Abstract
Purpose
To determine whether HPV DNA can be detected in the plasma of patients with HPV(+) oropharyngeal carcinoma (OP) and to monitor its temporal change during radiotherapy (RT).
Methods and Materials
We used PCR to detect HPV DNA in the culture media of HPV(+) SCC90, VU147T and the plasma of SCC90 and HeLa tumor bearing mice, non-tumor controls and those bearing HPV(-) tumors. We used real time quantitative PCR (qPCR) to quantify plasma HPV DNA in 40 HPV(+) OP, 24 HPV(-) head and neck cancer (HNC) patients and 10 non-cancer volunteers. Tumor HPV status was confirmed by p16INK4a staining and HPV16/18 PCR or HPV ISH. 14 patients had serial plasma samples for HPV DNA quantification during RT.
Results
HPV DNA was detectable in the plasma samples of SCC90- and HeLa-bearing mice but not in controls. It was detected in 65% of pretreatment plasma samples from HPV(+) OP patients using E6/7 qPCR. None of the HPV(-) HNC or non-cancer controls had detectable HPV DNA. Pretreatment plasma HPV DNA copy number correlated significantly with nodal metabolic tumor volume (assessed on FDG-PET). Serial measurements in 14 patients showed rapid decline in HPV DNA that became undetectable at RT completion. In 3 patients, HPV DNA rose to discernable level at the time of metastasis.
Conclusions
Xenograft studies indicated that plasma HPV DNA is released from HPV(+) tumors. Circulating HPV DNA is detectable in most HPV(+) OP patients. Plasma HPV DNA may be a valuable tool for identifying relapse.
Keywords: Human papillomavirus, oropharyngeal carcinoma, radiotherapy, plasma, circulating DNA
Introduction
Several studies have established a causal relationship between the human papillomavirus (HPV) infection and the development oropharyngeal carcinomas (OP).(1-3) Epidemiological studies have shown that the incidence of HPV-related OP is rising in the Western hemisphere, making it one of largest head and neck squamous cell carcinoma (HNSCC) subgroup.(4, 5) Prognostically, HPV(+) OP tumors fare significantly better than HPV(-) tumors. It has been proposed that treatment de-escalation for HPV(+) OP tumors merits study. Although the outcome for these patients is favorable, many still recur. Having a tumor specific maker for monitoring response and finding early recurrence during treatment de-escalation would be useful in OP patients.
Blood is the only fluid that is in direct contact with all organs and therefore offers an attractive non-invasive means of cancer surveillance. Since the first evidence showing that tumor-associated DNA can be detected in the serum of cancer patients (6), several studies have evaluated different types of tumor DNA as biomarkers for cancer surveillance.(7-12) The presence of viral DNA in viral-related tumors offers a distinct marker for detection in the blood. Epstein Barr Virus (EVB) DNA is often found in the plasma of nasopharyngeal carcinoma (NPC) patients, and it has been shown to be a sensitive and reliable marker for prognostication in NPC.(10, 11, 13) Several large studies have correlated pre-treatment circulating EBV DNA with stage progression and have shown that the post-treatment EBV DNA level was highly indicative of persistent tumor or early relapse.(14-18) Moreover, rises in circulating EBV DNA can precede clinical signs of recurrence for months, making it an inexpensive test for surveillance.(18) In contrast to NPC, the potential clinical application of circulating HPV DNA in OP cancer has not been investigated. Prior studies of circulating HPV DNA in cervical cancers have reported a pre-treatment detection rate ranging from 12% to 65% and the positivity rate is highly dependent on the patient population and the assay.(19-23) Only one study has evaluated circulating HPV DNA in HNSCC, using a combination of conventional PCR, southern blot hybridization and quantitative PCR (qPCR).(24) In this study, Capone et al. were able to detect E6/7 HPV DNA in 6 of 13 patients (46%) with HPV16(+) tumor. Since then, there has been no other publication on the role of plasma HPV DNA in HNSCC.
In this study, we ask if circulating HPV DNA is derived from HPV(+) HNSCC using xenograft models and whether it can be detected in the pretreatment plasma of patients with HPV(+) OP tumors by conventional and qPCR. In addition, we assessed the change in plasma HPV DNA levels in a subgroup of patients who had serial blood samples obtained during the course of chemoradiotherapy and at the time of relapse.
Materials and Methods
Patient selection
Between 6/1/2007 and 1/30/2010, 85 HNSCC patients participated in an institutional review board (IRB) approved biomarker study that allows for collection of blood and tumor tissue whenever available. Of these, we identified 40 patients with OP carcinomas that were positive for p16INK4a by immunohistochemical (IHC) staining at diagnosis and available tumor blocks at Stanford for additional HPV analysis, either by in-situ hybridization (ISH) or PCR. This group formed the HPV(+) cohort. Within this group, 14 patients had serial plasma samples collected during radiation treatment. For the HPV(-) cohort, we identified 24 patients with tumors that stained negative for p16INK4a and available tumor block for HPV status confirmation; these patients can have the primary tumor located either within or outside of the oropharynx. We also consented 10 non-cancer volunteers under a separate IRB approved study for blood samples.
Collected clinical information included age, gender, tumor site, tumor stage, nodal stage, overall stage (2002 AJCC staging system), tobacco and alcohol use. For tobacco use, patients were categorized as current users (those who quit <1 y before diagnosis), recently quit (those who quit <20 y before diagnosis), remotely quit (those who quit ≥20 y before diagnosis) or never users (those who smoked < one pack-year in their lifetime). For alcohol use, patients were classified as light (<1 drink/day), moderate (1-2 drink/day), and heavy drinker (>2 drink/day). Follow up information was collected for patients with HPV(+) tumor.
Metabolic tumor volume
The pretreatment metabolic tumor volume (MTV) was derived from the staging 18F-Deoxyglucose Positron Emission Tomography scan (FDG-PET). It was defined as the volume of hypermetabolic tissues within the tumor with a standard uptake value (SUV) >50% of the maximum SUV (also within the tumor).(14) These volumes were delineated using a semi-automatic custom software (MIMcontouring®, Cleveland, OH). MTV for the primary tumor (MTV-P) and involved nodes (MTV-N) were delineated independently and summed together as a total MTV (MTV-T). We have previously found that pretreatment MTV was prognostic for outcomes in HNSCC patients.(14)
p16INK4a Immunohistochemistry
Immunoperoxidase stains for p16INK4a (clone E6H4, Dako) were performed on 4μM-thick whole tumor sections as previously described.(25) p16INK4a staining was interpreted by a pathologist (CSK), who was blinded to the clinical data, and scored as follows: negative (weak cytoplasmic staining in < 5% of the cells), focally positive (focal strong nuclear and/or cytoplasmic staining in 5-80% of the cells) and diffusely positive (diffuse strong staining in >80% of the cells). (26-28)
Cell line
SCC90, a HPV16(+) oropharyngeal cancer (OP) cell line, was a gift from Dr. Robert Ferris (University of Pittsburgh). VU147T, another OP HPV16(+) cell line was a gift from Professor H. Joenje (VU Medical Center, Amsterdam). HeLa (HPV18+) and MiaPaCa-2 were obtained from the American Type Culture Collection. SAS, an HPV(-) oral cavity cancer cell line, was obtained from Japan Health Science Research Resources Bank. Cells were cultured in DMEM supplemented with 10% fetal bovine serum.
Xenograft study
All animal studies were approved by the Stanford University Administrative Panel on Laboratory Animal Care (APLAC). SCC90, HeLa, SAS and MiaPaCa-2 (all at 5 × 106 cells/injection) were implanted into the flanks of SCID mice. When tumors reached 500 mm3, 0.4 ml blood/mouse was collected for plasma samples, which were stored at -80°C until DNA extraction.
DNA extraction from plasma and tumor tissues
Collected blood samples from patients were centrifuged (3000 rpm 4°C for 10 min), aliquoted and stored at -80°C until DNA extra ction. Plasma DNA was extracted using the QIAamp Blood Mini Kit (QIAgen, Valencia, CA) per manufacturer's protocol. For paraffin-embedded tumors, the block was punched, de-paraffinized by xylene, and DNA was extracted following the instructions of QIA blood and tissue Kit (QIAgen).
Primer/probe information
The sequences for all primers and probes are listed in Table 1. (29),(30).
Table 1.
Sequences of primers and probes for conventional PCR and qPCR
| Gene Name | Forward Primer (5’ to 3’) | Reverse primer (5’ to 3’) | Probe (5’ to 3’) |
|---|---|---|---|
| Conventional PCR | |||
| HPV L1 | tttgttactgtggtagatacatc | gaaaaataaactgtaatcatattc | |
| HPV16 E6 1 | atgcaccaaaagagaactgc | ataatgtctatactcactaa | |
| HPV16 E6 2 | aatgtttcaggacccaca | catttatcacatacagcata | |
| HPV16 E7 1 | gatctctactgttatgagca | cacaattcctagtgtgccca | |
| HPV16 E7 2 | aattaaatgacagctcagag | ttaacaggtcttccaaagta | |
| HPV18 E6 1 | tactatggcgcgctttgagg | atacatttatggcatgcagc | |
| HPV18 E6 2 | atccaacacggcgaccctac | atgggtatactgtctctat | |
| HPV18 E7 1 | aagtatgcatggacctaagg | tgcttactgctgggatgcac | |
| HPV18 E7 2 | taaggcaacattgcaagaca | cacggacacacaaaggac | |
| human-β–Globin | caacttcatccacgttcacc | gaagagccaaggacaggtac | |
| mouse-β–Globin | acctgactgatgctgagaagg | cccttgaggctgtccaagtg | |
| qPCR | |||
| HPV16 E6/7 | gaaccgaaaccggttagtataa | atgtatagttgtttgcagctctgt | FAM-aggacccacaggagcgaccc-BHQ1 |
| HPV18 E6/7 | ggaccgaaaacggtgtatataa | cagtgaagtgttcagttcggt | CO560-atgtgagaaacacaccacaatactatggcgcg-BHQ1 |
| human-β–Globin | acacaactgtgttcactagc | caacttcatccacgttcacc |
Conventional HPV PCR – tumor and plasma
HPV DNA was detected with L1 G5+/6+ primer (L1 primer), and the amplification using a touchdown thermoprofile as described.(25) HPV16/18 E6, E7 nested primers were also used. β-globin primer was included in each assay to assess the quality of DNA extraction and amplification. Cycling conditions was described.(20)
Real-Time Quantitative HPV DNA PCR
Real-time PCR reactions were set up using the TaqMan PCR Master Mix (Roche, IN). HPV-16/18 E6/7 primers and probes were as described.(30) PCR primers and probes were synthesized by Biosearch Technologies (Novato, CA). β-globin was used as an internal positive control.
All PCR reactions were carried out in triplicate. A no template negative control was included in each analysis. DNA amplifications were carried out on ABI 9700 Sequence Detector (Applied Biosystems) which can detect multiple fluorescent dyes. Fluorescence data were analyzed with Sequence Detection System Software.
Statistics
Statistical analysis was performed using the Statview (Computing Resource Center, San Monica, CA) statistical software. The analysis of variance (ANOVA) and student T-test were used to compare plasma HPV DNA copy number between the different patient groups. Both the Spearman rank and the Pearson correlation tests were used to determine the relationship between plasma HPV DNA and continuous variables (e.g. MTV and tumor HPV DNA).
Results
Xenograft study
We used both the L1 G5+/6+ primer and HPV16/18 subtype specific nested E6/E7 primers for conventional PCR studies. Figure 1a shows strong detection of HPV DNA in the culture media of HPV16(+) SCC90 and VU147T HNSCC cell lines, indicating that these cells release DNA, presumably from cell death or lysis. We then implanted SCC90 and HeLa (HPV18+) cells in the flanks of SCID mice. VU147T cells failed to grow in mice. Mice without tumor implanted and mice bearing known HPV(-) cells (MiaPaCa2 and SAS) were used as negative controls. Figure 1b shows that 9 of 10 SCC90 and all HeLa tumor bearing mice had adequate DNA extraction from the plasma based on the β-globin amplification; of the 9 SCC90 mice, plasma HPV DNA was detectable in 8 (89%) with the L1 primers and in 7 (78%) with nested HPV16 E6/E7 primers. Of the 3 HeLa mice, plasma HPV DNA was detectable in all with both primers. However, plasma HPV DNA was not detectable in any of the control mice, indicating that the circulating DNA was derived directly from the implanted tumors that harbored HPV DNA.
Figure 1.
a. Detection of Human Papillomavirus16 DNA in the media of SCC 90 (Left) and VU147T (Right) by PCR (L1 and HPV16 E6/E7 primers). DNA from the lysates of SCC90, SiHa, VU147T cells and HPV16 plasmids were used as positive control and no template as negative control.
b. Detection of HPV DNA in the plasma of SCID mice, bearing HPV16(+) SCC90 (upper panel, lane 1-10) and HPV18(+) HeLa cells (lower panel, lane 6-8), but not in HPV(-) negative Miapaca2 (upper panel, lane 11-13), SAS cells (lower panel, lane 3-5) or non-tumor bearing mice (upper panel, lane 14-16). Conventional PCR was used with L1 and HPV16/18 E6/7 nested primers. βglobin confirmed adequate DNA extraction. DNA from HeLa cells and HeLa tumors was used as positive control and no template as negative control.
Validation of tumor HPV status in studied patients
We identified 40 OP patients with p16INK4a positive tumors and minimal smoking history as the putative HPV(+) group and 24 patients with p16INK4a negative HNSCC tumors (two OP, ten oral cavity, six hypopharyngeal, three laryngeal and three node positive cutaneous squamous cell carcinoma) as the putative HPV(-) control group. p16INK4a staining for all patients were re-reviewed and confirmed by two independent pathologists (CSK and BAP). All but one of the 40 p16INK4a positive OP tumors showed diffusely strong staining. Only one tumor showed focally positive pattern. None of the 24 previously classified p16INK4a negative tumors showed any staining.
We then evaluated HPV status in these 64 tumors by either conventional PCR using the L1 primers (N = 54) or in-situ hybridization (ISH) with HPVIII Family 16 probe (N = 10, Ventana, Tucson, AZ). We confirmed that all 40 p16INK4a positive OP tumors were HPV(+) and all 24 p16INK4a negative tumors were HPV(-). There was no discordance between p16INK4a staining and HPV status in this highly selected patient group. Table 2 shows the characteristics for the 40 patients with HPV(+) OP tumors. Consistent with prior reports, most of these patients were either non- or ex-smokers with small primary tumor and N2 nodal disease.
Table 2.
Characteristics of 40 patients with HPV(+) oropharyngeal carcinoma
| Characteristics | # pts (Total N= 40) | |
|---|---|---|
| Age | Median (Range) | 58.5 (42-85) |
| Gender | Male | 37 (92.5%) |
| Tobacco | Current | 6 |
| Recently quit | 7 | |
| Remotely quit | 7 | |
| Never | 20 | |
| Alcohol | <1 d/d | 26 |
| 1-2 d/d | 6 | |
| > 2 d/d | 8 | |
| Tumor subsite | Base of tongue | 18 |
| Tonsil | 20 | |
| Pharyngeal wall | 2 | |
| T-stage | T1 | 8 |
| T2 | 18 | |
| T3 | 10 | |
| T4 | 4 | |
| N-Stage | 0 | 4 |
| 2a | 5 | |
| 2b | 20 | |
| 2c | 10 | |
| 3 | 1 | |
| Stage | I | 1 |
| III | 4 | |
| IVA | 35 | |
| MTV (N = 37) | Tumor | 12.87 (2.8-94.6) |
| Median (Range) | Node | 13.45 (0-105.5) |
| Total | 32.14 (6.8-146.1) | |
| Tumor HPV DNA (copies/ng DNA, N = 27) | Median (range) | 89 (2-5693) |
| Plasma HPV DNA (copies/ml, N=40) | Median (range) | 222 (0-5500) |
In 27 HPV(+) tumors with enough tumor tissue for DNA extraction, the presence of E6/7 DNA was assessed by qPCR using HPV16/18 E6/7 specific primer/probe sets. Table 2 shows the range of E6/7 tumor DNA copy number per ng of extracted DNA for these patients; seven tumors had <10 copies/ng, seven had between 10-100 copies/ng and 13 had >100 copies/ng. There was no obvious relationship between tumor HPV DNA copy number and any clinical characteristics.
Detection of pretreatment circulating HPV DNA
After confirming the tumor HPV status, we isolated DNA from pretreatment plasma samples in all 64 HNSCC patients and 10 non-cancer controls. The presence of HPV in the plasma was assessed by conventional PCR using the L1 primer and HPV16/18 E6/7 nested primers. The plasma DNA copy number was then quantified by qPCR with HPV 16/18 E6/7 primer/probe sets. None of the 24 HPV(-) patients or the 10 non-cancer volunteers had HPV DNA detected in their blood. Figure 2a shows a conventional PCR gel of plasma HPV DNA in 4 representative patients with HPV16(+) tumors. The β-globin band shows that patient #2's sample was not analyzable due to failure to extract adequate DNA. In the other three patients with analyzable samples, we were able to detect plasma HPV DNA from two. Table 3 shows the sensitivity for the different primers in detecting HPV DNA in the blood. The L1 primer had the highest detection rate of 68%, followed by qPCR for E6/7 (65%). When less sensitive primers (nested E6 and E7) and conventional PCR were used, plasma HPV DNA was positive in only 42% of patients.
Figure 2.
a. Conventional PCR gel showing examples of detected plasma HPV DNA.
b. An example of a aPCR standard curve using HPV16 plasmid. The trend line was drawn for HPV16 plasmid plus genomic DNA.
c. Histogram showing the distribution of plasma HPV DNA copies/ml by the number of patients.
Table 3.
Detection rate of circulating HPV DNA based on tumor expression for different primers and PCR assays
| PCR type and Primer | HPV(+) Tumors |
| # Plasma (+)/# Tumors (+) (%) | |
| Conventional & L1 G5+/6+ primers | 27/40 (68%) |
| qPCR & HPV16/18 E6/7 primer/probe | 26/40 (65%) |
Next, we determined whether the volume of plasma used for DNA extraction influenced the qPCR detection rate. DNA was extracted from 0.2 ml and 0.6 ml plasma volume, respectively, from the first 24 HPV(+) OP patients and used for qPCR. The positive detection rate was significantly higher for the larger volume (17/24 for 0.6 ml versus 7/24 for 0.2 ml, p = 0.009). Therefore, all subsequent studies employed 0.6 ml of plasma. Figure 2b shows the typical standard curve generated using HPV16 and HPV18 plasmids. The addition of exogenous DNA did not affect the curve. The median and range of HPV DNA copy number per ml of plasma for the 40 HPV(+) OP patients are shown in table 2 and the histogram of copy number is shown in Figure 2c. In 26 patients with detectable HPV DNA, it was <500 copies/ml in 13 and >500 copies/ml in 13 patients.
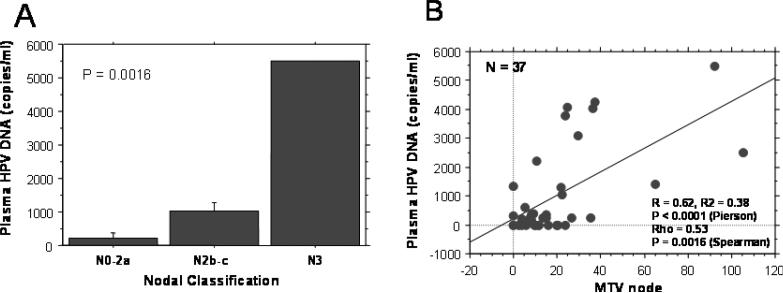
Relationship between pretreatment plasma HPV DNA copy number and clinical parameters
We evaluated the relationship between plasma HPV DNA copy number and other clinical/biologic parameters including tumor HPV DNA copy number, cigarette smoking, alcohol use, primary tumor subsite (base of tongue vs. other site), tumor classification, nodal classification, tumor and nodal metabolic volume (MTV). These data are shown in Table 4. The only parameters that achieved a significant correlation on univariate analysis were nodal classification (Figure 3a) and nodal MTV (Figure 3b). Tumor HPV copy number (p = 0.08), primary tumor subsite (higher for the base of tongue and oropharyngeal wall, p = 0.06) and age (higher for older patients, p = 0.07) were of borderline significance.
Table 4.
Relationship between plasma HPV DNA copy number and different clinical parameters
| Parameter | Patient number | Plasma HPV copy number +/-SE or Correlation coefficient (R and R2) | p-value | |
|---|---|---|---|---|
| Age | ≤ 58 | 20 | 554.2+/-220.7 | 0.09 |
| > 58 | 20 | 1361.4+/-404.3 | ||
| Continuous | 40 | R = 0.29, R2 = 0.09 | 0.07 | |
| Gender | Male | 37 | 1030.2+/-251.9 | 0.29 |
| Female | 3 | 64.7+/-64.7 | ||
| Cigarette use | Never | 20 | 923.8+/-323.1 | 0.97 |
| Former-recently quit | 7 | 777.9+/-430.1 | ||
| Former-remotely quit | 7 | 1147.3+/-605.2 | ||
| Current | 6 | 1060.0+/-890.1 | ||
| Alcohol Use | Light | 26 | 1050.4+/-303.5 | 0.34 |
| Moderate | 6 | 1425.8+/-822.6 | ||
| Heavy | 8 | 305.6+/-166.6 | ||
| T-classification | T1-2 | 26 | 903.0+/-267.7 | 0.76 |
| T3-4 | 14 | 1059.6+/-470.9 | ||
| N-Classification | N1-2A | 9 | 225.2+/-144.2 | 0.002 |
| N2B-C | 30 | 1026.1+/-262.7 | ||
| N3 | 1 | 5500 | ||
| Tumor Site | Base of tongue | 18 | 1432.3+/-408.0 | 0.06 |
| Pharyngeal wall | 2 | 1986.0+/-1792.0 | ||
| Tonsil | 20 | 425.2+/-217.0 | ||
| MTV | Primary | 37 | R = 0.12, R2 = 0.02 | 0.47 |
| Node | R = 0.62, R2 = 0.38 | <0.0001 | ||
| Primary + node | R = 0.51, R2 = 0.26 | 0.001 | ||
| Tumor | HPV copy number | 27 | R = 0.34, R2 = 0.12 | 0.08 |
MTV: metabolic tumor volume; SE: Standard error
Figure 3.
a. Relationship between nodal classification and plasma HPV DNA copy number.
b. Correlation between nodal metabolic tumor volume and plasma HPV DNA copy number.
Changes of HPV DNA copy number during chemoradiation
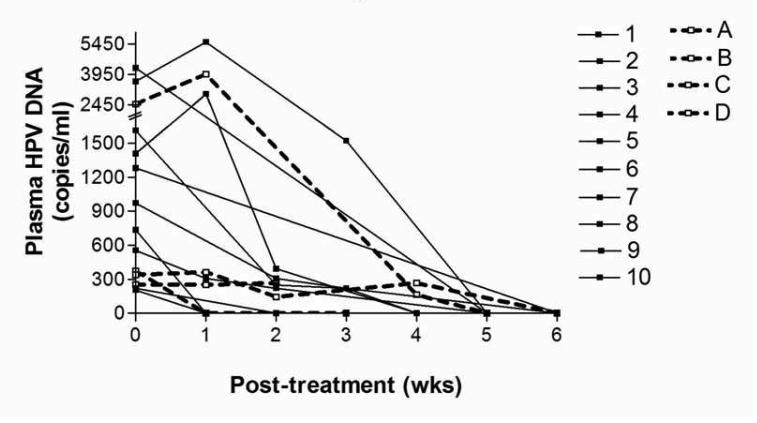
In 14 patients with detectable pretreatment plasma HPV DNA, we also obtained serial plasma samples (weekly or every other week) during the course concurrent chemoradiotherapy. We used qPCR to follow the change in plasma HPV DNA level. Four of these patients eventually relapsed: one locoregionally and 3 distantly in the lungs. Figure 4 shows the change in plasma HPV DNA over time during chemoradiotherapy for patients with and without subsequent relapse. There was a gradual decline in HPV DNA during therapy that became indiscernible by week six in all patients. Some patients showed a transient rise in HPV DNA during the first two weeks; however, the levels then decreased rapidly and became undetectable later on. There were no obvious differences in the rate of HPV DNA decline between the patients with eventual tumor relapse and those without.
Figure 4.
pPCR results showing declining of circulating HPV DNA copies/ml during chemoradiation in 14 patients with serial measurements. The dash lines (Patients A-D) represent patients who developed recurrence and the solid lines (Patients 1-10), those who did not recur.
We were able to obtain plasma sample at the time of relapse for the 3 patients who developed lung metastasis but not for the one patient with locoregional relapse. All three patients had removal of their lung tumors that proved to be metastatic squamous cell carcinoma, p16INK4a and HPV 16 positive. The time of relapse ranged from 12 to 22 months after the initial diagnosis. All 3 patients had detectable HPV DNA level in the plasma, ranging from 158-542 copies/ml, at the time of relapse. Of interest was a patient who had detectable HPV DNA in the blood (111 copies/ml) 4 months prior to the detection of lung metastasis. A surveillance chest CT 4 months later (required by a clinical trial that he was on) revealed a new lung nodule that proved to be HPV(+) metastatic squamous cell carcinoma on biopsy. The HPV DNA level at the time of the biopsy rose to 542 copies/ml.
Discussion
In this study, we have been able to detect circulating HPV DNA in the pretreatment plasma samples from the majority of HPV(+) OP cancer patients but in none of the HPV(-) HNSCC or non-cancer volunteers. Our xenograft studies confirmed that circulating HPV DNA was released from HPV(+) tumor cells.
Our plasma HPV DNA detection rate of 65% by qPCR is higher than previously reported for HNSCC and cervical cancer patients.(19-24, 31) By increasing the plasma volume from 0.2 to 0.6 ml, we were able to improve the yield of DNA extraction, which translated to a higher HPV DNA detection rate. However, the sensitivity of the assay still needs improvement. In addition, the circulating DNA copy number is lower than reported for EBV DNA in NPC.(16, 17, 32) The low copy number and the imperfect detection rate may be inherent to the fact that we are amplifying a single copy gene in the virus. Moreover, since HPV exists mainly as integrated rather than episomal form, the number of viral genome per tumor cell are likely lower than EBV, which exists mainly in episomal form. One strategy to improve detection sensitivity is to amplify a short repeat segment that is unique to the viral DNA. Such strategy has been used for EBV DNA and may be applicable for HPV.(10, 13, 18)
Due to our small sample collection, we were able to test only 24 non-HPV HNSCC and 10 non-cancer volunteers to assess the assay specificity. Although we were not able to detect any HPV DNA by either conventional or qPCR in these samples, we need to confirm these findings in a larger group of patients harboring HPV(-) tumors. More importantly, we need to test the assay on the plasma sample of people who are known to have HPV colonization in the oropharynx but do not have cancer. Prior analysis of circulating HPV DNA in cervical cancer patients indicated that the detection rate was dependent on tumor invasion and stage, with a higher detection rate noted for patients with invasive and late stage (stage III-IV) tumors than in those with carcinoma in situ.(20, 21, 33) At least two groups have reported zero HPV DNA detection rate in the blood of women with cervical HPV16 or 18 infection but without invasive cancer.(21, 34)
Our plasma HPV DNA detection rate in OP is much higher than those reported for cervical cancer that harbors the same HPV serotypes. We hypothesize that this discrepancy may be related to the fact that OP tumors are more likely to have nodal involvement than cervical cancer at diagnosis. Our data suggest a strong relationship between involved nodal volume and plasma HPV DNA levels. In fact, nodal MTV and nodal stage were the only parameters that correlated with plasma HPV DNA copy number on univariate analysis.
Treatment with combined chemoradiotherapy resulted in a rapid reduction and clearance of HPV DNA from the plasma in our pilot study of 14 patients with serial on-treatment samples. This is different from a prior study, which showed that HPV16 DNA is still detectable in the saliva of 11% of the patients with HPV16(+) tumors after therapy.(35) However, the rate of decline in plasma HPV DNA level during therapy in this small patient cohort was not sensitive enough to distinguish the patients who would eventually relapse from those who would not. This is in contrast with the story of circulating EBV DNA in NPC patients, where a persistently detectable post-treatment EBV DNA level was highly predictive of subsequent failure. A larger study with longer follow up is needed to confirm our findings.
Of interest is the detection of plasma HPV DNA at the time of relapse in 3 patients with lung metastasis. Two of the three patients had undetectable circulating HPV DNA at the completion of therapy and in the third patient, we did not have plasma sample for analysis at treatment completion. In all three patients, the circulating HPV DNA level rose to > 100 copies/ml at the time of metastasis. This suggests that plasma HPV DNA may serve as a non-invasive and inexpensive test to distinguish between lung metastasis from a HPV(+) OP and a new lung cancer. Although these data are intriguing, the patient group is quite small need to be confirmed in a much larger cohort.
In summary, using conventional and qPCR, we have been able to quantify circulating HPV DNA from the pretreatment plasma samples of HPV(+) OP patients. The DNA copy number correlated significantly with nodal classification and the metabolically active nodal volume. Serial measurements in a small number of patients indicated that plasma HPV DNA becomes undetectable with tumor response to chemoradiation. However, the rate of decline was indistinguishable between relapsing and non-relapsing patients. Intriguingly, in a few patients, plasma HPV level was measurable at the time of metastasis and may serve as an inexpensive marker in this setting. A larger study is necessary to validate the sensitivity and specificity of this assay and to determine whether post treatment DNA levels can be used to identify recurrence in these patients.
Acknowledgments
Supported by 1 R01 CA118582-05 (QTL, HC, SK, CSK, TK)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 2.D'Souza G, Kreimer AR, Viscidi R, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356(19):1944–56. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 3.Gillison ML, D'Souza G, Westra W, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100(6):407–20. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Engels EA, Anderson WF, et al. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26(4):612–9. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 5.Attner P, Du J, Nasman A, et al. The role of human papillomavirus in the increased incidence of base of tongue cancer. Int J Cancer. 2009 doi: 10.1002/ijc.24994. [DOI] [PubMed] [Google Scholar]
- 6.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–50. [PubMed] [Google Scholar]
- 7.Usadel H, Brabender J, Danenberg KD, et al. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002;62(2):371–5. [PubMed] [Google Scholar]
- 8.Shao ZM, Wu J, Shen ZZ, et al. p53 mutation in plasma DNA and its prognostic value in breast cancer patients. Clin Cancer Res. 2001;7(8):2222–7. [PubMed] [Google Scholar]
- 9.Anker P, Lyautey J, Lederrey C, et al. Circulating nucleic acids in plasma or serum. Clin Chim Acta. 2001;313(1-2):143–6. doi: 10.1016/s0009-8981(01)00666-0. [DOI] [PubMed] [Google Scholar]
- 10.Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59(6):1188–91. [PubMed] [Google Scholar]
- 11.Mutirangura A, Pornthanakasem W, Theamboonlers A, et al. Epstein-Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res. 1998;4(3):665–9. [PubMed] [Google Scholar]
- 12.Utting M, Werner W, Muller G, et al. A possible noninvasive method for the detection of bladder cancer in patients: microsatellite analysis of free DNA in urine and blood. Ann N Y Acad Sci. 2001;945:31–5. doi: 10.1111/j.1749-6632.2001.tb03861.x. [DOI] [PubMed] [Google Scholar]
- 13.Lo YM, Chan LY, Chan AT, et al. Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res. 1999;59(21):5452–5. [PubMed] [Google Scholar]
- 14.La TH, Filion EJ, Turnbull BB, et al. Metabolic tumor volume predicts for recurrence and death in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2009;74(5):1335–41. doi: 10.1016/j.ijrobp.2008.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin JC, Chen KY, Wang WY, et al. Detection of Epstein-Barr virus DNA the peripheral-blood cells of patients with nasopharyngeal carcinoma: relationship to distant metastasis and survival. J Clin Oncol. 2001;19(10):2607–15. doi: 10.1200/JCO.2001.19.10.2607. [DOI] [PubMed] [Google Scholar]
- 16.Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med. 2004;350(24):2461–70. doi: 10.1056/NEJMoa032260. [DOI] [PubMed] [Google Scholar]
- 17.Chan AT, Lo YM, Zee B, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst. 2002;94(21):1614–9. doi: 10.1093/jnci/94.21.1614. [DOI] [PubMed] [Google Scholar]
- 18.Le QT, Jones CD, Yau TK, et al. A comparison study of different PCR assays in measuring circulating plasma epstein-barr virus DNA levels in patients with nasopharyngeal carcinoma. Clin Cancer Res. 2005;11(16):5700–7. doi: 10.1158/1078-0432.CCR-05-0648. [DOI] [PubMed] [Google Scholar]
- 19.Wei YC, Chou YS, Chu TY. Detection and typing of minimal human papillomavirus DNA in plasma. Int J Gynaecol Obstet. 2007;96(2):112–6. doi: 10.1016/j.ijgo.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Sathish N, Abraham P, Peedicayil A, et al. HPV DNA in plasma of patients with cervical carcinoma. J Clin Virol. 2004;31(3):204–9. doi: 10.1016/j.jcv.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Kay P, Allan B, Denny L, et al. Detection of HPV 16 and HPV 18 DNA in the blood of patients with cervical cancer. J Med Virol. 2005;75(3):435–9. doi: 10.1002/jmv.20294. [DOI] [PubMed] [Google Scholar]
- 22.Liu VW, Tsang P, Yip A, et al. Low incidence of HPV DNA in sera of pretreatment cervical cancer patients. Gynecol Oncol. 2001;82(2):269–72. doi: 10.1006/gyno.2001.6289. [DOI] [PubMed] [Google Scholar]
- 23.Yang HJ, Liu VW, Tsang PC, et al. Quantification of human papillomavirus DNA in the plasma of patients with cervical cancer. Int J Gynecol Cancer. 2004;14(5):903–10. doi: 10.1111/j.1048-891X.2004.014528.x. [DOI] [PubMed] [Google Scholar]
- 24.Capone RB, Pai SI, Koch WM, et al. Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin Cancer Res. 2000;6(11):4171–5. [PubMed] [Google Scholar]
- 25.Kong CS, Narasimhan B, Cao H, et al. The relationship between human papillomavirus status and other molecular prognostic markers in head and neck squamous cell carcinomas. Int J Radiat Oncol Biol Phys. 2009;74(2):553–61. doi: 10.1016/j.ijrobp.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sano T, Oyama T, Kashiwabara K, et al. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol. 1998;153(6):1741–8. doi: 10.1016/S0002-9440(10)65689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keating JT, Cviko A, Riethdorf S, et al. Ki-67, cyclin E, and p16INK4 are complimentary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am J Surg Pathol. 2001;25(7):884–91. doi: 10.1097/00000478-200107000-00006. [DOI] [PubMed] [Google Scholar]
- 28.Kong CS, Balzer BL, Troxell ML, et al. p16INK4A immunohistochemistry is superior to HPV in situ hybridization for the detection of high-risk HPV in atypical squamous metaplasia. Am J Surg Pathol. 2007;31(1):33–43. doi: 10.1097/01.pas.0000213347.65014.ee. [DOI] [PubMed] [Google Scholar]
- 29.de Roda Husman AM, Walboomers JM, van den Brule AJ, et al. The use of general primers GP5 and GP6 elongated at their 3' ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76(Pt 4):1057–62. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz M, Scheungraber C, Herrmann J, et al. Quantitative multiplex PCR assay for the detection of the seven clinically most relevant high-risk HPV types. J Clin Virol. 2009;44(4):302–7. doi: 10.1016/j.jcv.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Dong SM, Pai SI, Rha SH, et al. Detection and quantitation of human papillomavirus DNA in the plasma of patients with cervical carcinoma. Cancer Epidemiol Biomarkers Prev. 2002;11(1):3–6. [PubMed] [Google Scholar]
- 32.Chan AT, Ma BB, Lo YM, et al. Phase II study of neoadjuvant carboplatin and paclitaxel followed by radiotherapy and concurrent cisplatin in patients with locoregionally advanced nasopharyngeal carcinoma: therapeutic monitoring with plasma Epstein-Barr virus DNA. J Clin Oncol. 2004;22(15):3053–60. doi: 10.1200/JCO.2004.05.178. [DOI] [PubMed] [Google Scholar]
- 33.Pornthanakasem W, Shotelersuk K, Termrungruanglert W, et al. Human papillomavirus DNA in plasma of patients with cervical cancer. BMC Cancer. 2001;1:2. doi: 10.1186/1471-2407-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kedzia H, Gozdzicka-Jozefiak A, Wolna M, et al. Distribution of human papillomavirus 16 in the blood of women with uterine cervix carcinoma. Eur J Gynaecol Oncol. 1992;13(6):522–6. [PubMed] [Google Scholar]
- 35.Agrawal Y, Koch WM, Xiao W, et al. Oral human papillomavirus infection before and after treatment for human papillomavirus 16-positive and human papillomavirus 16-negative head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14(21):7143–50. doi: 10.1158/1078-0432.CCR-08-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]