Abstract
Drug-drug interactions (DDIs) are major causes of serious adverse drug reactions. Most DDIs have a pharmacokinetic basis in which one drug reduces the elimination of a second drug, leading to potentially toxic drug levels. As a major organ of drug elimination, the kidney represents an important site for DDIs. Here, we screened a prescription drug library against the renal organic cation transporter OCT2/SLC22A2, which mediates the first step in the renal secretion of many cationic drugs. Of the 910 compounds screened, 244 inhibited OCT2. Computational analyses revealed key properties of inhibitors versus non-inhibitors, which included overall molecular charge. Four of six potential clinical inhibitors were transporter-selective in follow-up screens against additional transporters: OCT1/SLC22A1, MATE1/SLC47A1 and MATE2-K/SLC47A2. Two compounds showed different kinetics of interaction with the common polymorphism OCT2-A270S, suggesting a role of genetics in modulating renal DDIs.
Introduction
The vast majority of registered drugs are eliminated via the kidneys, either in the form of unchanged parent compound1 or after bioconversion to polar metabolites (Figure 1A). In a recent analysis of clinical elimination data for 391 drugs, Varma et al. showed that approximately 30% of the compounds were primarily excreted unchanged in the kidneys.1 In addition, hepatic metabolism is usually followed by renal excretion of the formed metabolites, as demonstrated by our own analysis of clinical mass balance data for more than three hundred drugs (Figure 1A). Consequently, inhibition of renal drug transport can result in significantly altered systemic levels of the parent drug molecule and of potentially active metabolites, with ultimate effects on the drug’s pharmacological and toxicological profile.2–6 Moreover, transport inhibitors may result in changes in the accumulation of drugs in the kidney leading to enhanced or reduced exposure and toxicity to the kidney. Such drug-drug interactions (DDIs) are typically detrimental and, therefore, sought to be avoided in drug development and therapy.
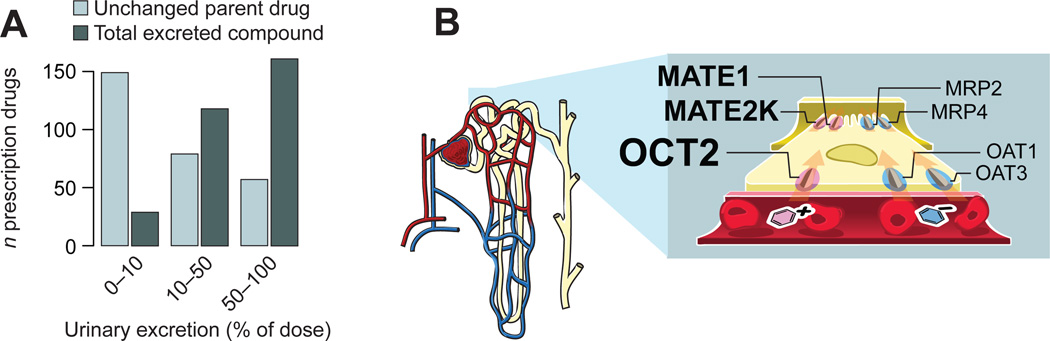
Figure 1. Renal excretion of drugs and metabolites.
(A) Urinary excretion profiles for prescription drugs. Data on the urinary recovery of 308 drugs was collected from published clinical mass balance studies. Light blue bars denote the renal recovery of unchanged parent drug. For approximately one-half of all analyzed compounds, renal excretion of the parent compound contributes significantly to the overall elimination (light blue bars, ≥10% urinary excretion of parent drug). Data on the total recovery of parent drug and metabolites (dark blue bars) show that renal elimination is an important pathway also for drugs that are metabolically converted, with more than 90% of all analyzed drugs being recovered in urine to a significant extent (dark blue bars, ≥10% urinary excretion of parent drug + metabolites). (B) Schematic illustration of key drug transporters expressed in renal proximal tubule epithelium. Cation transporters are on the left hand side and anion transporters on the right. The basolateral membrane facing the blood is nearest to the reader. The nephron is shown in the far left.
For organic cations, which make up the majority of currently used prescription drugs,7 the first step in renal secretion is mediated by the organic cation transporter 2 (OCT2, SLC22A2) (Figure 1B, which also shows the localization of other major renal drug transporters).8–9 OCT2 is responsible for the renal elimination of metformin,10–11 the most commonly prescribed treatment of type 2 diabetes, and is an important pharmacokinetic determinant for several cytostatic12 and antiretroviral agents.13
Over the past decades, major progress has been made in characterizing and predicting drug interactions with drug metabolizing enzymes, with the aim of avoiding potential DDIs. Recently, the limelight has shifted to drug transporter-mediated DDIs.2–6, 14–15 Accordingly, there is increased demand from regulatory agencies in the US16 and Europe17 to document transporter interactions of new chemical entities. However, in contrast to cytochrome P450 enzymes, probes that target specific transporters are scarce, hampering efficient evaluation of transporter effects on drug disposition. Systematic mapping of drug-transporter interactions on scales previously performed for drug metabolizing enzymes are thus warranted.
Here, we used a multi-tier fluorescent screening and computational modeling strategy to identify OCT2 inhibitors in a library of 910 prescription drugs to identify DDI liabilities, to develop specific probes for delineating drug transport processes, and to define the molecular properties leading to transporter binding.
Results
Identification of OCT2 inhibitors among prescription drugs
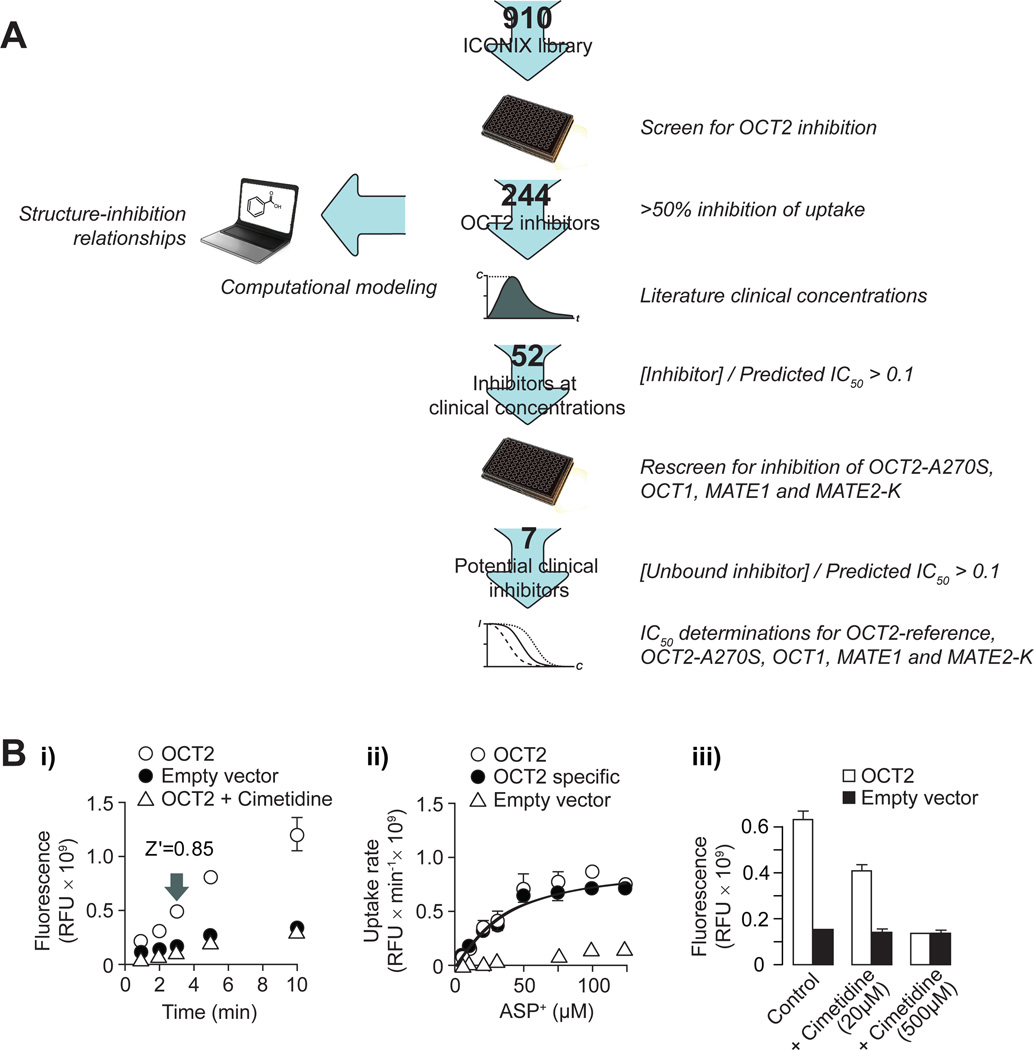
We screened a library of 910 prescription drugs and drug-like compounds using a high-throughput assay of renal organic cation transport (Figure 2). In the assay, uptake of the model OCT2 substrate 4-(4-(dimethylamino)styryl)-N-methylpyridinium (ASP+) into intact HEK293 cells stably expressing human OCT2 was measured fluorometrically. Substrate uptake was linear for >5 minutes (Figure 2Bi) with a Km of ASP+ transport of 36.4±6.8 µM (Figure 2Bii). Screening using 5 µM ASP+ and 3 minute incubation yielded a Z' factor of 0.85, indicating excellent assay performance. For validation purposes, the model OCT2 inhibitor cimetidine was included in all assay plates at concentrations of 20 µM and 500 µM, resulting in partial and complete inhibition of OCT2 mediated transport, respectively (Figure 2Biii).
Figure 2. Experimental and computational methods.
(A) Schematic of the workflow applied to identify OCT2 inhibitors. The number of compounds analyzed in each step is noted on the arrows. [Inhibitor]: total plasma concentration of inhibitor. [Unbound inhibitor]: concentration of inhibitor not bound to plasma proteins. (B) Uptake of ASP+ in HEK293 cells stably expressing OCT2. (Bi) Time course of ASP+ (5 µM) uptake without (open circles) or with 500 µM cimetidine (open triangles), and in cells transfected with an empty vector (closed circles). The Z' factor at the sampling time used for screening is indicated in the figure. (Bii) Concentration dependence of ASP+ uptake in OCT2 expressing cells (open circles) and empty vector transfected cells (open triangles). The OCT2 specific uptake (closed circles) was calculated by subtracting the non-specific uptake in empty vector transfected cells from that in the cells expressing OCT2. (Biii) Inhibitory effects of cimetidine on ASP+ (5 µM) uptake in OCT2 expressing cells (open bars) and in cells transfected with an empty vector (closed bars). Data are presented as mean ± s.d. (three separate samples from one representative experiment).
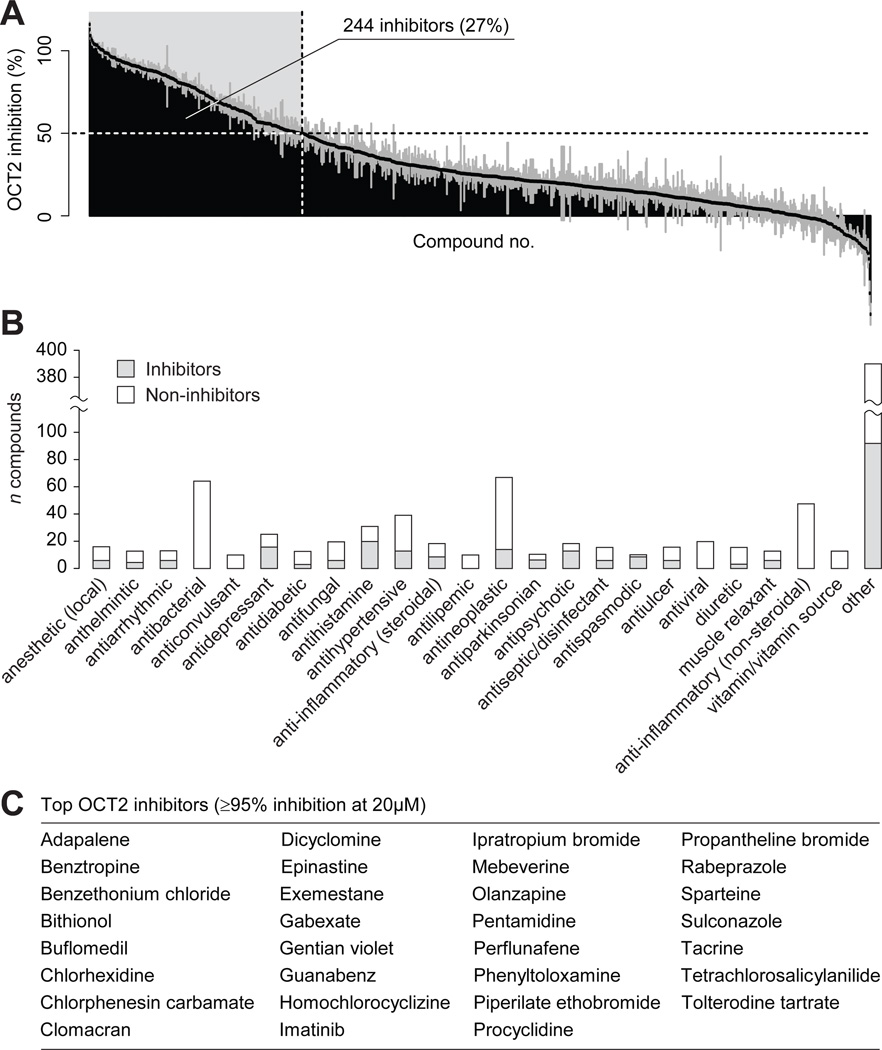
At 20 µM, 244 compounds decreased ASP+ transport by at least 50% (Figure 3A). OCT2 inhibitors were found across multiple pharmacological classes: in particular, the antidepressant, antihistamine, antiparkinsonian, antipsychotic and antispasmodic therapeutic classes were highly enriched in OCT2 inhibitors, with >60% of compounds in each of these therapeutic classes showing OCT2 inhibition potency (Figure 3B). Inhibitor activity was also common (>40%) in the local anesthetic, antiarrhythmic, steroid anti-inflammatory, antiseptic/disinfectant, antiulcer and muscle relaxant classes. Thirty-one inhibitors showed high potency towards OCT2 (≥95% inhibition) (Figure 3C).
Figure 3. Inhibitors of OCT2 identified in a screen of 910 prescription drugs and drug-like compounds.
(A) Overview of the results from the screening of OCT2 inhibition. Each bar represents one compound. 244 compounds resulting in at least 50% decreased uptake of ASP+ were classified as inhibitors (shaded in light gray). Data are presented as mean ± s.d. (samples in triplicate from one experiment). (B) Therapeutic classes of the screened compounds. Therapeutic classes with ≥ 10 members in the screening library are shown as individual bars; all other classes were combined (“other”). Shaded and white bars represent the number of OCT2 inhibitors and non-inhibitors in each class, respectively. (C) High-potency OCT2 inhibitors resulting in ≥ 95% inhibition at 20 µM, corresponding to estimated IC50 ≤ 1µM.
With the aim of identifying clinically relevant OCT2 inhibitors, we used the inhibitor activity measurements to estimate half-maximum inhibitory concentrations (IC50). These were then compared to plasma concentrations obtained after typical clinical doses. Fifty-two compounds were selected for further analyses on the basis of having Cmax / IC50 > 0.1 and being commercially available.
Specificity of OCT2 inhibition at clinical drug concentrations
The lack of clinical probes that target specific transporters is a severe obstacle for the mechanistic understanding of a drug’s pharmacokinetic properties. Accordingly, we determined the interaction of the 52 putative clinical OCT2 inhibitors against a panel of relevant renal and hepatic organic cation transporters (OCT1, MATE1 (SLC47A1), MATE2-K (SLC47A2)) and a common genetic polymorphism of OCT2, OCT2-A270S.
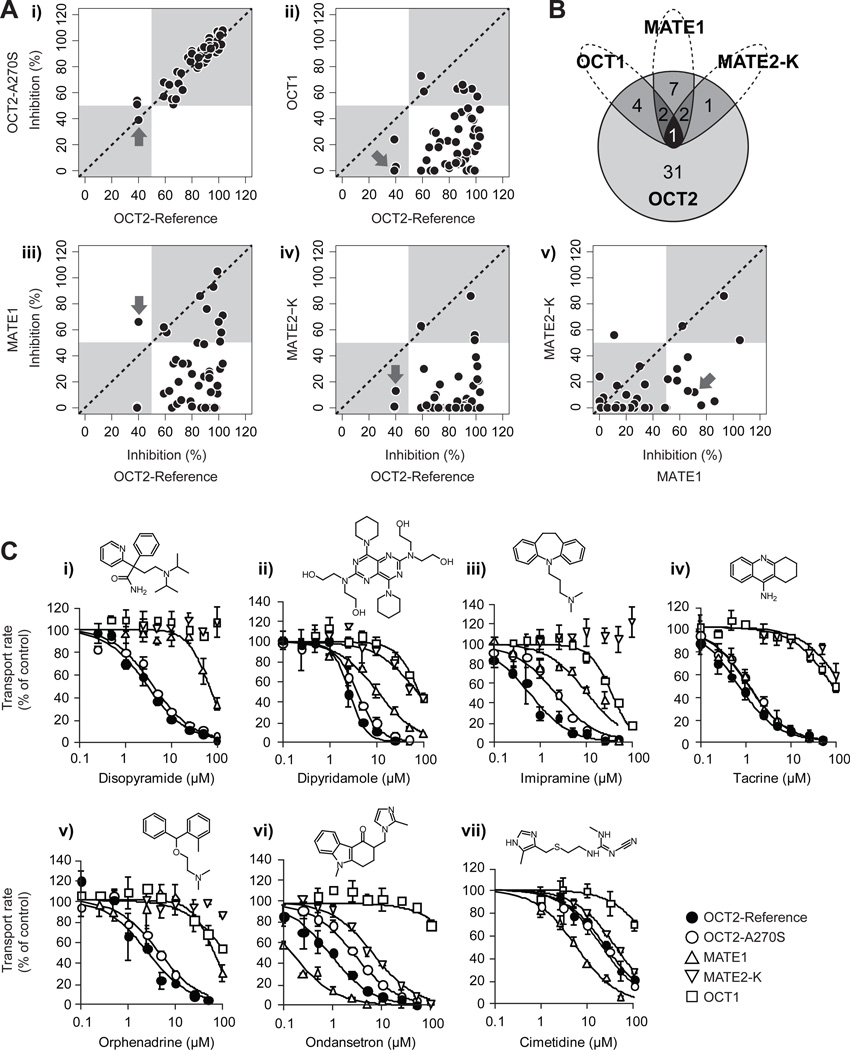
ASP+ was shown to be a suitable probe substrate for all evaluated transporters (Supporting Information, Figure S1). Rescreening against OCT2 confirmed all but three of the inhibitors from the initial screening, and the inhibition profile for the common genetic variant OCT2-A270S was well correlated with that of the reference protein, suggesting only minor effects of this genetic variant on inhibitors (Figure 4Ai). In contrast, despite a sequence identity of >70%, only 7 of the OCT2 inhibitors also affected the hepatic paralog OCT1 (Figure 4Aii; Figure 4B). A similar overlap was observed for the much more distantly related transporters MATE1 and MATE2-K (<10% sequence identity with OCT2), with 12 and 4 inhibitors in common with OCT2. Only one compound, the leukotriene antagonist zafirlukast, showed affinity for all four organic cation transporters.
Figure 4. Selectivity of OCT2 inhibitors for the polymorphic transporter, OCT2-A270S and other organic cation transporters.
(A) Correlation analyses between OCT2 inhibition and inhibition of OCT2-A270S, the hepatic homologue OCT1, and the apical organic cation transporters MATE1 and MATE2-K. The prototypical organic cation transport inhibitor cimetidine is indicated by the arrows. (B) Venn diagram showing the overlapping inhibitors for OCT2, OCT1, MATE1 and MATE2-K. (C) Selectivity of inhibition for putative clinical inhibitors of OCT2. The concentration dependent inhibition of ASP+ uptake is shown for HEK293 cells stably expressing OCT2-Reference (closed circles), OCT2-A270S (open circles), MATE1 (upward pointing triangles), MATE2-K (downward pointing triangles) and OCT1 (open squares). Data are presented as mean ± s.d. (three separate samples from one representative experiment).
For drugs tightly binding to plasma proteins, the free concentration in plasma is a better estimate of the drug that can interfere with OCT2 transporter function. Hence, in the next step, we increased the stringency of our selection criteria further, using unbound instead of total plasma concentrations. Figure 4C shows the concentration dependency of six compounds that exhibited transporter specific inhibition at clinical unbound plasma concentrations, together with that of the prototypical organic cation transport inhibitor cimetidine. Notably, cimetidine had considerably higher inhibition potency for MATE1 than for the other transporters in the panel, as did the antiemetic ondansetron, whereas disopyramide, imipramine and orphenadrine and to a lesser extent, dipyridamole, specifically inhibited OCT2. These compounds are thus potential candidates as selective clinical transporter inhibitors. Further, ondansetron, tacrine, dipyridamole and imipramine showed preferential inhibition of the renal transporters OCT2 and MATE1 compared to the hepatic OCT1, suggesting their use in delineating organic cation disposition on the organ level. Notably, imipramine and ondansetron had lower affinity towards the genetic variant OCT2-A270S than to the reference protein. Such selectivity differences are suggestive, since they imply a possibility of compounded effects of drug-induced inhibition and genetic modulation that may put certain subpopulations at an increased risk of drug-drug interactions.
Structural characteristics of OCT2 inhibitors
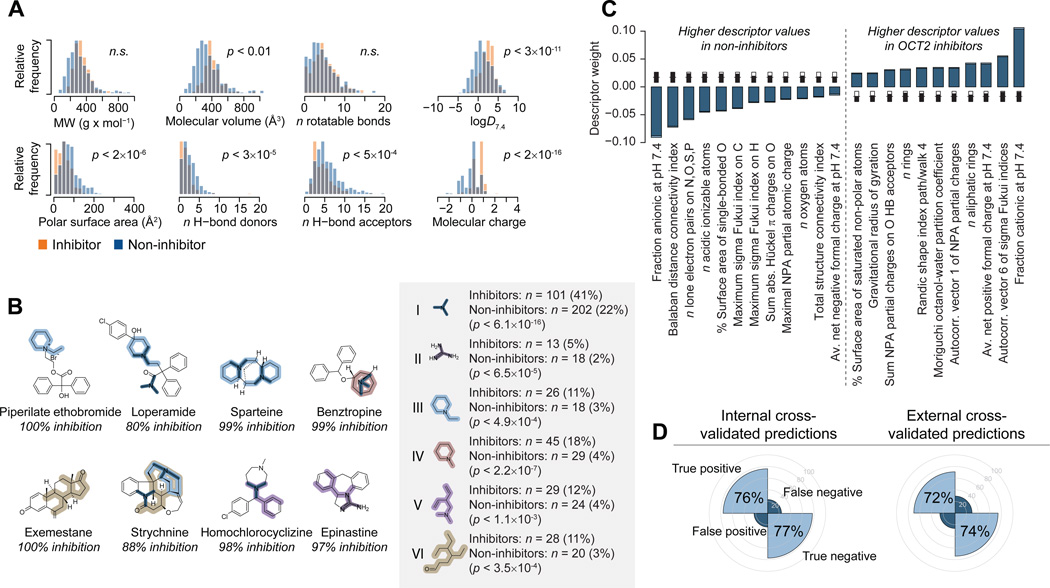
We leveraged the unique size of this transporter inhibition dataset to determine structural features that define ligand binding to OCT2. As a first step, we examined the distribution of key molecular properties in OCT2 inhibitors and non-inhibitors (Figure 5A). This revealed statistically significant differences in the molecular volume, polar surface area, the number of hydrogen bond donors and acceptors and, particularly, in the lipophilicity and average charge of molecules in these two groups.
Figure 5. Structural features of OCT2 inhibitors.
(A) Distribution of key physicochemical properties of OCT2 inhibitors (yellow) and non-inhibitors (blue). Differences were assessed with the two-sided unpaired Student’s t-test assuming unequal variances and are presented as uncorrected p-values. (B) Substructure analysis of OCT2 inhibitors. The inset shows substructure fragments enriched in inhibitors compared to the entire dataset, with absolute and relative frequencies of each fragment in inhibitors and non-inhibitors. Enrichment was assessed with the hypergeometric test and is presented as uncorrected p-values. Significantly enriched fragments are highlighted in example OCT2 inhibitors. (C) Molecular descriptors discriminating between inhibitors and non-inhibitors. Bars show the mean PLS regression coefficients from 500 cross-validated models (100 random cross-validation partitionings with five segments each); standard errors are shown in light blue. Descriptors with positive coefficients have higher values in inhibitors, and descriptors with negative coefficients have higher values in non-inhibitors. The descriptors shown were included in at least 50% of the final 500 models after optimization through iterative exclusion of uninformative descriptors; inset thermometer plots show in black the fraction of all models that include the descriptor. (D) Internal and external prediction results. The left-hand plot shows the fraction of true positive and true negative predictions (averaged over all 500 models) from the inner cross-validation loop, during which model parameters and descriptor selections are optimized. The right-hand plot shows predictions from the outer cross-validation loop (averaged over 100 random partitionings of the dataset). The latter thus represent unbiased external predictions.
An analysis of substructure fragments overrepresented in inhibitors also indicated the importance of positive charge for ligand binding to OCT2. A selection of motifs found disproportionally often in inhibitors are shown in Figure 5B, and include tertiary amine, guanidine, and N-methyl and N-ethyl piperidine fragments (I–IV). Also, several variations of a larger fragment containing a double bonded oxygen were identified as enriched (e.g., VI); these predominantly matched to a series of carbonyl substituted steroids, a majority of which inhibited OCT2 (63% of the compounds inhibited more than 50% of the transport at 20µM).
Substructures exclusively found in inhibitors tended to be fairly large and specific for smaller groups of homologous compounds. Examples include fragments specifically found in imidazole antifungals and in tricyclic antidepressants and in antihistamines (data not shown). Altogether, thirteen inhibitor-specific substructures accounted for a total of 60 (25%) of all OCT2 inhibitors. Each of these substructures matched at most seven inhibitors, demonstrating that activity is spread through multiple structural series, and suggesting that affinity for the transporter is likely caused by more general molecular properties that are not specific for a certain series of congeners.
Identification of molecular affinity determinants
To further elucidate the mechanisms of ligand binding to OCT2, we derived discriminant structure-activity models using partial least squares projection. We used a stringent double-loop cross-validation procedure to determine the most important molecular properties for discriminating between inhibitors and non-inhibitors (Supporting Information, Figure S3A) and to provide an unbiased estimate of model predictivity (Figure 5D; Supporting Information, Figure S3B and Table S2). The molecular properties with the largest influence on the models are shown in Figure 5C. Again, a positive overall molecular charge is clearly important for the inhibitory effect (see, e.g., the fraction cationic and the average net positive charge at pH 7.4, and the negative influence of descriptors related to negative charge). Also, lipophilicity (Moriguchi octanol-water partition coefficient) and, to a lesser extent, molecular size and shape (e.g., the gravitational radius of gyration and the total structure connectivity index) are important determinants of OCT2 inhibition.
Despite the clear importance of positive charge for inhibitory effect, many inhibitors are predicted to be primarily unionized at pH 7.4 (n = 73; 30% of all inhibitors), and a few carry a negative charge (n = 15; 6%). A corresponding spread in descriptor values within the inhibitor group is also apparent in lipophilicity, size and shape related descriptors (Figure 5A), further demonstrating that OCT2 inhibitors can come from widely varying structural families, and suggesting that different classes of inhibitors may bind to different sites in the protein.
Identifying subclusters of OCT2 inhibitors
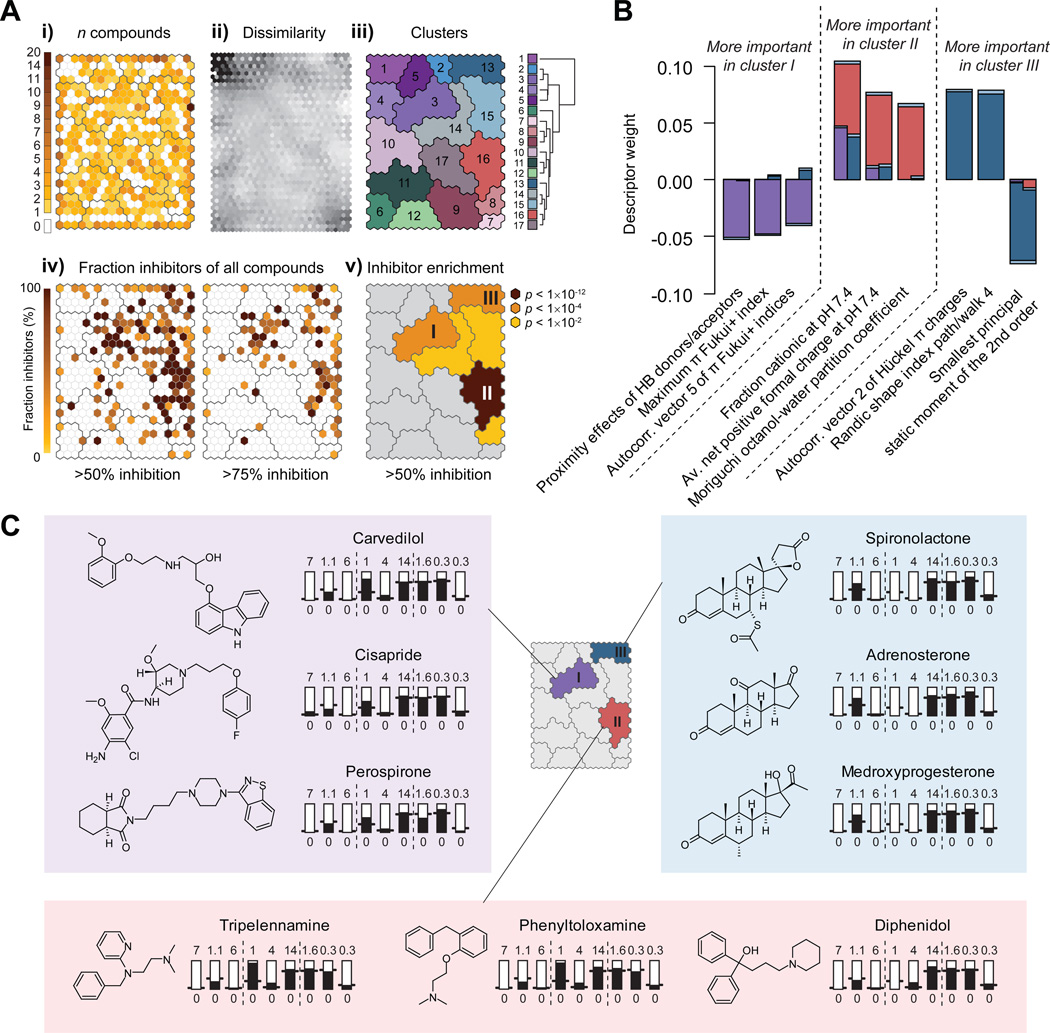
To examine if there was any underlying structure in the dataset that might explain the diversity of OCT2 inhibitors, we clustered all molecules in the dataset based on their similarity in molecular properties and visualized this as a self-organizing map (SOM) (Figure 6A). Each hexagon in the SOM is a cluster of related molecules, and similarity in molecular properties decreases with larger distances in the map. When visualizing the distances between adjacent hexagons (Figure 6Aii), larger clusters of similar molecules are evident; subsequent merging of hexagons that were not significantly different resulted in 17 final clusters of chemically similar compounds (Figure 6Aiii). While compounds of any activity are relatively uniformly distributed in the map (Figure 6Ai), the distribution of OCT2 inhibitors is shifted to the top-right part of the plot (Figure 6Aiv): 95% of all inhibitors are found in just 25% of the hexagons. Inhibitors primarily fall into three different groups; this pattern is more pronounced when a more stringent activity cutoff is applied (≥75% inhibition; Figure 6Aiv), and corresponds to three clusters significantly enriched with inhibitors (Figure 6Av).
Figure 6. Clustering of OCT2 inhibitory activity across the examined chemical library.
(A) Self-Organizing Map (SOM) clustering. Each hexagon is a cluster of similar molecules, and hexagons near each other are more similar than distant clusters. (Ai) Hexagons are colored according to the number of compounds they contain. (Aii) Hexagons are colored according to the similarity to their neighbors. Lighter color indicates greater similarity. (Aiii) Hexagons were merged into larger clusters if they were not significantly different (F-test p-value < 0.1). The hierarchical tree shows the chemical similarity relationships among the final clusters. (Aiv) Hexagons are colored according to the fraction of the compounds that inhibit at least 50% (left-hand SOM) or 75% (right-hand SOM) of OCT2-mediated transport. (Av) Enrichment of OCT2 inhibitors. Cluster colors are scaled to the negative log10 of the uncorrected hypergeometric test p-values. Clusters with enrichment p-values > 0.05 are colored grey. (B) Molecular descriptors specifically important for inhibitors in clusters I, II and III. The descriptors shown are the nine with the most significantly different PLS regression coefficients among separate models of OCT2 inhibitors in clusters I, II or III, respectively. Bars show mean PLS regression coefficients for cluster I (purple), II (red) and III (blue); standard errors are shown in light blue. (C) Structural differences between molecules in clusters I, II and III. Thermometer plots show molecular properties of prototypical members of each cluster, with descriptors in the same order as in (B).
We derived separate structure-activity relationships for each of these clusters. All compounds in the dataset were assigned to their nearest cluster (resulting in 261, 431 and 208 compounds in clusters I, II and III, respectively, of which 67, 127 and 49 inhibit OCT2), and models discriminating between inhibitors and non-inhibitors in each of these were developed (Supporting Information, Figure S4A). Similarities as well as differences are apparent upon closer examination of the most important molecular descriptors in the cluster-specific models (Figure 6B; Supporting Information, Figure S4A). Charge-related descriptors have an especially dominating influence in cluster II, and the lowest influence in cluster III. Size and shape-related descriptors play more pronounced roles in clusters I and III (Supporting Information, Figure S4A).
Prototypical inhibitors from each cluster are shown in Figure 6C. Perhaps most striking are the considerable differences in molecular size and flexibility among the clusters. In cluster I, inhibitors are generally more elongated than non-inhibitors, as judged by the positive influence of asphericity, the geometrical radius of gyration and the largest second-order principal static moment, and the negative influence of the medium second-order principal static moment (Supporting Information, Figure S4A). This is in contrast to inhibitors in clusters II and III, which tend to be smaller and more globular in shape. The size distributions in each cluster are visualized in the Supporting Information (Figure S4B), showing that the average inhibitor in cluster I is almost twice as long as other inhibitors. The majority of known transported substrates of OCT2 are in the size range of cluster II, and are assigned to this cluster based on their molecular properties (Supporting Information, Table S3).
Discussion
More than two million severe adverse drug reactions are estimated to occur in the United States each year, a quarter of which are attributable to interactions between co-administered drugs.18–20 An increasing number of studies report DDIs resulting from the inhibition of membrane transporters, adding to the complexity of drug safety assessment. To date, however, we lack the arsenal of selective probes necessary to mechanistically interpret transporter-mediated DDIs. Further, to our knowledge there have been no large screens of registered drugs to prospectively predict transporter based DDIs.
Here, we sequentially assessed inhibition of the renal organic cation transporter OCT2, counter-screened for inhibition of other major transporters, and prioritized hits based on their clinical plasma concentrations. Eighty-nine of the 244 identified OCT2 inhibitors are among the top 200 most prescribed drugs in the United States (IMS Health, accessed at http://www.rxlist.com). For several FDA-approved drugs OCT2 inhibition would occur near the therapeutically active plasma concentrations, meriting caution when co-administered with drugs whose renal excretion relies on OCT2 transport. Recently, the International Transporter Consortium proposed criteria that would trigger a clinical DDI study based on in vitro interaction studies with membrane transporters.21 Because of enormous interspecies differences in the tissue distribution and substrate and inhibitor specificity of transporters, the International Transporter Consortium recommended that clinical studies, rather than animal studies be conducted. Six drugs in our screen met these criteria for inhibition of OCT2, including cimetidine, which is the most extensively studied inhibitor of renal organic cation transport and the perpetrator in many of the reported clinical renal DDIs.5, 22–25 For inhibitors of OCT2, metformin was recommended as a model substrate in a clinical study. Clearly, based on our studies, such studies are warranted.
Using computational analyses, we showed that inhibitors of OCT2 are found across multiple structural and pharmacological classes and that inhibitor potency is not associated with substructures that define series of structurally related compounds, but rather to broader molecular properties like positive charge, lipophilicity, molecular size and flexibility. This is similar to the structure-inhibition relationship for the hepatic organic cation transporter OCT1, for which lipophilicity, positive atomic charge and hydrogen bond donors were predictive of inhibitors.26 Compounds from three distinct clusters that differ in molecular properties resulted in OCT2 inhibition, suggesting that the clusters may represent complementary inhibitory mechanisms. For example, known transported substrates of OCT2 are almost exclusively found in cluster II (Figure 6Av; Supporting Information, Table S3), suggesting competitive binding to the transport binding site of cluster II inhibitors; further, corticosterone, which is prototypical of the structures in our cluster III, results in an allosteric modulation of substrate binding to OCT2 and OCT1;27 and inhibitors in cluster I are distinct from the others in their larger size and flexibility, implying that inhibition may be caused by occlusion of the substrate binding site as suggested in crystallographic experiments for LeuT, a bacterial homologue of human SLC6 transporters.28–29 The pronounced importance of positive charge for the tentatively substrate-like inhibitors in cluster II is in line with the demonstrated tendency for substantial renal secretion of charged drugs.1 In contrast, lipophilicity is not a hallmark of actively secreted drugs;1, 30 its importance for the inhibitory endpoint studied here is however not surprising, given the well-established influence of hydrophobic interactions between ligands and proteins.
Notably, only 7 of the 52 compounds that inhibited OCT2 near the total clinical plasma concentration also inhibited its hepatic paralog OCT1. Because the two transporters are highly homologous especially in regions where circulating inhibitors are likely to bind, our data imply that the overall homology between OCT2 and OCT1 does not directly reflect homology in the binding site. For example, a single-residue switch in rabbit OCT2, from the OCT2 specific glutamate to the glutamine of OCT1, altered cimetidine binding to an OCT1-like phenotype.31 Our results also suggest that the affinity for OCT1 is consistently lower than for OCT2. A previous report of OCT1 inhibitors among registered drugs confirms this phenomenon (Supporting Information, Figure S2)26: the majority of the compounds that specifically inhibited OCT2 at 20µM also inhibit OCT1 at the higher concentration of 100µM. We suggest that the hepatic OCT1 transporter, which experiences high portal vein concentrations of orally absorbed compounds, through evolutionary mechanisms is less susceptible to circulating inhibitors.
Six drugs were identified that inhibit OCT2 at clinical concentrations of free drug, with inhibition potencies ranging from 0.6 to 23 µM (Table 1; Figure 4C). Of interest is that 5 of the potential in vivo probe inhibitors are eliminated exclusively (>95%) through metabolism (Supporting Information, Table S1). Previously, inhibitors of renal excretion of basic drugs were thought to be eliminated largely by the kidney. Our data suggest that drugs that are extensively metabolized may also be important inhibitors of renal secretion of basic drugs. Interestingly, we confirmed that cimetidine is a more potent inhibitor of the apically located MATE transporters than of OCT2 (Table 1; Figure 4C).32– 33 Our in vitro data for MATE1 and MATE2-K, in addition to those for OCT2 (Figure 4Cvii), suggest the necessity of conducting a clinical trial based on the proposed guidelines.21
Table 1.
Potency of putative clinical inhibitors in HEK293 cells expressing reference OCT2, the genetic variant OCT2-A270S, OCT1, MATE1 or MATE2-K.
| Compound | OCT2-Ref | OCT2-A270S | OCT1 | MATE1 | MATE2-K | Cmaxa (µM) |
PBb (%) |
|
|---|---|---|---|---|---|---|---|---|
| Disopyramide | IC50c (µM) | 2.9 | 3.9 | > 100 | 66 | > 100 | 7.95 | 50 |
| (Cmax,ud/ IC50) | (1.35) | (1.02) | (N. C.)e | (0.06) | (N. C.) | |||
| Dipyridamole | IC50 (µM) | 2.6 | 3.8 | 81 | 26 | 74 | 3.96 | 90a |
| (Cmax,u/ IC50) | (0.15)f | (0.10) | (0.005) | (0.02) | (0.01) | |||
| Imipramine | IC50 (µM) | 0.60 | 2.1 | 37 | 10 | > 100 | 0.71 | 90 |
| (Cmax,u/ IC50) | (0.12) | (0.03) | (0.003) | (0.01) | (N. C.) | |||
| Tacrine | IC50 (µM) | 0.68 | 1.2 | 83 | 1.1 | > 100 | 0.17 | 55 |
| (Cmax,u/ IC50) | (0.11) | (0.07) | (0.001) | (0.07) | (N. C.) | |||
| Orphenadrine | IC50 (µM) | 2.5 | 4.3 | > 100 | 65 | > 100 | 0.31 | 20 |
| (Cmax,u/ IC50) | (0.10) | (0.06) | (N. C.) | (0.004) | (N. C.) | |||
| Ondansetron | IC50 (µM) | 0.89 | 2.8 | > 100 | 0.15 | 6.9 | 0.13 | 73 |
| (Cmax,u/ IC50) | (0.04) | (0.01) | (N. C.) | (0.23) | (0.01) | |||
| Cimetidine | IC50 (µM) | 23 | 20 | > 100 | 5.7 | 39 | 11.9 | 19 |
| (Cmax,u/ IC50) | (0.41) | (0.47) | (N. C.) | (1.69) | (0.25) |
Cmax, therapeutic plasma concentration;
PB, plasma protein binding;
IC50, half-maximum inhibitory concentration;
Cmax,u, therapeutic unbound plasma concentration;
N. C., not calculated because of low inhibitor affinity.
OCT2 governs the entry of many circulating toxins into the tubular epithelium, including the neurotoxin 1-methyl-4-phenylpyridinium (MPP+), the DNA intercalator ethidium and the herbicide paraquat,8, 34–35 with the cellular uptake typically coupled to efflux into the urine via MATEs.34 Selective inhibition of OCT2 or MATEs would thus result in opposing effects on cellular toxin accumulation, decreasing and exacerbating renal toxicity, respectively. Accordingly, protection from cisplatin-induced nephrotoxicity was recently demonstrated in OCT2-deficient mice, and the reduced-function genetic variant OCT2-A270S was associated with decreased acute nephrotoxicity in cisplatin-treated patients.36 The selective inhibitors identified here may thus hold therapeutic potential as cytoprotectants in anticancer drug therapy. It is noteworthy that the co-administration of an inhibitor of organic anion transport, probenecid, to reduce the renal toxicity of the antiviral agent cidofovir is recommended in the official product label.37–38
In summary, we present the largest available resource of organic cation transporter inhibition data, identifying 244 OCT2 inhibitors among prescription drugs with six potential drugs that can be used as in vivo probes of renal transport or in protection against drug-induced nephrotoxicity.
Experimental Section
Collection of clinical mass balance data
Data for the urinary recovery of radiolabeled drugs were collected from Goodman and Gilman’s the Pharmacological Basis of Therapy7 and from literature references obtained from a PubMed search using the key words [‘mass balance’ OR ((‘renal’ OR ‘urine’ OR ‘urinary’) AND (‘excretion’ OR ‘elimination’ OR ‘clearance’ OR ‘recovery’))]. The final dataset included 308 unique drugs with data on the urinary recovery of parent drug and/or total radiolabel.
Reagents
4-(4-(Dimethylamino)styryl)-N-methyl-pyridinium iodide (ASP+) was purchased from Molecular Probes. The ICONIX compound screening library was obtained through the Small Molecule Discovery Center, University of California San Francisco, CA. Compounds used in the concentration-dependent and specificity screens were obtained from Sigma-Aldrich. All compounds were of analytical grade and of at least 95% purity.
Cell culture
Flp-In-human embryonic kidney (HEK-293-Flp-In) cell lines stably expressing human OCT1, OCT2, OCT2-A270S, MATE1 and MATE2-K were previously established in our laboratory.10, 34 Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 µg/ml streptomycin and 60 µg/ml hygromycin, at 37°C in a humidified atmosphere with 5% CO2.
Fluorescence uptake assay
Cells were seeded in black poly-D-lysine-coated 96-well plates (Greiner Bio-One, Frickenhausen, Germany) at 45000 cells/well 48 h prior to experiments. Before the assay, cells were washed twice in room temperature assay buffer (HBSS with 5.6 mM d-glucose, pH 7.4). Uptake was initiated by application of uptake buffer containing 1–125 µM of the fluorescent substrate ASP+ (with or without inhibitor) and incubated at room temperature. After 3 minutes, substrate uptake was stopped by aspirating the reaction mixture and washing the cells two times with HBSS with inhibitor (OCT2, OCT2-A270S and MATE1: 500µM cimetidine; OCT1: 500µM verapamil; MATE2-K: 250µM ondansetron). All compounds were analyzed in triplicate. The intensity of accumulated ASP+ fluorescence was measured using an Analyst AD plate reader (Molecular Devices, Sunnyvale, CA) with excitation and emission filters at 485 nm and 500–580 nm wavelength, respectively.
Transport kinetics were characterized by measuring the uptake of increasing substrate concentrations in transporter transfected cells and cells transfected with the empty vector. After subtracting out the non-specific transport, residual rates were fitted to the Michaelis-Menten equation: V = Vmax × S / (Km + S), where Vmax is the maximum transport rate, Km is the substrate concentration resulting in half-maximum uptake rate, and S is the concentration of ASP+, using GraphPad Prism version 5.01 (GraphPad Software Inc., San Diego, CA). Z' assay sensitivity factors were calculated according to Reference 39: Z' = 1 - (3 × sdsample + 3 × sdcontrol) / (meansample – meancontrol).
Transporter inhibition assay
Assay buffers were prepared by diluting 1 mM DMSO stock solutions with HBSS (pH 7.4) containing ASP+ (5 µM) to a final concentration of 20 µM (2% DMSO). For the screens against OCT1, OCT2, OCT2-A270S, MATE1 and MATE2-K, solid drug material obtained separately was used to prepare 2 mM DMSO stock solutions, which were diluted to a final concentration of 20 µM (1% DMSO). Non-specific transport was determined in separate wells on each assay plate using high concentrations of inhibitor: OCT2, OCT2-A270S and MATE1: 500 µM cimetidine; OCT1: 500 µM verapamil; MATE2-K: 250 µM ondansetron. In separate experiments, these conditions resulted in complete inhibition of transporter activity (Figure 2Biii; Figure 4Cvi). After subtracting out the non-specific transport, residual transport rates were used for further calculations.
Half-maximum inhibitory concentrations (IC50) were estimated from the screening inhibition measurements as: V = V0 / [1 + (I / IC50)], where V and V0 are the activity with and without inhibitor, respectively, and I is the inhibitor concentration of 20 µM. This approach is expected to give reasonable inhibitor affinity estimates when the screening concentration is within the linear part of the IC50 curve (approximately 20–80% inhibition, corresponding to IC50 values between 5 and 80 µM in the present study); this was subsequently confirmed by a good concordance between estimated and experimentally determined IC50 values (70% within 0.5 log unit, 100% within 1 log unit). The estimated IC50 values were compared with plasma concentration data collected from the literature, e.g., References 7 and 40–41.
Experimental IC50s were measured as the uptake of ASP+ in the presence of increasing concentrations of inhibitor. Data were fit using nonlinear regression to the equation V = V0 / [(1 + (I / IC50)n], where V and V0 are the ASP+ uptake rates in the presence and absence of the inhibitor, respectively, I is the inhibitor concentration and n is the slope.
Substructure fragment analysis
Substructure fragments enriched in OCT2 inhibitors were determined using MoFa version 2.2.42 Multiple runs were performed using alternative algorithm settings, matching or ignoring aromaticity; handling five- and six-membered rings as separate units or as individual atoms; and applying or omitting branch pruning with respect to perfect extensions. The statistical significance of substructure enrichment in inhibitors compared to all molecules was assessed using the hypergeometric test.
Molecular descriptor generation
Three-dimensional molecular structures were generated from SMILES representations using Corina version 3.0 (Molecular Networks, Erlangen, Germany), keeping the lowest energy conformation of a maximum of 100 alternative ring conformations, and were used as input for molecular descriptor calculation with DragonX version 1.4 (Talete, Milan, Italy), ADMETPredictor version 5.0 (SimulationsPlus, Lancaster, CA) and MAREA version 3.02.43 After removal of replicate molecular descriptors and descriptors having zero variance, 347 descriptors remained and were used for cluster analyses and as the starting point for structure-activity model development. Of the experimentally examined library, 10 compounds were excluded from further computational analyses because defined molecular structures were unavailable (e.g., teicoplanin) or because the structures contained non-parameterized features (e.g., strontium chloride).
Structure-activity modeling
Partial least squares discriminant analysis (PLS-DA) was used to develop computational models that differentiate between OCT2 inhibitors and non-inhibitors based on differences in molecular descriptor values. A double-loop cross-validation (CV) procedure was used to provide an unbiased estimate of the prediction accuracy (Supporting Information, Figure S3A).44 Variable selection was performed in two phases: first, the descriptors with lowest absolute PLS weight were iteratively removed until only the twenty-five most important ones remained; second, the same procedure was repeated, but descriptors were kept in the model if removal resulted in an inferior model. The entire double-loop procedure was repeated 100 times for different random partitionings of the dataset to enable calculation of confidence intervals of prediction accuracy estimates and model parameters. A permutation procedure was used to assess the statistical significance of the final models: the order of the dependent variable was randomly shuffled 1000 times, and the predictions of the permuted datasets were compared to those for the original dataset. All models gave predictions (as assessed by the area under the receiver operating characteristic) well outside the range of background probabilities from the randomized datasets (permutation p-values < 0.001; corresponding p-values assuming normally distributed background predictions < 10−37), showing that the model predictions are highly unlikely to be due to chance correlations.
Self-organizing map clustering
Compounds were clustered using the Self-Organizing Map (SOM) algorithm (SOM_PAK version 3.1; http://www.cis.hut.fi/research/som_lvq_pak.shtml), based on the similarity (Euclidean distances) of their molecular descriptor values (centered and scaled to unit variance). Twenty randomly seeded 18×25 hexagonal topology maps were calculated using a Gaussian neighborhood, a 10,000 step initial phase (rate: 0.5; 10 cell initial radius; linear decrease) and a 100,000 step final phase (rate: 0.02; 4 cell initial radius; linear decrease). The SOM with lowest average quantization error was used in subsequent analyses. Hexagons were subsequently merged into larger clusters (iteratively, in the order of merges in a hierarchical tree calculated from the hexagon mean vectors), if the between-to-within-cluster variance ratio of molecular properties was not significantly different (as judged by an F-test p-value < 0.1). Enrichment of OCT2 inhibitors in each of the final merged clusters was assessed using the hypergeometric test.
Supplementary Material
Acknowledgements
This research was supported by National Institutes of Health grants GM36780 and GM61390. We thank Steven Chen, UCSF Small Molecule Discovery Center, for help with assay automation and SimulationsPlus for providing access to the ADMETPredictor software.
Abbreviations
- DDI
drug-drug interaction
- HEK293
human embryonic kidney cell line
- MATE
multidrug and toxic compound extrusion
- OCT
organic cation transporter
- PLS-DA
partial least squares discriminant analysis
- SLC
solute carrier
- SOM
self-organizing map
Footnotes
Supporting Information Available:
Development and characterization of screening assays for additional major organic cation transporters; molecular structures and experimentally determined and computationally predicted OCT2 inhibition for all 910 evaluated compounds; comparison of inhibitor specificity between OCT2 and the hepatic paralog OCT1; computational modelling procedure and statistics for models of OCT2 inhibition; molecular properties of OCT2 inhibitors in each of the three inhibitor clusters; assignment of reported transported OCT2 substrates in the three ligand clusters. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Varma MV, Feng B, Obach RS, Troutman MD, Chupka J, Miller HR, El-Kattan A. Physicochemical determinants of human renal clearance. J. Med. Chem. 2009;52:4844–4852. doi: 10.1021/jm900403j. [DOI] [PubMed] [Google Scholar]
- 2.Cundy KC. Clinical pharmacokinetics of the antiviral nucleotide analogues cidofovir and adefovir. Clin. Pharmacokinet. 1999;36:127–143. doi: 10.2165/00003088-199936020-00004. [DOI] [PubMed] [Google Scholar]
- 3.Jaehde U, Sorgel F, Reiter A, Sigl G, Naber KG, Schunack W. Effect of probenecid on the distribution and elimination of ciprofloxacin in humans. Clin. Pharmacol. Ther. 1995;58:532–541. doi: 10.1016/0009-9236(95)90173-6. [DOI] [PubMed] [Google Scholar]
- 4.Hedaya MA, Elmquist WF, Sawchuk RJ. Probenecid inhibits the metabolic and renal clearances of zidovudine (AZT) in human volunteers. Pharm. Res. 1990;7:411–417. doi: 10.1023/a:1015835826114. [DOI] [PubMed] [Google Scholar]
- 5.Somogyi A, Stockley C, Keal J, Rolan P, Bochner F. Reduction of metformin renal tubular secretion by cimetidine in man. Br. J. Clin. Pharmacol. 1987;23:545–551. doi: 10.1111/j.1365-2125.1987.tb03090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fletcher CV, Henry WK, Noormohamed SE, Rhame FS, Balfour HH., Jr The effect of cimetidine and ranitidine administration with zidovudine. Pharmacotherapy. 1995;15:701–708. [PubMed] [Google Scholar]
- 7.Brunton L, Lazo J, Parker K, editors. Goodman and Gilman's The Pharmacological Basis of Therapeutics. 11th ed. New York, NY: McGraw-Hill, Inc.; 2005. [Google Scholar]
- 8.Gorboulev V, Ulzheimer JC, Akhoundova A, UlzheimerTeuber I, Karbach U, Quester S, Baumann C, Lang F, Busch AE, Koepsell H. Cloning and characterization of two human polyspecific organic cation transporters. DNA Cell Biol. 1997;16:871–881. doi: 10.1089/dna.1997.16.871. [DOI] [PubMed] [Google Scholar]
- 9.Motohashi H, Sakurai Y, Saito H, Masuda S, Urakami Y, Goto M, Fukatsu A, Ogawa O, Inui K. Gene expression levels and immunolocalization of organic ion transporters in the human kidney. J. Am. Soc. Nephrol. 2002;13:866–874. doi: 10.1681/ASN.V134866. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Li S, Brown C, Cheatham S, Castro RA, Leabman MK, Urban TJ, Chen L, Yee SW, Choi JH, Huang Y, Brett CM, Burchard EG, Giacomini KM. Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet. Genomics. 2009;19:497–504. doi: 10.1097/FPC.0b013e32832cc7e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, Owen RP, Sheardown SA, Yue L, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin. Pharmacol. Ther. 2008;83:273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang SZ, Lovejoy KS, Shima JE, Lagpacan LL, Shu Y, Lapuk A, Chen Y, Komori T, Gray JW, Chen X, Lippard SJ, Giacomini KM. Organic cation transporters are determinants of oxaliplatin cytotoxicity. Cancer Res. 2006;66:8847–8857. doi: 10.1158/0008-5472.CAN-06-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung N, Lehmann C, Rubbert A, Knispel M, Hartmann P, van Lunzen J, Stellbrink HJ, Faetkenheuer G, Taubert D. Relevance of the organic cation transporters 1 and 2 for antiretroviral drug therapy in human immunodeficiency virus infection. Drug Metab. Dispos. 2008;36:1616–1623. doi: 10.1124/dmd.108.020826. [DOI] [PubMed] [Google Scholar]
- 14.Neuvonen PJ, Niemi M, Backman JT. Drug interactions with lipid-lowering drugs: Mechanisms and clinical relevance. Clin. Pharmacol. Ther. 2006;80:565–581. doi: 10.1016/j.clpt.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 15.US Food and Drug Administration. Highlights of Prescribing Information: Tracleer (Bosentan) US FDA website [online] 2001 http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/021290s012lbl.pdf.
- 16.US Food and Drug Administration. Guidance for Industry. Drug Interaction Studies - Study Design, Data Analysis, and Implications for Dosing and Labeling. US FDA website [online] 2006 http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm072101.pdf.
- 17.European Medicines Agency. Guideline on the Investigation of Drug Interactions. EMEA web site [online] 2010 http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2010/05/WC500090112.pdf.
- 18.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients - A meta-analysis of prospective studies. JAMA. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 19.Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, Farrar K, Park BK, Breckenridge AM. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18,820 patients. Br. Med. J. 2004;329:15–19. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wienkers LC, Heath TG. Predicting in vivo drug interactions from in vitro drug discovery data. Nat. Rev. Drug Discov. 2005;4:825–833. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- 21.Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, Hoffmaster KA, Ishikawa T, Keppler D, Kim RB, Lee CA, Niemi M, Polli JW, Sugiyama Y, Swaan PW, Ware JA, Wright SH, Yee SW, Zamek-Gliszczynski MJ, Zhang L. Membrane transporters in drug development. Nat. Rev. Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abel S, Nichols DJ, Brearley CJ, Eve MD. Effect of cimetidine and ranitidine on pharmacokinetics and pharmacodynamics of a single dose of dofetilide. Br. J. Clin. Pharmacol. 2000;49:64–71. doi: 10.1046/j.1365-2125.2000.00114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng B, Obach RS, Burstein AH, Clark DJ, de Morais SM, Faessel HM. Effect of human renal cationic transporter inhibition on the pharmacokinetics of varenicline, a new therapy for smoking cessation: an in vitro-in vivo study. Clin. Pharmacol. Ther. 2008;83:567–576. doi: 10.1038/sj.clpt.6100405. [DOI] [PubMed] [Google Scholar]
- 24.Shiga T, Hashiguchi M, Urae A, Kasanuki H, Rikihisa T. Effect of cimetidine and probenecid on pilsicainide renal clearance in humans. Clin. Pharmacol. Ther. 2000;67:222–228. doi: 10.1067/mcp.2000.104018. [DOI] [PubMed] [Google Scholar]
- 25.Somogyi AA, Bochner F, Sallustio BC. Stereoselective inhibition of pindolol renal clearance by cimetidine in humans. Clin. Pharmacol. Ther. 1992;51:379–387. doi: 10.1038/clpt.1992.37. [DOI] [PubMed] [Google Scholar]
- 26.Ahlin G, Karlsson J, Pedersen JM, Gustavsson L, Larsson R, Matsson P, Norinder U, Bergstrom CA, Artursson P. Structural requirements for drug inhibition of the liver specific human organic cation transport protein 1. J. Med. Chem. 2008;51:5932–5942. doi: 10.1021/jm8003152. [DOI] [PubMed] [Google Scholar]
- 27.Gorboulev V, Shatskaya N, Volk C, Koepsell H. Subtype-specific affinity for corticosterone of rat organic cation transporters rOCT1 and rOCT2 depends on three amino acids within the substrate binding region. Mol. Pharmacol. 2005;67:1612–1619. doi: 10.1124/mol.104.008821. [DOI] [PubMed] [Google Scholar]
- 28.Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A Competitive Inhibitor Traps LeuT in an Open-to-Out Conformation. Science. 2008;322:1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Z, Zhen J, Karpowich NK, Law CJ, Reith MEA, Wang DN. Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures. Nat. Struct. Mol. Biol. 2009;16:652–657. doi: 10.1038/nsmb.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng B, LaPerle JL, Chang G, Varma MV. Renal clearance in drug discovery and development: molecular descriptors, drug transporters and disease state. Expert Opin. Drug Metab. Toxicol. 2010;6:939–952. doi: 10.1517/17425255.2010.482930. [DOI] [PubMed] [Google Scholar]
- 31.Zhang XH, Shirahatti NV, Mahadevan D, Wright SH. A conserved glutamate residue in transmembrane helix 10 influences substrate specificity of rabbit OCT2 (SLC22A2) J. Biol. Chem. 2005;280:34813–34822. doi: 10.1074/jbc.M506342200. [DOI] [PubMed] [Google Scholar]
- 32.Matsushima S, Maeda K, Inoue K, Ohta KY, Yuasa H, Kondo T, Nakayama H, Horita S, Kusuhara H, Sugiyama Y. The inhibition of human multidrug and toxin extrusion 1 is involved in the drug-drug interaction caused by cimetidine. Drug Metab. Dispos. 2009;37:555–559. doi: 10.1124/dmd.108.023911. [DOI] [PubMed] [Google Scholar]
- 33.Tsuda M, Terada T, Ueba M, Sato T, Masuda S, Katsura T, Inui K. Involvement of human multidrug and toxin extrusion 1 in the drug interaction between cimetidine and metformin in renal epithelial cells. J. Pharmacol. Exp. Ther. 2009;329:185–191. doi: 10.1124/jpet.108.147918. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Zhang S, Sorani M, Giacomini KM. Transport of paraquat by human organic cation transporters and multidrug and toxic compound extrusion family. J. Pharmacol. Exp. Ther. 2007;322:695–700. doi: 10.1124/jpet.107.123554. [DOI] [PubMed] [Google Scholar]
- 35.Lee WK, Reichold M, Edemir B, Ciarimboli G, Warth R, Koepsell H, Thevenod F. Organic cation transporters OCT1, 2, and 3 mediate high-affinity transport of the mutagenic vital dye ethidium in the kidney proximal tubule. Am. J. Physiol. Renal Physiol. 2009;296:F1504–F1513. doi: 10.1152/ajprenal.90754.2008. [DOI] [PubMed] [Google Scholar]
- 36.Filipski KK, Mathijssen RH, Mikkelsen TS, Schinkel AH, Sparreboom A. Contribution of organic cation transporteR2 (OCT2) to cisplatin-induced nephrotoxicity. Clin. Pharmacol. Ther. 2009;86:396–402. doi: 10.1038/clpt.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.US Food and Drug Administration. Highlights of Prescribing Information: Vistide (Cidofovir) US FDA website [online] 1999 http://www.accessdata.fda.gov/drugsatfda_docs/label/1999/020638s003lbl.pdf.
- 38.Lalezari JP, Kuppermann BD. Clinical experience with cidofovir in the treatment of cytomegalovirus retinitis. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1997;14:S27–S31. doi: 10.1097/00042560-199700001-00006. [DOI] [PubMed] [Google Scholar]
- 39.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 40.DiGregorio GJ, Barbieri EJ, editors. Handbook of commonly prescribed drugs with therapeutic, toxic and lethal levels. 19th ed. West Chester, PA: Medical Surveillance, Inc.; 2005. [Google Scholar]
- 41.Moffat AC, Osselton MD, Widdop B, Galichet LY, editors. Clarke's Analysis of Drugs and Poisons. 3rd ed. London, UK: Pharmaceutical Press; 2004. [Google Scholar]
- 42.Berthold MR, Hofer H, Borgelt C. Mining molecular fragments with mofa - Finding relevant substructures in sets of molecules. Abstr. Pap. Am. Chem Soc. 2003;225:U554–U554. [Google Scholar]
- 43.Matsson P, Bergstrom CA, Nagahara N, Tavelin S, Norinder U, Artursson P. Exploring the role of different drug transport routes in permeability screening. J. Med. Chem. 2005;48:604–613. doi: 10.1021/jm049711o. [DOI] [PubMed] [Google Scholar]
- 44.Freyhult E, Prusis P, Lapinsh M, Wikberg JE, Moulton V, Gustafsson MG. Unbiased descriptor and parameter selection confirms the potential of proteochemometric modelling. BMC Bioinformatics. 2005;6:50. doi: 10.1186/1471-2105-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barberi M, Merlin JL, Weber B. Sensitive determination of free and plasma protein-bound dipyridamole by high-performance liquid chromatography. J. Chromatogr. 1991;565:511–515. doi: 10.1016/0378-4347(91)80418-c. [DOI] [PubMed] [Google Scholar]
- 46.MacGregor TR, Sardi ED. In vitro protein binding behavior of dipyridamole. J. Pharm. Sci. 1991;80:119–120. doi: 10.1002/jps.2600800206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.