Abstract
Nucleosomes are the fundamental packing units of the eukaryotic genome. Understanding the dynamic structure of a nucleosome is a key to the elucidation of genome packaging in eukaryotes, which is tied to the mechanisms of gene regulation. CpG methylation of DNA is an epigenetic modification associated with the inactivation of transcription and the formation of a repressive chromatin structure. Unraveling the changes in the structure of nucleosomes upon CpG methylation is an essential step toward the understanding of the mechanisms of gene repression and silencing by CpG methylation. Here, we report single molecule and ensemble fluorescence studies to show how the structure of a nucleosome is affected by CpG methylation. The results indicate that CpG methylation induces tighter wrapping of DNA around the histone core accompanied by a topology change. These findings suggest that changes in the physical properties of nucleosomes induced upon CpG methylation may contribute directly to the formation of a repressive chromatin structure.
Nucleosomes are the fundamental packing units of the eukaryotic genome. Genes wrapped in nucleosomes are further compacted into chromatin and eventually into chromosome. The structural organization of the eukaryotic genome is associated with the regulated access to the genetic information and the protection of genes against physical damage1. A nucleosome core particle comprises ~147 bp DNA in a left-handed superhelical format with 1.65 turns around an octameric histone core containing two copies of H2A, H2B, H3, and H4 2. Covalent modifications of DNA and histones play important roles in regulating various genome activities.
Modifications of DNA that alter its physical properties will most likely affect the structure of nucleosomes3, 4. It has been suggested that the sequence dependent mechanics of wrapped nucleosomal DNA play a role in the positioning and dynamics of nucleosomes5. Recently, we reported that DNA methylation induces compaction and rigidity of the linker DNA region that may imply suppressed dynamics of nucleosome structure and consequently reduced gene accessibility6. DNA methylation typically takes place in a CpG dinucleotide context where the cytosine is methylated to 5-methylcytosine and it is associated with the formation of heterochromatin and silencing of genes critical for the regulation of growth and proliferation7, 8. Abnormal CpG methylation leads to fatal diseases and defective development. Most CpG-islands are hypomethylated in normal somatic tissues while most tumor types display hypermethylated CpG islands in tumor suppressor regions9–11. Methylation of CpG-islands found in promoter regions is important in gene repression during X-chromosome inactivation and in genomic imprinting12, 13.
Studies suggest that the extent of CpG methylation is closely tied to the modulated accessibility of genes by establishing and maintaining repressive or transcriptionally inactive structures of chromatin. CpG methylation has been implicated in the repressive chromatin mainly in two ways. Firstly, transcriptional repressors or related factors block transcription by binding to the methyl-CpG moiety14, 15. Secondly, the binding of transcription activation factors to DNA becomes inefficient due to the compaction and rigidity of the chromatin structure induced by CpG methylation16. Both hypotheses are supported by experimental evidence. However, a clear link between CpG methylation and the chromatin structure is yet to be unraveled. As nucleosomes are the fundamental building blocks of chromatin, the effects of CpG methylation on the structure of a nucleosome are at the core of this problem.
Here, we present effects of CpG methylation on the structure of the internal regions of nucleosomal DNA based on single molecule and ensemble averaging fluorescence measurements. Single molecule methods provide an efficient means to probe sub-population dynamics in a heterogeneous mixture of diverse species and states, which is particularly useful when the change of interest is small and easily lost during ensemble averaging. Utilizing this unique benefit of single molecule methods, we studied the changes in the structure of nucleosomes induced upon CpG methylation.
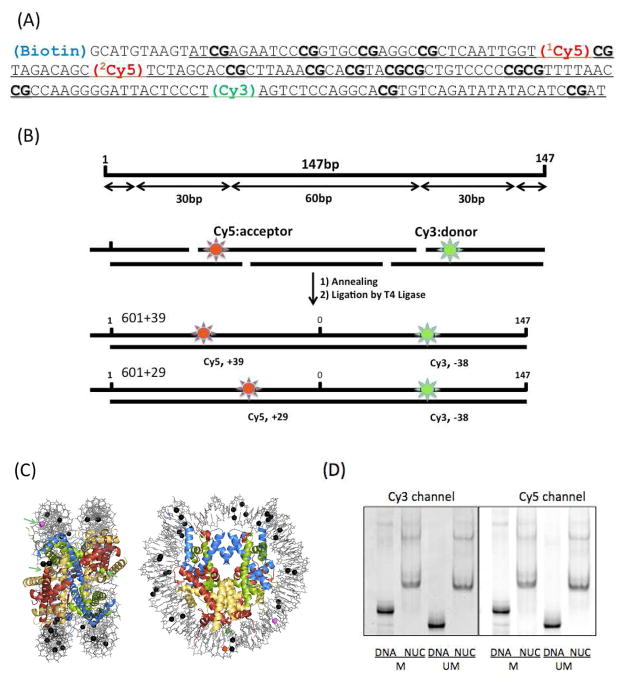
We designed two nucleosomal DNA constructs based on the Selex 601 sequence containing 15 CpG dinucleotides (Fig. 1A). The 601+39 DNA construct is labeled with Cy5 at the +39th nucleotide from the dyad and with Cy3 at the −38th nucleotide. The 601+29 DNA construct is labeled with Cy5 at the +29th nucleotide and with Cy3 at the −38th nucleotide (Fig. 1B & C). DNA was constructed by ligating oligonucleotides (IDT DNA, Coralville IA) some of which are already labeled with fluorophores or modified with methylation. Nucleosomes were reconstituted from the purified DNA constructs and Xenopus Laevis histones with yeast Nap117 and analyzed with native PAGE (5%) (Fig. 1D). Nucleosomes show one major band indicating homogenous positioning of histones along DNA. The methylated nucleosomal DNA displayed slow migration as compared to the unmethylated DNA, due to the increased mass with the additional 30 methyl groups. The nucleosomes assembled with the methylated DNA also show a lower mobility than the unmethylated nucleosomes.
Figure 1.
Nucleosomal DNA constructs based on the Selex 601 sequence and nucleosome reconstitution. (A) Selex 601 DNA sequence. Locations of Cy3 (FRET donor) and Cy5 (FRET acceptor) are marked. 1Cy5 is at the +39th base from the dyad (Cytosine, 74th) (601+39 DNA) and 2Cy5 is at the +29th base from the dyad (601+29 DNA). Cy3 is located at the −38th base from the dyad. Underlined sequence represents the double-strand DNA region. (B) The 147 bp DNA fragment comprises ~60 bp of (H3-H4)2 tetramer binding site at the center and ~30 bp of H2A-H2B dimer binding sites near the termini. (C) The locations of fluorophores (Cy3 (Green), 1Cy5 (Red), 2Cy5 (Purple)) and methylation (black) in a crystal structure (PDB: 3MVD) is shown in a sphere representation. (D) Unmethylated (UM) or methylated (M) nucleosomes were reconstituted with yeast nucleosome assembly protein 1 (Nap1) and analyzed on a 5% native polyacrylamide gel.
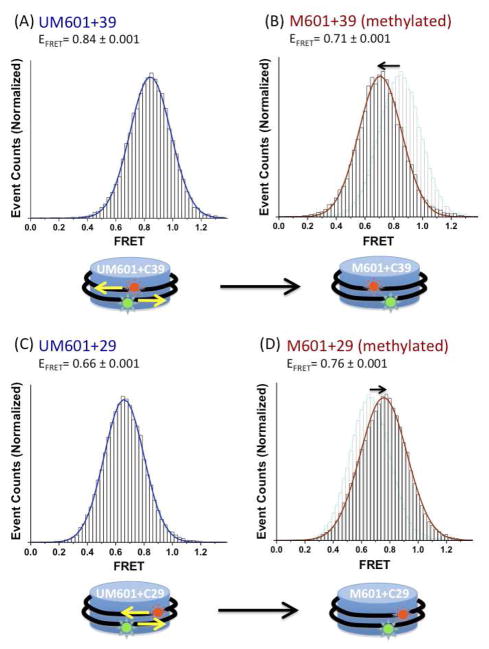
We immobilized the nucleosomes on a functionalized microscope slide via biotin-streptavidin conjugation and recorded the FRET signal from the individual nucleosomes. The FRET efficiency is approximated as Iacceptor/(Idonor+Iacceptor), where Idonor and Iacceptor denote the fluorescence intensities of the donor and the acceptor, respectively. Fig. S1 (Supporting Information) illustrates the typical FRET signals and Fig. 2 shows the FRET efficiency histograms of the two nucleosomes before and after CpG methylation.
FIGURE 2.
Effects of CpG methylation on the structure of assembled nucleosomes. FRET efficiency histograms of (A) unmethylated 601+39 nucleosomes, (B) methylated 601+39 nucleosomes, (C) unmethylated 601+29 nucleosomes, and (D) methylated 601+29 nucleosomes. The pale-colored histograms in (B) and (D) are the replicas of the histograms in (A) and (C), respectively.
Fig. 2 reveal that the nucleosomes show one major FRET efficiencies and the FRET efficiency of the M601+39 nucleosome (0.71) is lower than that of the unmethylated counterpart (UM610+39, 0.84), suggesting that the methylation affects the structure of a nucleosome. The decreased FRET efficiency of the methylated nucleosomes indicates the extent of DNA wrapping or the conformation of DNA is altered upon the methylation. As the increased distance between the two fluorophores in the 601+39 nucleosomes could result from either over- or under-wrapping of the DNA around the histone core, we further examined the FRET change with the 601+29 nucleosomes. The FRET efficiency of the UM601+29 nucleosome is 0.66, which is lower than 0.84 observed from the UM601+39 nucleosome, as expected according to the difference in the labeling positions of the fluorophores in the two constructs. The M601+29 nucleosomes display an increased FRET efficiency of 0.76. These results indicate that CpG methylation induces over-wrapping of DNA around the histone octamer in a nucleosome. Under- or over-wrapping of DNA would accompany a topology change as we previously reported on histone acetylated nucleosomes17. We examined the topology change of nucleosomal DNA upon CpG methylation by measuring fluorescence anisotropy of the FRET pair (Table 1). The results indicate a significant difference (7.0±1.5°) in the dipole alignment angle (β) between UM601+39 and M601+39 nucleosomes. Detailed information on the calculation is included in the Supporting Information. The decreased β from 46.7° to 39.7° upon the methylation should have resulted in increased FRET efficiency if the distance between the FRET pair did not change. The decreased FRET efficiency upon the methylation, therefore, confirms that the distance between the FRET pair must have been decreased. These results indicate that the topology of nucleosomal DNA changes upon CpG methylation, which accompanies over-wrapping.
Table 1.
Fluorescence anisotropy of unmethylated and methylated nucleosomes and the angle between the dipoles of the FRET pair.
| Excitation/Emission (nm) | 532/570 (Cy3) | 600/670 (Cy5) | 532/670 (Cy5 via FRET) | β (°) |
|---|---|---|---|---|
| UM601+39 | 0.33±0.01 | 0.33±0.01 | 0.07±0.01 | 46.7±0.97 |
| M601+39 | 0.34±0.01 | 0.33±0.01 | 0.13±0.01 | 39.7±1.12 |
We previously reported transient formation of a compact and rigid nucleosome structure induced by CpG methylation of 5S rDNA with M.SssI CpG methyltransferase6, which is supported by an earlier study that postulated stabilization of nucleosomal occupancy upon methylation18. This transient structure is represented by the shortening of the end-to-end distance in the linker DNA region, which suggests tight wrapping of DNA around the histone core. The current system enables us to investigate the structural changes in the internal regions of DNA upon CpG methylation. Multiple studies have reported that CpG methylation reduces the flexibility of free DNA19–21. For instance, C9pG10 methylation in a DNA dodecamer containing EcoRI restriction site reduces the flexibility of the phosphate-sugar backbone at the C9 position20. A 32mer nucleotide with the cAMP respective element (CRE) that has methylated nucleotides at the 8th and 16th positions showed increased stiffness possibly due to the restricted conformational space by the bulky methyl groups21.
As the rigidity of DNA is increased upon the methylation, we may expect that the nucleosomal DNA be less twisted in the methylated form. The change in the FRET efficiency upon the methylation indicates that CpG methylation induces over-wrapping of the internal regions of DNA around a histone octamer. Our results from the anisotropy measurements confirm the topology change upon CpG methylation, suggesting that the reduced extent of twist upon methylation causes the over-wrapping of DNA around the histone core. A Monte Carlo simulation study also suggests an under-winding of DNA with methylated CpG sequences resulting in an increased helical repeat from 10.5 to 11.0 bp/turn19. The increased helical repeat would imply that the histone content in a methylation rich region in the genome would be increased, which is beneficial to the formation of a repressive chromatin structure.
It has been reported that subtle rotational or translational changes of nucleosomal DNA in the promoter regions can play a role in gene regulation. For instance, the ability of a TATA-binding protein to bind to a nucleosomal target site depends on the rotational setting of the site22. A few base-pair shifted translational positioning of a nucleosome on the TATA box of a PHO5 gene displayed altered accessibility to the TATA box23. Therefore, the accessibility to a promoter region close to CpG islands can be sensitive to changes in DNA topology upon methylation, which may suggest another role for CpG hypermethylation in blocking promoter regions of tumor suppressors in cancerous cells10. The sequence used in our measurements has the GC content of 55% and the observed CpG/expected CpG of 1.29, which meet the classic definition of a CpG island24 except for that the length is shorter than 200bp.
Our study unraveled the effects of CpG methylation on the structure of nucleosomes: i) tighter wrapping of DNA around histones and ii) topology change of nucleosomal DNA, both of which can contribute to the formation of a repressive chromatin structure.
Supplementary Material
Acknowledgments
This research was funded by a Searle Scholar Award, a Henry and Dreyfus New faculty Award, and an NIH grant (R01GM097286) to TL.
Footnotes
Supporting Information. Figure S1 and supplementary methods are provided as the Supporting Information.
References
- 1.Kornberg RD, Klug A. Sci Am. 1981;244:52. doi: 10.1038/scientificamerican0281-52. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Anselmi C, Bocchinfuso G, De Santis P, Savino M, Scipioni A. Biophys J. 2000;79:601. doi: 10.1016/S0006-3495(00)76319-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Widom J. Quart Rev Biophys. 2001;34:269. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 5.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom AC, Field Y, Moore IK, Wang JPZ, Widom J. Nature. 2006;442:772. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH. J Am Chem Soc. 2010;132:1782. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein CB, Costa M. Mutat Res-Rev Mutat. 1997;386:163. doi: 10.1016/s1383-5742(96)00052-x. [DOI] [PubMed] [Google Scholar]
- 8.Bird A. Gene Dev. 2002;16:6. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 9.Bird AP. Nature. 1986;321:209. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M. Nat Rev Genet. 2007;8:286. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 11.Robertson KD. Oncogene. 2002;21:5361. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 12.Riggs AD, Pfeifer GP. Trends Genet. 1992;8:169. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 13.Takai D, Jones PA. Proc Natl Acad Sci USA. 2002;99:3740. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones PL, Veenstra GJC, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Nat Genet. 1998;19:187. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 15.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nature. 1998;393:386. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 16.Tate PH, Bird AP. Curr Opin Genet Dev. 1993;3:226. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 17.Lee JY, Wei S, Lee TH. J Biol Chem. 2011;286:11099. doi: 10.1074/jbc.M110.192047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA. Cancer Cell. 2007;12:432. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nathan D, Crothers DM. J Mol Biol. 2002;316:7. doi: 10.1006/jmbi.2001.5247. [DOI] [PubMed] [Google Scholar]
- 20.Geahigan KB, Meints GA, Hatcher ME, Orban J, Drobny GP. Biochemistry. 2000;39:4939. doi: 10.1021/bi9917636. [DOI] [PubMed] [Google Scholar]
- 21.Derreumaux S, Chaoui M, Tevanian G, Fermandjian S. Nucleic Acids Res. 2001;29:2314. doi: 10.1093/nar/29.11.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imbalzano AN, Kwon H, Green MR, Kingston RE. Nature. 1994;370:481. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Campa C, Politis P, Moreau JL, Kent N, Goodall J, Mellor J, Goding CR. Mol Cell. 2004;15:69. doi: 10.1016/j.molcel.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Gardiner-Garden M, Frommer M. J Mol Biol. 1987;196:261. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.