Abstract
Computationally designing protein-protein interactions with high affinity and desired orientation is a challenging task. Incorporating metal-binding sites at the target interface may be one approach for increasing affinity and specifying the binding mode, thereby improving robustness of designed interactions for use as tools in basic research as well as in applications from biotechnology to medicine. Here we describe a Rosetta-based approach for the rational design of a protein monomer to form a zinc-mediated, symmetric homodimer. Our metal interface design, named MID1 (NESG target ID OR37), forms a tight dimer in the presence of zinc (MID1-zinc) with a dissociation constant <30 nM. Without zinc the dissociation constant is 4 μM. The crystal structure of MID1-zinc shows good overall agreement with the computational model, but only three out of four designed histidines coordinate zinc. However, a histidine-to-glutamate point mutation resulted in four-coordination of zinc, and the resulting metal binding site and dimer orientation closely matches the computational model (Cα RMSD = 1.4 Å).
Keywords: computational protein interface design, protein-protein interaction, metal, zinc, cobalt, homodimer, de novo
Introduction
Protein-protein interactions are ubiquitous in biology, but the mechanisms of interaction are complex and only moderately understood. Rational design of new protein-protein interactions may improve our understanding of protein biophysics and may be used as a tool for developing novel competitive inhibitors, biosensors, network components, and protein therapeutics 1,2. While directed evolution is a powerful means to generate new binders, computational design may be used to sample more sequence space, to choose the binding location, and to choose the binding orientation for the interaction. Successful binding of a viral protein target with the desired location and orientation demonstrates that computational design is emerging as a new means to generate interactions for applied purposes 3. Other successes include redesigning existing interfaces 4–11, grafting known sidechain interaction motifs onto new protein scaffolds 12, producing modest-affinity binders de novo (Kd > 100 μM) 13,14, and combining computational design with directed evolution 15,16.
The computational approach to interface design remains very challenging due to sophisticated steric and chemical complementarity required for interaction, combined with the enormous conformational sampling required to optimize rigid body, backbone, and sidechain degrees of freedom. Furthermore, designed interactions are modeled with atomic-level detail, and slight modeling inaccuracies can result in no binding, weak binding, or binding in an unexpected orientation. For example, the work of Fleishman et al. required 73 yeast-displayed designs to identify two binders with Kd of ~2 μM 3, and the work of Karanicolas et al. led to an alternative binding orientation 16. With these challenges in mind, we use metal binding sites at a designed interface to drive association despite modeling inaccuracies, and also to achieve high affinity and orientation preference in a smaller and more tractable protein interface. Interactions between metals and histidine, cysteine, aspartate, and/or glutamate sidechains are stronger than protein-protein hydrogen bonds or van der Waals contacts. Thus, suboptimal hydrogen-bonding patterns or packing at the interface may be overcome by metal-binding interactions.
Metal-binding sites are an attractive computational design goal because coordination spheres are well-understood 17–20, protein-metal interactions are stronger than protein-protein contacts, and only a handful of mutations are required. Zinc has a well-established structural role in protein tertiary and quaternary structure of naturally occurring proteins 17,21–23, and engineering zinc binding sites was one of the earliest goals in computational protein design. Regan and co-workers and Hellinga and co-workers designed metal-binding sites in proteins twenty years ago 24–27. The field has matured to designing functional metalloproteins 28. Promoting quaternary structure using metal-binding sites has been explored in several variations by the DeGrado lab, including design of diiron- and porphyrin-mediated helical assemblies 29–32. The Tezcan group converted inter-molecular interactions observed in the crystalline state (crystal contacts) to solution-state interactions by placing histidines at the surface to form intermolecular zinc binding sites 33. This minimalist interface was then computationally optimized to achieve a metal-independent protein-protein interaction 34. Many designed metal-binding sites in proteins have been reported 35,36, and given a history of success in this endeavor, incorporating zinc binding sites at our designed interfaces may provide a foothold to computationally design a protein-protein interaction from scratch.
To test our strategy for metal-mediated protein interface design, we designed the surface of a monomer scaffold to symmetrically self-interact in a metal-mediated manner. The computational design protocol first uses RosettaMatch 37,38 to generate two-residue zinc binding sites on known monomeric scaffold proteins. These two-residue zinc binding sites on the monomer become tetrahedral four-residue zinc sites upon simulated dimerization, and symmetric sequence design with backbone minimization in Rosetta optimized the protein-protein contacts 39,40.
This symmetric zinc-mediated design approach may be used as a general strategy to control pharmacokinetic properties of injected protein therapeutics. As a compelling example, insulin is secreted as a zinc-mediated hexamer that becomes active only upon dissociation 41. Metal binding may help minimize the number of mutations of the active monomer required to achieve oligomerization. There are also practical incentives to design a homodimer. In the design stage, enforcing symmetry limits the vastness of conformational space, and furthermore, an interesting study proposes that a symmetric complex tends to be lower in energy than an asymmetric complex 42.
Our broader scientific goal is to develop computational methods for protein interface design, and the specific strategy discussed here features the design of a symmetric metal-mediated homodimer. Our success with this exploratory strategy is a step toward more advanced and reliable protein interface design methodology for diverse applications in medicine, biotechnology, and basic research.
Results
Computational Approach
To design a de novo metal-mediated protein interface (Figure 1), we first used RosettaMatch to design two-residue zinc binding sites on the surface of known monomeric protein scaffolds. Each scaffold surface was divided into patches of approximately 10 residues using a Rosetta application called SurfaceGroups. Each surface residue was treated as the center of a surface patch. For each surface patch, the residue positions were searched by RosettaMatch for “matches”, residue pairs that can be mutated to histidine or cysteine to coordinate a zinc ion with proper coordination distances and angles as well as histidine dihedrals (Figure S1). Upon enumeration of hundreds of thousands of two-residue zinc matches, a strict geometric evaluation was used to delete those matches with suboptimal geometry. This evaluation was performed using another Rosetta application called ZincMatchFilter, which sums deviations from ideal distances, angles, and dihedrals, normalized by standard-deviation values (Figure S1). The end-result of our RosettaMatch runs was 42,000 high-quality two-residue matches among more than 600 protein scaffolds (PDB codes given in Supplemental Information).
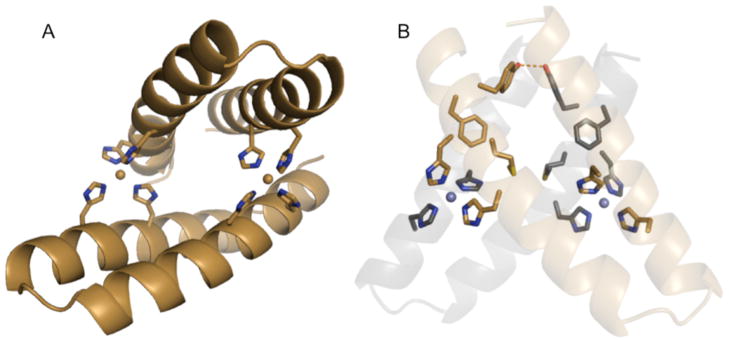
Figure 1. Computational model of MID1 (metal interface design 1).
A) The scaffold protein for this design is the 46-residue helix-turn-helix Rab4-binding domain of rabenosyn (PDB code 1YZM). The design is a symmetric homodimer with two interface zinc sites each coordinated by four histidines at i, i+4 positions on each helix. B) The protein-protein contacts feature a small hydrophobic core: Met38 interacts with the equivalent methionine in the opposing monomer; likewise, the Tyr41 hydroxyl groups are within hydrogen bonding distance of each other, and Phe42 packs against the opposing helix. In a top-down view, the top chain is tan, and the bottom chain is black.
In the second step of the protocol (Figure 2), all pairs of matches on a given scaffold were combinatorially enumerated and grafted onto the scaffold surface. In this case, the goal was to create a dimer with two metal sites at the interface. A symmetric zinc-mediated dimer was generated by duplicating the twice-grafted scaffold then rotating the second chain 180° about an axis bisecting and orthogonal to the zinc-zinc axis. The result of this rotation was a C2 symmetric dimer where match 1 on chain A coordinates zinc with match 2 on chain B, and match 2 on chain A coordinates zinc with match 1 on chain B. The rotation ignored backbone clashes, and although the geometry within the individual two-residue matches remained constant, the tetrahedral arrangement of match pairs was typically far from ideal. Thus, to relieve backbone clashes and improve the tetrahedral angles about zinc, the second chain was incrementally rotated about the zinc-zinc axis to maintain symmetry while also exploring one degree of freedom for rigid body alignment. In the event that an alignment featured no backbone clashes and good tetrahedral geometry, this complex was output as a designable starting structure. Designable starting structures were infrequent, but the number of designable starting structures was still large due to the extensive number of match pairs combined with a fine-grained rotational search. Among 600 scaffolds, 500,000 designable starting structures were identified using our Rosetta protocol, named SymMetalInterface_TwoZN_setup.
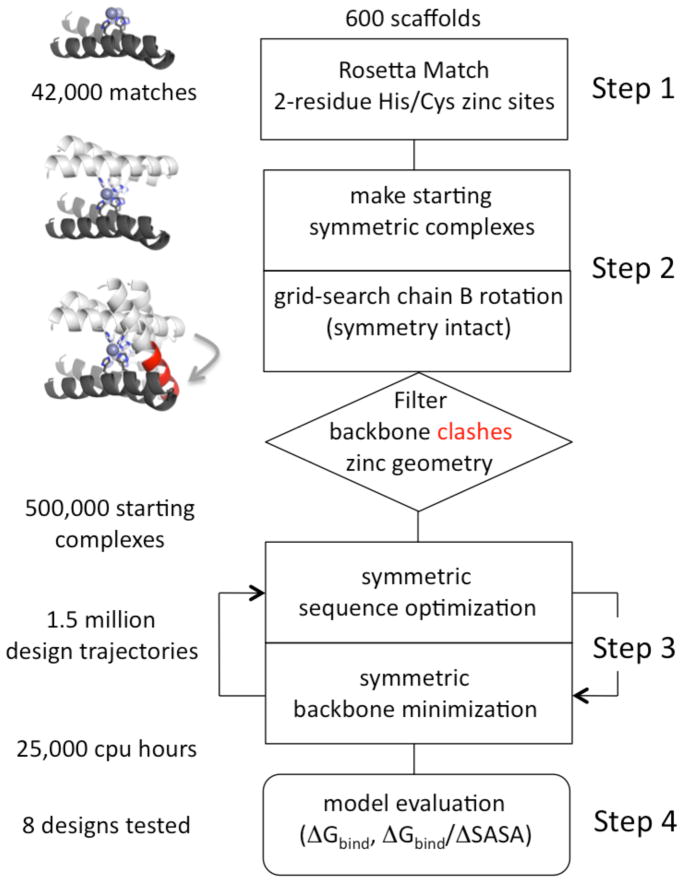
Figure 2. Flow chart of the protocol for the design of the symmetric metal-mediated interface.
In Step 1, 600 monomer scaffold surfaces were scanned for two-residue cysteine/histidine zinc binding sites using the RosettaMatch algorithm. In Step 2, all pairs of two-residue zinc binding sites for a given scaffold were grafted onto the surface, and the monomer was converted to a C2-symmetric dimer by rotation (see Methods). The second chain was rotated about the zinc-zinc axis to maintain symmetry while grid-searching the rigid-body alignment for relief of clashes and proper zinc coordination geometry. In Step 3, symmetric interface design was iterated with symmetric backbone minimization using Monte-Carlo simulated annealing. In Step 4, a large number of design models were filtered based on two primary metrics: computed binding energy, excluding contribution from zinc (ΔGbind) and binding energy per unit of interface surface area (ΔGbind/ΔSASA).
All designable starting structures were inputs for the symmetric interface design step (SymMetalInterface_TwoZN_design), which iterated three times between sequence optimization and backbone minimization. During sequence optimization, bias was given to the native amino acid to limit the number of mutations. These simulations required ~1 minute per starting structure, ~10,000 cpu hours total, due to the restriction of binding orientation by the zinc binding sites, which were constrained during these simulations. Design models were evaluated using a Rosetta application called Interface Analyzer, which computes binding energy (ΔGbind), interface surface area (ΔSASA, Å2), and binding energy density (ΔGbind/ΔSASA). Design models with ΔGbind better than −20 Rosetta energy units (R.e.u.) (ignoring contributions from metal binding) and ΔGbind/ΔSASA better than −0.015 R.e.u./Å2 were considered. Other metrics included packing quality, number of unsatisfied hydrogen bonds at the interface, and zinc-coordination geometry. We also considered the number of mutations and diversity in the residue composition of the zinc binding sites (histidine/cysteine combinations). In the end, we chose eight designs to experimentally test. Four of these designs contain two zinc binding sites as described, and four contain one zinc binding site; these four were designed using a slightly different setup protocol for Step 2, shown in Figure 2 (see Computational Methods).
The eight designs tested had diverse characteristics (Table S1, and Figure S2). Hydrophobicity: four interfaces were predominantly hydrophobic, two interfaces were predominantly polar, and two interfaces were mixed hydrophobic and polar. Zinc binding sites: five were 4-Cys, two were 4-His, and one was 2-Cys/2-His. Secondary structure: three were helical, two were helical with loops, one was sheet-to-sheet, one was primarily loop, and one was mixed with loops, strands, and helices. Size of interface: the largest interface tested was 2430 Å2 with 18 mutations and the smallest was 1230 Å2 with 10 mutations.
Biophysical Characterization
Eight designs were expressed as C-terminal fusions to 6xHis-MBP tags to promote expression and solubility. Six out of eight designs either suffered from poor expression or formed higher-order oligomers (Supplemental Table S2). The design derived from the Rab4-binding domain of rabenosyn (PDB code 1YZM) – a 46-residue helix-turn-helix scaffold – migrated during size-exclusion chromatography with the hydrodynamic radius expected for the 6xHis-MBP-fusion protein. The design model has two 4-histidine zinc binding sites, each histidine coordinates zinc with its ε2 nitrogen, and the δ1 nitrogens are solvent exposed (Figure 1A). Met38, Tyr41, and Phe42 on both chains interact to form a small hydrophobic core with the significant protein-protein contacts in the design model; Met38 interacts with the equivalent methionine in the opposing monomer; likewise, the Tyr41 hydroxyl groups are within hydrogen bonding distance of each other, and Phe42 packs against the opposing helix (Figure 1B). This design is named MID1, for metal interface design 1.
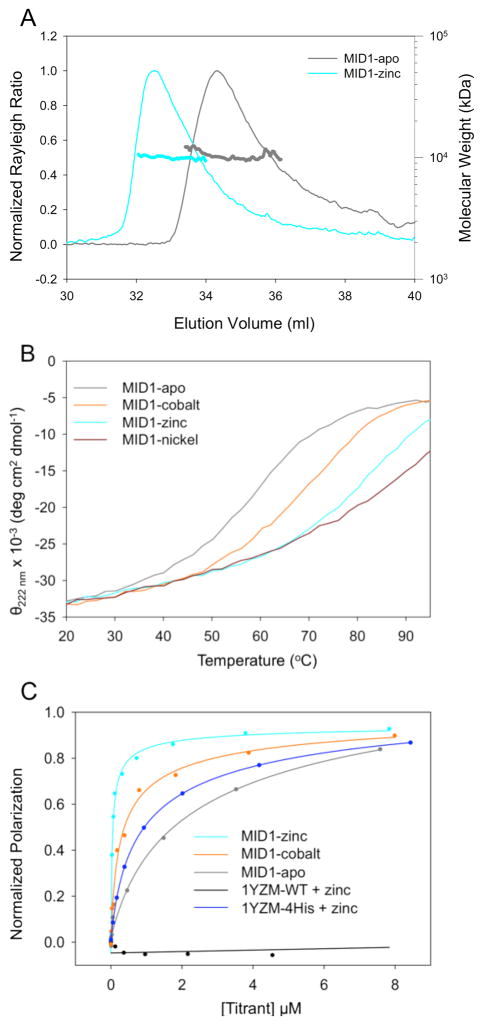
MID1 was cleaved from the 6xHis-MBP fusion domain by TEV proteolysis, and the oligomeric state of MID1 was characterized by size exclusion chromatography (Figure S3) and multiple angle light scattering (MALS, Figure 3A). The theoretical size of the dimer is 10.6 kDa, and MALS indicated a molar mass of 10.5 kDa and 10.0 kDa for MID1 without metal (MID1-apo) and MID1 with zinc (MID1-zinc), respectively. Thus, MID1 forms a dimer at high micromolar concentrations with and without zinc. For evidence of metal binding, thermally-induced unfolding in the presence and absence of various metals was monitored by circular dichroism. The midpoint of thermal unfolding (Tm) of MID1 (57°C) was similar to 1YZM wild-type (56°C). The Tm of reversible unfolding of MID1 increased in response to equimolar additions of cobalt (70°C), zinc (81°C), and nickel (87°C) (Figure 3B), suggesting metal-binding events. Divalent metal ions often bind to the surface of proteins and mediate non-specific interactions. In that respect, it is noteworthy to mention that the Tm of MID1 was not affected when manganese, calcium, magnesium, or iron was added.
Figure 3. Biophysical characterization of MID1.
A) Multiple angle light scattering data indicate that MID1-apo and MID1-zinc exist as a dimer. B) Thermal denaturation monitored by circular dichroism indicates that cobalt, zinc, and nickel increase the melting temperature (Tm) from 57°C to 70°C, 81°C, and 87°C, respectively. C) Fluorescence polarization titration experiments provide estimates of the dimer dissociation constant, Kd, in the presence and absence of metal. Fluorescently-tagged MID1 is diluted to 10 nM to promote a starting monomer state, and unlabeled MID1 is titrated in the presence or absence of metal; dimerization increases the fluorescence polarization. To estimate Kd, the binding curves were fit using an equation describing a homodimer equilibrium interaction (Supplemental Information). Cobalt and zinc improve the apparent Kd from 4300 nM to 410 nM and <30 nM, respectively. The wild-type scaffold 1YZM shows no dimerization, and 1YZM with the four designed histidines (1YZM-4His) binds with a Kd of 1400 nM in the presence of 12 μM zinc.
The equilibrium dissociation constant for MID1 dimerization was measured using fluorescence polarization in the presence and absence of metal. MID1 was recloned with a C-terminal glycine-cysteine extension for fluorescent dye conjugation. MID1-GC-Bodipy was diluted to 10 nM in the starting sample, and unlabeled MID1 was titrated in the absence or presence of 12 μM metal ion. An increase in polarization was observed, indicating the formation of a higher-molecular-weight complex. Titration curves were fit to a homodimer equilibrium model (Supplemental Methods) to obtain estimates for the dissociation constant, Kd. The Kd’s for MID1-apo, MID1-cobalt, and MID1-zinc are 4300 nM, 410 nM, and <30 nM, respectively (Figure 3C) – zinc binding leads to a >200-fold increase in binding affinity. With histidines for zinc coordination but without the designed protein-protein contacts (1YZM-4His), binding occurred with a Kd of 1400 nM (Figure 3C). Thus, zinc binding alone allows weak association (Kd = 1400 nM), protein-protein interaction alone allows weak association (Kd = 4300 nM), but a combination of zinc binding and protein-protein interactions allows tight binding (Kd < 30 nM).
Structural Validation
Biophysical characterization of MID1 indicated that we had successfully designed a metal-mediated high-affinity protein-protein interaction using a computational approach. In order to obtain structural information of MID1-zinc by NMR spectroscopy, we nominated the protein as a community outreach target of the Protein Structure Initiative (PSI) and collaborated with the Northeast Structural Genomics Consortium (NESG: http://www.nesg.org; NESG target ID OR37). We observed only 26 out of the 46 expected polypeptide backbone amide peaks in 2D [15N, 1H]-HSQC (Figure S4). Furthermore, only 17 of those could be assigned, that is, Gln6, Gln7, the polypeptide segment comprising residues 18 to 31 and the C-terminal residue Asp46. Residues 18 to 31 are located in the helical hairpin and are not part of the designed interface (Figure S4). Prediction of helical polypeptide segments using the chemical shift index (CSI 43) confirms that the helical hairpin is present in solution. However, a total of 29 NH moieties, including those of the interface, are affected by conformational dynamics to an extent that either broadens lines beyond detection, or prevents resonance assignment.
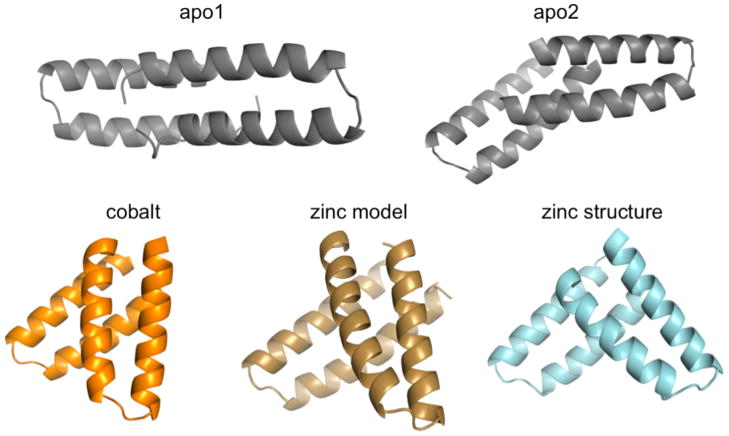
The lack of resonance assignments precluded solving the structure of MID1-zinc by NMR, so we then used X-ray crystallography to determine the three-dimensional structure of MID1 for comparison to the model that we obtained by computational methods. We determined six crystal structures – MID1-apo1, MID1-apo2, MID1-cobalt, and MID1-zinc, MID1-H12E-zinc, and MID1-H35E-zinc, each using diffraction data to 1.28 Å resolution or higher (data collection and refinement statistics in Table S3). MID1-apo1 and MID1-apo2 crystallized in the same buffer condition but in different crystal forms, with MID1-apo1 containing one molecule in the asymmetric unit, and MID1-apo2 containing two molecules in the asymmetric unit. In both structures, the backbone of the helical hairpin is very similar to that in the 1YZM scaffold, indicating that the designed mutations did not significantly alter the tertiary structure (Figure S5). The two structures show two possible binding modes for MID1-apo dimerization, with the caveat that small interfaces (~1000 Å2 in this case) can be difficult to distinguish from crystal contacts 44, i.e., interactions that would not be observed free in solution. In both interfaces, designed sidechains form unanticipated hydrophobic contacts and hydrogen bonds (Figure S6). The dimeric structures of MID1-apo1 and MID1-apo2 do not resemble the computational model (Figure 4): RMSD upon alignment of 78 equivalent helical Cα backbone atoms is 8.6 Å and 7.8 Å, and the crossing angle between the two chains is different from the model by 74° (Table 1).
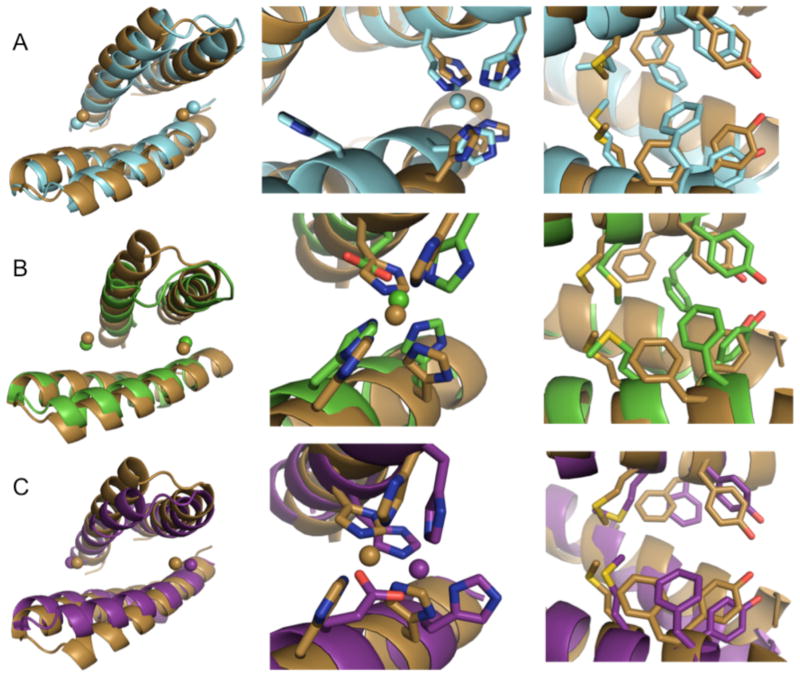
Figure 4. Binding orientation of MID1-apo1/2, MID1-zinc, MID1-cobalt.
The crystal structure of MID1-apo (gray) shows two dimer orientations, neither were predicted. The presence of zinc (cyan) and cobalt (orange) promotes an orientation that resembles the zinc model (tan). RMSD values for global alignment and dimer crossing angle comparisons are given in Table 1.
Table 1.
Parameters describing homodimer binding orientation
| RMSD alignment to model (Å)1 | Symmetry deviation (Å)1,2 | Dimer crossing angle difference3 | |
|---|---|---|---|
| Model | -- | 0.02 | 0° |
| MID1-apo1 | 8.6 | 3.98 | N/A |
| MID1-apo2 | 7.8 | 2.28 | 74° |
| MID1-cobalt | 2.2 | 0.12 | 17° |
| MID1-zinc | 2.4 | 0.33 | 19° |
| MID1-H12E-zinc | 1.4 | 0.47 | 8° |
| MID1-H35E-zinc | 2.5 | 0.33 | 7° |
Calculated using helical Cα positions. This removed the flexible termini, as well as the alternative turn conformation in chain B of MID1-H12E (apparent in Figure 5B, left) due to sterics of crystal packing.
Sdev < 0.2 Å is considered symmetric.
Dimer crossing angles were computed as described in the Methods section.
Comparing the MID1-apo structures to the MID1-zinc crystal structure, we observe a zinc-dependent reorientation of the MID1 dimer that closely resembles the MID1-zinc design model (Figure 4, Figure 5A). The zinc atoms are in the designed positions (Figure 5A, left), the structure aligns to the design model with an RMSD of 2.5 Å for 78 equivalent Cα backbone atoms representing helical positions only, and the crossing angle between chains differs from the model by only 19° (Table 1). Thus, zinc binding promotes the intended binding orientation. The zinc-mediated dimer was modeled symmetrically, and although it is not perfectly symmetric in the crystal structure, the MID1-zinc dimer is much more symmetric than the MID1-apo dimers – symmetry deviation is 0.33 Å with zinc, 3.98 Å and 2.28 Å without zinc (Table 1). For reference, symmetry deviation of 0.2 Å is considered symmetrical 45. Despite these similarities, we found in the crystal structure that only 3 out of 4 histidines coordinate each zinc (Figure 5A, center). H35 does not coordinate zinc; the coordination sphere is instead completed by carboxylates from either the C-terminal aspartate of a symmetry-related molecule (Figure S7A) or a tartrate molecule from the crystallization buffer (Figure S7B). Furthermore, all four histidines were predicted to coordinate zinc with the ε2 nitrogen, but the structure shows one histidine coordinates zinc with the δ1 nitrogen (Figure 5A, center, Supplementary Table S4). The observed zinc-coordination arrangement may be responsible for the small deviation in binding orientation and deviations in atomic-level protein-protein interface contacts. As designed, Met38, Tyr41, and Phe42 interact at the interface. However, the hydroxyls on Tyr41 are not within hydrogen bonding distance, and Phe42 on chain A makes direct contacts with Phe42 on chain B, an interaction that is not present in the design model (Figure 5A, right).
Figure 5. Comparison of the MID1-zinc model to A) MID1-zinc, B) MID1-H12E-zinc, and C) MID1-H35E-zinc crystal structures.
Left panel: the global alignment of zinc-bound dimers, spheres indicate zinc ions. Center panel: the observed zinc-coordination geometry compared to the model. Right panel: the observed interface sidechain contacts compared to the model.
We hypothesized that repairing the zinc binding site to achieve four-coordination would improve the agreement of the dimer structure with the computational model. To repair the zinc binding site, the four histidines were individually mutated to glutamate (H12E, H16E, H35E, H39E) to form 3-His, 1-Glu zinc binding sites. We chose glutamate instead of aspartate because the sidechain length closely recapitulates the ε2 nitrogen position. Crystallization trials led to structures of MID1-H12E-zinc and MID1-H35E-zinc. In fact, MID1-H12E does display four-coordination of zinc; the H12E glutamate mutation causes H35 to participate in zinc binding. The zinc binding site is accurately modeled (Figure 5B, center), and the overall orientation closely resembles the design model, featuring a crossing angle difference of only 8° and a helical Cα RMSD of only 1.4 Å (Table 1 and Figure 5B, left). Looking at the sidechain interactions, the location of Met38 is partially accurate and Tyr41 does make a hydrogen bond as predicted, however Phe42 is again not modeled correctly (Figure 5B, right). Phe42 is not accurately modeled due to helix unwinding, an unpredicted change to the backbone (Figure S9).
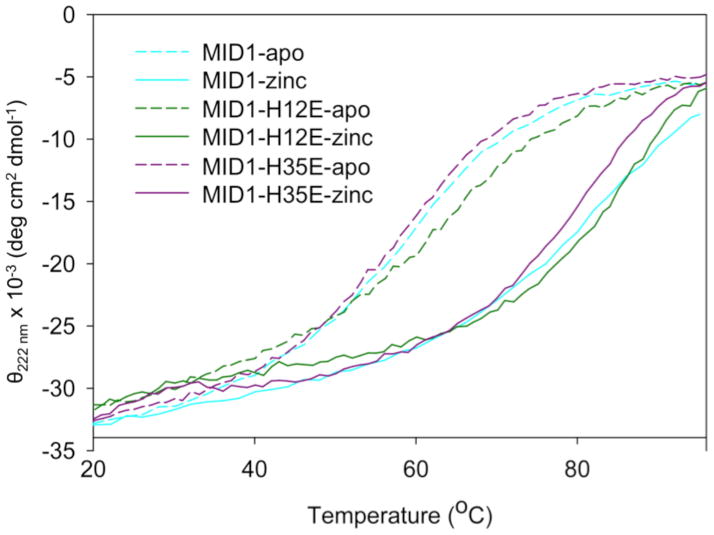
The other point mutant that crystallized, MID1-H35E, also displays four-coordination of zinc, where H35 previously did not participate in zinc binding. This zinc binding site does not agree with the original model as well as MID1-H12E (Figure 5C, center); however, as predicted, Phe42 only contacts the opposing helix, not its symmetric counterpart (Figure 5C, right). Although the H12E and H35E mutations add an additional zinc coordination bond (Figure S10), zinc binding does not thermostabilize these mutants more than the original MID1 design (Figure 6).
Figure 6. Circular dichroism thermal denaturation of MID1, MID1-H12E, and MID1-H35E with and without zinc.
Despite adding an additional zinc coordination bond, the H12E and H35E mutations do not result in increased thermostability in response to zinc.
As a second approach to recover the H35 coordination of zinc, we crystallized MID1 bound to cobalt (MID1-cobalt). Cobalt prefers six-coordination, and the MID1-cobalt crystal structure shows that all four histidines do coordinate cobalt with the ε2 nitrogen, and the octahedral coordination sphere is completed by the C-terminal aspartate, which wraps around to participate in the metal binding (Figure S8, center).
Thus, we have observed four different metal-bound structures of the MID1 dimer: MID1-zinc, MID1-H12E-zinc, MID1-H35E-zinc, and MID1-cobalt. In all four cases the overall binding orientation is similar as dictated by the two metal sites, but variations in sidechain packing and helix crossing angles suggest that there is some plasticity to the designed interface (Figure 7). In addition to observing varied sidechain packing in the four crystal structures, the absence of NMR peaks for residues at the dimer interface in the 2D [15N, 1H]-HSQC of MID1 indicates that residues are exchanging between multiple environments on the NMR timescale. Although sidechain positions showed variability and deviated from the prediction, these residues were still critical for affinity (compare MID1-zinc and 1YZM-4His + zinc in Figure 3C), likely due to the increased hydrophobicity of the design (Figure S11). Taken together these results suggest that atomic-level design of sidechain-sidechain interactions was not critical in this metal-mediated approach to interface design. Instead, providing roughly complementary nonpolar surfaces may have been sufficient. This less-stringent approach to interface design may improve the success rate while also maintaining affinity and orientation specificity due to metal binding. Similar observations have been made by Tezcan and co-workers when using metal to template helix bundle assemblies. They found that surface metal sites were sufficient to drive metal-mediated oligomerization at high protein concentrations (>100 μM) 33, and that subsequent computational redesign of surrounding residues to be more hydrophobic promoted oligomerization at lower protein concentrations 34.
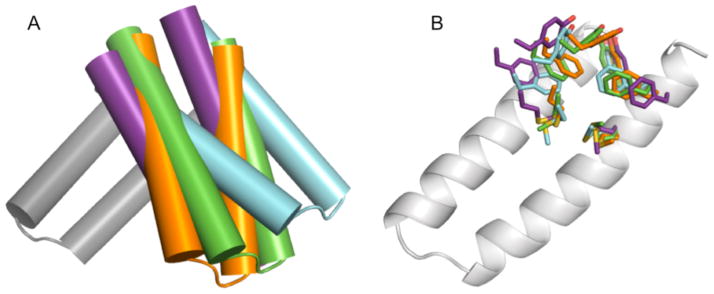
Figure 7. Superimposition of four metal-bound MID1 crystal structures.
MID1-zinc (cyan), MID1-cobalt (orange), MID1-H12E-zinc (green), and MID1-H35E-zinc (purple) were aligned by chain A to the wild-type 1YZM scaffold (gray) to compare the relative orientation of chain B. A) Cylinder representations of helices show slightly different binding orientations for each dimer. B) Sidechain placements are slightly different for each dimer. These minor differences suggest that the MID1 dimer interface may have plasticity and exchange among multiple orientations.
Discussion
We have described the computational design of a metal-mediated protein-protein interaction, and this result is unique in several ways. First, we simultaneously engineered the metal-binding site and the protein-protein contacts, as opposed to the metal-templating strategy utilized by Salgado et al., where first, metal-binding sites were incorporated, followed by crystal structure determination of the resulting low-affinity complex and its use in rationally redesigning protein-protein contacts in a stepwise manner 34. Second, we rationally designed toward a certain binding mode and were able to obtain crystal structures to test the accuracy of our model – structure determination of a complex can be an elusive step in characterizing de novo designed interactions 13,14. Third, we achieved the desired orientation – the work by Karanicolas et al. demonstrates that alternative orientations can degrade the accuracy of designed interactions 16. Finally, we achieved high affinity (Kd < 30 nM) using only computational design, without subsequently optimizing the interaction with directed evolution techniques – when Fleishman et al. tested 73 computational designs, the two best hits had Kd’s > 2000 nM, and evolution by yeast display was required to achieve high affinity 3.
Our strategy to use metal binding to promote protein interaction was chosen to address two major pitfalls caused by modeling inaccuracies, weak binding and nonspecific binding orientation. Our strategy was successful: in the absence of metal, the MID1 design dimerized only weakly and with two types of nonspecific orientations. In the presence of metal, the desired binding orientation was achieved with high affinity, despite minor discrepancies at the atomic level between the computational model and the crystal structure. Metal binding can thus improve robustness of computationally designed interactions.
A compelling reason for performing protein design is to rigorously test our understanding of protein energetics, and by performing multiple rounds of design coupled with experimental validation it may be possible to improve understanding. The crystal structure of MID1 showed that only three of the four histidines participated in metal binding. This result prompted us to reexamine the composition of zinc binding sites in the PDB. Although histidines at positions i and i+4 on a helix make up a common zinc coordination motif, a zinc binding site with four histidine ligands is a rare occurrence. Our own survey of the Protein Data Bank (Supplemental Methods) revealed that only 7 out of 1705 zinc binding sites have a 4-histidine arrangement with no instances of an all-helical 4-histidine arrangement. By contrast, there are 185 instances of 3-histidine 1-aspartate/glutamate arrangements (Supplemental Table S5). Having a negatively charged ligand, such as aspartate or glutamate, to coordinate the metal2+ ion may be more energetically favorable than four neutral ligands. Interestingly, in both the MID1-zinc and MID1-cobalt crystal structures, we observe a strong preference for carboxylate coordination to complete the primary coordination sphere of a metal2+ ion – all four metal ions are coordinated by a carboxylate group (Figure S7). The Tezcan group also intended to engineer 4-histidine zinc binding sites but observed a 3-His/1-Asp coordination sphere instead 33. These findings prompted us to repair the MID1 metal binding site by mutating one of the histidines to glutamate. Indeed, crystal structures of MID1-H12E-zinc and MID1-H35E-zinc showed the desired tetrahedral coordination of zinc and the binding orientation of MID1-H12E-zinc was very close to our original design model. These results suggest that future designs should feature zinc sites with 3-histidines and 1-aspartate/glutamate, or 2-histidines and 2-aspartate/glutamates.
Including aspartates and glutamates when designing zinc binding sites should also allow our interface design protocol to be applied to more scaffolds. Including aspartates and glutamates during RosettaMatch simulations more than doubles the number of potential two-residue zinc matches on a protein surface (59 vs. 228 on the 1YZM scaffold), and because matches are paired combinatorially, having more two-residue zinc binding sites would greatly increase the number of match pairs (1,711 vs. 25,878 on the 1YZM scaffold) and the number of designable starting structures. Thus, this approach should be useful for designing dimers even when limited to a single scaffold.
We also experimentally tested designs with 4-cysteine sites, the most common arrangement for zinc coordination (Supplemental Table S5); however, these designs did not lead to successful dimers but instead formed higher-order oligomers. While we could not determine if oligomer formation was due to misfolding or nonspecific association, we may have been at high risk of nonspecific association when using cysteines, which are hydrophobic, to coordinate zinc at a hydrophobic interface. Previous studies have shown that homodimeric interfaces tend to be more hydrophobic than heterodimeric interfaces 46, though perhaps more polar character would help us avoid high-order nonspecific oligomerization in future designs.
In conclusion, we have computationally designed a metal-mediated homodimer with high affinity and orientation preference. We have gained important insights to guide future design efforts; an aspartate/glutamate should accompany histidines in the coordination sphere, all-atom interface design may not be necessary with this approach, and designs with 4-cysteine zinc sites should have more polar character. We will continue to pursue the design of symmetric metal-mediated protein-protein interactions because this strategy could be a powerful approach to modulate the pharmacokinetics of injected protein therapeutics. If activity is observed for a monomer, then metal-mediated oligomerization could be a means to increase retention and lead to prolonged activity of the therapeutic protein.
Computational Methods
Identifying two-residue zinc binding sites
The computational design protocol is summarized in the flowchart shown in Figure 2. The first step in the protocol was to design two-residue zinc binding sites on scaffold-protein surfaces using RosettaMatch. RosettaMatch 38,47 searches residue positions on a fixed backbone for sidechains that can satisfy geometric requirements of an input “transition state”. For a zinc binding site, the transition state is composed of a zinc atom and tetrahedrally arranged virtual atoms. Residue types were limited to cysteine and histidine. Ideal geometries were as follows: distances were 2.33 Å and 2.05 Å for sulfur-zinc and nitrogen-zinc coordination bonds, respectively. Angles vertexed at zinc were 109°, angles vertexed at histidine nitrogens were 125°, and angles vertexed at cysteine sulfurs were 109°. Histidine dihedrals were measured from zinc-nitrogen-carbon-carbon for either Nε2 or Nδ1 coordination. Ideal Nε2 dihedrals were 180° and ideal Nδ1 dihedrals were 0° (Figure S1). These values were specified using a zinc transition-state parameter file and a geometric constraint file (Supplemental Methods). RosettaMatch also requires inputs for residue positions to search, so the surface of all scaffolds was divided into patches that were searched independently. Each surface residue (<16 neighboring residues) was treated as the center of a patch, and the patch included all of its surface neighbors (Cβ-Cβ distance < 10 Å). Despite the geometric constraints that are inputs for RosettaMatch, the output matches are not all geometrically accurate. Output matches were thus filtered according to the desired distance, angle, and dihedral measurements using a Rosetta application called ZincMatchFilter. Deviations from the ideal measurement were normalized by standard deviations, so a score <2.0 for a two-residue match is within standard deviation, on average. Furthermore, two-cysteine matches at consecutive residue positions were omitted because we did not observe this motif in natural zinc sites. Command lines and inputs for SurfaceGroups, RosettaMatch, and ZincMatchFilter are given in the Supplemental Methods.
Generating starting structures with two zinc ions per interface
Step 2 in the Rosetta protocol named SymMetalInterface_TwoZN_setup was summarized in the Computational Approach section of the Results section. Additionally, in the two-zinc protocol, dimers with zinc-zinc distances < 10 Å apart were thrown out. The remaining were rotated by 5° increments about the zinc-zinc axis, thus exploring 72 orientations per 2-residue match pair.
Generating starting structures with one zinc ion per interface
To design interfaces containing one zinc instead of two, we used a different protocol named SymMetalInterface_OneZN_setup for generating starting structures. One two-residue match was grafted onto the scaffold surface, this chain was duplicated, and the duplicated chain was rotated 180° about an axis that passes through the zinc and is parallel to the nitrogen-nitrogen vector (if histidines form the metal-binding site). This rotation results in a square-planar arrangement about zinc. To achieve two different tetrahedral arrangements, the second chain was rotated by ±90° about an axis that passes through zinc and bisects the previously mentioned nitrogen-nitrogen pair. Thus, two symmetric tetrahedral starting structures per match were generated. Constraints of 2-by-2 residue zinc binding sites leave little freedom for rigid-body searching. However, alignment was optimized while keeping the tetrahedral geometry within one standard deviation (15° per angle). A combinatorial grid-search of +10°, 0°, and −10° rotations about three orthogonal axes intersecting at the zinc position (giving 27 orientations per starting dimer) were explored, and orientations without backbone clashes were accepted as designable starting structures.
Symmetric protein interface design
Symmetry-definition files describe the symmetry of a complex, and a script within Rosetta (make_NCS.pl) was used to create symmetry-definition files for all designable starting structures. A symmetric interface was designed for all designable starting structures using Monte-Carlo simulated-annealing iterated with gradient-based backbone minimization using the protocols SymMetalInterface_OneZN_design and SymMetalInterface_TwoZN_design. Symmetry is maintained by assigning identical torsion angles to the sidechains and backbones of symmetry-related residue positions 39,40. Sequence positions were allowed for design only if the Cβ sidechain atom was within 10 Å of a Cβ atom on the opposing chain, and the native amino acid was given a bonus weight of 1.5 to limit mutations. To prevent worsening of zinc binding geometry due to backbone minimization, the distances, angles and dihedrals in the zinc binding sites were harmonically constrained. Each constraint was given a weight of 1.0. Simulations were performed using the UNC Topsail supercomputing cluster.
Design filtering and evaluation
To evaluate and rank a large number of design models, binding energy was computed as the energy of the complex minus the energy of both separated chains. For speed of calculation, the separated partners were neither repacked nor minimized. Energetic contributions from zinc were not considered in this calculation. Interfaces with ΔGbind < −20 Rosetta energy units (R.e.u.) were kept. Evaluating solely on computed binding energy biases toward large interfaces, so binding energy per Å2 of interface surface area (binding energy density, R.e.u./Å2) was also a critical metric. Interfaces with binding-energy density < −0.015 were kept. With a more manageable list of designs, other metrics were considered, such as final zinc binding geometry, packing quality 48 (packing score > 0.5), and the number of unsatisfied hydrogen bonds at the interface (<= 6). When making final selections for designs to test, we opted for variety in the zinc ligands (histidine versus cysteine), and favored designs that required relatively few mutations. In the end, we tested four designs from the two-zinc protocol, and four designs from the one-zinc protocol (Supplemental Table S1).
Symmetry and crossing angle analysis
Symmetry deviation was calculated as previously described 45. If A and B are two atoms in one chain, and A′ and B′ are the corresponding atoms in the other chain, then symmetry deviation (Sdev) was calculated as the average of |distance(A, B′) – distance(A′, B)| for all helical Cα carbons. Only helical positions were considered to avoid consideration of flexible termini and a crystal packing artifact in chain B of MID1-H12E.
To compare the dimeric crossing angle in our MID1 crystal structures with that in the model, first a vector was computed for each helix. This vector was defined as the vector between the center of the first four Cα atoms and the center of the last four Cα atoms of the helix. For each pair of helix vectors (chain A helix 1 with chain B helix 1, chain A helix 1 with chain B helix 2, chain A helix 2 with chain B helix 1, and chain A helix 2 with chain B helix 2), an angle between the two vectors and vector scalar product was calculated: . Each of the four computed angles was subtracted from the corresponding angle from the zinc model, and a root mean square deviation was obtained. The values for the root mean square deviation from the model are reported in Table 1.
Experimental Methods
Gene synthesis and cloning, protein expression and purification
Genes were ordered from GenScript, USA optimized for expression in E. coli with an N-terminal BamHI restriction site, a C-terminal stop codon, and a C-terminal SalI restriction site. The pQE-80L expression vector was altered by adding an N-terminal 6x-His tag and an MBP fusion with a TEV protease cleavage site (pQE-H6MBP). Insertion of genes into the pQE-H6MBP vector was confirmed by DNA sequencing analysis. Plasmids were transformed into BL21(DE3) pLysS cells for gene expression. Cells were grown at 37°C in LB broth containing 67 mg/L ampicillin to OD600 = 0.6–0.8 and were then induced with 0.3 mM IPTG. Growth continued at 18°C for 16 hours. Cell pellets were resuspended in lysis buffer containing 20 mM Tris-HCl, 100 mM NaCl, 10% (v/v) glycerol, 0.5 mM DTT, 0.5 mM PMSF, and 1 mM benzamidine. Following sonication, 2 units of RNase and DNase were added for a 20-minute incubation at room temperature followed by centrifugation at 15,000 rpm for 20 minutes (Sorvall RC-5B Plus series). The cleared lysate was subjected to immobilized-metal affinity chromatography (IMAC) using a Ni-NTA HisTrap HP column (GE Healthcare) equilibrated with 20 mM Tris, pH 8.0, 100 mM NaCl, 25 mM imidazole (His-column loading buffer). 1 mM DTT and EDTA were added to the eluted protein, and TEV proteolysis (0.05 mg/ml TEV) occurred overnight at 4°C with gentle rotation. The protein was dialyzed against His-column loading buffer and subjected to IMAC a second time to remove the cleaved 6xHis-MBP fusion. The flow-through was supplemented with 1 mM DTT and 1 mM EDTA to eliminate endogenously bound metal ions and maintain reducing conditions. The flow-through was concentrated for size exclusion chromatography on a Superdex-75 column (GE Healthcare, HiLoad 16/60 prep grade). Appropriate fractions were combined and concentrated (Amicon Ultra, Millipore). Purity was assessed using SDS-PAGE, and protein concentrations were estimated by absorbance at 280 nm using theoretical molar extinction coefficients 49.
Multiple angle light scattering (MALS)
Protein sample of MID1 and MID1-zinc (12 mg/ml) in buffer containing 20 mM MOPS, pH 6.9, 100 mM NaCl, and 0.2 g/L sodium azide was injected onto a 25-ml Superdex-75 size exclusion column (GE Healthcare) connected to a multi-angle light scattering instrument (DAWN HELEOS II, Wyatt Technologies) and a refractometer (OPTILAB rEX, Wyatt Technologies). For both runs, a single elution peak was analyzed using the ASTRA software package (Wyatt Technologies), giving a molecular weight calculated based on light scattering and refractive index.
Circular dichroism
Circular dichroism data were collected on a JASCO J-815 CD spectrometer with the temperature controlled by a JASCO Peltier device and water bath. Experiments were performed in a 1-mm cuvette at 25 μM protein concentration in a buffer containing 10 mM MOPS pH 6.9, 25 mM NaCl, and 0.5 mM TCEP. Additions of metal were equimolar to the protein concentration, 25 μM. Far-UV scans from 190–250 nm of MID1 confirmed the helical character of MID1. Thermal denaturation of MID1 was monitoring at a wavelength of 222 nm to provide estimates of the melting temperature, Tm. The temperature ramp-rate was 3°C/min, and data points were taken every 1°C. Data are reported in units of mean residue ellipticity:
Fluorescence polarization
Fluorescence polarization was used to obtain apparent dissociation constants (Kd) of MID1-apo, MID1-zinc, and MID1-cobalt. The C-terminus of MID1 was extended with a glycine-cysteine (MID1-GC) for conjugation of the thiol-reactive fluorophore Bodipy (507/545)-iodoacetamide (Molecular Probes). MID1-GC was buffer-exchanged into 50 mM Tris-Cl, pH 7.5 followed by addition of 1 mM TCEP. A 20 mM stock solution of Bodipy dissolved in DMSO was added to 6-fold molar excess of MID1-GC, and labeling proceeded overnight at 4°C with gentle inversion. The reaction was stopped by adding 50 mM BME, followed by centrifugation to remove free dye. Any remaining free dye was removed by desalting using a PD10 column (GE Healthcare) with buffer containing 50 mM Tris-Cl, pH 7.5 and 5 mM BME. The labeling efficiency was determined by UV/Vis absorption using an extinction coefficient of 69,000 M−1cm−1 (at 508 nm) for Bodipy. Labeling efficiency of 90% was achieved.
Binding assays were performed using a SPEX FluoroLog-3 instrument (Jobin Yvon Horiba, Edison, NJ). To observe counts greater than 100,000 per second for a 10 nM MID1-Bodipy sample, a cuvette of 1 cm pathlength (3 ml volume) was used with slits opened to 10 nm. Excitation and emission wavelengths were 508 and 545 nm. During the titrations, each polarization reading was taken as an average of three measurements with 0.1 second integration, and readings were taken in triplicate. Data were analyzed to obtain apparent Kd values using a homodimer equilibrium-binding model (Supplemental Methods).
X-Ray Crystallography
For crystallization, MID1 was expressed and purified as described above and stored at 4°C in 100 mM ammonium acetate, pH 7.0 at a concentration of 20 mg/ml prior to crystallization. For crystallization of MID1-zinc, MID1-H12E-zinc, MID1-H35E-zinc, or MID1-cobalt, zinc sulfate or cobalt chloride were added to this MID1 stock solution at equimolar concentration with the protein. Crystals were obtained by vapor diffusion at 20°C from hanging drops with ratios of protein to crystallization solution of 2:1, 1:1 or 1:2 and with drop sizes of 0.3μl to 2μl. Crystals generally appeared within five days and grew to final sizes of up to 300 × 100 × 100 μm.
Prior to data collection, crystals were plunged into liquid nitrogen directly from the crystallization drop, except for MID1-zinc and MID1-H12E-zinc crystals, which were first dipped into LV cryo oil (Mitegen). All datasets were collected at beamlines 23IDB or IDD (GM/CA-CAT) at the Advanced Photon Source (APS), Argonne National Laboratory (ANL), Argonne, IL. Data were processed using the program HKL2000 50. Structures were determined by molecular replacement with the program Phaser 51, using the coordinates of a truncated, wild-type scaffold (PDB code 1YZM) as the search model. Refinement was carried out using the program Phenix consisting of conjugate-gradient minimization and refinement of individual anisotropic atomic displacement parameters, interspersed with manual revisions of the models using the program Coot 52.
For data collection and refinement statistics and a list of residues that could not be located in the electron density see Supplemental Table S3.
NMR Spectroscopy
Uniformly 13C,15N-labeled MID1-zinc was generated for NMR studies. An overnight culture of transformed BL21(DE3)pLysS cells was used to inoculate 6 liters of LB broth containing ampicillin. Cells were grown to an OD600 of 0.6 and were pelleted by centrifugation at 4000 rpm for 20 minutes (Sorvall RC-3 series, Thermo Scientific). The supernatant was removed, and cells were resuspended in 2 liters M9 minimal media containing 15N-ammonium chloride (1 g/L) and 13C-glucose (2 g/L). After a recovery period (shaking for 1 hour at 37°C), expression was induced with 0.3 mM IPTG and proceeded overnight at 18°C. The purification procedure previously described yielded uniformly 13C,15N-labeled MID1-zinc. The final yield was a 1.0 mM solution in a Shigemi NMR tube (buffer: 10 mM MOPS pH 6.9, 25 mM NaCl, 10% D20).
NMR spectra were recorded at 25°C on a Varian INOVA 750 spectrometer equipped with a cryogenic probe. A 2D [15N,1H]-HSQC spectrum was recorded with 1.3 hours of measurement time, and two through-bond correlation G-matrix Fourier transform (GFT) NMR experiments 53,54 complemented by 3D HNNCO 55 were performed for assignment of the polypeptide backbone and 13Cb resonances (total measurement time: ~21 hrs). The spectra were processed and analyzed with the programs PROSA 56 and CARA 57, respectively. Unambiguous sequence-specific backbone (1HN, 15N, 13Ca, and 13C′) and 13Cb resonance assignments were obtained only for Gln 6, Gln 7, the polypeptide segment comprising residues 18 to 31 and Asp 46 by using (4,3)D HNNCabCa/CabCa(CO)NHN and 3D HNNCO.
Supplementary Material
Table 2.
Summary of crystal formation: crystallization buffer, solvent content, space group, cell parameters, and resolution.
| structure | crystallization buffer | solvent content | space group | molecules in asymmetric unit | Bragg spacing (dmin) |
|---|---|---|---|---|---|
|
MID1-apo1 PDB code XXXX |
0.1 M MES, pH 5.97 30% (v/v) PEG 600 7.5% (w/v) PEG 1000 5% (v/v) glycerol |
22% | P212121 | 1 | <0.9 Å |
|
MID1-apo2 PDB code XXXX |
0.1 M MES pH 6.0 30% (v/v) PEG 600 5% (w/v) PEG 1000 10% (v/v) glycerol |
22% | P212121 | 2 | 1.2 Å |
|
MID1-cobalt PDB code XXXX |
0.1 M MES, pH 6.0 30% (v/v) PEG 200 10% (w/v) PEG 3000 |
29% | P1 | 8 | 1.2 Å |
|
MID1-zinc PDB code XXXX |
0.1 M sodium citrate, pH 5.5 0.08 M K/Na tartrate |
30% | P212121 | 2 | 1.1 Å |
|
MID1-H12E-zinc PDB code XXXX |
0.1 M bicine, pH 9.0 27% (w/v) PEG 3350 |
27% | P212121 | 2 | 1.0 Å |
|
MID1-H35E-zinc PDB code XXXX |
0.1 M sodium acetate, pH 4.0 15% (w/v) PEG 600 10% isopropanol 5% (v/v) glycerol |
28% | P212121 | 2 | 1.0 Å |
Acknowledgments
This work was funded by the NIH grant GM073960 and by the National Science Foundation graduate research fellowship (2009070950 to B.D.). We thank Jenny Williams and ITS Research Computing at UNC for their important assistance in running extensive design simulations on the Topsail supercomputing cluster. We thank Dr. Ashutosh Tripathy of the UNC Macromolecular Interaction Facility for MALS data collection and analysis, and Dr. Greg Young of the UNC Biomolecular NMR Lab. We thank Andrew Leaver-Fay for help with RosettaMatch and Steven Lewis for helping to write the Rosetta applications SurfaceGroups and ZincMatchFilter. We thank SER-CAT for resources provided for the crystallography studies, and we thank NESG for resources provided for the NMR studies in this work.
Footnotes
Supporting Information Available. Figures: ideal zinc-coordination geometry; ribbon diagrams of the eight tested designs; size exclusion chromatograms; 2D [15N, 1H]-HSQC spectra with partial resonance assignments; crystal structures of MID1-apo1, MID1-apo2, MID1-zinc, and MID1-cobalt; QUILT analysis of MID1 hydrophobicity. Tables: computed parameters for eight designs; experimental end-results for eight designs; crystallography statistics; zinc binding geometries; zinc ligand statistics. Other: PDB scaffold list, Rosetta command lines and input, fluorescence polarization homodimer equilibrium fitting equation.
References
- 1.Karanicolas J, Kuhlman B. Curr Opin Struct Biol. 2009;19:458–63. doi: 10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandell DJ, Kortemme T. Nat Chem Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- 3.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Science. 2011;332:816–21. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reina J, Lacroix E, Hobson SD, Fernandez-Ballester G, Rybin V, Schwab MS, Serrano L, Gonzalez C. Nat Struct Biol. 2002;9:621–7. doi: 10.1038/nsb815. [DOI] [PubMed] [Google Scholar]
- 5.Shifman JM, Mayo SL. J Mol Biol. 2002;323:417–23. doi: 10.1016/s0022-2836(02)00881-1. [DOI] [PubMed] [Google Scholar]
- 6.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Mol Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 7.Havranek JJ, Harbury PB. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 8.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. J Mol Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 9.Bolon DN, Grant RA, Baker TA, Sauer RT. Proc Natl Acad Sci U S A. 2005;102:12724–9. doi: 10.1073/pnas.0506124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoryan G, Reinke AW, Keating AE. Nature. 2009;458:859–64. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sammond DW, Bosch DE, Butterfoss GL, Purbeck C, Machius M, Siderovski DP, Kuhlman B. J Am Chem Soc. 2011;133:4190–2. doi: 10.1021/ja110296z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Zhu X, Liang H, Cao A, Chang Z, Lai L. Proc Natl Acad Sci U S A. 2007;104:5330–5. doi: 10.1073/pnas.0606198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang PS, Love JJ, Mayo SL. Protein Sci. 2007;16:2770–4. doi: 10.1110/ps.073125207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jha RK, Leaver-Fay A, Yin S, Wu Y, Butterfoss GL, Szyperski T, Dokholyan NV, Kuhlman B. J Mol Biol. 2010;400:257–70. doi: 10.1016/j.jmb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guntas G, Purbeck C, Kuhlman B. Proc Natl Acad Sci U S A. 2010;107:19296–301. doi: 10.1073/pnas.1006528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karanicolas J, Corn JE, Chen I, Joachimiak LA, Dym O, Peck SH, Albeck S, Unger T, Hu W, Liu G, Delbecq SG, TMCPS, Liu DR, Baker D. Mol Cell. 2011;42:250–60. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auld DS. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 18.Lee YM, Lim C. J Mol Biol. 2008;379:545–53. doi: 10.1016/j.jmb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Alberts IL, Nadassy K, Wodak SJ. Protein Sci. 1998;7:1700–16. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rulisek L, Vondrasek J. J Inorg Biochem. 1998;71:115–27. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 21.Cunningham BC, Mulkerrin MG, Wells JA. Science. 1991;253:545–548. doi: 10.1126/science.1907025. [DOI] [PubMed] [Google Scholar]
- 22.Krishna SS, Majumdar I, Grishin NV. Nucleic Acids Research. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis AM, Berg JM. Journal of the American Chemical Society. 2009;131:11492–11497. doi: 10.1021/ja9028928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regan L, Clarke ND. Biochemistry. 1990;29:10878–83. doi: 10.1021/bi00501a003. [DOI] [PubMed] [Google Scholar]
- 25.Hellinga HW, Caradonna JP, Richards FM. J Mol Biol. 1991;222:787–803. doi: 10.1016/0022-2836(91)90511-4. [DOI] [PubMed] [Google Scholar]
- 26.Hellinga HW. Fold Des. 1998;3:R1–8. doi: 10.1016/S1359-0278(98)00001-7. [DOI] [PubMed] [Google Scholar]
- 27.Klemba M, Gardner KH, Marino S, Clarke ND, Regan L. Nat Struct Biol. 1995;2:368–73. doi: 10.1038/nsb0595-368. [DOI] [PubMed] [Google Scholar]
- 28.Lu Y, Yeung N, Sieracki N, Marshall NM. Nature. 2009;460:855–62. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summa CM, Rosenblatt MM, Hong JK, Lear JD, DeGrado WF. J Mol Biol. 2002;321:923–38. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 30.Marsh EN, DeGrado WF. Proc Natl Acad Sci U S A. 2002;99:5150–4. doi: 10.1073/pnas.052023199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korendovych IV, Senes A, Kim YH, Lear JD, Fry HC, Therien MJ, Blasie JK, Walker FA, Degrado WF. J Am Chem Soc. 2010;132:15516–8. doi: 10.1021/ja107487b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bender GM, Lehmann A, Zou H, Cheng H, Fry HC, Engel D, Therien MJ, Blasie JK, Roder H, Saven JG, DeGrado WF. J Am Chem Soc. 2007;129:10732–40. doi: 10.1021/ja071199j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salgado EN, Faraone-Mennella J, Tezcan FA. J Am Chem Soc. 2007;129:13374–5. doi: 10.1021/ja075261o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salgado EN, Ambroggio XI, Brodin JD, Lewis RA, Kuhlman B, Tezcan FA. Proc Natl Acad Sci U S A. 2010;107:1827–32. doi: 10.1073/pnas.0906852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh D, Pecoraro VL. Current Opinion in Chemical Biology. 2005;9:97–103. doi: 10.1016/j.cbpa.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Matthews JM, Loughlin FE, Mackay JP. Current Opinion in Structural Biology. 2008;18:484–490. doi: 10.1016/j.sbi.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. PLoS One. 2011:6. doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Rothlisberger D, Baker D. Protein Sci. 2006;15:2785–94. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andre I, Bradley P, Wang C, Baker D. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das R, Andre I, Shen Y, Wu YB, Lemak A, Bansal S, Arrowsmith CH, Szyperski T, Baker D. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18978–18983. doi: 10.1073/pnas.0904407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunn MF. Biometals. 2005;18:295–303. doi: 10.1007/s10534-005-3685-y. [DOI] [PubMed] [Google Scholar]
- 42.Andre I, Strauss CE, Kaplan DB, Bradley P, Baker D. Proc Natl Acad Sci USA. 2008;105:16148–52. doi: 10.1073/pnas.0807576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wishart DS, Sykes BD. J Biomol NMR. 1994;4:171–80. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 44.Bahadur RP, Zacharias M. Cell Mol Life Sci. 2008;65:1059–72. doi: 10.1007/s00018-007-7451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andre I, Strauss CEM, Kaplan DB, Bradley P, Baker D. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16148–16152. doi: 10.1073/pnas.0807576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhanhua C, Gan JG, Lei L, Sakharkar MK, Kangueane P. Bioinformation. 2005;1:28–39. doi: 10.6026/97320630001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. PLoS One. 2011;6:e19230. doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheffler W, Baker D. Protein Sci. 2010;19:1991–5. doi: 10.1002/pro.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gill SC, von Hippel PH. Analytical Biochemistry. 1989;182:319–26. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallographica Section D-Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S, Szyperski T. J Biomol NMR. 2004;28:117–30. doi: 10.1023/B:JNMR.0000013827.20574.46. [DOI] [PubMed] [Google Scholar]
- 54.Atreya HS, Szyperski T. Proc Natl Acad Sci U S A. 2004;101:9642–7. doi: 10.1073/pnas.0403529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavanagh J. Protein NMR spectroscopy: principles and practice. Academic Press; 2007. [Google Scholar]
- 56.Güntert P, Dötsch V, Wider G, Wüthrich K. Journal of Biomolecular NMR. 1992;2:619–629. [Google Scholar]
- 57.Keller R. The Computer Aided Resonance Assignment Tutorial. Cantina Verlag; Zürich: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.