Cheng Y, Putt M, Feldman M, Ziober B, et al. (2005). Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene 24(7), 1244–1251.
Roepman P, Wessels LF, Kettelarij N, Kemmeren P, Miles AJ, Lijnzaad P, Tilanus MG, Koole R, Hordijk GJ, van der Vliet PC, et al. (2005). An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet 37(2), 182–186.
Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, and Cordon-Cardo C (2006). Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol 24(5), 778–789.
Schlingemann J, Habtemichael N, Ittrich C, Toedt G, Kramer H, Hambek M, Knecht R, Lichter P, Stauber R, and Hahn M (2005). Patient-based cross-platform comparison of oligonucleotide microarray expression profiles. Lab Invest 85(8), 1024–1039.
Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, Shyr Y, Murphy BM, Cmelak AJ, Burkey BB, et al. (2006). Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res 12(3 Pt 1), 701–709.
Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA Jr, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, et al. (2005). Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol 167(6), 1763–1775.
Stransky N, Vallot C, Reyal F, Bernard-Pierrot I, de Medina SG, Segraves R, de Rycke Y, Elvin P, Cassidy A, Spraggon C, et al. (2006). Regional copy number-independent deregulation of transcription in cancer. Nat Genet 38(12), 1386–1396.
Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN, et al., (2004). Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet 154(1), 27–35.
Supplementary Results
Predictive Performance of Published Gene Expression Models in HNSCC Patients
We sought to determine whether two recently published predictive models of radiation response in patients could predict outcome in our data sets. We evaluated whether the Weichselbaum et al. [21] IFN related DNA damage resistance signature found to predict efficacy of adjuvant chemo/radiotherapy in breast cancer patients was predictive in our samples. We applied the IFN-related DNA damage resistance signature model to the HNSCC data sets described previously, and this did not successfully stratify patients according to response in either ROC (Rickman data set; Wilcoxon rank-sum test, P = .5) or Kaplan-Meier (evaluating multiple points on ROC as dichotomizing cutoffs) analyses (Rickman data set; best overall survival Kaplan-Meier χ2, P = .131; best progression-free survival Kaplan-Meier χ2, P = .258) or multivariate Cox proportional hazards regression models (Rickman data set; overall survival, P = .470; progression-free survival, P = .61).
The second model was one describing a 10-gene radiosensitivity index (RSI, high index = radioresistance) [22]. We evaluated this RSI model on the BLA-40 and Rickman data sets. RSI was not significantly correlated with SF2 in the BLA-40 cell lines (Pearson correlation = 0.183, one-sided cor.test P = .132; Spearman correlation = 0.174, one-sided cor.test P = .144). For the Rickman data set, RSI was not significantly different between survivors and deceased or between patients who relapsed and patients who did not relapse, for the entire set of patients treated with radiation (with or without chemotherapy) or for the set of patients treated with radiation alone.
Characteristics and Network Analysis of the 41 Genes in the GEM
Genes corresponding to probes in the model were identified and GO information obtained from the PANTHER database (Table W3). No molecular function classification (Figure W1A) was found for 15 of the 41 genes, whereas no biologic process classification (Figure W1B) was found for 16 of the 41 genes in the model. No GO terms are significantly overrepresented among the genes for which such terms are available (hypergeometric test). However, biologic process terms that are represented multiple times include immunity and defense (five genes), transport (four genes), cell proliferation and differentiation (three genes), and induction of apoptosis (three genes). Gene expression and protein synthesis, activation, and destruction terms are also represented multiple times: mRNA transcription regulation (two genes), protein biosynthesis (three genes), protein folding (two genes), protein phosphorylation (three genes), and proteolysis (four genes). The most common GO Biological Process classes were unclassified, transport, immunity, and defense and proteolysis, whereas the GO Molecular Function classes were unclassified, nucleic acid binding, ribosomal proteins, and transcription factors.
Supplementary Material
Abbreviations
- PPIB
cyclophilin B
- COXEN
Coexpression Extrapolation
- GEM
gene expression model
Footnotes
This work was supported in part by National Institutes of Health grants DK069264 to P.D.W., CA06294 to M.D.S. and W.A.B., HL081690 to J.K.L., CA075115 to D.T., and U19AI067773 and US Department of Energy DE-FG02-07ER46336 to S.A.A.
This article refers to supplementary materials, which are designated by Tables W1 to W4 and Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Vineis P, Alavanja M, Buffler P, Fontham E, Franceschi S, Gao YT, Gupta PC, Hackshaw A, Matos E, Samet J, et al. Tobacco and cancer: recent epidemiological evidence. J Natl Cancer Inst. 2004;96:99–106. doi: 10.1093/jnci/djh014. [DOI] [PubMed] [Google Scholar]
- 2.Boffetta P, Hecht S, Gray N, Gupta P, Straif K. Smokeless tobacco and cancer. Lancet Oncol. 2008;9:667–675. doi: 10.1016/S1470-2045(08)70173-6. [DOI] [PubMed] [Google Scholar]
- 3.Riesterer O, Milas L, Ang KK. Use of molecular biomarkers for predicting the response to radiotherapy with or without chemotherapy. J Clin Oncol. 2007;25:4075–4083. doi: 10.1200/JCO.2007.11.8497. [DOI] [PubMed] [Google Scholar]
- 4.Tofilon PJ, Camphausen K. Molecular targets for tumor radiosensitization. Chem Rev. 2009;109:2974–2988. doi: 10.1021/cr800504x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sengelov L, von der Maase H. Radiotherapy in bladder cancer. Radiother Oncol. 1999;52:1–14. doi: 10.1016/s0167-8140(99)00090-0. [DOI] [PubMed] [Google Scholar]
- 6.Kaufman DS, Shipley WU, Griffin PP, Heney NM, Althausen AF, Efird JT. Selective bladder preservation by combination treatment of invasive bladder cancer. N Engl J Med. 1993;329:1377–1382. doi: 10.1056/NEJM199311043291903. [DOI] [PubMed] [Google Scholar]
- 7.Choi N, Baumann M, Flentjie M, Kellokumpu-Lehtinen P, Senan S, Zamboglou N, Kosmidis P. Predictive factors in radiotherapy for non-small cell lung cancer: present status. Lung Cancer. 2001;31:43–56. doi: 10.1016/s0169-5002(00)00156-2. [DOI] [PubMed] [Google Scholar]
- 8.Kitagawa Y, Sano K, Nishizawa S, Nakamura M, Ogasawara T, Sadato N, Yonekura Y. FDG-PET for prediction of tumour aggressiveness and response to intra-arterial chemotherapy and radiotherapy in head and neck cancer. Eur J Nucl Med Mol Imaging. 2003;30:63–71. doi: 10.1007/s00259-002-0978-z. [DOI] [PubMed] [Google Scholar]
- 9.Vidyasagar MS, Maheedhar K, Vadhiraja BM, Fernendes DJ, Kartha VB, Krishna CM. Prediction of radiotherapy response in cervix cancer by Raman spectroscopy: a pilot study. Biopolymers. 2008;89:530–537. doi: 10.1002/bip.20923. [DOI] [PubMed] [Google Scholar]
- 10.Osman I, Sherman E, Singh B, Venkatraman E, Zelefsky M, Bosl G, Scher H, Shah J, Shaha A, Kraus D, et al. Alteration of p53 pathway in squamous cell carcinoma of the head and neck: impact on treatment outcome in patients treated with larynx preservation intent. J Clin Oncol. 2002;20:2980–2987. doi: 10.1200/JCO.2002.06.161. [DOI] [PubMed] [Google Scholar]
- 11.Wangsa D, Ryott M, Avall-Lundqvist E, Petersson F, Elmberger G, Luo J, Ried T, Auer G, Munck-Wikland E. Ki-67 expression predicts loco-regional recurrence in stage I oral tongue carcinoma. Br J Cancer. 2008;99:1121–1128. doi: 10.1038/sj.bjc.6604633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheridan MT, O'Dwyer T, Seymour CB, Mothersill CE. Potential indicators of radiosensitivity in squamous cell carcinoma of the head and neck. Radiat Oncol Investig. 1997;5:180–186. doi: 10.1002/(SICI)1520-6823(1997)5:4<180::AID-ROI3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 13.Minna JD, Girard L, Xie Y. Tumor mRNA expression profiles predict responses to chemotherapy. J Clin Oncol. 2007;25:4329–4336. doi: 10.1200/JCO.2007.12.3968. [DOI] [PubMed] [Google Scholar]
- 14.Williams PD, Lee JK, Theodorescu D. Genomancy: predicting tumour response to cancer therapy based on the oracle of genetics. Curr Oncol. 2009;16:56–58. doi: 10.3747/co.v16i1.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres-Roca JF, Eschrich S, Zhao H, Bloom G, Sung J, McCarthy S, Cantor AB, Scuto A, Li C, Zhang S, et al. Prediction of radiation sensitivity using a gene expression classifier. Cancer Res. 2005;65:7169–7176. doi: 10.1158/0008-5472.CAN-05-0656. [DOI] [PubMed] [Google Scholar]
- 16.Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, Reimers M, Chen Y, Scudiero DA, Weinstein JN, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 17.Eschrich SA, Pramana J, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, Rocha-Lima C, Kelley S, Calvin DP, et al. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. 2009;75:489–496. doi: 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong YF, Selvanayagam ZE, Wei N, Porter J, Vittal R, Hu R, Lin Y, Liao J, Shih JW, Cheung TH, et al. Expression genomics of cervical cancer: molecular classification and prediction of radiotherapy response by DNA microarray. Clin Cancer Res. 2003;9:5486–5492. [PubMed] [Google Scholar]
- 19.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, Su AW, Shaikh AY, Roach P, Kreike B, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghadimi BM, Grade M, Difilippantonio MJ, Varma S, Simon R, Montagna C, Fuzesi L, Langer C, Becker H, Liersch T, et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol. 2005;23:1826–1838. doi: 10.1200/JCO.2005.00.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumur CI, Ladd AC, Wright HV, Penberthy LT, Wilkinson DS, Powers CN, Garrett CT, DiNardo LJ. Genes involved in radiation therapy response in head and neck cancers. Laryngoscope. 2009;119:91–101. doi: 10.1002/lary.20005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Broek GB, Wildeman M, Rasch CR, Armstrong N, Schuuring E, Begg AC, Looijenga LH, Scheper R, van der Wal JE, Menkema L, et al. Molecular markers predict outcome in squamous cell carcinoma of the head and neck after concomitant cisplatin-based chemoradiation. Int J Cancer. 2009;124:2643–2650. doi: 10.1002/ijc.24254. [DOI] [PubMed] [Google Scholar]
- 23.Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, Grimshaw A, Theodorescu D. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci USA. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams PD, Cheon S, Havaleshko DM, Jeong H, Cheng F, Theodorescu D, Lee JK. Concordant gene expression signatures predict clinical outcomes of cancer patients undergoing systemic therapy. Cancer Res. 2009;69:8302–8309. doi: 10.1158/0008-5472.CAN-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith SC, Baras AS, Lee JK, Theodorescu D. The COXEN principle: translating signatures of in vitro chemosensitivity into tools for clinical outcome prediction and drug discovery in cancer. Cancer Res. 2010;70:1753–1758. doi: 10.1158/0008-5472.CAN-09-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher RA. The use of multiple measurements in taxonomic problems. Ann Eugen. 1936;7:179–188. [Google Scholar]
- 27.Venables WN, Ripley BD. Modern Applied Statistics with S. New York, NY: Springer; 2002. [Google Scholar]
- 28.Akaike H. New look at statistical—model identification. IEEE Trans Autom Control Ac. 1974;19:716–723. [Google Scholar]
- 29.Reimers N, Kasper HU, Weissenborn SJ, Stutzer H, Preuss SF, Hoffmann TK, Speel EJ, Dienes HP, Pfister HJ, Guntinas-Lichius O, et al. Combined analysis of HPV-DNA, p16 and EGFR expression to predict prognosis in oropharyngeal cancer. Int J Cancer. 2007;120:1731–1738. doi: 10.1002/ijc.22355. [DOI] [PubMed] [Google Scholar]
- 30.Shonka DC, Jr, Shoushtari AN, Thomas CY, Moskaluk C, Read PW, Reibel JF, Levine PA, Jameson MJ. Predicting residual neck disease in patients with oropharyngeal squamous cell carcinoma treated with radiation therapy: utility of p16 status. Arch Otolaryngol Head Neck Surg. 2009;135:1126–1132. doi: 10.1001/archoto.2009.153. [DOI] [PubMed] [Google Scholar]
- 31.McKenna WG, Weiss MC, Bakanauskas VJ, Sandler H, Kelsten ML, Biaglow J, Tuttle SW, Endlich B, Ling CC, Muschel RJ. The role of the H-ras oncogene in radiation resistance and metastasis. Int J Radiat Oncol Biol Phys. 1990;18:849–859. doi: 10.1016/0360-3016(90)90407-b. [DOI] [PubMed] [Google Scholar]
- 32.McKenna WG, Weiss MC, Endlich B, Ling CC, Bakanauskas VJ, Kelsten ML, Muschel RJ. Synergistic effect of the v-myc oncogene with H-ras on radioresistance. Cancer Res. 1990;50:97–102. [PubMed] [Google Scholar]
- 33.Cosaceanu D, Budiu RA, Carapancea M, Castro J, Lewensohn R, Dricu A. Ionizing radiation activates IGF-1R triggering a cytoprotective signaling by interfering with Ku-DNA binding and by modulating Ku86 expression via a p38 kinase-dependent mechanism. Oncogene. 2007;26:2423–2434. doi: 10.1038/sj.onc.1210037. [DOI] [PubMed] [Google Scholar]
- 34.Karin M, Yamamoto Y, Wang QM. The IKK NF-κB system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed KM, Li JJ. NF-κB-mediated adaptive resistance to ionizing radiation. Free Radic Biol Med. 2008;44:1–13. doi: 10.1016/j.freeradbiomed.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habraken Y, Piette J. NF-κB activation by double-strand breaks. Biochem Pharmacol. 2006;72:1132–1141. doi: 10.1016/j.bcp.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 37.Magne N, Toillon RA, Bottero V, Didelot C, Houtte PV, Gerard JP, Peyron JF. NF-κB modulation and ionizing radiation: mechanisms and future directions for cancer treatment. Cancer Lett. 2006;231:158–168. doi: 10.1016/j.canlet.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 38.Arlt A, Schafer H. NFκB-dependent chemoresistance in solid tumors. Int J Clin Pharmacol Ther. 2002;40:336–347. doi: 10.5414/cpp40336. [DOI] [PubMed] [Google Scholar]
- 39.Fischer G, Tradler T, Zarnt T. The mode of action of peptidyl prolyl cis/trans isomerases in vivo: binding vs. catalysis. FEBS Lett. 1998;426:17–20. doi: 10.1016/s0014-5793(98)00242-7. [DOI] [PubMed] [Google Scholar]
- 40.Kofron JL, Kuzmic P, Kishore V, Colon-Bonilla E, Rich DH. Determination of kinetic constants for peptidyl prolyl cis-trans isomerases by an improved spectrophotometric assay. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- 41.Melchior A, Denys A, Deligny A, Mazurier J, Allain F. Cyclophilin B induces integrin-mediated cell adhesion by a mechanism involving CD98-dependent activation of protein kinase C-delta and p44/42 mitogen-activated protein kinases. Exp Cell Res. 2008;314:616–628. doi: 10.1016/j.yexcr.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Lin SY, Yang J, Everett AD, Clevenger CV, Koneru M, Mishra PJ, Kamen B, Banerjee D, Glod J. The isolation of novel mesenchymal stromal cell chemotactic factors from the conditioned medium of tumor cells. Exp Cell Res. 2008;314:3107–3117. doi: 10.1016/j.yexcr.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herman M, Weinstein T, Korzets A, Chagnac A, Ori Y, Zevin D, Malachi T, Gafter U. Effect of cyclosporin A on DNA repair and cancer incidence in kidney transplant recipients. J Lab Clin Med. 2001;137:14–20. doi: 10.1067/mlc.2001.111469. [DOI] [PubMed] [Google Scholar]
- 44.Sugie N, Fujii N, Danno K. Cyclosporin-A suppresses p53-dependent repair DNA synthesis and apoptosis following ultraviolet-B irradiation. Photodermatol Photoimmunol Photomed. 2002;18:163–168. doi: 10.1034/j.1600-0781.2002.00765.x. [DOI] [PubMed] [Google Scholar]
- 45.Ahlers C, Kreideweiss S, Nordheim A, Ruhlmann A. Cyclosporin A inhibits Ca2+-mediated upregulation of theDNArepair enzyme DNA polymerase β in human peripheral blood mononuclear cells. Eur J Biochem. 1999;264:952–959. doi: 10.1046/j.1432-1327.1999.00700.x. [DOI] [PubMed] [Google Scholar]
- 46.Herman M, Ori Y, Chagnac A, Weinstein T, Korzets A, Zevin D, Malachi T, Gafter U. DNA repair in mononuclear cells: role of serine/threonine phosphatases. J Lab Clin Med. 2002;140:255–262. doi: 10.1067/mlc.2002.127738. [DOI] [PubMed] [Google Scholar]
- 47.Neijenhuis S, Verwijs-Janssen M, Kasten-Pisula U, Rumping G, Borgmann K, Dikomey E, Begg AC, Vens C. Mechanism of cell killing after ionizing radiation by a dominant negative DNA polymerase β. DNA Repair, (Amst) 2009;8:336–346. doi: 10.1016/j.dnarep.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Vens C, Dahmen-Mooren E, Verwijs-Janssen M, Blyweert W, Graversen L, Bartelink H, Begg AC. The role of DNA polymerase β in determining sensitivity to ionizing radiation in human tumor cells. Nucleic Acids Res. 2002;30:2995–3004. doi: 10.1093/nar/gkf403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vermeulen C, Verwijs-Janssen M, Begg AC, Vens C. Cell cycle phase dependent role of DNA polymerase β in DNA repair and survival after ionizing radiation. Radiother Oncol. 2008;86:391–398. doi: 10.1016/j.radonc.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 50.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 51.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 53.Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 54.Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, Young J, Dembele D, Thibault C, Muller D, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–2498. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 55.Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, Wolf H, Orntoft TF. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- 56.Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Rickman DS, Millon R, De Reynies A, Thomas E, Wasylyk C, Muller D, Abecassis J, Wasylyk B. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27:6607–6622. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 2.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update—from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shonka DC, Jr, Shoushtari AN, Thomas CY, Moskaluk C, Read PW, Reibel JF, Levine PA, Jameson MJ. Predicting residual neck disease in patients with oropharyngeal squamous cell carcinoma treated with radiation therapy: utility of p16 status. Arch Otolaryngol Head Neck Surg. 2009;135:1126–1132. doi: 10.1001/archoto.2009.153. [DOI] [PubMed] [Google Scholar]
- 4.Titus B, Frierson HF, Jr, Conaway M, Ching K, Guise T, Chirgwin J, Hampton G, Theodorescu D. Endothelin axis is a target of the lung metastasis suppressor gene RhoGDI2. Cancer Res. 2005;65:7320–7327. doi: 10.1158/0008-5472.CAN-05-1403. [DOI] [PubMed] [Google Scholar]
- 5.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, Orntoft TF. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 7.Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, Safo MK, Theodorescu D. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–3813. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- 8.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 9.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 11.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher RA. The use of multiple measurements in taxonomic problems. Ann Eugen. 1936;7:179–188. [Google Scholar]
- 15.Venables WN, Ripley BD. Modern Applied Statistics with S. New York: Springer; 2002. [Google Scholar]
- 16.Reimers N, Kasper HU, Weissenborn SJ, Stutzer H, Preuss SF, Hoffmann TK, Speel EJ, Dienes HP, Pfister HJ, Guntinas-Lichius O, et al. Combined analysis of HPV-DNA, p16 and EGFR expression to predict prognosis in oropharyngeal cancer. Int J Cancer. 2007;120:1731–1738. doi: 10.1002/ijc.22355. [DOI] [PubMed] [Google Scholar]
- 17.Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiat Res. 1990;122:86–94. [PubMed] [Google Scholar]
- 18.Nomura T, Yamamoto H, Mimata H, Shitashige M, Shibasaki F, Miyamoto E, Nomura Y. Enhancement by cyclosporine A of taxol-induced apoptosis of human urinary bladder cancer cells. Urol Res. 2002;30:102–111. doi: 10.1007/s00240-002-0239-4. [DOI] [PubMed] [Google Scholar]
- 19.Obata Y, Yamamoto K, Miyazaki M, Shimotohno K, Kohno S, Matsuyama T. Role of cyclophilin B in activation of interferon regulatory factor-3. J Biol Chem. 2005;280:18355–18360. doi: 10.1074/jbc.M501684200. [DOI] [PubMed] [Google Scholar]
- 20.Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, Theodorescu D. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 21.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, Su AW, Shaikh AY, Roach P, Kreike B, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eschrich SA, Pramana J, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, Rocha-Lima C, Kelley S, Calvin DP, et al. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. 2009;75:489–496. doi: 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, Reimers M, Chen Y, Scudiero DA, Weinstein JN, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.