Summary
Aging and age-related diseases can be viewed as the result of the lifelong accumulation of stress insults. The identification of mutant strains and genes which are responsive to stress and can alter longevity profiles provides new therapeutic targets for age-related diseases. Here we reported that a Drosophila strain with reduced expression of ribose-5-phosphate isomerase (rpi), EP2456, exhibits increased resistance to oxidative stress and enhanced lifespan. In addition, the strain also displays higher levels of NADPH. The knockdown of rpi in neurons by double-stranded RNA interference recapitulated the lifespan extension and oxidative stress resistance in Drosophila. This manipulation was also found to ameliorate the effects of genetic manipulations aimed at creating a model for studying Huntington’s disease by overexpression of polyglutamine in the eye, suggesting that modulating rpi levels could serve as a treatment for normal aging as well as for polyglutamine neurotoxicity.
Keywords: ribose-5-phosphate isomerase, pentose phosphate pathway, neuron, oxidative stress, longevity, polyglutamine toxicity, Drosophila
Introduction
Aging is a universal, progressive, deleterious process, and also a major cause of many diseases (Miller 2009). The identification of mutants which can prolong lifespan can help uncover genetic mechanisms involved in regulating aging and may lead to new medical interventions to postpone aging and treat age-related diseases (Finkel 2005; Fontana et al. 2010). Several conserved mechanisms which affect aging have been identified including: the modulation of caloric intake by caloric restriction (CR), changing exposure to reactive oxygen species (ROS), altering the expression of target of rapamycin (TOR) and the insulin/IGF-1 pathways (Lu & Finkel 2008; Fontana et al. 2010; Kenyon 2010).
Increased longevity is often associated with higher resistance to different environmental stress (Arking et al. 1991). The manipulation of stress-responsive genes can be utilized to extend lifespan. For instance, Drosophila lifespan is increased by overexpression of the antioxidant Cu-Zn superoxide dismutase (SOD) (Parkes et al. 1998; Sun & Tower 1999), or the heat shock protein (HSP) gene hsp70, hsp27, hsp26, and hsp22 (Tatar et al. 1997; Morrow et al. 2004; Wang et al. 2004). Understanding what genes can modulate responses to stress and alter longevity can be applied to develop novel therapies. Mimicking some consequences of caloric restriction, the administration of Sir2 agonists extends lifespan in metazoans (Wood et al. 2004). Increased expression of SIRT1, a human homologue of SIR2, or administration of resveratrol both promote the survival of neuronal cells, protect against Alzheimer disease (Kim et al. 2007). Resveratrol improves the health and survival of mice fed high-fat diets (Baur et al. 2006). Therefore, the selection for elevated tolerance to stress can be used to screen for new genes or mutants to identify targets for lifespan extension and potential disease therapies (Wang et al. 2004; Liao et al. 2008; Liu et al. 2009).
The pentose phosphate pathway (PPP) is an important cellular defense system against oxidative stress. This pathway helps to remove excessive ROS resulting from mitochondrial oxidative phosphorylation. The major function of PPP is to generate nicotinamide adenine dinucleotide phosphate (NADPH) to provide more reduced form of glutathione to counteract the damaging effects of ROS. Long-lived flies contain higher glucose-6-phosphate dehydrogenase (G6PD) activity, a rate limiting enzyme in pentose phosphate pathway (Luckinbill et al. 1990). In addition, Hsp27 can increase G6PD activity (Preville et al. 1999), and Hsp27 overexpression transgenic flies extend lifespan and exhibit better resistance to oxidative stress (Wang et al. 2004; Liao et al. 2008). Modulation of G6PD expression regulates NADPH levels and protects neuronal cells against nitrosative stress-induced apoptosis (Garcia-Nogales et al. 2003). Transgenic mice with neuronal expression of G6PD displayed neuroprotective action against oxidative stress (Mejias et al. 2006). In addition, it was reported that PPP plays an important role in promoting neuronal survival upon exposure to reactive oxygen and nitrogen species (Bolanos & Almeida 2010). Recently it was shown that G6PD overexpression transgenic flies with elevated NADPH levels enhance lifespan and increase tolerance to oxidative stress (Legan et al. 2008). These data suggest a correlation of PPP activity with oxidative stress response and lifespan.
Neurodegenerative diseases are devastating progressive conditions which usually occur at late-age. Oxidative damage is one of the major causes of many progressive neurodegenerative diseases such as Huntington’s disease (Trushina & McMurray 2007). Huntington’s disease is a neurodegenerative disease in which with expanded CAG repeats cause a form of polyglutamine toxicity in neuronal cells. Several different Drosophila models for polyglutamine diseases were established to look for suppressor genes to alleviate polyglutamine toxicity by overexpressing toxic polyglutamine encoding constructs in the eye and the resulting rough eye used to identify enhancers or suppressors of the effect (Warrick et al. 1998; Kazemi-Esfarjani & Benzer 2000; Sang et al. 2005). A number of genes able to rescue polyglutamine toxicity were identified, including CBP, hsp70, dhdj1, and hsp27 (Warrick et al. 1999; Kazemi-Esfarjani & Benzer 2000; Taylor et al. 2003; Liao et al. 2008). Some of the genes that rescued the polyglutamine toxicity in the fly eyes were also found to promote longevity, like hsp70, hsp27, and CBP (Tatar et al. 1997; Liao et al. 2008; Zhang et al. 2009). Thus, in some cases genes which enhance lifespan can also modulate polyglutamine toxicity and thus be useful for preventing neurodegeneration.
Here we reported that a Drosophila strain EP2456 with reduced expression of ribose-5-phosphate isomerase (rpi) expresses higher level of NADPH and exhibits increased resistance to oxidative stress and enhanced lifespan. Furthermore tissue specific knockdown of rpi in neurons by double-stranded RNA interference recapitulated the lifespan extension and oxidative stress resistance in Drosophila. It also rescued the rough eye morphology resulting from polyglutamine toxicity. These data provide a new approach for postponing aging and suggest a possible treatment for neurodegenerative diseases.
Results
Reduced ribose-5-phosphate isomerase expression in neurons enhances lifespan and increases resistance to oxidative stress in Drosophila
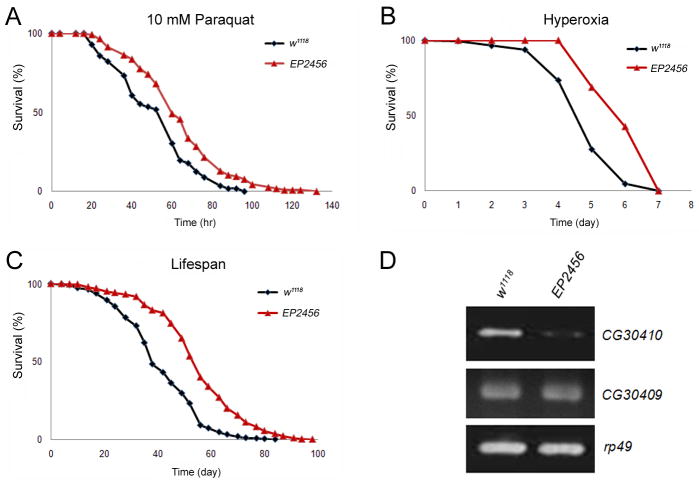
Enhanced longevity is usually associated with better resistance to environmental stresses. We performed a screen for enhanced stress resistance, to identify genes which extend lifespan in Drosophila (Wang et al. 2004; Liao et al. 2008; Liu et al. 2009). Here, we report the characterization of EP2456, a strain which exhibits enhanced resistance to paraquat and hyperoxia-induced oxidative stress respectively with 27% and 31% increase in mean survival time relative to the control w1118 (Fig. 1A, 1B, and supplemental Table 1). EP2456 is also long-lived with 39% increase in mean lifespan compared to w1118 (Fig. 1C, S. Table 1).
Fig. 1. Mutant fly EP2456 with reduced expression of rpi displays increased resistance to oxidative stress and enhanced lifespan.
EP2456 (red triangle) exhibits elevated resistance to (A) 10 mM paraquat induced oxidative stress (P<0.05) and (B) hyperoxia (95% O2) induced oxidative stress (P<0.01) compared to the control w1118 (black diamond). (C) EP2456 displays increased mean lifespan (P<0.001) compared to w1118 at 25°C. (D) Reduced expression of CG30410 (rpi) was detected in EP2456 compared to w1118, but no differences in the expression of the neighboring CG30409 between w1118 and EP2456. The expression of rp49 was used as a control.
To find the gene responsible for these phenotypes, we used semi-quantitative PCR to measure the level of expression of transcripts in the region near the P-element insertion, CG30409 and CG30410, relative to w1118 (Fig. 1D). Expression of CG30410 was reduced by 80% in the mutant, while CG30409 remained unchanged. Since CG30410 encodes a ribose-5-phosphate isomerase (rpi) which is involved in regulating NADPH levels via the pentose phosphate pathway and NADPH is involved in combating oxidative stress, these results are consistent with the downregulation of rpi affecting oxidative stress response.
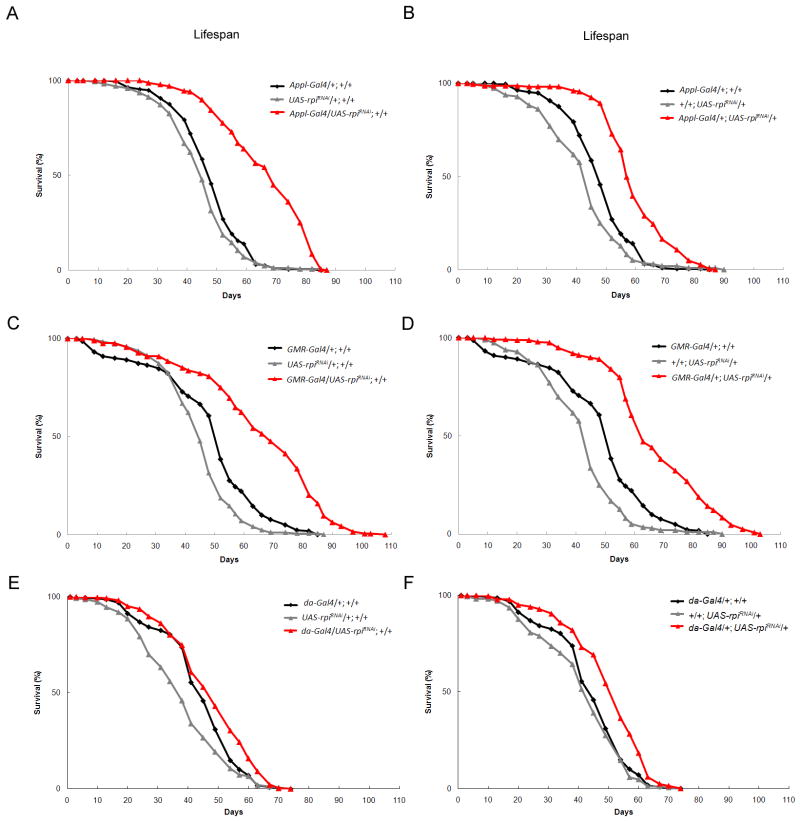
To determine whether decreased rpi expression extends lifespan, we generated independent Gal4 driven transgenic RNA knockdown lines, UAS-rpiRNAi with the insertion on either the second or third chromosome. Since it is known that neuronal tissue is particularly sensitive to oxidative stress, we compared the effects of reducing rpi expression in the neuronal tissue and eye to that of reducing the expression in the whole animal. The knockdown of rpi by tissue specific drivers Appl-Gal4 and GMR-Gal4 resulted in increased resistance to oxidative stress. However these effects were not seen with the ubiquitous driver da-Gal4 (S. Table 2). The flies with the pan-neuronal knockdown of rpi by Appl-Gal4, displayed a significant increase in mean lifespan ranged from 25% to 38% compared to the controls (Fig. 2A, 2B, and S. Table 3). Interestingly, using the predominantly eye specific GMR-Gal4 also resulted in a 35% and 38% in mean lifespan extension (Fig. 2C, 2D, and S. Table 3). These effects were confirmed using five independent UAS-rpiRNAi transgenic lines driven by Appl-Gal4 or GMR-Gal4 (results summarized in supplemental table 4). Ubiquitous knockdown of rpi by da-Gal4 resulted in modest enhancement of longevity of only 8% to 14% relative to the controls (Fig. 3E, 3F, and S. Table 3). It is possible that the knockdown of rpi by da-Gal4 in the other tissues beside neurons nullifies the beneficial effect on lifespan and oxidative tolerance. Interestingly, adult specific knockdown of rpi using the RU486-induced neuronal Gal4 driver, elav-GS Gal4 did not result in lifespan extension (supplemental Fig. 1). This is similar to what was observed by Simonsen et al. where lifespan extension was detected in the atg8a overexpression flies driven by Appl-Gal4 but not able by elav-Gal4 driver (Simonsen et al. 2008). Whether the age-dependent or the cell-type specific expression differences or both of the two different neuronal Gal4 drivers contributing to the differences in the lifespan results awaits further experiments to clarify. Together, the results indicate that the tissue-specific knockdown of rpi in neurons increases resistance to oxidative stress and extends lifespan in Drosophila.
Fig. 2. The RNA interference knockdown expression of rpi in neurons extends lifespan in Drosophila.
The independent transgenic fly lines with knockdown of rpi (A, B) by Appl-Gal4 (red triangle) or (C, D) by GMR-Gal4 (red triangle) exhibit dramatically increased lifespan than the control flies (black diamond and grey triangle). (E, F) The transgenic fly lines with knockdown of rpi ubiquitously by da-Gal4 (red triangle) only display little increased lifespan compared to the control flies (black diamond and grey triangle).
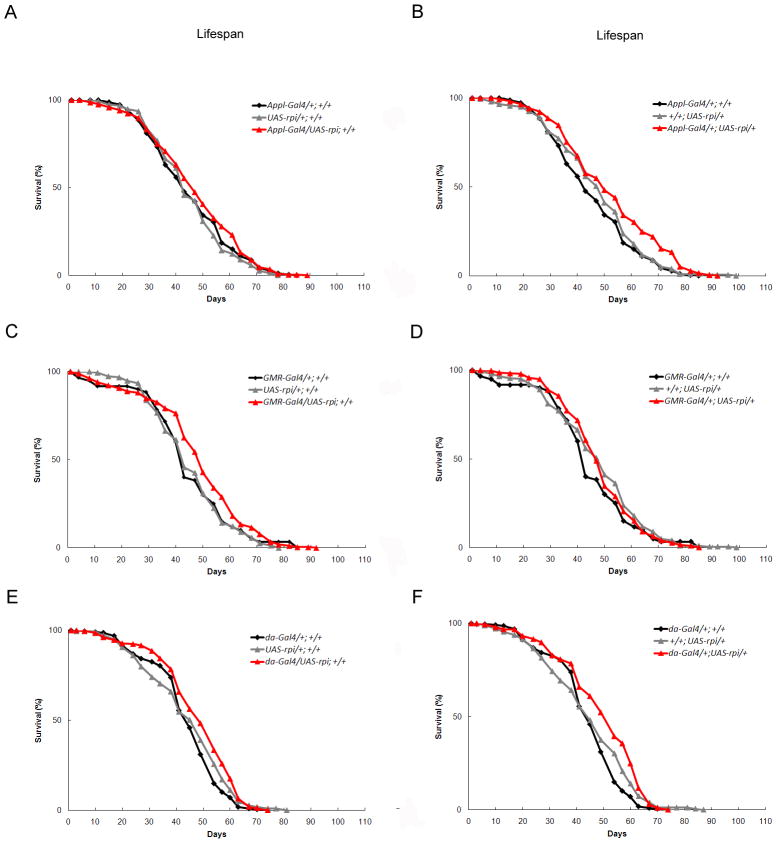
Fig. 3. The trangenic flies with overexpression of rpi do not have shortened lifespan.
(A to F) The rpi overexpression transgenic flies with UAS-rpi either on the second or third chromosome driven by Appl-Gal4, GMR-Gal4, and da-Gal4 (red triangle) do not show altered lifespan compared to their control flies (black diamond, grey triangle).
Overexpression of rpi does not shorten lifespan nor aggravates response to oxidative stress in Drosophila
Since the knockdown of rpi in neurons extends lifespan, we examined whether overexpression of rpi decreases lifespan and enhances susceptibility to oxidative stress. Independent Gal4 responsive transgenic lines were generated to measure the effect of overexpressing rpi. Transgenic lines, containing the full-length rpi cDNA, UAS-rpi were crossed with the same Gal4 drivers used previously, and their progeny were tested for lifespan and oxidative stress. None of the transgenic flies overexpressing rpi showed decreased lifespan (Fig. 3A, B, C, D, E, F, S. Fig. 1, and S. Table 5 and 7), nor were they sensitive to oxidative stress (data not shown). In fact, certain specific cross combinations resulted in marginal increases in mean lifespan upon Gal4 induction. The data revealed that overexpression of rpi does not aggravate the response to oxidative stress nor decrease lifespan in Drosophila.
Knockdown expression of rpi increases glucose-6-phosphate dehydrogenase activity and the levels of NADPHand the reduced form of glutathione
We proposed that reduced rpi could lead to elevated levels of NADPH, since rpi is involved in the pentose phosphate pathway and thus in producing more NADPH for the generation of the reduced form of glutathione (GSH). The knockdown of rpi may result in excess ribulose-5-phosphate to be metabolized via non-oxidative phase of PPP and feedback to glucose-6-phosphate to generate more NADPH (Bolanos & Almeida 2010). Therefore, we measured the NADPH levels in both the mutant and the control flies. A two-fold increased level of NADPH was measured in EP2456 compared to w1118 (Fig. 4A). We further examined the NADPH levels in the flies with the knockdown of rpi either by Appl-Gal4 or GMR-Gal4 driver as well as their control flies. We also detected increased levels of NADPH in the flies with the knockdown of rpi compared to the controls (Fig. 4B, C).
Fig. 4. Increased NADPH levels are detected in the longevity flies with lowered rpi expression.
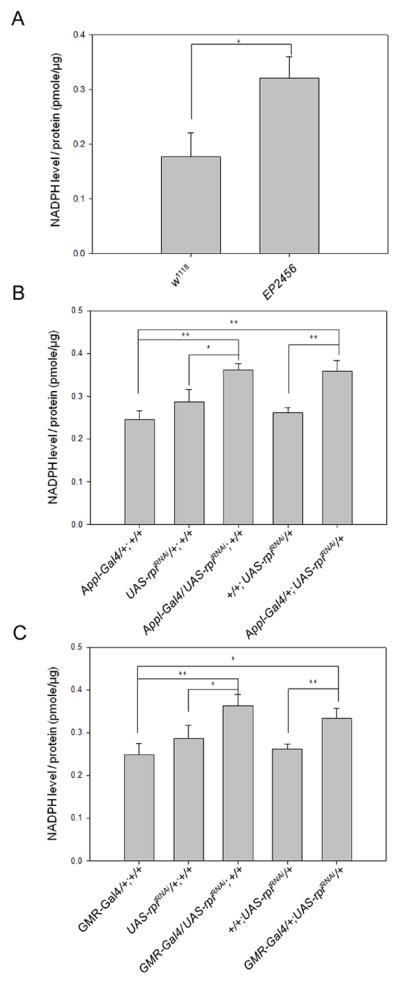
The NADPH level of each fly line was determined and normalized by its protein concentration. Each bar represents mean ± SEM (n = 3) (unit: pmole/ug). *P < 0.05; **P < 0.01 (A) w1118 (0.18 ±0.03) versus EP2456 (0.32±0.02). (B) Appl-Gal4/+;+/+ (0.25±0.01), UAS-rpiRNAi/+;+/+ (0.27±0.02), Appl-Gal4/UAS-rpiRNAi;+/+ (0.36±0.01), +/+;UAS-rpiRNAi/+ (0.26±0.01), Appl-Gal4/+;UAS-rpiRNAi/+ (0.36±0.02). (C) GMR-Gal4/+;+/+ (0.25±0.02), GMR-Gal4/UAS-rpiRNAi;+/+ (0.36±0.02), GMR-Gal4/+;UAS-rpiRNAi/+ (0.33±0.01).
Overexpression of G6PD was reported to extend lifespan in Drosophila (Legan et al. 2008), and G6PD is the rate-limiting enzyme of the PPP. To determine if G6PD was altered in our strains, we observed both young and old EP2456 flies and observed increased levels of G6PD in both (S. Fig. 2). We also measured G6PD activity and the levels of reduced GSH in Drosophila S2 cells and found that rpi-knockdown resulted in a 20% increase of G6PD activity and a 25% enhanced level of GSH compared to the untreated cells (S. Fig. 3). Together, it suggests that reducing rpi results in elevated levels of NADPH as a result of changes in the pentose phosphate pathway.
The knockdown of rpi attenuates the polyglutamine toxicity induced Drosophila rough eyes, which depends on G6PD and transaldolase expressions
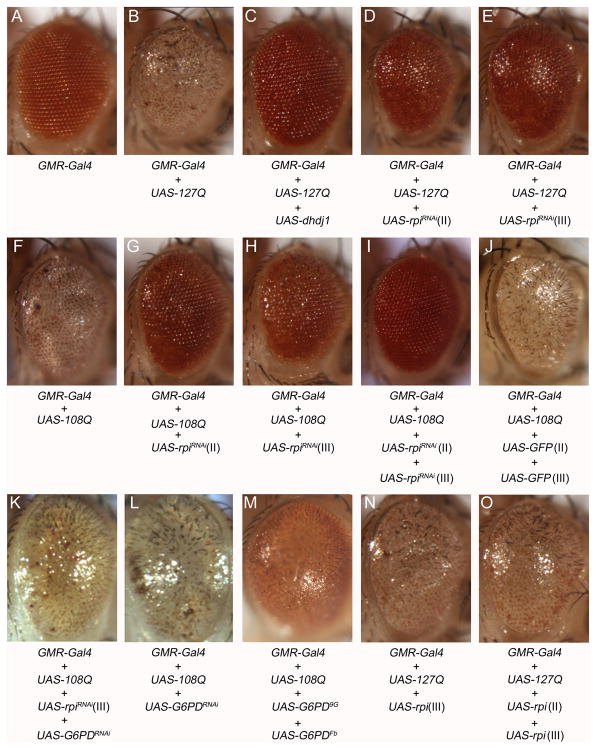
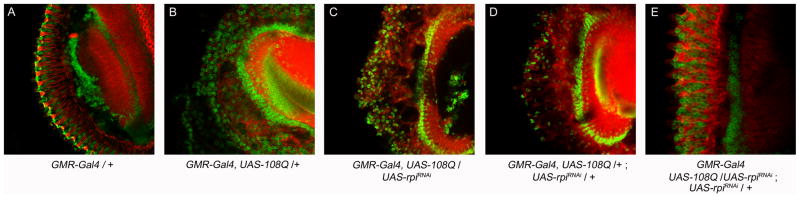
Since rpi knockdown flies appear to have a neuroprotective effect against oxidative damage, we tested whether this manipulation would protect against other types of neural toxicity such as polyglutamine toxicity. The GMR-GAL4>UAS-127Q (Kazemi-Esfarjani & Benzer 2000) flies and GMR-Gal4>108Q (Sang et al. 2005) expressing a long tract of glutamines displayed a rough eye phenotype and loss of pigment distribution (Fig. 5B, F). We tested whether rpi knockdown could rescue the effect by simultaneously expressing the polyQ and the rpi knockdown in the eye using GMR-Gal4. The knockdown of rpi dramatically rescued the phenotype of the rough eye and loss of pigment in both GMR-GAL4>UAS-127Q and GMR-GAL4>UAS-108Q flies (Fig. 5D, E, G, H). The effect was comparable with the result by the expression of dhdj1 (Fig. 5C). These results show that rpi knockdown is effective at blocking polyQ toxicity resulting from two independently generated lines in two different genetic backgrounds. In addition, the knockdown of rpi was shown to be dose dependent since two copies of UAS-rpiRNAi exhibit significantly better rescue of the eye phenotype compared to only one copy of UAS-rpiRNAi (Fig. 5G, H, I). To exclude the possibility that the rescue observed resulted from a titration of Gal4 proteins by the extra UAS sequences introduced, we crossed GMR-Gal4, UAS-108/Cyo with the fly strains harboring two copies of UAS-GFP (UAS-GFP;UAS-GFP) and none of their offspring shows any rescued eye phenotype (Fig. 5J).
Fig. 5. The knockdown of rpi rescues the polyglutamine-induced rough eyes, and the overexpression of rpi does not further deteriorate the rough eye phenotype.
The eye phenotypes resulting from different combinations of different transgenes of 127Q, 108Q, rpi, G6PD, GFP constructs are shown from A to O. (A) One copy of GMR-Gal4 displayed normal eye appearance as a control. (B) The expression of UAS-127Q by GMR-Gal4 exhibits rough eye phenotype. (C) The expression of UAS-hdj1 rescues the rough eye phenotype (Kazemi-Esfarjani & Benzer 2000), which was used as a positive control. (D, E) The RNAi knockdown of rpi by the transgene on the second (II) or the third (III) chromosome rescues the 127Q-induced rough eye. (F) The expression of UAS-108Q shows similar rough eye phenotype. (G, H) The RNAi knockdown of rpi also rescues the 108Q-induced rough eye. (I) The knockdown of rpi with two copies of UAS-rpiRNAi rescues the rough eye much better than a single copy of UAS-rpiRNAi. (J) The 108Q-induced rough eye cannot be rescued by two copies of UAS-GFP. (K) Knockdown of G6PD blocks the rescue by rpi knockdown on 108Q-induced rough eye. (L) Knockdown of G6PD does not degenerate the rough eye. (M) Overexpression of two copies of UAS-G6PD partially rescues the rough eye. (N) Overexpression of one copy of UAS-rpi does not further worsen the 127Q-induced rough eye. (O) The expression of two copies of UAS-rpi still does not deteriorate the rough eye.
Since reducing rpi expression elevates NADPH level and increases expression of G6PD, we asked whether the rpi mediated rescue of the polyglutamine toxicity depends on the expression of G6PD. We found that reducing G6PD levels by knockdown abolished the rescue mediated by rpi knockdown (Fig. 5K). However, the knockdown of G6PD in GMR-Gal4>UAS-108Q does not further deteriorate 108Q-induced rough eyes (Fig. 5L). Overexpression of two copies of UAS-G6PD can partially rescue the 108Q-induced rough eyes (Fig. 5M). The results suggest that G6PD expression mediates the effects of rpi potentially by its effect on NADPH levels.
Since our hypothesis is that rpi knockdown results in a shunt of ribulose-5-phosphate back to glucose-6-phosphate via the non-oxidative phase by a series of enzymes including transaldolase, thus the rescue by rpi knockdown of polyQ toxicity should depend on transaldolase activity. Indeed, the knockdown of transaldolase expression also blocks the rescue by the knockdown of rpi expression of 108Q toxicity (S. Fig. 4) As seen using SEM at higher magnification reduced transaldolase expression suppresses the rescue of 108Q toxicity by rpi knockdown (S. Fig 4). This indicates that the rescue by the knockdown of rpi requires transaldolase activity. Disruption of transaldolase activity has been shown to lower NADPH level (Qian et al. 2008). All these results support the conclusion that the enhancement of lifespan, resistance to oxidative stress, and alleviation of polyglutamine toxicity resulting from knockdown of rpi are mediated via increased G6PD activity and NADPH level.
Overexpression of rpi does not exacerbate the polyglutamine toxicity
Since the knockdown of rpi suppressed polyglutamine toxicity, we tested whether overexpression of rpi would enhance polyglutamine toxicity. Overexpression of rpi did not show any further deterioration of the rough eyes in the GMR-GAL4>UAS-127Q flies (Fig. 5N) even when two copies of UAS-rpi were used (Fig. 5O). This result correlates with the data from lifespan and stress assays, indicating that overexpression of rpi does not result in any significant negative effect on the phenotypes measured
The knockdown of rpi improves the abnormal retinal ultra-structure by polyglutamine toxicity
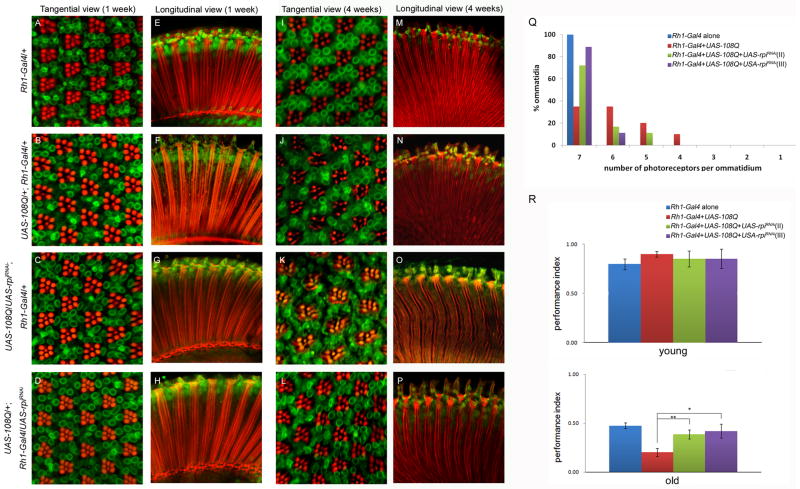
The compound eye of a fruitfly is composed of approximately 760 ommatidia. Each ommatidium contains 8 photoreceptor cells (R1–R8), supporting cells, pigment cells, and a cornea. Every photoreceptor cell which is responsible for phototransduction possesses the cell body and the rhabdomere. Expressing polyglutamine in the eye causes toxicity to neuronal cells, leading to dysfunction and cell death in selected neurons. Although knockdown of rpi effectively rescued the rough eye and pigment distribution under polyglutamine toxicity, we asked whether it can improve the integrity of neurons in fly eyes. We examined the ultra-structure of retina in the GMR-Gal4>UAS-108Q with and without the knockdown of rpi by whole mount staining (Sang & Ready 2002). A longitudinal view of the retina in GMR-Gal4>UAS-108Q revealed that the internal structure was disrupted and the cells had degenerated, resulting in large vacuoles not seen in the control flies (GMR-Gal4; Fig. 6A, B). Conversely, the retina of the flies expressing 108Q and also having reduced rpi levels showed improved structural integrity and more F-actin staining by pholloidin (Fig. 6C, D), the vacuoles however still existed showing that the rescue is not complete. As expected when two copies of UAS-rpiRNAi were used the rescue of the phenotype was better (Fig. 6 C, D, E), as was seen by observing the external eye morphology (Fig. 5G, H, I). Thus, the knockdown of rpi in GMR-Gal4>UAS-108Q not only rescued the external rough eye and the eye pigment but also improved the internal ultra-structural disorder of retina.
Fig. 6. The knockdown of rpi improves the internal structure of the fly eyes under polyglutamine toxicity.
The structures of the 4-day-old fly eyes were assessed by the staining of phalloidin to indicate rhabdomeres (red) and anti-lamin to nuclear membrane (green). (A) The control fly with one copy of GMR-Gal4 displays normal integrity of rhabdomere structure. (B) The expression of UAS-108Q under GMR-Gal4 exhibits massive cell loss and collapsed structure with vacuoles. (C, D) The knockdown of rpi either on the second or third chromosome shows more rhabdomere staining, though the vacuoles still exist. (E) The knockdown of rpi with two copies of UAS-rpiRNAi dramatically improves the structure of rhabdomere and decreases the size of vacuoles.
The knockdown of rpi attenuates adult-onset polyglutamine toxicity and ameliorates the damaged phototaxis in aged Drosophila
Many polyglutamine diseases are progressive and late-onset. However, GMR-Gal4 driver expresses Gal4 in the early development throughout adult stage. To mimic the late-onset event and check if the knockdown of rpi still can rescue it, we used Rh1-Gal4 driver, which expresses Gal4 in the late pupa stage, to express UAS-108Q alone or in combination with UAS-rpiRNAi. At one week of age no photoreceptor degeneration was observed in Rh1-Gal4>UAS-108Q flies (Fig. 7B, F) or in the knockdown of rpi, Rh1-Gal4>UAS-108Q, UAS-rpiRNAi (Fig. 7C, D, G, H), relative to the control Rh1-Gal4 driver alone (Fig. 7A, E). This reveals that 108Q toxicity is not observable in the one-week flies. Four-week old control fly Rh1-Gal4 showed normal ommatidium morphology, an ordered array of R1-R6 photoreceptor neurons surrounding the central R7 cell and the intact continuous structure of rhabdomeres as those in the one-week old Rh1-Gal4 (Fig. 7A,E, I, M). In age matched Rh1-Gal4>UAS-108Q flies, the photoreceptor neurons degenerated and the rhabdomeres were fragmented (Fig. 7J, N). Knockdown of rpi significantly improved rhabdomere structure and rescued the photoreceptor neurons of each ommatidium (Fig. 7 K, L, O, P). The percentage of the intact ommatidia containing seven photoreceptors were significantly improved from 38% to 72% - 90% in the flies with the knockdown of rpi in Rh1-Gal4>UAS108Q;UAS-rpiRNAi compared to Rh1-Gal4>UAS108Q alone (Fig. 7Q). These data demonstrate that rpi knockdown also relieves adult-onset polyglutamine toxicity.
Fig. 7. The knockdown of rpi suppresses adult-onset polyglutamine toxicity in aged Drosophila.
Phalloidin staining to rhabdomere (red) and anti-lamin staining to nuclear membrane (green) of the eyes of the 1-week young and 4-week old flies with different transgenes were shown from A to P. The images from A to D (1-week) and I to L (4-week) are tangential view of the retina, and those from E to H (1-week) and M to P (4-week) are longitudinal view of the retina. (A, E, I, M) Both the 1-week and 4-week control flies Rh1-Gal4/+ show normal structures of ommatidia which consist of an ordered array of R1-R6 photoreceptor neurons surrounding the central R7 cell, and a well lineup of rhabdomeres. (J, N) Severe degeneration of R1-R6 photoreceptor neurons and the disintegrated structures of rhabdomeres were observed only in the eyes of 4-week old Rh1-Gal4>UAS-108Q, (B, F) but not in those of the 1-week young ones. (C, D, G, H) The knockdown of rpi either on the second or third chromosome improved the structure of photoreceptor neurons and rhabdomeres under Rh1-Gal4>UAS108Q. (Q) The knockdown of rpi mitigates the photoreceptor neurons loss caused by polyglutamine toxicity. The distribution of the number of photoreceptor neurons in ommatidia under the expression of different transgenes was shown. (R, the upper panel) The one-week young flies show no differences in the phototaxis function among the fly lines under Rh1-Gal4. (R, the lower panel) The knockdown of rpi significantly rescues the impaired phototaxis function in the old flies under adult-onset polyglutamine toxicity.
To determine if the rescued eyes also function better, we measured phototaxis of the young and old Rh1-Gal4>UAS108Q flies with or without the knockdown of rpi. Since the Rh1 mediated Gal4 expression starts in the late pupa stage, there is no difference of the phototaxis activity among the young flies under 108Q with or without the knockdown of rpi (Fig. 7R, the upper panel). However, as flies aged polyglutamine toxicity reduced the phototaxis activity from 48% to 20%. The effect of polyglutamine toxicity on phototaxis was largely rescued by the rpi knockdown back to 39% and 42% respectively (Fig. 7R, the lower panel). Together, knockdown of rpi not only rescued the abnormal structure of ommatidia but also functionally amended the damaged phototaxis activity by polyglutamine toxicity.
Discussion
In this study, we report that reduction in the expression of rpi can lead to enhanced longevity, resistance to oxidative damage and protect against polyglutamine induced neurodegeneration. The reduced expression of rpi by RNAi in neuronal tissue and eyes also provided significant lifespan extension and better resistance to oxidative stress in Drosophila. The results demonstrated that the specific tissues like neuronal tissue and perhaps the photoreceptor neurons in the eyes may be the target sites of the beneficial effect of knockdown expression of rpi. Neurons have been proposed to be important for modulating longevity and stress resistance (Wolkow 2002; Tatar et al. 2003; Garelick & Kennedy 2010). Overexpression of superoxide dismutase (SOD) in motorneurons increased resistance to oxidative stress and enhanced lifespan in Drosophila melanogaster (Parkes et al. 1998). Activation of JNK signaling in neurons but not in muscle had better resistance to oxidative stress and extended lifespan (Wang et al. 2003). Neuronal expression of hsp26 and hsp27 elevated lifespan and resistance to oxidative stress (Liao et al. 2008). Mutations that affected the function of specific sensory neurons influenced longevity in C. elegans and Drosophila (Alcedo & Kenyon 2004; Libert et al. 2007).
We observed that overexpression of rpi did not have any adverse effect on the lifespan and oxidative stress resistance. This may be due to the fact that the enzyme does not catalyze the rate-limiting step in the pathway it controls and that therefore effects are only observed when the enzyme levels are reduced to the point at which flux through the pathway is altered. Additional reports have shown that inhibition of short neuropeptide F (sNPF) expression in neurons by RNAi silencing extended lifespan but overexpression of sNPF did not shorten the lifespan in Drosophila (Lee et al. 2008), suggesting that critical levels of gene expression are needed to promote longevity changes, however that changes in only one direction may have effects on longevity.
Many progressive neurodegenerative diseases are caused by oxidative damage that leads to neuronal cell death. Polyglutamine toxicity has been shown to be involved in a number of progressive neurodegenerative disorders. It has been shown that polyQ can induce intracellular ROS and cause cell death (Wyttenbach et al. 2002). Overexpression of Cu-Zn superoxide dismutase significantly increased lifespan and oxidative stress resistance (Parkes et al. 1998; Sun & Tower 1999). Neuronal expression of hsp27 not only increases tolerance to oxidative stress and lifespan, it also ameliorates Parkinsonism climbing disorder and mild polyglutamine toxicity (Liao et al. 2008). Our finding that the reducing rpi expression in neurons leads to higher levels of NADPH and enhanced oxidative stress resistance and reduced sensitivity to polyglutamine toxicity suggests a new strategy for neuronal protection. RPI isomerizes ribulose 5-phosphate to ribose 5-phophate in the non-oxidative phase of pentose phosphate pathway that facilitates synthesis of the nucleotides and nucleic acids. A recent report suggested that the lack of activity of 6-phophofructo-2-kinase/fructose 2,6-bisphosphatase isoform 3 (PFKFB3) in neurons causes glucose-6-phosphate (G6P) to be metabolized predominantly via PPP to generate NADPH against nitrosative stress (Herrero-Mendez et al. 2009; Bolanos & Almeida 2010). The knockdown of rpi may allow the excessive ribulose-5-phosphate to be metabolized to become fructose-5-phosphate (F6P) by non-oxidative phase of PPP (Wamelink et al. 2008). The lack of PFKFB3 activity in neurons may re-distribute the recycled F6P back to G6P for the re-entry of PPP and generate more NADPH in oxidative phase of PPP. Therefore, we hypothesize that lowered expression of rpi in neurons may allow the ribulose-5-phosphate via F6P to be recycled back into G6P and result in more NADPH to cope with oxidative stress. The oxidized glutathione (GSSG) needs a glutathione reductase which requires NADPH as a co-factor to direct the regeneration of reduced form of glutathione (GSH). The reduced GSH is the antioxidant used to protect cells from oxidative stress (Masella et al. 2005). Exposure of cells to the antioxidant N-Acetyl-L-cysteine (NAC) or GSH protected against polyglutamine-induced cell death (Wyttenbach et al. 2002).
In our studies, mRNA levels of G6PD, a rate-limiting enzyme for the generation of NADPH, was increased in both young and old flies with reduced rpi. In addition, both the G6PD activity and the reduced form of glutathione were increased by knockdown of rpi in Drosophila S2 cells. Likewise overexpression of G6PD increases resistance to oxidative stress and enhances lifespan (Legan et al. 2008). This may explain the mechanism through which lowered rpi expression in neurons enhances resistance to oxidative stress and suppression of polyglutamine toxicity. Interestingly, the rescue effect by the knockdown of rpi on 108Q toxicity was abrogated by the knockdown of G6PD (Fig. 5K). On the other hand, expression of two copies of UAS-G6PD can partially rescue 108Q toxicity (Fig. 5M). Together, it supports the notion that the effect by rpi knockdown depends on the expression of G6PD.
In summary, the knockdown of rpi in neurons provides enhanced resistance to oxidative stress, extends lifespan, and alleviates polyglutamine toxicity in Drosophila by at least one of the mechanisms to increase NADPH level through elevated G6PD activity. The development of new compounds which suppress ribose-5-phosphate isomerase enzyme activity may serve a new avenue for postponement of aging, a therapy for polyglutamine neurodegenerative diseases, and even on other age-related diseases like cancers.
Experimental procedures
Fly strains and maintenance
All flies were raised on standard fly food at 25°C, 65% humidity, 12hr light/dark cycle incubator. The fly lines used in the experiment EP2456, w1118, GMR-Gal4, Appl-Gal4, da-Gal4, GMR-Gal4/CyO;UAS-127Q were originally derived from Dr. Seymour Benzer’s lab (Caltech, USA). The RU486-induced elav-GeneSwitch (elav-GS) Gal4 line was provided by Dr. Pei-Yu Wang, where it was from Dr. Keshishian H. at Yale University. EP2456 and all the Gal4 drivers (except elav-GS here) used in the lifespan assay were back-crossed with w1118 ten times and generated as the homozygous lines used in the experiment. The fly lines, GMR-Gal4, UAS-108Q/Cyo and UAS-108Q;Rh1-Gal4 and Rh1-Gal4, were provided by Dr. Tzu-Kang Sang at NTHU. Transgenic RNAi line UAS-G6PDRNAi (#3337) targeting glucose-6-phosphate dehydrogenase (G6PD, CG12529) and UAS-tal RNAi (#106308) targeting transaldolase (tal, CG2827) were purchased from VDRC. The G6PD transgenic overexpression lines UAS-G6PD9G and UAS-G6PDFb were kindly provided by Dr. William Orr (Legan et al. 2008).
Oxidative stress and lifespan assays
Four-days old were collected and sexed and kept on the standard fly food with 25 flies per vial overnight. For paraquat assay, the flies were fed with 10 mM paraquat in 5% sucrose solution and the number of deaths were counted six hours until all the flies were dead. For hyperoxia assay, the flies were kept in a chamber with 95% oxygen adjusted by ProOx 110 (BioSpherix, Ltd.) and counted daily untill all dead. Student t-test was used to calculate P value to determine statistical significance. For lifespan assay, the flies were maintained in a 25°C/65% humidity-controlled incubator, transferred to new food every 3 -4 days untill all were dead. Log-rank test was used to calculate P value to determine statistical significance. At least three independent measurements were performed for each experiment.
Generation of the transgenic flies to express either the full-length coding sequence or the double-stranded RNA of ribose-5-phosphate isomerase
To generate UAS-rpi transgenic flies, the full-length coding sequence (CDS) of rpi (CG30410, which contains no intron) was amplified from the genomic DNA from Canton S by PCR using forward primer (5′-CAGCTCGAGTTAACCACATGTTATCTGGGTT-3′) and reverse primer (5′-GTCACTAGTAACTCCGCCCA AGAACTACTTG-3′), and the purified CDS was subcloned into XhoI/SpeI sites of the transgenic vector pINDY6 (Wang et al. 2004). To generate UAS-rpiRNAi transgenic flies, the 338-bp partial CDS fragment, which was analyzed by BLAST to avoid the off-target effects, was amplified by PCR with the primer set (5′-TCAGAATTCGGCGGTGGACCAGTGGGTGACTGA-3′ and 5′-TGAGAATTCAGTCGGCCACCACGATGAAGTGCTT-3′), the purified partial CDS was subcloned into EcoRI site of SympUAST vector (Giordano et al. 2002). The constructs were first verified by DNA sequencing to confirm that no point mutation derived from PCR amplification were introduced, and later used to generate the multiple independent transgenic lines. The ability of UAS-rpi to overexpress and UAS-rpiRNAi to knockdown the expression of rpi upon Gal4 induction was verified by RT-PCR and the folds of changes were listed in the supplemental table 6. All primer sequence is available upon request.
NADPH measurement
Ten flies of each specific strain were frozen at -80°C in a freezer. The frozen flies were washed with cold phosphate buffer saline (PBS) and extracted with cold NADP/NADPH extraction buffer. All the sample preparation was always kept on ice and according to the protocol in NADP/NADPH quantitation kit (BioVision, Cat.# K347-100). The values of the samples at O.D. 450 nm were measured and used to calculate NADPH concentration of each sample according the standard curve. The NADPH value of each sample was normalized by its own protein concentration. At least three independent measurements were carried out. Student t-test was used to calculate P value to determine statistical significance.
Whole-mount retina immunostaining
The procedure was adapted from the method by Sang and Ready (Sang & Ready 2002). The fly eyes of each sample at specific time points were fixed in 4% paraformaldehyde in PBS for 1 hour and washed three times in PBST(PBS plus 0.3% Triton X-100) with 10 minute each time. Then the eyes were incubated with TRITC-phalloidin (Sigma) to stain F-actin and incubated with 1:20 diluted anti-lamin monoclonal antibodies (ADL67.10, Developmental Studies Hybridoma Bank) in PBST plus 5% goat serum at 4°C overnight. After the eyes were washed three times in PBST, they were incubated with 1:100 diluted FITC-conjugate anti-mouse IgG (Jackson ImmunoResearch) for 4 hours, and washed three times in PBST, mounted with the mounting medium (VECTASHIELD), and examined by the confocal microscopy with Zeiss 510.
Phototaxis assay
The 4-days old male flies of each strain were collected at a density of 30 flies per vial, maintained on the regular fly food, and transferred to new fly food every 3 to 4 days to age the flies to the specific time for the whole mount retina staining and the phototaxis assay (Benzer 1967). In a darkroom, a 10-watt fluorescent lamp was used as a light source. Approximately 100 flies of the same strain were pooled into a test tube, and placed in the countercurrent apparatus, and left to relax for 2 minutes, then banged down in the apparatus and the empty tube side was placed toward to the light source and given 10 seconds for flies to react, and slide the tubes to collect the flies walking toward to the light. Performance index (PI) was calculated by the number of the flies walked to the light side divided by the total number of flies. Three independent measurements were performed. Student t-test was used to calculate P value to determine statistical significance.
Supplementary Material
Acknowledgments
We thank Drs. William Ja and Jui-Chou Hsu for the critical reading and suggestion for the manuscript. We also thank Dr. William Orr to provide UAS-G6PD transgenic fly lines and share the preliminary information for personal communication. We appreciate Dr. Pei-Yu Wang and Dr. Haig Keshishian to share elav-GS Gal4 line. We thank the funding support from National Science Council (97-2311-B-007-004-MY3), Chang-Tsing Collaboration grant (99N2430E1), NTHU Booster grant (99N2903E1) to Dr. Horng-Dar Wang. The grants from National Health Research Institute (NHRI-EX100-9925SC) and NSC (98-2314-B-182A-082-MY3) to Dr. Chao-Yung Wang, and the National Institutes of Health grant (1R15AG027749) to Dr. Ted Brummel are acknowledged. We are indebted for the partial funding support from the Brian Research Center by Dr. Ann-Shyn Chiang at NTHU, the assistance from Fly Core Taiwan by Dr. Chau-Ti Ting, and the transgenic fly support by Dr. Y. Henry Sun at Academia Sinica.
Footnotes
Author Contributions:
C-T Wang, Y-C Chen, Y-Y Wang, M-H Huang, T-L Yen, H Li, C-J Liang, T-K Sang,, and S-C Cho carried out the experiments and analyzed the data; H-D Wang and C-H Yuh designed the experiments, analyzed the data, and together with C-Y Wang and T. J. Brummel discussed the data and wrote the manuscript.
References
- Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Arking R, Buck S, Berrios A, Dwyer S, Baker GT., 3rd Elevated paraquat resistance can be used as a bioassay for longevity in a genetically based long-lived strain of Drosophila. Dev Genet. 1991;12:362–370. doi: 10.1002/dvg.1020120505. [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S. BEHAVIORAL MUTANTS OF Drosophila ISOLATED BY COUNTERCURRENT DISTRIBUTION. Proc Natl Acad Sci U S A. 1967;58:1112–1119. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos JP, Almeida A. The pentose-phosphate pathway in neuronal survival against nitrosative stress. IUBMB Life. 2010;62:14–18. doi: 10.1002/iub.280. [DOI] [PubMed] [Google Scholar]
- Finkel T. Radical medicine: treating ageing to cure disease. Nat Rev Mol Cell Biol. 2005;6:971–976. doi: 10.1038/nrm1763. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Nogales P, Almeida A, Bolanos JP. Peroxynitrite protects neurons against nitric oxide-mediated apoptosis. A key role for glucose-6-phosphate dehydrogenase activity in neuroprotection. J Biol Chem. 2003;278:864–874. doi: 10.1074/jbc.M206835200. [DOI] [PubMed] [Google Scholar]
- Garelick MG, Kennedy BK. TOR on the brain. Exp Gerontol. 2010 doi: 10.1016/j.exger.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano E, Rendina R, Peluso I, Furia M. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics. 2002;160:637–648. doi: 10.1093/genetics/160.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Mendez A, Almeida A, Fernandez E, Maestre C, Moncada S, Bolanos JP. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol. 2009;11:747–752. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P, Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, Kim AK, You KH, Tatar M, Yu K. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol. 2008;10:468–475. doi: 10.1038/ncb1710. [DOI] [PubMed] [Google Scholar]
- Legan SK, Rebrin I, Mockett RJ, Radyuk SN, Klichko VI, Sohal RS, Orr WC. Overexpression of glucose-6-phosphate dehydrogenase extends the life span of Drosophila melanogaster. J Biol Chem. 2008;283:32492–32499. doi: 10.1074/jbc.M805832200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao PC, Lin HY, Yuh CH, Yu LK, Wang HD. The effect of neuronal expression of heat shock proteins 26 and 27 on lifespan, neurodegeneration, and apoptosis in Drosophila. Biochem Biophys Res Commun. 2008;376:637–641. doi: 10.1016/j.bbrc.2008.08.161. [DOI] [PubMed] [Google Scholar]
- Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, Pletcher SD. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- Liu YL, Lu WC, Brummel TJ, Yuh CH, Lin PT, Kao TY, Li FY, Liao PC, Benzer S, Wang HD. Reduced expression of alpha-1,2-mannosidase I extends lifespan in Drosophila melanogaster and Caenorhabditis elegans. Aging Cell. 2009;8:370–379. doi: 10.1111/j.1474-9726.2009.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Finkel T. Free radicals and senescence. Exp Cell Res. 2008;314:1918–1922. doi: 10.1016/j.yexcr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckinbill LS, Riha V, Rhine S, Grudzien TA. The role of glucose-6-phosphate dehydrogenase in the evolution of longevity in Drosophila melanogaster. Heredity. 1990;65(Pt 1):29–38. doi: 10.1038/hdy.1990.66. [DOI] [PubMed] [Google Scholar]
- Masella R, Di Benedetto R, Vari R, Filesi C, Giovannini C. Novel mechanisms of natural antioxidant compounds in biological systems: involvement of glutathione and glutathione-related enzymes. J Nutr Biochem. 2005;16:577–586. doi: 10.1016/j.jnutbio.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Mejias R, Villadiego J, Pintado CO, Vime PJ, Gao L, Toledo-Aral JJ, Echevarria M, Lopez-Barneo J. Neuroprotection by transgenic expression of glucose-6-phosphate dehydrogenase in dopaminergic nigrostriatal neurons of mice. J Neurosci. 2006;26:4500–4508. doi: 10.1523/JNEUROSCI.0122-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA. Cell stress and aging: new emphasis on multiplex resistance mechanisms. J Gerontol A Biol Sci Med Sci. 2009;64:179–182. doi: 10.1093/gerona/gln072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- Preville X, Salvemini F, Giraud S, Chaufour S, Paul C, Stepien G, Ursini MV, Arrigo AP. Mammalian small stress proteins protect against oxidative stress through their ability to increase glucose-6-phosphate dehydrogenase activity and by maintaining optimal cellular detoxifying machinery. Exp Cell Res. 1999;247:61–78. doi: 10.1006/excr.1998.4347. [DOI] [PubMed] [Google Scholar]
- Qian Y, Banerjee S, Grossman CE, Amidon W, Nagy G, Barcza M, Niland B, Karp DR, Middleton FA, Banki K, Perl A. Transaldolase deficiency influences the pentose phosphate pathway, mitochondrial homoeostasis and apoptosis signal processing. Biochem J. 2008;415:123–134. doi: 10.1042/BJ20080722. [DOI] [PubMed] [Google Scholar]
- Sang TK, Li C, Liu W, Rodriguez A, Abrams JM, Zipursky SL, Jackson GR. Inactivation of Drosophila Apaf-1 related killer suppresses formation of polyglutamine aggregates and blocks polyglutamine pathogenesis. Hum Mol Genet. 2005;14:357–372. doi: 10.1093/hmg/ddi032. [DOI] [PubMed] [Google Scholar]
- Sang TK, Ready DF. Eyes closed, a Drosophila p47 homolog, is essential for photoreceptor morphogenesis. Development. 2002;129:143–154. doi: 10.1242/dev.129.1.143. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Sun J, Tower J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol Cell Biol. 1999;19:216–228. doi: 10.1128/mcb.19.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Tatar M, Khazaeli AA, Curtsinger JW. Chaperoning extended life. Nature. 1997;390:30. doi: 10.1038/36237. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Taye AA, Campbell C, Kazemi-Esfarjani P, Fischbeck KH, Min KT. Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Dev. 2003;17:1463–1468. doi: 10.1101/gad.1087503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushina E, McMurray CT. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 2007;145:1233–1248. doi: 10.1016/j.neuroscience.2006.10.056. [DOI] [PubMed] [Google Scholar]
- Wamelink MM, Struys EA, Jakobs C. The biochemistry, metabolism and inherited defects of the pentose phosphate pathway: a review. J Inherit Metab Dis. 2008;31:703–717. doi: 10.1007/s10545-008-1015-6. [DOI] [PubMed] [Google Scholar]
- Wang HD, Kazemi-Esfarjani P, Benzer S. Multiple-stress analysis for isolation of Drosophila longevity genes. Proc Natl Acad Sci U S A. 2004;101:12610–12615. doi: 10.1073/pnas.0404648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev Cell. 2003;5:811–816. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Paulson HL, Gray-Board GL, Bui QT, Fischbeck KH, Pittman RN, Bonini NM. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- Wolkow CA. Life span: getting the signal from the nervous system. Trends Neurosci. 2002;25:212–216. doi: 10.1016/s0166-2236(02)02133-1. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Wyttenbach A, Sauvageot O, Carmichael J, Diaz-Latoud C, Arrigo AP, Rubinsztein DC. Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum Mol Genet. 2002;11:1137–1151. doi: 10.1093/hmg/11.9.1137. [DOI] [PubMed] [Google Scholar]
- Zhang M, Poplawski M, Yen K, Cheng H, Bloss E, Zhu X, Patel H, Mobbs CV. Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS Biol. 2009;7:e1000245. doi: 10.1371/journal.pbio.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.