Abstract
By performing homology modeling, molecular docking, and molecular dynamics (MD) simulations, we have developed three-dimensional (3D) structural models of M5 muscarinic acetylcholine receptor (mAChR) and two complexes for M5 mAChR binding with antagonists SVT-40776 and Solifenacin in the environment of lipid bilayer and solvent water. According to the simulated results, each of the antagonists is orientated horizontally in the binding pocket formed by transmembrane helices 2, 3, and 5 to 7. The cationic head group of each of the antagonists interacts wtih a negatively charged residue, Asp110, through electrostatic and hydrogen bonding interactions. The simulated results also reveal some significant difference between the binding modes of SVT-40776 and Solifenacin. In particular, SVT-40776 is persistently hydrogen-bonded with the side chain of residue Tyr458, whereas Solifenacin cannot form a similar hydrogen bond with residues around its carbonyl group. Such significant difference in the binding structures is consistent with the fact that SVT-40776 has a much higher binding affinity (Kd = 0.4 nM) with M5 mAChR than that of Solifenacin (Kd = 31 nM) with the same reeptor. The calculated binding free energy change (−2.3 ± 0.3 kcal/mol) from Solifenacin to SVT-40776 is in good agreement with the experimentally derived binding free energy change (−2.58 kcal/mol), suggesting that our modeled M5 mAChR structure and its complexes with the antagonists are reliable. The new structural insights obtained from this computational study are expected to stimulate future, further biochemical and pharmacological studies on the detailed structures of M5 and other subtypes of mAChRs.
Introduction
The muscarinic acetylcholine receptors (mAChRs) mediate many actions of neurotransmitter acetylcholine in the central and peripherial nervous systems. These receptors have been found to play very important and diverse roles in many physiological processes such as cardiovascular, motor, attention, learning, memory, pain, and sleep.1,2,3,4 Different from the nicotinic acetylcholine receptors (nAChRs), which are ligand-gated ion channels formed by five subunits, mAChRs are G-protein coupled receptors (GPCRs), which operate through G-proteins to alter second messager systems involving adenylate cyclase and phospholipase Cβ. The mAChRs belong to the class-A GPCRs and have five distinct subtypes (M1 to M5). The M5 mAChR is expressed primarily at the substantia nigra, ventral tegmental area, cerebral cortex, and the striatum area of the central nervous system (CNS).1,2 Recent studies revealed that M5 mAChR was involved in the regulation of striatal dopamine release and in rewarding brain stimulations.5,6,7 It was also found that M5 mAChR-knockout mice had reduced morphine-induced locomotion, establishing that M5 mAChR is a potential target for the treatment of drug abuse.8,9,10,11 Therefore, it has become a high priority to understand the molecular moechanisms for M5 mAChR’s agonizing and antagonizing processes, and develop novel and selective M5 ligands as potential therapeutics.
The first highly potent ligand of M5 mAChR was derived from a series of 5-trifluoromethoxy N-benzyl Isatins and found to be a positive allosteric modulator.12 Some other compounds were also developed to be high-affinity antagonists of mAChRs, but these compounds did not show no significant selectivity toward M5 receptor.13, 14, 15,16 In order to design highly potent and M5 mAChR-selective agonists or antagonists, one needs to understand the molecular determinants for the M5 mAChR-ligand binding, and then perform structure-based ligand design. For this purpose, one first needs to develop a reasonable and reliable structural model of M5 mAChR-ligand complex. Due to the difficulty in protein expression and crystalization of membrane proteins, there is no avaliable high-resolution crystal structure for any subtype of mAChR from any species. However, recent progresses of structure-function studies on mAChRs and some other class-A GPCRs have paved the way of fully understanding the structure-function relationships of mAChRs.11,17,18,19,20,21 For example, the X-ray crystal structures for different conformational states of rhodposin22,23,24 from different species have provided fundamental insights into the molecular details on the receptor activation mechanism. High-resolution X-ray crystal structures of human β2 adrenergic receptor25,26,27 bound with inverse agonist revealed significant structural difference for both the transmembrane helices and the second extraceullar loop (EL-2) when comapred with the structures of rhodopsin. The crystal structure of turkey β1 adrenergic receptor(β1-AR) 28 bound with high-affinity antagonist represents a possible extracellularly-open conformational state for the adrenergic receptors. In this X-ray structure of β1-AR, the antagonist cyanopindolol (CYP) was located inside a binding pocket formed by transmembrane (TM) helices 3, and 5 to 7. The tert-butylamino group (cationic head) of CYP intereacted with negatively charged side chain of residue Asp121 from TM3 of β1-AR through both electorstatic and hydrogen-bonding interactions. The crystal structure of human A2A adenosine receptor29 bound with the selective antagonist demonstrated that the EL-2 and the antagonist binding site of adenosine receptors are quite different from those of rhodopsin and the adrenergic receptors. The differences in receptor structures and the ligand-binding sites observed from these crystal structures indicate that each subfamily of GPCRs may have different inherent strutural flexibility and, therefore, adopt different conformational states under different incoming stresses. The latest information from the X-ray crystal structures for CXCR4 receptor30 bound with antagonists further support the viewpoint about the structural flexibility and functional diversity of GPCRs. This background indicates that it is a challenge to reliably study the structural mechanisms for other GPCRs. Special attention needs to be paid on the selection of a template for the construction of a structural model of any other sepcific GPCR. For the available X-ray structures of bovine rhodopsin, the native agonist (i.e. retinal) is covalently bound with the side chain of residue Lys296 of the receptor. The sequence identity between bovine rhodopsin and mAChRs are quite low (mostly <20%). Thus, the structure of bovine rhodopsin is not a suitable template for use in homology modeling of mAChRs binding with antagonists. Earlier homology models of mAChRs based on the X-ray structure of bovine rhodopsin did not account for the possible difference at the ligand binding site.31,32,33,34 Thus, the modeled atomic contacts between the mAChR and ligand are not reliable. Phylogenetically, mAChRs are more closely related to adrenergic receptors than either adenosine receptors or the CXCR4 receptor.17,23 Currently, the antagonist-bound β1-AR is the most reasonable and appropriate template to model the structures of mAChRs for the purpose of studying the receptor-antagonist binding. The combined use of the X-ray structure of β1-AR-antagonist complex and current computational modeling has enabled us to construct a reasonable 3D model of M5 mAChR.
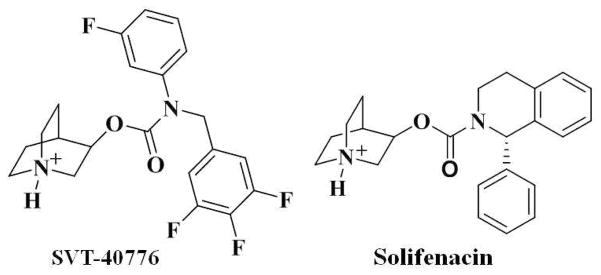
The present study aimed to develop a reasonable structural model of M5 mAChR and understanding its binding with antagonists. For this purpose, the 3D structure of human M5 mAChR was modeled, focusing on its binding with two representative antagonists (Scheme 1), (R)-quinudin-3-yl 3-fluorophenyl(3,4,5-trifluorobenzyl)carbamate (SVT-40776) and (1S,3′R)-quinuclidin-3′-yl 1-phenyl-1,2,3,4-tetrahydroisoquinoline-2-carboxylate (Solifenacin).13,15 Based on the energy-minimized structure of M5 mAChR, the protonated species of these two antagonists were docked into the binding site. The intermolecular interactions between M5 mAChR and the antagonists were analyzed based on the trajectoris of subsequent molecular dnyamics (MD) simulations. We found that SVT-40776 can form an additional hydrogen bond with the side chain of residue Tyr458 of M5 mAChR, which is consistent with the fact that SVT-40776 has a higher affinity than that of Solifenacin with the same receptor. The new structural insights obtained from this computational study should be valuable for future design further biochemical and pharmacological studies on M5 and other subtypes of mAChRs.
Scheme 1.
Computational Methods
Homology modeling and structural optimization
To study the binding of SVT-40776 and Solifenacin with M5 mAChR in atomic detail, a homology model of human M5 mAChR was built based on the X-ray crystal structure of turkey β1-AR (PDB entry of 2VT4 at 2.7Å resolution, B chain)28 by using the Protein Modeling module of Discovery Studio (version 2.5.5, Accelrys, Inc. San diego, CA). The amino acid sequence of human M5 mAChR was directly extracted from the uniprot.org (access number: P08912). The sequence alignment was generated by using ClusterW with the Blosum scoring function.35,36 The best alignment was selected according to both the alignment score and the reciprocal positions of the conserved residues among all the subtypes of mAChRs and adrenergic receptors. These include the DRY motif near the intracellular end of transmembrane 3 (TM3), the NPxxY(x)6F motif from TM7, and the position of disulfide bond between the Cys103 near the extracellular end of TM3 and the Cys183 of EL-2. The first 27 residue at the N-terminal, the last 20 residue at the C-terminal and the third intracellular loop (IL-3, residues from #225 to #435) of M5 mAChR were omited due to the lack of the corresponding homolog sequence in the template. The alignment was first performed by using the default values of the parameters in program ClusterW,35 e.g. the minimum sequence length of aligning was set as 10, and then was improved by adjusting the values of the parameters in order to obtain a higher alignment score. During the alignment process, the most important conserved motifs (i.e. the DRY motif from TM3, and the NPxxY(x6)F motif from TM7) were strictly reserved from the previous round of alignment to the next round of alignment, while the newly aligned conserved regions were also added for further alignment. The sequence identity for the regions except the omitted ones reached 34.3% and the sequence homology became 61.7% for the final alignment with 277 residues of β1-AR. We also tried to use another scoring function, i.e. the PAM matrix.35 According to the sequence alignment based on the PAM matrix, the sequence identity was 31.2%, lower than that for the sequence alignment based on the Blosum scoring function. The use of the PAM matrix could not align the conserved residue Cys183 of M5 mAChR with Cys199 of β1-AR. Based on the criteria of sequence identity and the reciprocal positions of the conserved residues, we finally elected to use the sequence alignment generated by using the Blosum scoring function. The coordinates of the heavy atoms in the conserved regions were directly transformed from the template structure (i.e. the X-ray crystal structure of β1-AR28), whereas the non-equivalent residues were mutated from the template to the corresponding ones of M5 mAChR. The side chains of the non-conserved residues were relaxed during the process of homology modeling in order to remove the possible steric overlap or hindrance with the neighboring conserved residues.
In order to simulate the actual physiological environment, the initial M5 mAChR model was inserted into a pre-equilibrated POPC lipid bilayer and then solvated by two layers of water molecules at each side of lipid bilayer. The area per lipid (AL) for this starting lipid bilayer was calcualted as the area of xy plane of the simulation box divided by the number of lipid molecules per layer. The all-atom model of the POPC bilayer was generated by using the membrane plugin of the VDW software37 and the initial size of the membrane was expanded to be large enough to encompass the target protein. The geometry of POPC molecule was optimized by performing ab initio electronic structure calculation at the HF/6-31G* level using Gaussian03 program.38 The HF/6-31G* calculation was also performed to determine the restrained electrostatic potential (RESP)-fitted charges for POPC molecules. The relative orientation of M5 mAChR in the lipid bilayer was determined by referring the similar orientation of the structure β1-AR,28 i.e. the helix 8 in parallel with the intracellular surface of the membrane. When inserting, any POPC molecule was removed if it had more than 50% of its non-hydrogen atoms within a distance of 2.5 Å to any non-hydrogen atoms of M5 mAChR. The solvent layers were added by using the LEaP module of Amber11 program suite.39 The protein together with the lipid bilayer was solvated in a rectangular box of TIP3P water molecules40 with a minimum solute wall distance of 10 Å. Standard protonation states at physiological environment (pH ~7.4) were set to all ionizable residues of M5 mAChR, and the position of the proton was properly set on Nδ1 atom of residue His478. Additional 11 Cl− ions were added in the solvent as counter ions to neutralize the system. The system composed of 42,911 atoms, including 132 POPC molecules and 6,892 water molecules.
After the whole system was set up, a series of energy minimizations (geometric optimizations) were carried out by using the Sander module of Amber11 program suite39 uisng the conjugate gradient energy-minimization method with a non-bonded cutoff of 10 Å. The first 2,000 steps of the energy minimization was done for the backbone of M5 mAChR while the side chains were fixed, and then the next 40,000 steps for the lipid molecules, water molecules, and the side chain atoms of M5 mAChR. In order to get the solute (M5 mAChR) better solvated, the subsequent MD simulations and energy minimizations were performed on the environment (i.e. the lipid molecules, water molecules, and the counter ions). First, 1.0 ns MD simulations were performed on the water molecules with NTV ensemble at T = 300 K. The environment was energy-minimized for 20,000 steps followed by a 2.0 ns MD simulation on the lipid molecules with anisotropic pressure coupling (i.e. the parameter ntp = 2 in the input file). After these MD simulations, the environment and side chains of M5 mAChR were energy-minimized for 20,000 steps. Finally, the system was energy-minimized for 6,000 steps for all atoms, and a convergence criterion of 0.001 kcal mol−1 Å−1 was achieved.
Molecular docking and MD simulation
Based on the structural model of M5 mAChR obtained in the present study, the binding modes of the receptor with two antagonists (Scheme 1) were explored through molecular docking by using AutoDock 3.0.5 program.41 Previous site-directed mutation on Asp105 of M1 mAChR and mutation on Asp103 of M2 mAChR11 showed that these negatively charged residues are critical for the ligand binding. This negatively charged residue from TM3 of M1 or M2 receptor corresponds to residue Asp121 from TM3 of β1-AR,28 and to residue Asp110 of M5 mAChR. Based on the sequence comaprison between M5 mAChR and β1-AR, and based on the fact that both SVT-40776 and Solifenacin are competitive antagonists,13,15 it is reasonable to assume that the binding site of M5 mAChR for these typical antagonists should be around residue Asp110 from TM3. The atomic charges of the protonated antagonists were also determined as the restrained electrostatic potential (RESP) charges determined by using the standard RESP procedure implemented in the Antechamber module of the Amber11 program39 following the electronic structure and electrostatic potential calculations at the HF/6-31G* level. During the docking process, a conformational search was performed using the Solis and Wets local search method,42 and the Lamarkian genetic algorithm (LGA)41 was applied to deal with the M5 mAChR-antagonist interactions. Among a series of docking parameters, the grid size was set to be 60 × 60 × 60 and the grid space was the default value of 0.375 Å. The possible binding site of M5 mAChR for these two antagonists was first roughly defined as a similar site of β1-AR structure for the antagonist cyanopindolol,28 i.e. the cavity around transmembrane helices 2, 3, and 5 to 7. The binding site was then hunted by changing the center and the size of the docking grid. All the complex candidates were evaluated and ranked in terms of the binding energy by using the standard energy score function implemented in the docking program and, finally, the geometric matching quality through visual checking. For each antagonist, the initial binding structure was selected from the docked candidates, of which the protonated head group (Scheme 1) was orientated toward the negatively charged side chain of Asp110 of M5 mAChR. The selected initial structure of M5 mAChR-SVT-40776 complex was ranked #1 in the clustering histogram, with a positional root-mean squeare deviation (RMSD) of 1.63 Å from the average structure of the same cluster. The selected initial structure of M5 mAChR-Solifenacin complex was also ranked #1 in clustering histogram, with the RMSD value of 0.80 Å.
In order to further relax the selected initial M5 mAChR-antagonist complex structures, MD simulations were performed by using the Sander module of Amber11.39 MD simulations were performed also for the purpose of making the EL-2 more reasonably packed on the extracellular surface of the complex structure. The whole system was gradually heated to 300 K by weak-coupling method43 and equilibrated for about 2.0 ns. Throughout the MD simulations, a 10 Å non-bonded interaction cutoff was used and the non-bonded list was updated every 25 steps. The particle mesh Ewald (PME) method44 was applied to treat long-range electrostatic interactions. The lengths of covalent bonds involving hydrogen atoms were fixed with the SHAKE algorithm,45 enabling the use of a 2-fs time step to numerically integrate the equations of motion. Finally, the production MD was kept running about 4.0 ns with a periodic boundary condition in the NTP ensemble at T = 300 K with Berendsen temperature coupling and at P = 1 atm with anisotropic molecule-based scaling.43
Calculation of binding free energy change
We employed a molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) method46 to estimate the binding free energy change from one inhibitor to another for the same receptor. According to the MM-PBSA method, the free energy of a ligand binding with the M5 receptor, ΔGbind, is calculated from the difference between the free energy of the receptor-ligand complex (GR-L) and the sum of the free energies of the unbound receptor (GR) and ligand (GL) as Eq.(1):
| (1) |
ΔGbind was evaluated as the sum of the changes in the MM gas-phase binding energy (ΔEMM), solvation free energy (ΔGsolv), and entropic contribution (−TΔS).
| (2) |
| (3) |
| (4) |
| (5) |
The MM binding energies were calculated with the Sander module of the Amber program. Electrostatic solvation free energy (ΔGPB) was calculated by the finite-difference solution to the Poisson-Boltzmann (PB) equation implemented in the Delphi program. 47, 48 The MSMS program49 was used to calculate the solvent-accessible surface area (SASA) for the estimation of the nonpolar solvation energy (ΔGnp) using Eq.(5) with parameters γ = 0.00542 kcal/Å2 and β = 0.92 kcal/mol.
The entropic contribution, −TΔS, to the binding free energy was calculated using a local program (a stand-alone program) developed in our own laboratory. The computational procedure used to evaluate the −TΔS was the same as that described in our recent publications.50,51,52,53 As we described previously, the entropy contribution is divided into two parts: solvation entropy (ΔSsolv) and conformational entropy (ΔSconf):
| (6) |
The solvation entropy is gained by solvent water molecules on being displaced from the active site by the ligand during binding and was calculated by using the parameters established previously. 54 The contribution to the binding free energy from the conformational entropy change is proportional to the number (ΔNrot) of the lost rotatable bonds during the binding:
| (7) |
in which w is the scaling factor which may be slightly different for different receptors. Nevertheless, our previous studies50–53 consistently revealed that w = 0.6434 (the smallest value) to 0.8452 (the largest value). In the present study, we simply used the middle value of the range, i.e. w = 0.7434 ± 0.1009, as the final result is not very sensitive to the w value within the range of the w values (see below).
It has been demonstrated55 that the MM-PBSA method is reliable for calculating the relative binding free energies between various ligands with a same receptor via Eq.(8), although it might not be very accurate for calculating the absolute binding free energies with a membrane protein.
| (8) |
Most of the MD simulations were performed on a supercomputer (i.e. the Dell X-series Cluster with 384 nodes or 4,768 processors) at University of Kentucky Center for Computational Sciences. Some other modeling and computations were carried out on SGI Fuel workstations in our own lab.
Results and Discussion
Structural model of M5 mAChR
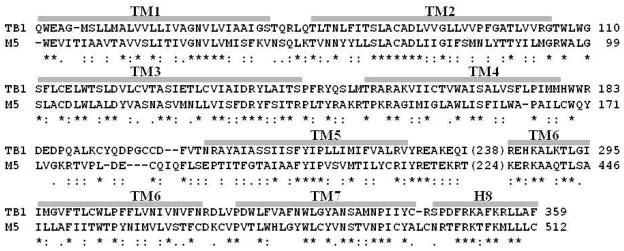
The amino acid sequence alignment (Figure 1) between human M5 mAChR and the turkey β1-AR shows that 8 regions with high homology can be assigned to 8 α-helices. These include the 7 transmembrane helices (TM1 to TM7) and a small α-helix H8 consisting of residues Asn498 to Cys512, which is supposed to locate on the membrane cytoplasm interface. The finally obtained sequence alignment is depicted in Figure 1. Our structural model of M5 mAChR based on the sequence alignment depicted in Figure 1 is significantly different from the previously reported M5 mAChR model.31,33,34 For example, the start and the end positions of each TM in our current model are different from the corresponding positions in the previous reported M5 mAChR model31 using the crystal structure of rhodopsin as the template.22 In particular, the TM3 consists of 35 residues in our model, but it only consists of 21 residues in the previously reported model.31 Based on the reported crystal structures of class-A GPCRs,18,19,25,26,27,28,29,30 the TM3 was significantly longer than that of rhodopsin.22,24 This information supports our new model developed by using the turkey β1-AR as the template and the sequence alignment depicted in Figure 1.
Figure 1.
Aligned sequence of M5 mAChR with the template turkey β1-AR. The identified 7 transmembrane (TM) helices and helix 8 are labeled above the sequence. The numbers in parenthesis are the positions for the last residue of TM5. The stars refer to the identical residues, the double filled periods refer to the conservative substitutions, and the filled perigods to the variable conservative substitutions.
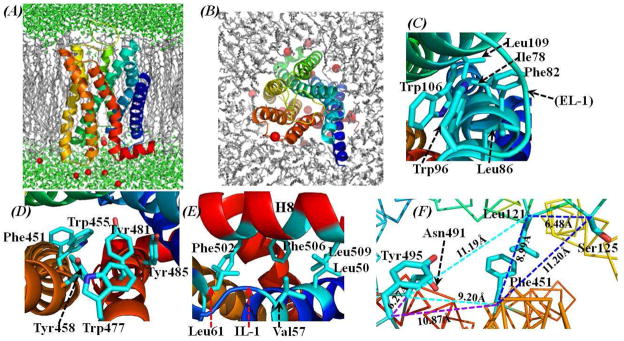
As modeled, the assembly of these helices in a lipid bilayer environment (Figure 2) is structurally organized similar to the template β1-AR.28 The area per lipid (AL) in the lipid bilayer was estimated to be ~63.1 Å2, which is very close to that (~63.0 Å2) of the most commonly used POPC lipid bilayers described in the literature.56 In order to track how M5 mAChR interacted with the surrounding lipid molecules, we counted the total number of contacts (Ncontact) between the non-hydrogen atoms of the lipid bilayers and the non-hydrogen atoms of the M5 receptor for each snapshot of the MD trajectory. The criterion (cutoff) for the inter-atomic contact was set as 5.0 Å, and the tracked results are provided in Supporting Information (Figure S1). The tracked Ncontact increased slightly during the first 2.0 ns of the MD simulation on the lipid molecules, and fluctuated during the next 2.0 ns (the equilibration stage) of the MD simulation for the M5 mAChR-antagonist complex structure (see below for further discussion). The tracked Ncontact kept constant, with an average Ncontact value of 1019, during the final 4.0 ns of the MD simulation for the production stage (see below for further discussion). The changes of Ncontact along the MD trajectory (Figure S1 in Supporting Information) suggest that the packing between the lipid molecules and the M5 mAChR was gradually improved and equilibrated during the MD simulation. The whole model of M5 mAChR appears to be in a conformation opening toward the extracellular side. The energy-minimized M5 mAChR structure (Figure 2) has a positional root-mean-square deviation (RMSD) of 0.23 Å for Cα atoms from the initial structural model. Relative to the template β1-AR structure,28 the RMSD for Cα atoms of the 7 TMs was only 0.52 Å. These small structural deviations suggest a high fidelity for the structure prediction of M5 mAChR. There are some proline-induced kinks at the conserved positions along the span of transmembrane segments. These positions are Pro205 from TM5, Pro457 from TM6, and Pro492 from TM7 (Figure 1). These kinks could enable the structural rearrangements required for the activation of G-protein effectors. As observed from the energy-minimized structure of M5 mAChR, most of the hydrophobic side chains of the 7 TMs are orientated toward the surrounding lipid molecules, stabilizing the transmembrane structure of the receptor, while the helix-helix packing is featured by inter-helical polar contacts and hydrogen bonding interactions. Specifically, a cluster of hydrophobic resides, being composed of residues Ile78, Phe82, and Leu86 from TM2 and Trp106 and Leu109 from TM3 (Figure 2C), is located near the extracellular end of these two helices. This cluster of hydrophobic residues is also close to the aromatic residue Trp96 of EL-1, making the TM2, TM3, and EL-1 tightly aggregated. Another cluster of aromatic residues locates close to the extracellular end of TM6 and TM7 and is formed by residues Phe451, Trp455, Tyr458, Trp477, Tyr481, and Tyr485 (Figure 2D). These residues are stacked in parallel, while all the hydroxyl groups of residues Tyr458, Tyr481, and Tyr485 point to the internal cavity among TM2, TM3, TM5, TM6, and TM7. Residues Phe502, Phe506, and Leu509 from H8 interact with the hydrophobic residues Leu50 and Val57 from TM1 and Leu61 from IL-1 (Figure 2E), making H8 closely packed with the neighboring TM1 and the surrounding lipid molecules. Such specific local interactions must help to stabilize the whole receptor in the membrane environment. The X-ray structure of β1-AR28 showed that its EL-2 formed a short α-helix and defined the entrance of the ligand-binding pocket. Unlike the EL-2 of β1-AR, the EL-2 of M5 mAChR cannot form any regular secondary structure, indicating that this loop might be easy to adapt to the outside stimulations.
Figure 2.
Energy-minimized structural model of human M5 mAChR in the physiological environment. (A) View along the normal of the membrane. The M5 mAChR protein is represented as ribbon in rainbow color, lipid molecules in grey, Cl− ions in read spheres, and water molecules in green. (B) Top view of M5 mAChR protein and the lipid molecules. Water molecules are not shown. (C) A hydrophobic packing by residues Ile78, Phe82, and Leu86 from TM2, Trp106 and Leu109 from TM3, and Trp96 from EL-1; (D) An aromatic cluster by residues Phe451, Trp455, and Tyr458 from TM6 and Trp477, Tyr481, and Tyr485 from TM7; (E) Interactions between residues Phe502, Phe506, and Leu509 from H8, Leu50 and Val57 from TM1, and Leu61 from IL-1; (F) Cluster of triple residues. There are three groups of these triple residues, i.e. Leu121-Ser125-Phe374, Leu121-Phe451-Asn491, and Phe451-Asn491-Tyr495. Each of these triple-residues groups could form a Zn2+ binding site if they were mutated to residue Histidine and adjust local conformation. The distances among the Cα atoms of each group of triple residues are labeled and represented by lines in different colors. (C) and (D) are in the similar orientation as that of (B), whereas (E) and (F) are the top view from the intracellular side. All residues are colored by the atom type, and the hydrogen atoms are omitted for clarity.
Our energy-minimized M5 mAChR model is considerably different from the previously reported M5 mAChR models.31,32,33,34 The major difference exists not only in the starting position and the length of each helix, but also in the assembly of the 7 TMs. These differences between our current model and the previous models are mainly due to the fact that the more reasonable template28 was used in our M5 mAChR structure modeling. As noted above, the template (i.e. the structure of β1-AR) used to build our model is phylogenetically closer to mAChRs, whereas the previous models31,32,33,34 were constructed based on the crystal structure of phylogenetically distant bovine rhodopsin.22 Histidine-substitution studies on M1 mAChR20 revealed that the triple mutations as Leu116His/Phe374His/Asn414His (the M1 numbering, and the same thereafter for residues of M1 mAChR) or Leu116His/Ser120His/Phe374His or Phe374His/Asn414His/Tyr418His led to high-affinity binding of the inactive receptor with Zn2+ ion, suggesting that these residues formed a network of intramolecular interactions. As these residues are highly conserved not only in mAChRs, but also throughout the class-A GPCRs,17,23 it is reasonable to assume that the M5 mAChR also retains such network of intramolecular interactions. Comparing the sequence of the M5 receptor with that of M1 mAChR, residues Leu121 and Ser125 from TM3, Phe451 from TM6, and Asn491 and Tyr495 from TM7 of M5 mAChR are at the corresponding positions of those residues in M1 mAChR. A careful inspection on the structural model of M5 mAChR in the present study revealed that the side chains of these residues all point to the central cavity between TM3, TM5, TM6, and TM7 (Figure 2F). The distances among the Cα atoms of residues Leu121, Ser125, and Phe374 are all close to 11.0 Å (Figure 2F, blue colored lines). The similar distances were found for the Cα atoms among the triple residues Leu121-Phe451-Asn491 (Figure 2F, cyan colored lines) and the triple residues Phe451-Asn491-Tyr495 (Figure 2F, magenta colored lines). As observed in our M5 mAChR model structure, the side chains of these triple-residues would approach toward each other inside the central cavity if they were mutated to residue Histidine. A Zn2+-binding site will be formed by the side chains of any of these triple residues after possible local conformational adjustment, and the whole receptor structure will probably be kept inactive. The usage of a better structural template28 and the qualitative consistency with the observations of Histidine-substitution studies20 have made our current M5 mAChR structural model more reliable than the previously reported M5 mAChR models.31,32,33,34
Binding mode of M5 mAChR with antagonists
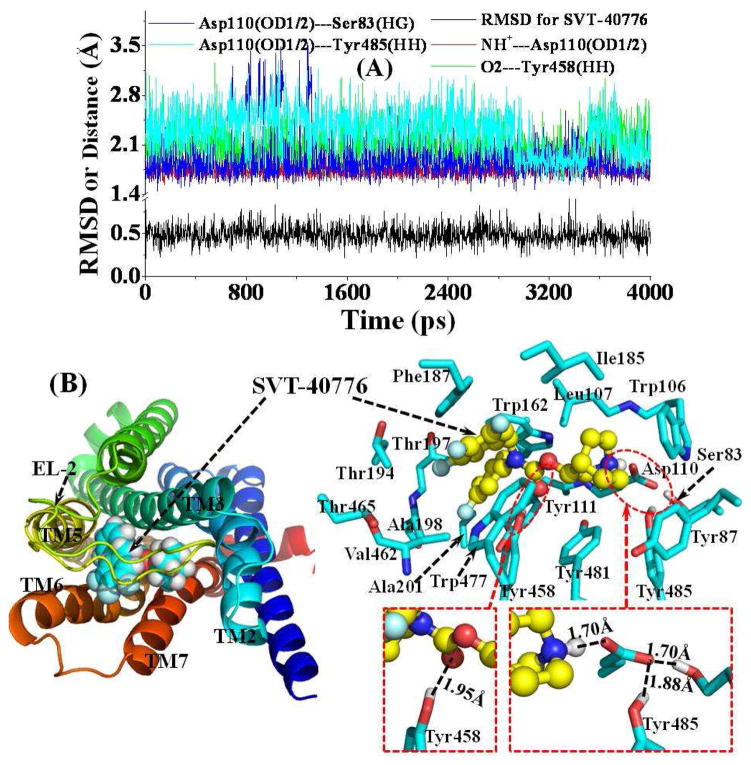
The antagonist-binding site in M5 mAChR is located near the extracellular end of TM3, TM5, TM6, and TM7. As shown in Figure 3, TM2 and TM4 are also partially involved in the formation of the antagonist-binding site. In addition, the antagonist-binding site is partially covered by EL-2. In a typical structure of the MD-simulated M5 mAChR-SVT-40776 complex (Figure 3B) at the snapshot of the MD trajectory at 4.0 ns (Figure 3A), the SVT-40776 molecule is orientated horizontally inside the binding pocket (Figure 3B). The cationic head of SVT-40776 is anchored around the negatively charged side chain of residue Asp110 from TM3, interacting with each other through electrostatic attraction and strong hydrogen bonding (Figure 3B). As tracked from the MD simulations (Figure 3A), the average shortest distance for the hydrogen bond through the proton at the cationic head of SVT-40776 and the negatively charged atoms at the side chain of Asp110 is 1.75 Å. Meanwhile, residue Asp110 is also hydrogen-bonded with the side chain of Ser83 from TM2 (blue curve in Figure 3A) and the side chain of Tyr485 from TM7 (cyan curve in Figure 3A). The cationic head of SVT-40776 is also closely packed with residues Trp106 and Leu107 from TM3, Tyr481 from TM7, and Ile185 from EL-2. Residue Tyr87 from TM2 is also within 5 Å from the cationic head of SVT-40776. The carbonyl oxygen of SVT-40776 is always hydrogen-bonded with the hydroxyl group on the side chain of Tyr458 from TM6 with an average distance of 2.08 Å throughout the entire MD simulations (green curve of Figure 3A). One of the fluoro-substituted phenyl group of SVT-40776 is packed in parallel with the underneath Tyr111 from TM3 and the above Phe187 from EL-2. It is also packed perpendicularly with the side chain of Trp162 from TM4. This fluoro-substituted phenyl group of SVT-40776 is also surrounded by residues Thr194, Thr197, Ala198, and Ala201 from TM5. The fluorine atom at the meta-position of the phenyl group of SVT-40776 has no specific hydrogen-bonding interaction with the surrounding residues as observed from the MD-simulated complex structure (Figure 3B). However, the three fluoro substitutes on another phenyl group of SVT-40776 are all solvent-exposed, probably hydrogen bonding with surrounding water molecules. One of these three fluoro substitutes is also close to the side chain of Ser465 from TM6, but without direct contact, indicating that they interact indirectly through water molecules. In addition, the three fluoro-substituted phenyl groups of SVT-40776 are packed in parallel with the aromatic side chain of residue Trp477 from TM7 and Phe187 from EL-2.
Figure 3.
(A) Tracked positional RMSD for SVT-40776 from its original coordinates and tracked distances along the MD trajectory. NH+---Asp110(OD1/2) represents the shortest distance between the proton at the cationic head of SVT-40776 and the negatively charged atoms (OD1 or OD2) on the side chain of residue Asp110; O2---Tyr458(HH) is the distance for the hydrogen bond formed by the carbonyl oxygen of SVT-40776 with the Tyr458 side chain; Asp110(OD1/2)---Ser83(HG) represents the distance for the hydrogen bond formed by the Asp110 side chain and the Ser83 side chain; Asp110(OD1/2)---Tyr485(HH) is the distance for the hydrogen bond formed by the Asp110 side chain and the Tyr485 side chain. (B) Top view of representative structure for M5 mAChR-SVT-40776 complex, taken from the last snapshot of the MD simulation. The coloring scheme for the complex structure is the same as that used in Figure 2B, and the SVT-40776 is shown as spheres (left panel) or ball-and-stick (right panel). Residues within 5 Å of SVT-40776 are labeled and shown in stick. The hydrogen bonding interaction between the Asp110 side chain and the cationic head of SVT-40776, the hydrogen bonding interaction between the carbonyl oxygen of SVT-40776 and the Tyr458 side chain, and the hydrogen bonding interactions among side chains of Asp110, Ser83, and Tyr485 are shown in dashed lines along with the labeled distances.
The residue Asp110 is totally conserved across all subtypes of mAChRs. The same Asp residue at the similar position in M1 and M2 mAChRs has been indicated to play a vital role in the binding of agonist acetylcholine by Asp/Glu mutation.11 Residues Tyr111, Tyr458, Tyr481, and Tyr485 are also conserved for mAChRs, and the residues at the corresponding positions in M3 mAChR were found to be important for the binding of agonists studied by cross-linking strategy.57 It would be interesting to design further biological studies including site-directed mutagenesis in the future to verify the computationally predicted binding mode of SVT-40776 in this study.
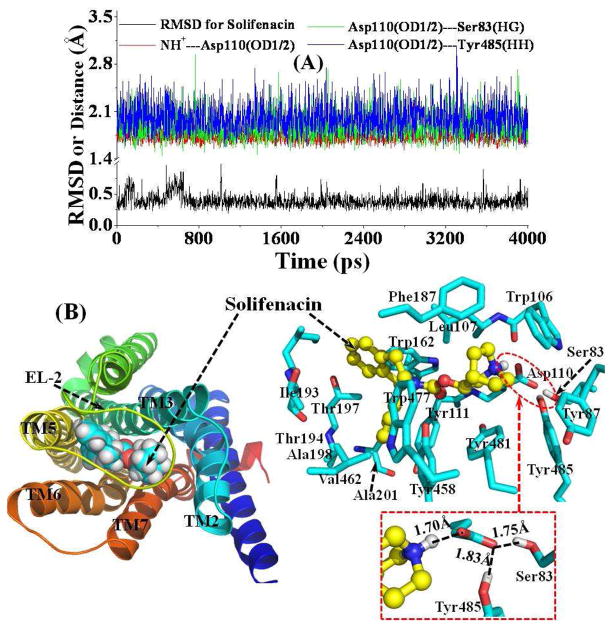
Figure 4 depicts the tracked important distances concerning the intermolecular interactions in the MD-simulated structural model of M5 mAChR-Solifenacin complex. The mode of binding of Solifenacin with M5 mAChR is generally similar with that of SVT-40776 (Figure 3). The detailed atomic interactions between Solifenacin and M5 mAChR are featured as electrostatic, hydrogen bonding, and hydrophobic contacts, as shown in Figure 4 for a typical structure of the MD-simulated complex at the 4.0 ns snapshot. The proton at the cationic head of Solifenacin is also hydrogen-bonded with the side chain of Asp110 with an average distance of 1.75 Å during the MD simulation (Figure 4A). For a major difference between the M5 mAChR-Solifenacin binding and the M5 mAChR-SVT-40776 binding, Phe187 is packed with the cationic head group of Solifenacin, whereas Ile185 is packed with the cationic head group of SVT-40776. For another significant difference, there is no hydrogen-bonding interaction between the carbonyl oxygen of Solifenacin and the surrounding residues including Tyr458 (Figure 4B), whereas a persistent hydrogen bond is formed between the carbonyl oxygen of SVT-40776 and the side chain of Tyr458 (Figure 3). This key structural difference in intermolecular interactions may be helpful to understand why the binding affinity of SVT-40776 is much higher than that of Solifenacin with the same receptor. As measured,13,15 the dissociation constant (Kd) of Solifenacin with M5 mAChR was 31 nM and the Kd of SVT-40776 was 0.4 nM. The phenyl group on the tail of Solifenacin is located at the similar sub-site as that of SVT-40776, and the tail of Solifenacin is loosely packed with the side chains of residues Val462 and Trp477.
Figure 4.
(A) Plots of the tracked positional RMSD for Solifenacin from its original coordinates and tracked distances. NH+---Asp110(OD1/2) represents the shortest distance between the proton at the cationic head of Solifenacin and the negatively charged atoms (OD1 or OD2) on the side chain of residue Asp110; Asp110(OD1/2)---Ser83(HG) represents the distance for the hydrogen bond formed by the Asp110 side chain and the Ser83 side chain; Asp110(OD1/2)---Tyr485(HH) is the distance for the hydrogen bond formed by the Asp110 side chain and the Tyr485 side chain. (B) Top view of representative structure for M5 mAChR-Solifenacin complex, taken from the last snapshot of the MD simulations. The coloring scheme for the complex structure is the same as that used in Figure 2B, and the Solifenacin is presented as spheres (left panel) or ball-and-stick (right panel). Residues within 5 Å of Solifenacin are labeled and shown in stick. The hydrogen bonding interaction between the Asp110 side chain and the cationic head of Solifenacin, and the hydrogen bonding interactions among side chains of Asp110, Ser83, and Tyr485 are shown in dashed lines along with the labeled distances.
For a better understanding of the binding affinity difference between Solifenacin and SVT-40776, the MM-PBSA calculations were performed to estimate the binding free energy change, i.e. ΔΔGbind in Eq.(8), from Solifenacin to SVT-40776 for the binding with M5 mAChR by using the energy-minimized receptor-ligand binding structures depicted in Figures 3 and 4. Based on the MM-PBSA calculations, we obtained ΔΔEbind = −1.4 kcal/mol and −TΔΔS = −0.9 ± 0.3 kcal/mol corresponding to w = 0.7434 ± 0.1009. These data suggest that both the enthalpy and entropy favor SVT-40776 binding with the receptor. Thus, according to Eq.(8), the MM-PBSA calculations led to the computational prediction of ΔΔGbind = −2.3 ± 0.3 kcal/mol. For comparison, we may also convert the experimentally measured dissociation constant (Kd = [R][L]/[R-L]) to the binding free energy, denoted by ΔGbind(expt), by using the well-known thermodynamic equation, i.e. ΔGbind = RTlnKd at T = 298.15 K. Based on the experimental data, i.e. Kd = 0.4 nM for SVT-40776 and Kd = 31 nM for Solifenacin,13,15 we have ΔΔGbind(expt) = RTlnKd(SVT-40776) − RTlnKd(Solifenacin) = −2.58 kcal/mol. Our computationally estimated ΔΔGbind value of −2.3 ± 0.3 kcal/mol is in good agreement with the experimentally derived ΔΔGbind(expt) value of −2.58 kcal/mol. The good agreement between the computational and experimental binding free energy differences suggests that the modeled complex structures for M5 mAChR binding with the antagonists are reliable.
Conclusions
The present computational modeling, molecular docking, and molecular dynamics simulations have led us to develop a reasonable 3D structural model of M5 mAChR and understand its binding with antagonists SVT-40776 and Solifenacin. Our modeled 3D structures of M5 mAChR in complex with the antagonists have provided valuable structural insights concerning how M5 mAChR interacts with its antagonists at atomic level. In common, the cationic head group of both antagonists interacts with residue Asp110 from TM3 through electrostatic attraction and hydrogen bonding, and the cationic head is also packed tightly with the surrounding aromatic residues from TM3 and TM7. However, the carbonyl oxygen of SVT-40776 is persistently hydrogen-bonded with residue Tyr458, whereas there is no hydrogen-bonding interaction between the carbonyl oxygen of Solifenacin and the surrounding residues. This significant difference in intermolecular interactions contributes to the observed difference in the binding affinity between these two antagonists with the same receptor. The good agreement between the calculated binding free energy change (−2.3 ± 0.3 kcal/mol) from Solifenacin to SVT-40776 and the experimentally derived binding free energy change (−2.58 kcal/mol) suggests that our modeled M5 mAChR structure and its complexes with the antagonists are reliable. The new structural insights obtained from this computational study should be valuable for future design of further biochemical and pharmacological studies on the detailed structures of M5 and other subtypes of mAChRs.
Supplementary Material
Acknowledgments
This work was supported by NIH (DA025948 and DA030667). The authors also acknowledge the Center for Computational Sciences (CCS) at the University of Kentucky for supercomputing time on a Dell Supercomputer Cluster consisting of 388 nodes or 4,816 processors.
Footnotes
Supporting Information Available. Additional figure (Figure S1) for the tracked number of contacts between the M5 mAChR and the surrounding lipid bilayer molecules. This material is available free of charge via the Internet http://pubs.acs.org.
References
- 1.Bonner TI, Young AC, Brann MR, Buckley NJ. Cloning and expression of the human and rat m5 muscarinic acetylcholine receptor genes. Neuron. 1988;1:403–410. doi: 10.1016/0896-6273(88)90190-0. [DOI] [PubMed] [Google Scholar]
- 2.Eglen RM, Choppin A, Dillon MP, Hegde S. Muscarinic receptor ligands and their therapeutic potential. Curr Opin Chem Biol. 1999;3:426–432. doi: 10.1016/S1367-5931(99)80063-5. [DOI] [PubMed] [Google Scholar]
- 3.Bymaster FP, McKinzie DL, Felder CC, Wess J. Use of M1-M5 muscarinic receptor knockout mice as novel tools to delineate the physiological roles of the muscarinic cholinergic system. Neurochem Res. 2003;28:437–442. doi: 10.1023/a:1022844517200. [DOI] [PubMed] [Google Scholar]
- 4.Jonkam C, Zhu Y, Jacob S, Rehberg S, Kraft E, Hamahata A, Nakano Y, Traber LD, Herndon DN, Traber DL, Hawkins Hk, Enkhbaatar P, Cox RA. Muscarinic receptor antagonist therapy improve acute pulmonary dysfunction after smoke inhalation injury in sheep. Crit Care Med. 2010;38:2339–2344. doi: 10.1097/CCM.0b013e3181f8557b. [DOI] [PubMed] [Google Scholar]
- 5.Yeomans J, Forster G, Blaha C. M5 muscarinic receptors are needed for slow activation of dopamine neurons and for rewarding brain stimulation. Life Sci. 2001;68:2449–2456. doi: 10.1016/s0024-3205(01)01038-4. [DOI] [PubMed] [Google Scholar]
- 6.Forster GL, Yeomans JS, Takeuchi J, Blaha CD. M5 muscarinic receptors are required for prolonged accumbal dopamine release after electrical stimulation of the pons in mice. J Neurosci. 2002;22:RC190. doi: 10.1523/JNEUROSCI.22-01-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendor J, Lizardi-Ortiz J, Westphalen RI, Hemmings HC, Jr, Sulzer D, Flajolet M, Greengard P. AGAP1/AP-3-dependent endocytic recycling of M5 muscarinic receptors promotes dopamine release. The EMBO J. 2010;29:2813–2826. doi: 10.1038/emboj.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araya R, Noguchi T, Yuhki M, Kitamura N, Higuchi M, Saido TC, Seki K, Itohara S, Kawano M, Tanemura K, Wess J, Yamada M. Loss of M5 muscarinic acetylcholine receptors leads to cerebrovascular and neuronal abnormalities and cognitive deficits in mice. Neurobiol Dis. 2006;24:334–344. doi: 10.1016/j.nbd.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Steidl S, Yeomans JS. M5 muscarinic receptor knockout mice show reduced morphine-induced locomotion but increased locomotion after cholinergic antagonism in the ventral tegmental area. J Pharm Expt Ther. 2009;328:263–275. doi: 10.1124/jpet.108.144824. [DOI] [PubMed] [Google Scholar]
- 10.Raffa RB. The M5 muscarinic receptor as possible target for treatment of drug abuse. J Clin Pharm Ther. 2009;34:623–629. doi: 10.1111/j.1365-2710.2009.01059.x. [DOI] [PubMed] [Google Scholar]
- 11.Hulme E, Curtis CAM, Page KM, Jones PG. The role of charge interactions in muscarinic agonist binding, and receptor-response coupling. Life Sci. 1995;56:891–898. doi: 10.1016/0024-3205(95)00025-2. [DOI] [PubMed] [Google Scholar]
- 12.Gridges TM, Marlo JE, Niswender CM, Jones CK, Jadhav SB, Gentry PR, Plumley HC, Weaver CD, Conn PJ, Lindsley CW. Discovery of the first highly M5-preferring muscarinic acetylcholine receptor ligand, an M5 positive allosteric modulator derived from a series of 5-trifluoromethoxy N-benzyl isatins. J Med Chem. 2009;52:3445–3448. doi: 10.1021/jm900286j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohtake A, Saitoh C, Yuyanma H, Ukai M, Okutsu H, Noguchi Y, Hatanaka T, Suzuki M, Sasamata M, Miyata K. Pharmacological characterization of a new antimuscarinic agent, sulifenacin succinate, in comparison with other antimuscarinic agents. Biol Pharm Bull. 2007;30:54–58. doi: 10.1248/bpb.30.54. [DOI] [PubMed] [Google Scholar]
- 14.Jones LH, Randall A, Napier C, Trevethick M, Sreckovic S, Watson J. Design and synthesis of a fluorescent muscrinic antagonist. Bioorg Med Chem Lett. 2008;18:825–827. doi: 10.1016/j.bmcl.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Salcedo S, Davalillo S, Cabellos J, Lagunas C, Balsa D, Pérez-del-Pulgar S, Ballarín M, Fernández AG. In vivo and in vitro pharmacological characterization of SVT-40776, a novel M3 muscarinic receptor antagonist, for the treatment of overactive bladder. Brit J Pharmacol Soc. 2009;156:807–817. doi: 10.1111/j.1476-5381.2008.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casarosa P, bouyssou T, Germeyer S, Schnapp A, Gantner F, Pieper M. Preclinical evaluation of long-acting muscarinic antagonists: comparison of tiotropium and investigational drugs. J Pharmcol Expt Ther. 2009;330:660–668. doi: 10.1124/jpet.109.152470. [DOI] [PubMed] [Google Scholar]
- 17.Congreve M, Langmead CJ, Mason JS, Marshall FH. Progress in structure based drug design for G protein-coupled receptors. J Med Chem. 2011;54:4283–4311. doi: 10.1021/jm200371q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaakola VP, Ijzerman AP. The crystallographic structure of the human adenosine A2A receptor in a high-affinity antagonist-bound state: implications for GPCR drug screening and design. Curr. Opion. Struct. Biol. 2010;20:401–414. doi: 10.1016/j.sbi.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Shukla AK, sun JP, Lefkowitz RJ. Crystallizing thinking about the β2-adrenergic receptor. Mol Pharmacol. 2008;73:1333–1338. doi: 10.1124/mol.108.045849. [DOI] [PubMed] [Google Scholar]
- 20.Lu ZL, Hulme EC. A network of conserved intramolecular contacts defines the off-state of the transmembrane switch mechanism in a seven-transmembrane receptor. J Biol Chem. 2000;275:5682–5686. doi: 10.1074/jbc.275.8.5682. [DOI] [PubMed] [Google Scholar]
- 21.Bokoch MP, Zou Y, Rasmussen SGF, Liu CW, Nygaard R, Rosenbaum DM, Fung JJ, Choi HJ, Thian FS, Kobilka TS, Puglisi JD, Weis WI, Pardo L, Prosser RS, Mueller L, Kobilka BK. Ligand-specific regulation of the extracellular surface of a G–protein coupled receptor. Nature. 2010;463:108–114. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 23.Mustafi D, Palczewski K. Topology of class A G protein-coupled receptors: insights gained from crystal structures of rhodopsins, adrenergic and adenosine receptors. Mol Pharmacol. 2009;75:1–12. doi: 10.1124/mol.108.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci U S A. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen SGF, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–388. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 26.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EYT, Velasquez J, Kuhn P, Stevens RG. A specific cholesterol binding site is established by the 2.8Å structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tates CG, Schertler GF. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–492. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. The 2. 6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedretti A, Vistoli G, Marconi C, Testa B. Muscarinic receptors: a comparative analysis of structural features and binding modes through homology modeling and molecular docking. Chem Biodiv. 2006;3:481–501. doi: 10.1002/cbdv.200690052. [DOI] [PubMed] [Google Scholar]
- 32.Avlani VA, Gregory KJ, Morton CJ, Parker MW, Sexton PM, Christopoulos A. Critical role for the second extracellular loop in the binding of both orthosteric and allosteric G protein-coupled receptor ligands. J Biol Chem. 2007;282:25677–25686. doi: 10.1074/jbc.M702311200. [DOI] [PubMed] [Google Scholar]
- 33.Peng JYC, Vaidehi N, Hall SE, Goddard WA., III The predicted 3D structures of the human M1 muscarinic acetylcholine receptor with agonist or antagonist bond. Chem Med Chem. 2006;1:878–890. doi: 10.1002/cmdc.200600047. [DOI] [PubMed] [Google Scholar]
- 34.Vistoli G, Pedretti A, Dei S, Scapecchi S, Marconi C, Romanelli MN. Docking analyses on human muscarinic receptors: Unveiling the subtypes peculiarities in agonist binding. Bioorg Med Chem. 2008;16:3049–3058. doi: 10.1016/j.bmc.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henikoff S, Henikoff JG. Amino-acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphrey W, Dalke A, Schulten K. VMD-Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 38.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision A1. Gaussian, Inc; Pittsburgh, PA: 2003. [Google Scholar]
- 39.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo L, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Liu L, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER. Vol. 11. University of California; San Francisco: 2010. [Google Scholar]
- 40.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 41.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 42.Solis FJ, Wets RjB. Minimization by random search techniques. Maths Opera Res. 1981;6:19–30. [Google Scholar]
- 43.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 44.Darden T, York D, Pedersen L. Particle mesh Ewald—an Nlog(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 45.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 46.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 47.Gilson MK, Sharp KA, Honig BH. Calculating electrostatic interactions in biomolecules: Method and error assessment. J Comput Chem. 1987;9:327–335. [Google Scholar]
- 48.Jayaram B, Sharp KA, Honig B. The electrostatic potential of B-DNA. Biopolymers. 1989;28:975–993. doi: 10.1002/bip.360280506. [DOI] [PubMed] [Google Scholar]
- 49.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 50.Pan Y, Gao D, Zhan CG. Modeling the catalysis of anti-cocaine catalytic antibody: Competing reaction pathways and free energy barriers. J Am Chem Soc. 2008;130:5140–5149. doi: 10.1021/ja077972s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu H, Goren AC, Zhan CG. Characterization of the structures of phosphodiesterase 10 binding with adenosine 3′,5′-monophosphate and guanosine 3′,5′-monophosphate by hybrid quantum mechanical/molecular mechanical calculations. J Phys Chem B. 2010;114:7022–7028. doi: 10.1021/jp911527y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, Zhan X, Xiong Y, Liu J, Zhan CG. Fundamental reaction pathway and free energy profile for hydrolysis of intracellular second messenger adenosine 30,50-cyclic monophosphate (cAMP) catalyzed by phosphodiesterase-4. J Phys Chem B. 2011;115:12208–12219. doi: 10.1021/jp205509w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li D, Huang X, Han K, Zhan C-G. Catalytic mechanism of cytochrome P450 for 5′-hydroxylation of nicotine: fundamental reaction pathways stereoselectivity. J Am Chem Soc. 2011;133:7416–7427. doi: 10.1021/ja111657j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raha K, Merz KM. Large Scale Validation of a Quantum Mechanics Based Scoring Function: Predicting the Binding Affinity and the Binding Mode of a Diverse set of Protein-Ligand Complexes. J Med Chem. 2005;48:4558–4575. doi: 10.1021/jm048973n. [DOI] [PubMed] [Google Scholar]
- 55.Huang X, Gu HH, Zhan CG. Mechanism for cocaine blocking the transport of dopamine: insights from molecular modeling and dynamics simulations. J Phys Chem. 2009;113:15057–15066. doi: 10.1021/jp900963n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sapay N, Tieleman DP. Combination of the CHARMM27 force field with united-atom lipid force fields. J Comput Chem. 2011;32:1400–1410. doi: 10.1002/jcc.21726. [DOI] [PubMed] [Google Scholar]
- 57.Han SJ, Hamdan FF, Kim SK, Jacobson KA, Bloodworth LM, Li B, Wess J. Identification of an agonist-induced conformational change occurring adjacent to the ligand-binding pocket of the M3 muscarinic acetylcholine receptor. J Biol Chem. 2005;280:34849–34858. doi: 10.1074/jbc.M506711200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.