Abstract
The function of RNA depends on its ability to adopt complex and dynamic structures, and the incorporation of site-specific crosslinking probes is a powerful method for providing distance constraints that are valuable in RNA structural biology. Here we describe a new RNA-RNA crosslinking strategy based on Pt(II) targeting of specific phosphorothioate substitutions. In this strategy cis-diammine Pt(II) complexes are kinetically recruited and anchored to a phosphorothioate substitution embedded within a structured RNA. Substitution of the remaining exchangeable Pt(II) ligand with a nucleophile supplied by a nearby RNA nucleobase results in metal-mediated crosslinks that are stable during isolation. This type of crosslinking strategy was explored within the catalytic core of the Hammerhead ribozyme (HHRz). When a phosphorothioate substitution is installed at the scissile bond normally cleaved by the HHRz, Pt(II) crosslinking takes place to nucleotides G8 and G10 in the ribozyme active site. Both of these positions are predicted to be within ~8 Å of a phosphorothioate-bound Pt(II) metal center. Crosslinking depends on Mg2+ ion concentration, reaching yields as high as 30%, with rates that indicate cation competition within the RNA three-helix junction. Crosslinking efficiency depends on accurate formation of the HHRz tertiary structure, and crosslinks are not observed for RNA helices. Combined these results show promise for using kinetically inert Pt(II) complexes as new site-specific crosslinking tools for exploring RNA structure and dynamics.
Introduction
Posttranscriptional RNA processes underlie gene regulation in all forms of life. The complex and highly regulated ‘ribonome’ includes a growing list of noncoding RNAs whose regulatory functions may involve RNA-protein or small-molecule interactions, and accompanying changes in structure.1–3 Underlying these functions is the ability of the RNA biopolymer to fold into complex architectures. These dynamic structures scaffold molecular recognition as well as platform biomolecular catalysis, and give rise to RNA’s diverse roles in biology.
With modern genomic tools, the discovery of important RNA sequences has outpaced the determination of corresponding molecular structures.4,5 Current computational methods are fairly successful at predicting helical regions, even in very large RNAs. While higher-order structure prediction for large RNAs is gaining significant success,5 de novo structure prediction benefits greatly from experimentally-determined constraints either as input or for verification. Such constraints are also critical in validating RNA structure models obtained from biophysical methods including X-ray crystallography, cryo-EM, and small-angle X-ray scattering.6–8
Crosslinking reagents, chemical modification and hydroxyl-radical cleavage experiments are all used to obtain three-dimensional distance constraints for large RNAs.9–13 Selectively reactive crosslinking reagents have proven to be very informative.10–13 Through reaction with exogenous small molecules,10 synthetic installation of UV-sensitive nucleobase analogues,11 or reactive 2′-ribose substituents,12 or by simply capitalizing on the inherent photoreactivity of endogenous nucleobases,2a,11c,13 it is possible to create covalent crosslinks between nearby positions within a structured RNA. Subsequent identification of crosslinked partners establishes spatial relationships in large RNA and ribonucleoprotein (RNP) complexes with nucleotide-level precision. Major hurdles in the use of crosslinking reagents arise from the challenge of balancing reactivity and selectivity, along with the accompanying variability in yields.
The nucleic acid coordination properties of square-planar Pt(II) complexes14 make them attractive candidates as crosslinking reagents and have been well characterized due to the clinical success of cisplatin (cis-diamminedichloro Pt(II)) and related DNA-targeting Pt(II) antitumor drugs.15 Generally, Pt(II) complexes form kinetically inert adducts with soft biomolecular nucleophiles, and do so particularly well with the N7 position of purine bases in RNA and DNA, as well as with cysteine thiolate and histidine imidazole ligands in proteins. Pt(II)-complexes with two exchangeable coordination sites form stable bifunctional chelates between neighboring positions on a biomolecule. This well-characterized reactivity has inspired previous efforts to use Pt(II) complexes to probe DNA and RNA conformation,16 biomolecular contacts in ribonucleoprotein complexes,17 and recently, solvent exposure of nucleotides within the ribosome.18 The results from these studies show that Pt(II) reagents readily form inter- and intrastrand adducts in RNAs, and may do so in regions of complex structure. In order to capitalize on this reactivity, we sought to develop a targeted Pt(II)-based crosslinking strategy for RNA structure analysis.
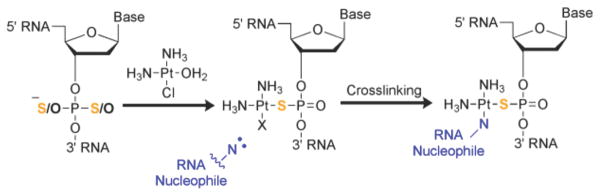
In developing a new Pt(II)-based crosslinking system we sought to bias kinetic targeting of Pt(II) metal complexes such that they were directed to a specific and predetermined position within a structured RNA (Scheme 1).19 Previous reports by Elmroth, Lippard and coworkers have shown that phosphorothioate substitutions (exchange of one of the non-bridging in the RNA or DNA phosphodiester backbone for a sulfur atom) are capable of recruiting Pt(II) complexes19 3–4 fold faster than equivalent, single-stranded GG targets.20 Similarly promising were early reports by Orgel and coworkers exploring the use of phosphorothioate and cysteamine substitutions in creating Pt(II) crosslinks at the termini of DNA duplexes.21 Building on these investigations we envisioned using phosphorothioate substitutions to recruit and anchor Pt(II) complexes to selected positions within pre-folded RNAs. A second ligand exchange reaction would then result in crosslinking between the selected site and nearby RNA ligands (Scheme 1), which could be identified by RNA mapping techniques. This method relies on phosphorothioate substitutions that are readily available from synthetic sources or can be biosynthetically incorporated. The resulting crosslinks represent proximity between phosphodiester and nucleobase moieties, and complement information available from photoaffinity methods that crosslink nucleobases. It is possible that the small cationic Pt(II) compounds may mimic natural metal ion cofactors, targeting electrostatic pockets in complex RNA structures while causing minimal structural perturbations.
Scheme 1.
Pt(II)-phosphorothioate RNA crosslinking.
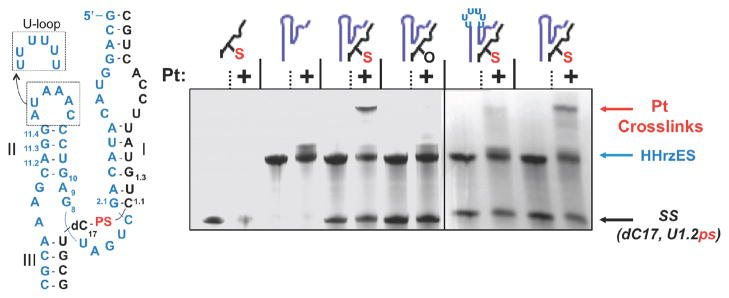
We chose a well-characterized RNA model in order to test this Pt(II)-crosslinking technique. The Hammerhead ribozyme is a catalytic RNA motif with a conserved core sequence that is predicted in untranslated regions of the genomes of many organisms, including humans.22–24 In plant viroids, the embedded self-cleaving HHRz sequence functions to cleave transcripts during rolling circle replication. For mechanistic studies in vitro, the HHRz has been adapted as a two-stranded construct in which a hammerhead ‘enzyme’ strand cleaves a complementary ‘substrate’ strand in trans. The early discovery and broad biological distribution of the HHRz have prompted its use as a model system for understanding chemical aspects of RNA catalysis.25 The functional core of the HHRz is formed at the junction of three helical regions of RNA and its arrangement is influenced by tertiary contacts formed between a flanking loop and receptor (Figure 1).23,26 Recent crystal structures of the HHRz have revealed a complex core architecture in which nucleobase and ribose moieties are seemingly well-aligned to catalyze the site-specific phosphotransesterification reaction carried out by the ribozyme.27 While significant attention has been directed at determining the detailed catalytic mechanism of the HHRz, the chemical roles played by specific nucleobase and sugar functionalities and, in particular, metal ion species in facilitating catalysis have been under active investigation.28 For this reason, the scissile bond of the HHRz represented an interesting location at which to test a new metal-mediated crosslinking strategy.
Figure 1.
Pt(II)-induced crosslinking of the Hammerhead ribozyme. a) The sequence of HHRzES-SS(dC17,C1.1ps) ribozyme used in this study (left) consists of an ’enzyme strand’ (ES, blue) and phosphorothioate substituted ’substrate strand’ (SS, black).’PS’ (red) denotes the phosphorothioate substitution installed at the HHRz cleavage site and U-loop depicts a non-folding mutant. On the right, a denaturing gel depicting crosslinks formed by modified HHrz components. Gel lanes from left to right (− or + platinum): (i) isolated PS-modified SS (ii) isolated HHRz enzyme strand, (iii) the PSsubstituted HHRzES-SS ribozyme, (iv) a non-PS-substituted HHrzES-SS ribozyme, (v) a PS-substituted HHHrzES-SS ribozyme with a folding-deficient U-loop mutation, (vi) a repeat experiment using the PS-substitute HHRzES-SS ribozyme. Conditions (see Methods): 40 μM RNA in 1mM Mg(NO3)2, 100 mM NaNO3, 10 mM Na2PO4, pH 7.0, 16 h, 37°C, ± 3 equivalents activated [cis-diammine Pt(II)] analyzed by 20% dPAGE and stained with methylene blue.
Results and Discussion
Pt(II)-PS crosslinks in the Hammerhead ribozyme active site
The properties of Pt(II) as a targeted RNA crosslinking agent were investigated using a two-piece HHrz construct comprised of ‘enzyme’ (HHRzES) and ‘substrate’ (SS) strands (Figure 1). When annealed in the presence of divalent cations, this complex catalyzes cleavage of the phosphodiester bond between C17 and C1.1. HHRz catalysis has been proposed to be facilitated by coordination of a metal ion to the pro-Rp oxygen at this phosphodiester bond,29,23,26 and so we were optimistic that a phosphorothioate substitution at this bond might capture a Pt(II) probe and produce crosslinks to nearby soft ligands. Pt(II) crosslinking studies were therefore performed with a C17-C1.1 phosphorothioate (PS) substitution (mixed Rp and Sp diastereomers), along with an inhibitory 2′-deoxy C17 modification to prevent cleavage during the study (SS(dC17, C1.1ps), Figure 1).
Initial crosslinking experiments used ionic conditions that promote global folding of the ribozyme (1 mM Mg2+, 100 mM Na+)30 and only three equivalents of cis-[Pt(NH3)2(OH2)Cl]+ in order to reduce off-target platination. In these conditions, interstrand crosslinks were indeed observed as higher molecular-weight products (Figure 1) that contain both HHRz strands (data not shown). Importantly, these products were not observed in the absence of the C1.1 phosphorothioate substitution, demonstrating the requirement for and likely involvement of the Pt(II)-thioate interaction in forming intermolecular crosslinks. Crosslinking from the C1.1 phosphorothioate position was confirmed through mild alkali hydrolysis mapping11,16c.31 of isolated crosslinks (Figure S1). In these experiments, the crosslinked site is identified by a gap in the RNA hydrolysis ladder that occurs 3′ to the crosslinked position and is due to formation of higher molecular-weight fragments that contain the crosslinked species. In an untreated sample, limited hydrolysis of the 5′-end labeled phosphorothioate-containing substrate strand shows the expected gap due to removal of the 2′-OH at dC17, but hydrolysis products beyond that position are observed. By contrast, in the crosslinked sample the hydrolysis ladder does not extend beyond dC17 due to the crosslink formed by its 3′-phosphorothioate and the 43-nucleotide enzyme strand (Figure S1).
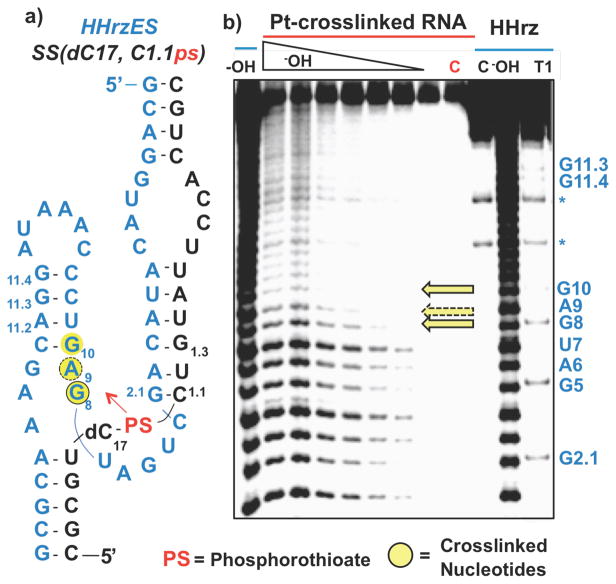
In order to identify the RNA nucleotide(s) trapped by the targeted Pt(II)-thioate probe, crosslinks generated using a 5′ end-labeled HHRz ES were isolated and similarly mapped. The resulting hydrolysis ladder shows a significant loss of products 3′ to U7, identifying G8 as a major site of Pt(II)-induced crosslinking (Figure 2b). These data are taken to indicate that Pt(II) is chelated between a C1.1 phosphorothioate and the N7 atom of G8. Interestingly, with increasing hydrolysis time, a second fall-off in cleavage intensity following A9 is also observed, indicating that Pt(II)-crosslinking to the N7 atom of G10.1 also takes place.
Figure 2.
a) HHRz secondary structure with crosslinked nucleobases in yellow. b) Cleavage products produced by alkali hydrolysis of the Pt-crosslinked HHRzES-SS(dC17, C1.1ps) complex formed using 5′ end-labeled HHRzES. Control lanes (blue overline): C: 5′ endlabeled HHRzES T1: G-specific sequence ladder generated by partial nuclease digestion with RNase T1 -OH: Reference alkali hydrolysis ladder. Platinum crosslinked lanes (red overline): C: isolated, Ptcrosslinked HHRz-SS(dC17, C1.1ps) –OH lanes: isolated, Ptcrosslinked HHRz-SS(dC17, C1.1ps) treated using alkali hydrolysis conditions for increasing amounts of time. Yellow arrows designate platinum crosslinking sites as indicated by loss of hydrolysis products 3′ to the site.
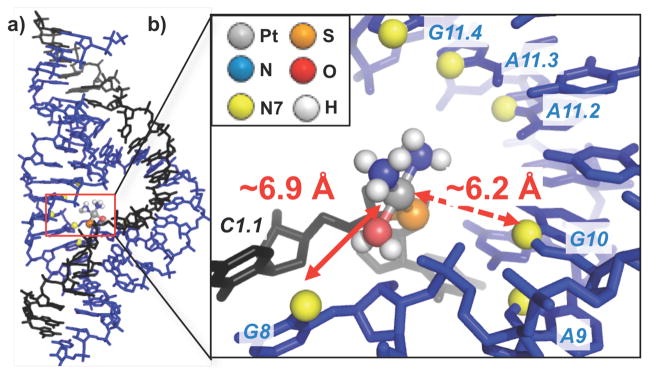
In order to understand the structural context for the Pt(II)-phosphorothioate/nucleobase crosslinks observed in this system, we used a HHRz crystal structure reported by Scott and coworkers27 to model the Pt(II) moiety and estimate distances to the observed crosslinking sites. Using HHRz structure PDB 2OEU27 (nearly identical to the sequence used in this study) we constructed a [Pt(NH3)2(OH2)] fragment bound to either the Rp (Figure 3) or Sp (Figure S2) stereoisomer of a C1.1 phosphorothioate. Holding the RNA structure static, molecular mechanics calculations were used to minimize conformational strain and steric clash between Pt(II) probe and the RNA. The resulting models for both phosphorothioate isomers show the N7 atoms of multiple purines, including G8 and G10.1 in the core, to be located ~6–11 Å away from the Pt(II) ion. In this seemingly target-rich coordination environment, the specificity of crosslinking to the G8 and G10.1 positions is intriguing. The steric influence from nonexchangeable cis-ammine ligands may have some influence; lowest energy structures of both Pt(II)-bound Rp and Sp isomers, calculated from several different starting geometries, all show the Pt(II) ion to be directed toward the interior of the ribozyme’s active site with the cis-diammine ligands oriented distal to the G8/G10.1 face. Once a [Pt(NH3)2]-aqua species is bound to the embedded phosphorothioate, crosslinking is expected to occur through an associative mechanism32 and may favor the N7 atoms of G8 or G10.1 due to additional factors, such as local structural flexibility and geometric alignment. Thus, based on comparison with predictions from crystallographic data, this Pt(II)-thioate crosslinking approach results in crosslinks between a phosphorothioate-bound Pt(II) complex and nucleobase ligands that are within ~6–8 Å. Further specificity may be conferred due to properties such as specific phosphorothioate isomer, steric interactions of the Pt(II)-ammine ligands, and local flexibility.
Figure 3.
a) Crystal structure of the HHRz27 (pdb: 2OEU) with modeled Pt(II) reagent. b) Structural model of Pt(NH3)2(OH2) bound to the Rp stereoisomer of a C1.1 phosphorothioate. Distances from Pt(II) to G8, the major site of PtII crosslinking, and additional site G10.1 are indicated. Distances to other purine N7 sites and model built with the Sp stereoisomer are given in Figure S2. Models generated as described in Methods.
Kinetics of Pt(II)-RNA crosslinking
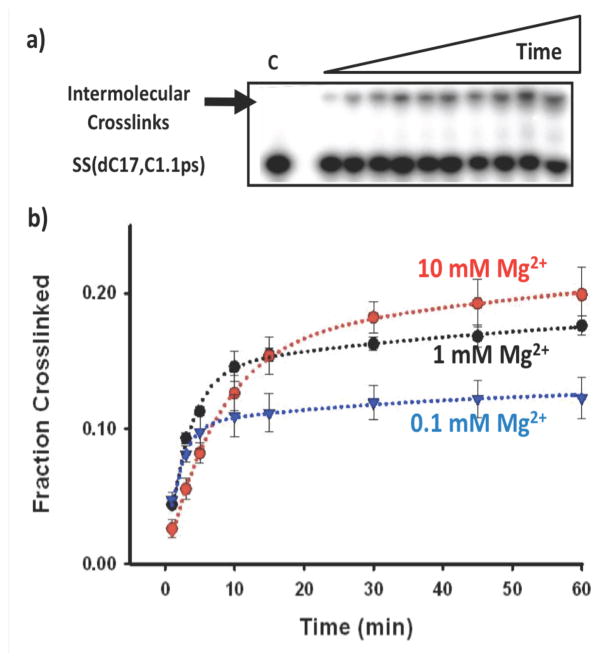
In order to better understand the influence of RNA structure and solution conditions, we characterized the kinetics of HHRz crosslinking at three Mg2+ concentrations and also tested the crosslinking efficiency of related RNA constructs. Crosslinking kinetics experiments were carried out by monitoring product formation over times ranging from 1 min to 8 h (Figure 3a). In all cases, crosslinking was 80% complete within twenty minutes of adding the Pt(II) reagent. This relatively fast timescale and the match with expectations based on crystallography, in conjunction with the control experiments described below, support the conclusion that the crosslinked product reflects near-native structure and is not the result of a large Pt(II)-induced conformational change in the RNA.33
The kinetic traces for crosslinked product formation were best fit using biphasic rate equations (Figures 3b and 3c) that reflect two noninteracting populations. The majority population undergoes relatively fast crosslinking, with Mg2+-dependent rate constants that decrease with increasing Mg2+ concentration. As described below, these rates are likely reporting on coordination of cis-[Pt(NH3)2(OH2)Cl]+ to the RNA site, followed by a fast crosslinking step. Based on the consistent rate of the slower population of 0.01–0.02 min−1 we suspect that this population reflects a rate-limiting step involving Pt(II) aquation equilibria.34
For the majority and faster population, in 1 mM Mg2+, crosslinking takes place with an observed rate of 0.34 ± 0.04 min−1 (t1/2= ~ 2 min). For purposes of comparison with other studies, a second-order rate constant can be estimated as 47 M−1 sec−1 35 Based on comparison with values from previous studies of Pt(II)-oligonucleotide interactions,20 this relatively fast reaction likely reflects initial binding of [Pt(II)(NH3)2OH2]+ to the embedded phosphorothioate ligand.This value is ~50-fold faster than previously reported rate constants for platination of phosphorothioate substitutions in 2- to 16-mer single-stranded DNAs, measured in similar conditions. The observation that the crosslinking rate measured for the HHRz is faster than the previously measured reactions monitoring platination of phosphorothioates in unstructured DNA oligonucleotides suggests that initial Pt(II)-sulfur coordination is the rate-limiting step in the HHRz crosslinking reaction, and that the electrostatics of structured RNA enhance this rate in the HHRz. Platination and crosslinking of 41-mer hairpin-like RNAs, closer in size to the 63-mer HHRz but lacking a phosphorothioate substitution, was previously measured to be ~2 M−1sec−1 under nearly identical ionic conditions.16c For comparison to the present study, this value can be multiplied by a ‘phosphorothioate factor’ of 3–4, which is the reported kinetic preference for platination of phosphorothioates versus nucleobase targets in single-stranded DNAs. The resulting estimate of ~8 M−1sec−1 for the RNA hairpins is still 5–6 times slower than observed here for a phosphorothioate embedded in the 3-helix junction of the HHRz. Taken together, the relatively fast crosslinking rates found in these studies seem to reflect the properties of the HHRz tertiary structure in recruiting metal ions. This hypothesis fits well with previous studies including computational modeling of the electrostatic topography of other 3-helix junction motifs.36
A rate-limiting step of Pt(II)-RNA (phosphorothioate) coordination would predict sensitivity to competing cations. Consistent with this, the rates of HHRz crosslinking decrease with increasing ionic strength. Opposing trends of decreased rates but increased crosslinking yields are observed with added Mg2+ (Figure 4 and Table 1) and likely reflect the dual influences of Mg2+ in both folding the HHRz and in metal ion competition for partitioning in the RNA ionic atmosphere. Mg2+ promotes global folding around the 3-helix junction of the HHRz with a K1/2 ~ 1 mM Mg2+ in 0.1 M NaCl,30 suggesting that the increase to 30% crosslinking yield in higher Mg2+ is due in part to higher population of globally folded molecules. In order to further address the tertiary organization necessary to crosslink the HHRz we characterized Pt(II) crosslinking using a “docking-deficient” HHRz mutant. In the HHrz(Uloop) construct (Figure 1), replacement of the helix II 5′-CAAAUA-3′ terminal loop with a series of U nucleotides interferes with accurate tertiary folding of the ribozyme and presumably organization of the molecule’s catalytic core.26,30a,37 A significantly lower yield of crosslinked product is observed for this folding-deficient construct (Figure 1). This finding, along with the Mg2+-dependent increase in crosslink yields of the native HHRz (Table 1) supports the interpretation that Pt(II) crosslinking in the core of the HHRz is dependent on accurate tertiary folding.
Figure 4.
Kinetic characterization of Pt(II)-crosslinking in the HHRzES-SS(dC17,C1.1ps) ribozyme construct. a) Typical autoradiograph obtained during kinetic studies demonstrating the appearance of crosslinked products over time. b) Kinetic traces obtained from crosslinking reactions carried out at varing Mg2+ concentrations. Conditions as in Figure 1 with Mg2+ concentration varied as indicated.
Table 1.
Pt(II)-phosphorothioate RNA crosslinking rate constants and yields. Conditions: 40 μM RNA in 1mM Mg(NO3)2, 100 mM NaNO3, 10 mM Na2PO4, pH 7.0, 120 μM [Pt(NH3)2(OH2)Cl]+, 37°C.
| [Mg2+] | Fast kobs (min−1) | Fast k2, calc (M−1s−1) | Slow kobs (min−1) | Slow k2, calc (M−1s−1) | Average Yield |
|---|---|---|---|---|---|
| 0.1 mM | 0.55 ± 0.19 | 76 ± 26 | 0.02 ± 0.02 | 3 ± 2 | 13% |
| 1 mM | 0.34 ± 0.04 | 47 ± 5 | 0.01 ± 0.01 | 1 ± 1 | 20% |
| 10 mM | 0.14 ± 0.01 | 19 ± 1 | 0.01 ± 0.01 | 1 ± 1 | 30% |
Structural determinants for Pt(II)-RNA crosslink formation
We sought to further examine the range of structural contexts in which Pt(II)-phosphorothioate crosslinking could take place. Pt(II) treatment of phosphorothioate substitutions at other positions in the HHRz as well as in unstructured oligonucleotides resulted in RNA cleavage at these sites (Figure S3). The presence of RNA cleavage productss with 2′-3′ cyclic phosphate termini suggests that Pt(II) coordination to a phosphorothioate substitution is capable of activating nucleophilic attack of the adjacent ribose 2′-OH. Thus, 2′-deoxy substitutions were used in conjunction with phosphorothioate substitutions in these studies (Scheme 1). Interestingly, although crosslinks were formed with a phosphorothioate at the HHRz cleavage site, movement of the phosphorothioate to the adjacent C1.1-U1.2 linkage did not result in intermolecular crosslinking (Figure S4). HHrz activity is tolerant to phosphorothioate substitution at this location,28a,38 suggesting minimal perturbation to the local geometry. Although crosslinks were not observed, [Pt(NH3)2] coordination to the SS(dC17, U1.2ps) substrate strand was confirmed using MALDI-MS, suggesting that the lack of crosslinking at this site was not due to a loss of Pt(II) coordination to the target phosphorothioate (data not shown). Molecular modeling of the HHRz-SS(dC17, U1.2ps) construct indicates that G8 and G10.1 may be >8 Å from a PS-coordinated Pt(II) probe (Figure S2). At this site, there is also the potential for a Pt(II) probe to form intramolecular adducts with the neighboring G1.3 nucleotide,39 an interaction that would preclude intermolecular crosslinking. The potential to form crosslinks between strands within A-form helices was then tested using both SS(dC17, U2.1ps) and SS(dC17, C1.1ps) RNAs and their complements (Figure S5). Crosslinks were not observed between strands in these base-paired models, as was previously found in attempts to use cis-diammine Pt(II) to crosslink internal phosphorothioates in fully based-paired, B-form DNA helices.21 As in DNA, intermolecular crosslinking between phosphorothioates and purine nucleobases on different strands of A-form RNA helices may be prohibited from crosslinking by this reagent by longer distances (~>10 Å) as well as increased rigidity. Because the phosphorothioate substitution embedded in the SS(dC17, C1.1ps) duplex is not adjacent to a G or A nucleotide, it seems unlikely that crosslinking in this construct is inhibited by the formation of intrastrand Pt(II)adducts. These findings all indicate that Pt(II) crosslinking may be selective for tertiary interactions and reflect proximity in near-native structures in complex RNAs.
Reversibility of Pt(II)-RNA crosslinks
For some applications, a desirable aspect of an RNA crosslinking reagent may be reversibility, facilitating sequence analysis following isolation of crosslinked species. Previous work has shown that thiourea reverses Pt(II)-cysteine-RNA crosslinks in RNA-protein complexes17a and Pt(II)-purine adducts in structured RNAs.40 As expected, incubation with thiourea also reverses the otherwise stable Pt(II)-phosphorothioate RNA crosslinks formed in the HHrz (Figure S6). Crosslink reversal would leave a deoxy-phosphorothioate linkage on one strand, which may be identified by PS-selective cleavage. These properties would be useful for high-throughput applications in larger systems.
Relationship of Pt(II) ligands to native metal sites
The properties of square-planar cis-diammine Pt(II) complex differs from Mg2+, the most metal most likely used by functional RNAs, in size, coordination preference, and geometric constraints. Thus, while the Pt(II) agent may efficiently form crosslinks in the electrostatic pockets that are characteristic of native metal sites in RNAs, the specific ligands involved may differ from those expected for Mg2+. The majority of Mg2+ sites that have been detected by X-ray crystallography involve at least one inner-sphere coordination to a phosphodiester non-bridging oxygen atom, as is biased in this crosslinking approach with a Pt(II)-thiophosphate linkage.41,42 Crosslinking, however, is biased by the strong preference of Pt(II) for purine imino N, or other thio groups that might be present in modified systems. By contrast, Mg2+ often interacts with non-phosphate ligands via hydrogen-bonding by a Mg2+-aqua ligand. Metal ion substitution studies in RNA have shown that transition metals may occupy the same site as does Mg2+, but with slight shifts in ligands that reflect these metals’ coordination preferences.42,43 In the present case, metal coordination to the scissile phosphate of the HHRz has been predicted based on biochemical and computational studies. Current models suggest that G10.1, the minor crosslinked product, provides an additional ligand to this active-site metal at some point in the reaction pathway.23,29,44 To date, however, there has been no suggestion of metal coordination, functional or otherwise, to the G8 nucleobase despite its proximity. Efficient HHRz activity requires base-pairing at the G8:C3 position, and different base-pairs reduce activity by 2–4 orders of magnitude.28a,45,46 Thus, the Pt(II)-induced crosslink between a scissile phosphorothioate and G8 may reflect proximity, but a relationship between metal coordination to G8 and activity remains to be evaluated.
Summary and Perspective
In summary, we have described a new RNA crosslinking strategy based on the targeting of a kinetically inert Pt(II) complex to a specific phosphorothioate-substituted position within a structured RNA. Once anchored to this position the Pt(II) probe selectively forms exchange-inert crosslinks to nearby nucleotides. In this example we embedded a phosphorothioate substitution within the catalytic core of the Hammerhead ribozyme, a three-helix junction RNA motif. Based on comparison with recent crystallographic structures and biochemical data, the resulting major crosslink to G8 and minor crosslink to G10.1, are predicted to be based on ≤~8 Å proximity within a near-native structure, as is desirable for a crosslinking agent. Crosslinks occur with yields of ~30% at higher Mg2+ concentrations and are not observed in global-folding mutants, or in model RNA duplexes. Accumulated biochemical and crystallographic evidence supports metal coordination to the N7 position of G10.1 in ground-state structures, but the potential for the Pt(II) crosslink to reflect a native metal ion interaction with G8 requires further evaluation. In all cases, the specific crosslinks formed by this method may be influenced by steric or electronic encumberances imposed by the Pt(II) ligand set and/or by local RNA structural dynamics. We anticipate that this method may be of particular use for identification of RNA junction regions, where local electrostatics will enhance the kinetics of Pt(II) coordination and non-canonical RNA structures may provide local flexibility. For detailed investigations, stereopure phosphorothioate populations47 and isomeric Pt(II) reagents hold potential for structure probing at a more precise level. As a more general application, the availability of methods for global phosphorothioate incorporation and subsequent identification, developed for NAIM (Nucleotide Analogue Interference Mapping) techniques,48 suggests extension of Pt(II) crosslinking to high-throughput methods of RNA structure elucidation. Overall, Pt(II)-phosphorothioate and other targeted Pt(II) crosslinking methods have potential to add to the expanding repertoire of techniques being used to probe the structure of the transciptome.
Supplementary Material
Acknowledgments
We thank Kory Plakos for assisting with Mg2+-dependent kinetic studies, W. Luke Ward for helpful discussions, and the Center for Advanced Materials Characterization at Oregon for support of MALDI-MS facilities. Funding from the NIH (GM 058096) and the University of Oregon are gratefully acknowledged.
Footnotes
Supporting Information Available: Methods, MALDI-MS and dPAGE data, and molecular modeling data mentioned in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Sharp PA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]; (b) Atkins JF, Gestetland RF, Cech TR, editors. RNA Worlds. Cold Spring Harbor Perspectives in Biology, Cold Springs Harbor Laboratory Press; Cold Springs Harbor, New York: 2011. [Google Scholar]; (c) Mattick JS. Nat Rev Genetics. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 2.(a) Darnell RB. Wiley Int Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz JA, Westhof E. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 4.(a) Schroeder S. J Virology. 2009;83:6326–6334. doi: 10.1128/JVI.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Jossinet F, Ludwig TE, Westhof E. 2007;10:279–285. doi: 10.1016/j.mib.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 5.(a) Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Curr Op Struct Bio. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]; (b) Westhof E, Masquida B, Jossinet F. Cold Spring Harbor Perspectives in Biology. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Das R, Karanicolas J, Baker D. Nat Meth. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Flores SC, Altman RB. RNA. 2010;16:1769–1778. doi: 10.1261/rna.2112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Reues FE, Garst AD, Batey RT. Methods Ezymol. 2009;469:119–139. doi: 10.1016/S0076-6879(09)69006-6. [DOI] [PubMed] [Google Scholar]; (b) Moers BHM. Methods. 2009;47:168–176. doi: 10.1016/j.ymeth.2008.09.006. [DOI] [PubMed] [Google Scholar]; (c) Holbrook SR, Kim SH. Biopolymers. 1997;44:3–21. doi: 10.1002/(SICI)1097-0282(1997)44:1<3::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 7.Mitra K, Frank J. Annu Rev Biophys Biomol Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]; (b) Baird NJ, Ludtke SJ, Khan H, Chiu W, Pan T, Sosnik TR. J Am Chem Soc. 2010;132:16352–16353. doi: 10.1021/ja107492b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Jurica MS. Curr Opin Struct Biol. 2008;18:315–320. doi: 10.1016/j.sbi.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang YX, Zuo XB, Wang JB, Yu P, Butcher SE. Methods. 2010;52:180–191. doi: 10.1016/j.ymeth.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Herschlag D, editor. Methods in Enzymology. Academic Press; London: 2009. Biophysical, Chemical and Functional Probes of RNA Structure, Interactions and Folding: Part A; p. 468. [DOI] [PubMed] [Google Scholar]; (b) Weeks KM. Curr Op Struct Bio. 2010;20:1–10. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Das R, Kudaravalli M, Jonikas M, Laederach A, Fong R, Schwans JP, Baker D, Piccirilli JA, Altman RB, Herschlag D. Proc Natl Acad Sci. 2008;205:4144–4149. doi: 10.1073/pnas.0709032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efimov VA, Fedyunin SV, Chakhmakhcheva OG. Russian J Bioorg Chem. 2010;36:49–72. [Google Scholar]
- 11.(a) Harris ME, Christian EL. In: Handbook of RNA Biochemistry. Hartmann RK, Bindereif A, Schon A, Westhof E, editors. Wiley; 2008. pp. 374–384. [Google Scholar]; (b) Juzumiene D, Shapakina T, Kirillov S, Wollenzein P. Methods. 2001;25:333–343. doi: 10.1006/meth.2001.1245. [DOI] [PubMed] [Google Scholar]; (c) Lambert D, Heckman JE, Burke JM. Biochemistry. 2006;45:7140–7147. doi: 10.1021/bi052457x. [DOI] [PubMed] [Google Scholar]
- 12.(a) Hummel CF, Pincus MR, Brandt-Rauf PW, Frei GM, Carty RP. Biochemistry. 1987;26:135–146. doi: 10.1021/bi00375a020. [DOI] [PubMed] [Google Scholar]; (b) Thomas JM, Perrin DM. J Am Chem Soc. 2008;46:15467–15475. doi: 10.1021/ja804496z. [DOI] [PubMed] [Google Scholar]; (c) Sigurdsson ST, Tuschl T, Eckstein F. RNA. 1995;1:575–583. [PMC free article] [PubMed] [Google Scholar]; (d) Cohen SB, Cech TR. J Am Chem Soc. 1997;119:6259–6268. [Google Scholar]; (e) Stage-Zimmerman TK, Uhlenbeck OC. Nat Struct Biol. 2001;8:863–867. doi: 10.1038/nsb1001-863. [DOI] [PubMed] [Google Scholar]
- 13.Brimacombe Rl, Stiege W, Kyriatsoulis A, Maly P. Methods Enz. 1988;164:287–309. doi: 10.1016/s0076-6879(88)64050-x. [DOI] [PubMed] [Google Scholar]
- 14.(a) Lippert B. In: Progress in Inorganic Chemistry. Lippard SJ, editor. Vol. 37. Wiley; New York: 1989. pp. 1–97. [Google Scholar]; (b) Chapman EG, Hostetter A, Osborn M, Miller A, DeRose VJ. In: Metal Ions In Life Sciences: Structural and Catalytic Roles of Metal Ions in RNA. Sigel A, Sigel H, Sigel RKO, editors. Royal Society of Chemistry; Cambridge, UK: 2011. [PubMed] [Google Scholar]
- 15.(a) Kelland L. Nat Rev Cancer. 2007;7:573–583. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]; (b) Wang D, Lippard SJ. Nat Rev Drug Disc. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 16.(a) Escaffre M, Chottard JC, Bombard S. Nucl Acids Res. 2002;30:5222–5228. doi: 10.1093/nar/gkf672. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Nuc Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bombard S, Kozelka J, Favre A, Chottard JC. Eur J Biochem. 1998;252:25–35. doi: 10.1046/j.1432-1327.1998.2520025.x. [DOI] [PubMed] [Google Scholar]; (c) Hostetter AA, Chapman EG, DeRose VJ. J Am Chem Soc. 2009;131:9250–9257. doi: 10.1021/ja809637e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Tukalo MH, Kubler MD, Kern D, Mougel M, Ehresmann C, Ebel JP, Ehresmann B, Giegé R. Biochemistry. 1987;26:5200–5208. doi: 10.1021/bi00390a045. [DOI] [PubMed] [Google Scholar]; (b) Moine H, Bienaimé C, Mougel M, Reinbolt J, Ebel JP, Ehresmann C, Ehresmann B. FEBS Letters. 1988;228:1–6. doi: 10.1016/0014-5793(88)80572-6. [DOI] [PubMed] [Google Scholar]; (c) Metz-Boutigue MH, Reinbolt J, Ebel JP, Ehresmann C, Ehresmann B. FEBS Letters. 1989;245:194–200. doi: 10.1016/0014-5793(89)80220-0. [DOI] [PubMed] [Google Scholar]; (d) Rasmussen NJ, Wikman FP, Clark BFC. Nuc Acids Res. 1990;18:4883–4889. doi: 10.1093/nar/18.16.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Yusupova G, Reinbolt J, Wakao H, Laalami S, Grunberg-Manago M, Romboy P, Ehresmann B, Ehresmann C. Biochemistry. 1996:352987–2984. doi: 10.1021/bi9519415. [DOI] [PubMed] [Google Scholar]
- 18.(a) Rijal K, Chow CS. Chem Comm. 2009:107–109. doi: 10.1039/b816633a. [DOI] [PubMed] [Google Scholar]; (b) Hostetter AA, Osborn MF, DeRose VJ. ACS Chem Biol. doi: 10.1021/cb200279p. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strothkamp KG, Lippard SJ. Proc Natl Acad Sci USA. 1976;73:2536–2540. doi: 10.1073/pnas.73.8.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Elmroth SKC, Lippard SJ. J Am Chem Soc. 1994;116:3633–3634. [Google Scholar]; (b) Elmroth SKC, Lippard SJ. Inorg Chem. 1995;34:5234–5243. [Google Scholar]; (c) Kjellström J, Elmroth SKC. Inorg Chem. 1999;38:6193–6199. doi: 10.1021/ic990622p. [DOI] [PubMed] [Google Scholar]
- 21.(a) Chu BCF, Orgel LE. Nuc Acids Res. 1989;17:4783–4798. doi: 10.1093/nar/17.12.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chu BF, Orgel LE. Nuc Acids Res. 1990;18:5163–5171. doi: 10.1093/nar/18.17.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chu BF, Orgel LE. DNA and Cell Bio. 1990;9:71–76. doi: 10.1089/dna.1990.9.71. [DOI] [PubMed] [Google Scholar]; (d) Gruff E, Orgel LE. Nuc Acids Res. 1991;19:6849–6854. doi: 10.1093/nar/19.24.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Nucl Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Uhlenbeck OC. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 23.Blount KF, Uhlenbeck OC. Annu Rev Biophys Biomol Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 24.de la Peña M, García Robles RNA. 2010;16:1943–1950. doi: 10.1261/rna.2130310.Seehafer C, Kalweit A, Steger G, Gräf S, Hammann C. RNA. 2011;17:21–28. doi: 10.1261/rna.2429911.Humans: de la Peña M, García Robles I. EMBO Reports. 2010;11:711–716. doi: 10.1038/embor.2010.100.
- 25.Rios AC, Tor Y. Curr Op Chem Bio. 2009;13:660–668. doi: 10.1016/j.cbpa.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.(a) Osborne EM, Schaak JE, DeRose VJ. RNA. 2005;11:187–196. doi: 10.1261/rna.7950605. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ward LW, DeRose VJ. RNA. in press. [Google Scholar]
- 27.(a) Martick M, Lee TS, York DM, Scott WG. Chemistry and Biology. 2008;15:332–342. doi: 10.1016/j.chembiol.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Martick M, Scott WG. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.(a) Nelson JA, Uhlenbeck O. RNA. 2008;14:605–615. doi: 10.1261/rna.912608. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Leclerc F. Molecules. 2010;15:5389–5407. doi: 10.3390/molecules15085389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Karbenstein K, Peracchi A, Beigelman L, Herschlag D. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- 30.(a) Kim NK, Murali A, DeRose VJ. J Am Chem Soc. 2005;127:14134–14135. doi: 10.1021/ja0541027. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Boots JL, Canny MD, Azimi E, Pardi A. RNA. 2008;14:2212–2122. doi: 10.1261/rna.1010808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(a) Huggins W, Shapkina T, Wollenzien P. RNA. 2007;13:2000–2011. doi: 10.1261/rna.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Behlen LS, Sampson JR, Uhlenbeck OC. Nucleic Acids Res. 1992;20:4055–4059. doi: 10.1093/nar/20.15.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Butcher SE, Burke JM. Biochemistry. 1994;33:992–999. doi: 10.1021/bi00170a018. [DOI] [PubMed] [Google Scholar]
- 32.Crabtree RH. The Organometallic Chemistry of the Transition Metals. 3. Wiley and Sons; New York: 2001. [Google Scholar]
- 33.Al-Hashimi HH, Walter NG. Curr Op Struct Bio. 2008;18:321–29. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hindmarsh K, House DA, Turnbull MM. Inorg Chim Acta. 1997;257:11–18. [Google Scholar]
- 35.A second-order rate constant for the initial fast reaction phase was estimated by dividing the observed rate by the initial concentration of Pt(II). This procedure is only valid under pseudo-first-order conditions, which were not established in these particular studies. Previous studies of phosphorothioate-Pt(II) kinetics (ref. 20b) and Pt(II)-RNA kinetics (ref. 16c), performed under similar conditions, did establish pseudo-first-order behavior. In the present case, however, the heterogeneity of the Pt(II) reagent with respect to aquation, and the potential for additional Pt(II)-RNA adducts that are not observed as final crosslinked product, are noted as unaccounted variables.
- 36.(a) Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Proc Natl Acad Sci USA. 2002;96:9077–9082. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mohanty U, Spasic Al, Kim HD, Chu S. J Phys Chem B. 2005;109:21369–21374. doi: 10.1021/jp050005o. [DOI] [PubMed] [Google Scholar]
- 37.Kim N-K, Bowman MK, DeRose VJ. J Am Chem Soc. 2010 doi: 10.1021/ja101317g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osborne EM, Ward WL, Ruehle MZ, DeRose VJ. Biochemistry. 2009;48:10654–10664. doi: 10.1021/bi900614v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kratochwil NA, Parkinson JA, Sacht C, Murdoch PS, Brown T, Sadler PJ. Euro J Inorg Chem. 2001:2743–2746. [Google Scholar]
- 40.Chapman EG, DeRose VJ. J Am Chem Soc. 2010;132:1946–1952. doi: 10.1021/ja908419j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein DJ, Moore PB, Steitz TA. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.(a) Pyle AM. J Biol Inorg, Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]; (b) DeRose VJ, Burns S, Kim NK, Vogt M. Comprehensive Coordination Chemistry. I. Elsevier; St. Louis: 2003. pp. 787–813. [Google Scholar]
- 43.Shi H, Moore PB. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee TS, Giambaşu GM, Sosa CP, Martick M, Scott WG, York DM. J Mol Bio. 2009;388:195–206. doi: 10.1016/j.jmb.2009.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson JA, Uhlenbeck OC. RNA. 2008;14:43–54. doi: 10.1261/rna.717908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee TS, York DM. J Am Chem Soc. 2010;132:13505–13518. doi: 10.1021/ja105956u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frederiksen JK, Piccirilli J. Methods Enzym. 2009;468:289–309. doi: 10.1016/S0076-6879(09)68014-9. [DOI] [PubMed] [Google Scholar]
- 48.Suydam IT, Strobel SA. Methods Enzym. 2009;468:4–30. doi: 10.1016/S0076-6879(09)68001-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.