Abstract
Angelman syndrome is a severe neurodevelopmental disorder caused by deletion or mutation of the maternal allele of the ubiquitin protein ligase E3A (Ube3a)1–3. In neurons, the paternal allele of Ube3a is intact but epigenetically silenced4–6, raising the possibility that Angelman syndrome could be treated by activating this silenced allele to restore functional UBE3A protein7,8. Using an unbiased, high-content screen in primary cortical neurons from mice, we identified twelve topoisomerase I inhibitors and four topoisomerase II inhibitors that unsilence the paternal Ube3a allele. These drugs included topotecan, irinotecan, etoposide, and dexrazoxane (ICRF-187). At nanomolar concentrations, topotecan upregulated catalytically active UBE3A in neurons from maternal Ube3a-null mice. Topotecan concomitantly downregulated expression of the Ube3a antisense transcript that overlaps the paternal copy of Ube3a9–11. These results suggest that topotecan unsilences Ube3a in cis by reducing transcription of an imprinted antisense RNA. When administered in vivo, topotecan unsilenced the paternal Ube3a allele in several regions of the nervous system, including neurons in the hippocampus, neocortex, striatum, cerebellum and spinal cord. Paternal expression of Ube3a remained elevated in a subset of spinal cord neurons for at least twelve weeks after cessation of topotecan treatment, suggesting transient topoisomerase inhibition can have enduring effects on gene expression. While potential off-target effects remain to be investigated, our findings suggest a therapeutic strategy for reactivating the functional but dormant allele of Ube3a in patients with Angelman syndrome.
No effective therapies exist for Angelman syndrome (AS)—an imprinting disorder caused by mutations or deletions in the maternal allele of Ube3a1–3. Ube3a is biallelically expressed in most tissues of the body; however, in rodents and humans, most neurons express Ube3a only from the maternally-inherited allele4,12–14. This unique epigenetic pattern of regulation suggested that it might be possible to unsilence the dormant paternal Ube3a allele in neurons7,8.
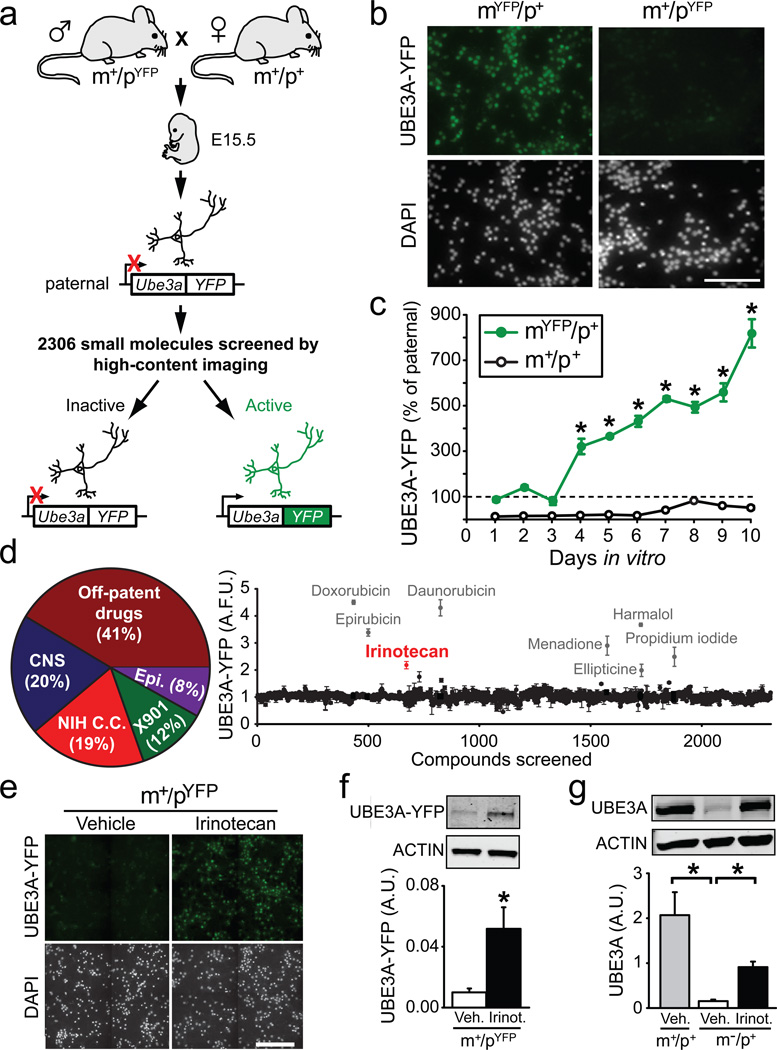
To test this possibility, we developed a 384-well high-content screen using primary mouse cortical neurons from Ube3a-Yellow Fluorescent Protein (Ube3a-YFP) knockin mice15, and searched for drug-like molecules that could unsilence the paternal Ube3a-YFP allele (Fig. 1a). This screen was based on our observation that the imprinting of Ube3a-YFP was maintained in vitro in cultured embryonic cortical neurons. Notably, Ube3a-YFP expression was undetectable (silenced) in cultured neurons when paternally inherited (Ube3am+/pYFP), but was expressed when maternally inherited (Ube3amYFP/p+) (Fig. 1b), with expression increasing from 4 to 10 days in vitro (DIV) (Fig. 1c). This significant difference between maternal and paternal UBE3A-YFP protein levels provided a large screening window and a Z’-factor score of 0.58 (determined by statistically comparing antibody-enhanced fluorescence intensities and variations between maternal and paternal UBE3A-YFP signals at DIV10), making our high-content platform suitable for unbiased screening.
Figure 1. A small-molecule screen identifies a topoisomerase inhibitor that unsilences the paternal allele of Ube3a in neurons.
a, High-content screen flowchart. E15.5 cortical neurons with a paternally inherited Ube3a-YFP allele were cultured in 384-well plates and treated with small molecules from DIV7–DIV10. Active compounds that unsilence the paternal Ube3a-YFP allele were detected with antibody-enhanced fluorescence and high-content imaging. b, High-content imaging of DIV7 neurons that inherited Ube3a-YFP maternally (mYFP/p+) or paternally (m+/pYFP). Nuclei were stained with DAPI. Scale bar = 50 µm. c, Mean ± s.e.m. levels of UBE3A-YFP fluorescence in neurons cultured from maternal Ube3a-YFP (mYFP/p+) or wild-type (m+/p+) mice, normalized to levels in age-matched neurons cultured from paternal Ube3a-YFP mice (m+/pYFP). Two-way ANOVA revealed main effects of genotype, duration, and a genotype-duration interaction (P<0.001); Bonferroni post hoc test examined comparisons between maternal and paternal Ube3a-YFP mice from DIV4 to DIV10, *P<0.001; n=2–6 culture wells/day. d, Pie chart depicting categories of the 2,306 screened compounds and graph summarizing presumptive UBE3A-YFP in arbitrary fluorescence units (A.F.U.) after small molecule treatments. Small molecules that were subsequently found to be autofluorescent (Supplementary Fig. 2) are depicted in gray. The initial screen identified one active compound, irinotecan (red). e, High magnification view of wells treated with vehicle (0.2% DMSO) or 10 µM irinotecan for 72 hr. Neuron density and health is similar in vehicle- and irinotecan-treated cells as evidenced by counterstaining with the nuclear marker DAPI. Scale bar = 100 µm. f,g Western blots of cultured cortical neurons probed with GFP or UBE3A antibodies, respectively, ± irinotecan (10 µM for 72h). f, *P<0.05; two-tailed t-test, n=3/group. g, *P<0.001, one-way ANOVA with Bonferroni post hoc, n=7–8/group. All data are presented as means ± s.e.m.
To perform the screen, we cultured Ube3am+/pYFP neurons for 7 days and then treated these neurons with compounds (10 µM for 72 hours) from multiple small molecule libraries (Fig. 1d, Supplementary Fig. 1). In total, we screened 2,306 small molecules in quadruplicate, normalizing values to vehicle-treatment (0.2% DMSO) (for full methods see Supplementary Methods; for list of all compounds tested see Supplementary Table 1). While methylation and other epigenetic marks are thought to control imprinting of Ube3a9,16–18, to our surprise, none of the commonly used compounds that target the epigenome, including chromatin remodeling drugs and DNA methyltransferase inhibitors, unsilenced the paternal Ube3a-YFP allele. A number of compounds were identified as false positives (gray compounds in Fig. 1d) due to their intrinsic fluorescence (Supplementary Fig. 2). Our initial screen identified one compound—irinotecan, an FDA-approved camptothecin-based topoisomerase type I inhibitor. Irinotecan lacked intrinsic fluorescence and upregulated UBE3A-YFP fluorescence (Fig. 1d,e and Supplementary Fig. 3). Irinotecan (10 µM) also upregulated paternal UBE3A-YFP protein (Fig. 1f) and endogenous UBE3A protein (Fig. 1g) in neuronal cultures from Ube3am+/pYFP and Ube3am–/p+ mice (AS model mice13), respectively.
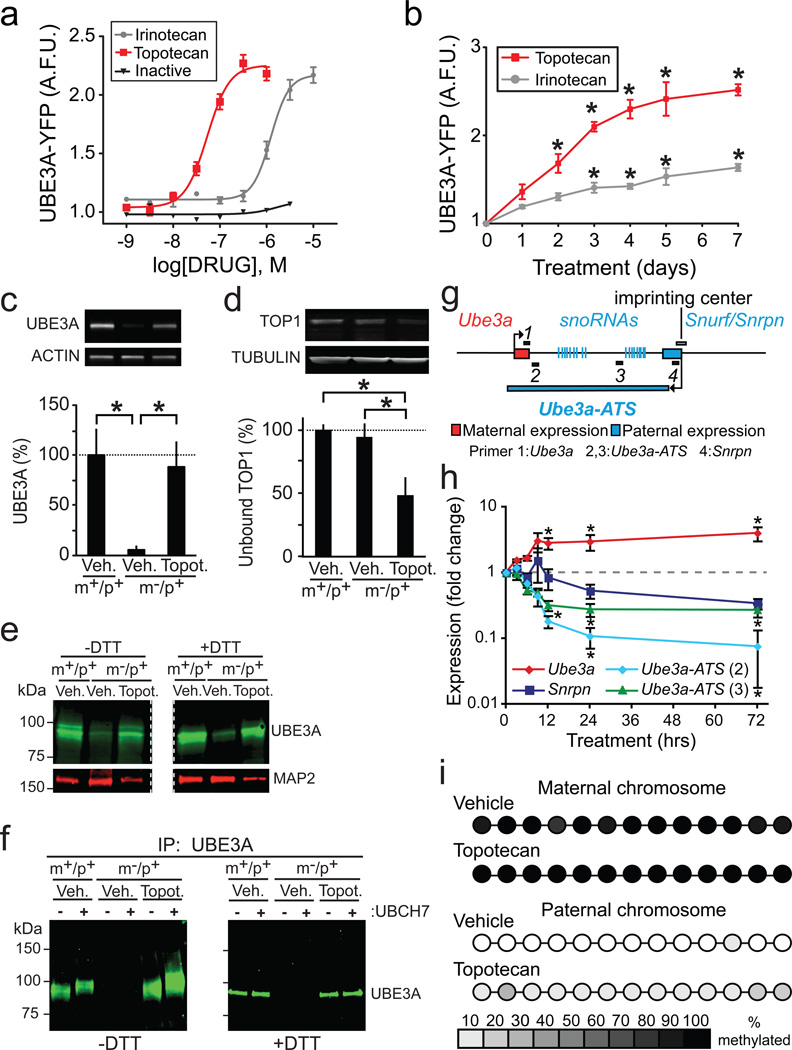
Many topoisomerase I inhibitors, including irinotecan and the related FDA-approved drug topotecan, are derived from the natural product camptothecin (CPT)19. To explore structure activity relationships, we tested CPT analogs and other topoisomerase inhibitors (Fig. 2a; Supplementary Figs. 4–10), all of which lack inherent fluorescence (Supplementary Fig. 3). We found that irinotecan and topotecan upregulated paternal UBE3A-YFP in a dose-and time-dependent manner in cultured neurons, with topotecan being 20× more potent than irinotecan (Fig. 2a,b; Supplementary Fig. 11). In contrast, an inactive analog of CPT (lactam E-ring-CPT) that does not inhibit topoisomerases20 failed to unsilence the paternal Ube3a-YFP allele (Fig. 2a; Supplementary Fig. 4). Ten additional topoisomerase I inhibitors unsilenced Ube3a-YFP in a dose-dependent manner, including CPT analogs and structurally distinct indenoisoquinolines (Table 1 and Supplementary Figs. 4–7). Furthermore, four structurally distinct topoisomerase II inhibitors (etoposide, dexrazoxane, ICRF-193, and amsacrine) also unsilenced the paternal Ube3a-YFP allele (Table 1 and Supplementary Figs. 8–10). Thus, our data with 16 topoisomerase inhibitors and one inactive analog strongly suggest that inhibition of topoisomerase I or II can unsilence the paternal Ube3a allele.
Figure 2. Topotecan unsilences the paternal allele of Ube3a and the unsilenced protein is catalytically active.
a, Dose-response curves for unsilencing paternal Ube3a-YFP. Inactive = lactam E ring-camptothecin. n=4/data point. b, UBE3A-YFP levels in neurons from Ube3am+/pYFP mice increase with duration of topotecan (300 nM) or irinotecan (1 µM) treatment. *P<0.05, oneway ANOVA with Bonferroni post hoc tests relative to day zero, n=4–8/group. A.F.U.=arbitrary fluorescence units. c, Western blots and quantification of UBE3A and the loading control actin. *P<0.05, one-way ANOVA with Bonferroni post hoc tests, n=4/group. d, Quantification of unbound TOP1 and representative western blots. β-tubulin was used as a loading control. One-way ANOVA with Bonferroni post hoc tests, *P<0.05; n=3/group. e, Western blot from vehicle- and topotecan-treated neurons from wild-type (m+/p+) and maternal Ube3a-deficient (m–/p+) mice. f, Western blots examining UBE3A ubiquitin-thioester formation following immunoprecipitation with an anti-UBE3A antibody and in vitro ubiquitination in the presence or absence of the ubiquitin conjugating enzyme (E2), UBCH7. All data are presented as means ± s.e.m. g, Schematic demonstrating location of 4 primer sets used to probe mRNA expression shown in h. h, Normalized mRNA levels in cultured Ube3am–/p+ neurons following vehicle or 300 nM topotecan. Expression is given as a ratio of expression in drug treated cells to vehicle treated cells, normalized to the housekeeping gene RPL22. *P<0.05 compared to 0 hr, Kruskal-Wallis one-way ANOVA followed by post hoc tests, n=4–5 cultures/data point. i, Schematic summarizing methylation status of the Snrpn promoter region on the maternal and paternal chromosome following treatment with vehicle or 300 nM topotecan (see complete primer 1 data set in Supplementary Fig.12). Average methylation status is indicated using a grayscale.
Table 1.
Efficacies and potencies of topoisomerase inhibitors for unsilencing the paternal allele of Ube3a-YFP in cultured neurons.
| Compound | Potency EC50 (nM) |
Efficacy Emax (fold over vehicle) |
|---|---|---|
| Camptothecin derivatives | ||
| 7-Ethyl-Camptothecin (7-Ethyl-C PT) | 7.2 ± 2.3 | 1.70 ± 0.04 |
| 7-Ethyl-10-H ydroxy-CPT | 11 ± 3.2 | 1.99 ± 0.06 |
| 10-H ydroxy-CPT | 14 ± 5.7 | 1.82 ± 0.08 |
| Belotecan (CKD 602) | 19 ± 4.4 | 1.88 ± 0.05 |
| Camptothecin (CPT) | 21 ± 3.8 | 2.11 ± 0.05 |
| Topotecan* | 54 ± 3.4 | 2.25 ± 0.05 |
| Rubitecan (9-Nitro-CPT) | 62 ± 18 | 2.09 ± 0.09 |
| Irinotecan* | 994 ± 13 | 2.17 ± 0.05 |
| Silatecan (DB67) | 2,244 ± 171 | 1.65 ± 0.05 |
| Lactam E ring-CPT (inactive) | inactive | inactive |
| Indenoisoquinoline derivatives | ||
| NSC725776 | 10 ± 1.6 | 1.76 ± 0.03 |
| NSC706744 | 11 ± 3.2 | 1.84 ± 0.07 |
| NSC724998 | 14 ± 2.2 | 1.69 ± 0.03 |
| Podophyllotoxin derivative | ||
| Etoposide* | 1,600 ± 980 | 1.68 ± 0.04 |
| Bis-dioxopiperazine derivatives | ||
| ICRF-193 | 205 ± 70 | 2.21 ± 0.09 |
| Dexrazoxane (ICRF-187)* | 2,0470 ± 1,450 | 1.82 ± 0.05 |
| Aminoacridine derivative | ||
| Amsacrine | 27 ± 5.2 | 1.74 ± 0.06 |
FDA-approved compounds
We focused our remaining studies on the topoisomerase I inhibitor topotecan because it is approved for use in humans, it unsilenced Ube3a in the low nanomolar range, and topotecan (300 nM, 72 h) restored UBE3A protein to wild-type levels in cultured neurons from Ube3am–/p+ mice (Fig. 2c).
Many topoisomerase inhibitors, including topotecan, covalently link topoisomerases to DNA, forming stable DNA-enzyme complexes that are separable from free topoisomerase enzymes19. Since topotecan inhibits topoisomerase I (TOP1) and Top1 is expressed at high levels in the developing and adult brain19,21, we focused our subsequent analysis on this enzyme. We found that topotecan (300 nM, 72 h) significantly reduced the amount of free TOP1 (Fig. 2d) in cultured neurons, indicating that topotecan engages its known molecular target at doses that unsilence the paternal Ube3a allele.
UBE3A is a HECT (homology to E6 carboxyl terminus) domain E3 ligase that forms a thioester-ubiquitin intermediate in the presence of E1 and E2 enzymes22. This thioester-ubiquitin intermediate is required for HECT domain E3 ligases to mono- and polyubiquitinate their substrates23. Interestingly, we noticed that topotecan (300 nM, 72 hr) upregulated UBE3A protein in Ube3am–/p+ cultures along with a higher molecular weight form (resolved after running gels for longer times; Fig. 2e). This high molecular weight band was also seen in wild-type (Ube3am+/p+) cultures and was lost upon addition of the reducing agent dithiothreitol (DTT) (Fig. 2e). These data suggest that the unsilenced paternal copy of UBE3A is catalytically active and competent to form a thioester-ubiquitin intermediate, just like wild-type, maternal-derived UBE3A23.
To further demonstrate that UBE3A was catalytically active, we immunoprecipitated UBE3A from cultured wild-type and Ube3am–/p+ neurons (+/− topotecan), then tested these samples for a gel-mobility-shift in the presence or absence of the ubiquitin E2 UBCH724. Both wild-type (maternal-derived) and topotecan-unsilenced (paternal-derived) UBE3A underwent mobility shifts in the presence of UBCH7 plus free ubiquitin that were abolished by DTT (Fig. 2f). This observation indicates the mobility shift was due to addition of covalent ubiquitin, and demonstrates that topotecan can upregulate a functional UBE3A enzyme.
Ube3a is repressed in cis by a large antisense transcript (Ube3a-ATS) that overlaps the paternal allele of Ube3a (Fig. 2g)9,10. Ube3a-ATS is expressed exclusively from the paternal allele as a result of allele-specific methylation of an imprinting center that overlaps the Ube3a-ATS and Snurf/Snrpn transcription start site25. We next sought to determine if topotecan regulated Ube3a expression through changes in Ube3a-ATS expression or altered methylation at the imprinting center. We found that topotecan upregulated expression of Ube3a in cultured neurons from Ube3am–/p+ mice while concomitantly downregulating expression of Ube3a-ATS and Snrpn (Fig. 2h). However, topotecan did not alter methylation at the imprinting center (Fig. 2i, Supplementary Fig. 12). Taken together, these data suggest that topotecan unsilences paternal Ube3a by reducing transcription of a regulatory antisense RNA without appreciably affecting genomic methylation at the imprinting center.
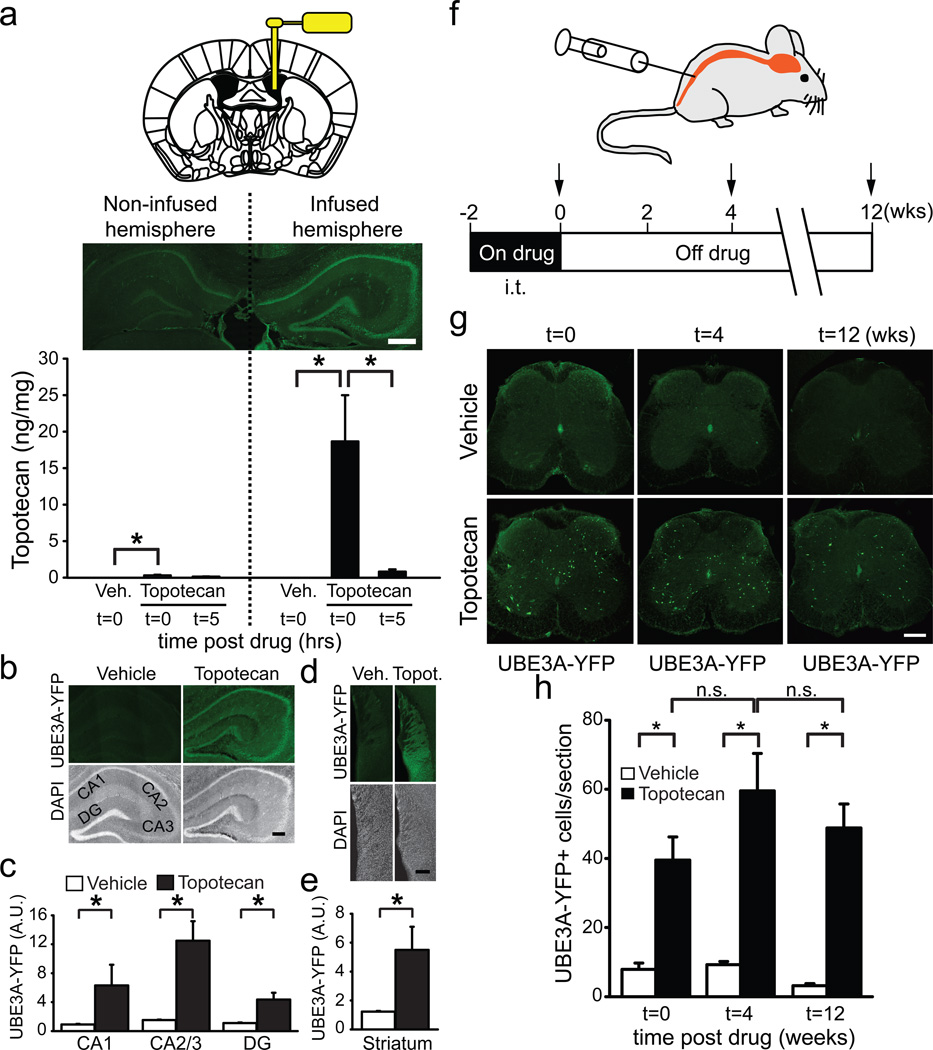
We then sought to determine if topotecan could unsilence the paternal Ube3a allele in vivo. We first identified a dose that was well tolerated, meaning there were no significant decreases in body weight between the beginning and end of the drug treatments (Supplementary Fig. 13). We then administered topotecan (3.74 µg/h) unilaterally into the lateral ventricle of Ube3am+/pYFP or Ube3am-/p+ mice by intracerebroventricular (i.c.v.) infusion for two weeks and then sacrificed the mice either immediately or 5 hr after drug cessation. Strikingly, topotecan unsilenced paternal Ube3a in the hippocampus, striatum, and cerebral cortex of the infused hemisphere, but had only a modest effect on the contralateral (non-infused) hemisphere with no effect in the cerebellum (Fig. 3a–e, Supplementary Figs. 14–15). Pharmacokinetic analyses demonstrated that a significant amount of topotecan was detectable in the infused hemisphere immediately following treatment whereas low levels were present in the contralateral (non-infused) hemisphere and in cerebellum (Fig. 3a, Supplementary Fig. 14). However, a higher dose of topotecan (21.6 µg/h for five days) did unsilence the paternal allele of Ube3a in Purkinje neurons of the cerebellum (Supplementary Fig. 16). No significant difference in topotecan levels was detected in blood between drug- and vehicle-treated mice (data not shown). Topotecan concentrations significantly declined five hours after cessation of i.c.v. drug delivery (Fig. 3a), indicating that topotecan does not persist and is rapidly removed/metabolized in the brain. Taken together, these pharmacokinetic and pharmacodynamic data suggest that the degree to which topotecan unsilences the paternal Ube3a allele is directly correlated with drug concentrations in the brain. Moreover, our data indicate topotecan has the potential to unsilence the paternal Ube3a allele throughout the nervous system.
Figure 3. Topotecan enduringly unsilences the paternal allele of Ube3a in vivo.
a, Unilateral delivery of topotecan (i.c.v.) using a mini-osmotic pump into the lateral ventricle of Ube3am+/pYFP mice in vivo. Two weeks of topotecan infusion (3.74 µg/h) unsilenced the paternal Ube3a-YFP allele in the hippocampus of the infused hemisphere near the site of drug delivery, while only modestly unsilencing Ube3a-YFP in the contralateral (non-infused) hemisphere. Scale bar = 500 µm. Pharmacokinetic analyses measuring topotecan levels in the infused and non-infused hemisphere immediately (t=0) or five hours (t=5) after cessation of drug delivery. *P<0.05, one-way ANOVA with Bonferroni post hoc test, n=5–9/group. b, Representative sections and c, quantification of optical intensity of UBE3A-YFP in hippocampal regions (CA1, CA2/3, and dentate gyrus=DG) of the topotecan-infused hemisphere or the hemisphere of vehicle-treated mice. *P<0.05, Mann-Whitney rank sum test, n=5/group. d, Representative sections and e, quantification of paternal UBE3A-YFP in the striatum following i.c.v. infusion of topotecan. *P<0.05, two-tailed t test, n=4/group. f, Schematic depicting schedule for i.t. delivery of topotecan (50 nmol/day for 10 of 14 days) and endpoints (arrows) immediately, 4 weeks, and 12 weeks after cessation of drug. g,h Topotecan (i.t.) increased the number of UBE3A-YFP-positive spinal neurons compared to vehicle, and the unsilencing of Ube3a-YFP was maintained for at least 12 weeks. *P<0.001, one-way ANOVA with Bonferroni post hoc test, n=7–8/group.
Genomic imprinting is thought to be established only during prescribed germline and embryonic periods of development and imprinted genes typically remain epigenetically regulated throughout life26. Thus, we next sought to determine if topotecan had transient or long-lasting effects on paternal Ube3a expression. To test this possibility, we turned to an intrathecal (i.t.) delivery protocol (Fig. 3f) because topotecan (Fig. 3g) and irinotecan (not shown) unsilenced paternal Ube3a in a sparse population of lumbar spinal neurons, allowing us to quantify all UBE3A-YFP-positive neurons. Moreover, i.t. delivery has been used to deliver topotecan to the brain in humans27. We found that topotecan (50 nmol/5 µL i.t. once daily, for 10 of 14 days) was well tolerated (Supplementary Fig. 13) and significantly increased the number of paternal UBE3A-YFP-positive cells in the lumbar spinal cord of mice (Fig. 3g,h, Supplementary Fig. 17a). The vast majority (>93%) of these UBE3A-YFP-positive cells were NeuN+ neurons (Supplementary Fig. 17a,b), indicating topotecan unsilences Ube3a primarily in neurons in vivo. Moreover, the unsilenced paternal UBE3A-YFP protein was expressed at levels comparable to maternal UBE3A-YFP controls (Supplementary Fig. 17c). Remarkably, the number of UBE3A-YFP-positive spinal cord neurons remained elevated 12 weeks following cessation of drug treatment (Fig. 3g,h), much longer than the elimination of topotecan from tissue (Fig. 3a). These results indicate that topotecan can enduringly unsilence paternal Ube3a in a subset of spinal neurons and suggest that a single course of drug treatment has the capacity to permanently modify expression of Ube3a.
In conclusion, we found that topoisomerase inhibitors can unsilence the paternal allele of Ube3a and the paternally-derived protein is functional. These findings suggest that topoisomerase inhibitors have the potential to rescue molecular, cellular, and behavioral deficits associated with loss of UBE3A7,13,28. Ube3a expression is modestly upregulated in the brain of Top2b knockout mice29, providing genetic support that topoisomerases regulate Ube3a expression. We also found that topotecan reduced expression of paternal Snrpn and Ipw, a genomic region whose deletion is associated with Prader-Willi syndrome7,17. To what extent topotecan regulates expression of other genes in the brain, including Igf2r, Kcnq1, and Gnas imprinted gene clusters, and whether long-term treatments with topoisomerase inhibitors produce a Prader-Willi-like condition are unknown. However, topotecan and irinotecan are approved for use in patients with cancer, penetrate into the central nervous system, and are well-tolerated when administered chronically to both adult and pediatric patients27,30. Importantly, patients treated with topoisomerase inhibitors do not display symptoms associated with Prader-Willi syndrome. Ultimately, our study indicates that topoisomerase inhibitors regulate gene expression through a transcriptional mechanism and could be used to treat symptoms associated with AS.
Methods Summary
All animal procedures were approved by the University of North Carolina at Chapel Hill Animal Care and Use Committee. We used male and female Ube3a-YFP knockin mice15, Ube3am–/p+ mice13, and their age-matched, wild-type controls. High-content screening was performed on a BD Pathway 855 system. UBE3A-YFP was detected for drug screening with an anti-GFP antibody (Novus Biologicals, NB600-308; 1:1000) because intrinsic YFP fluorescence levels were low in cultured neurons and tissue sections. All data are presented as mean ± s.e.m., with sample sizes (n) shown in figures or stated in text. Statistical analyses were performed using SigmaPlot 11 (Systat Software). Normality tests (Shapiro-Wilk) and equal variance tests were run and passed (P>0.05) before parametric statistical analyses were run.
Full methods accompany this paper as supplementary material.
Supplementary Material
Acknowledgements
We thank Drs. Art Beaudet and Yong-hui Jiang for providing Ube3a-YFP and Ube3am–/p+ mice; Thorfinn Riday and Ji Eun Han for assistance in i.c.v. mini-osmotic pump infusion; Andrew Burns for assistance in maintaining mouse colonies; Dr. Vladimir Gukassyan for help with the Surveyor and confocal imaging systems; Kirk McNaughton for help with tissue sectioning; Dr. William Zamboni for providing belotecan, rubitecan, and silatecan; and William Janzen and the Center of Integrative Chemical Biology and Drug Discovery for providing the epigenetic library. B.D.P., M.J.Z., and B.L.R. were supported by the Simons Foundation Autism Research Initiative (SFARI) and by the Angelman Syndrome Foundation. B.D.P. and M.J.Z. were supported by the NIMH. B.D.P. was support by the National Eye Institute (R01EY018323) and NC TraCS (50KR41016). M.J.Z. was supported by the National Institute of Neurological Disorders and Stroke (NINDS) (R01NS060725, R01NS067688). B.L.R. was supported by NIH HHSN-271-2008-00025-C, the NIMH Psychoactive Drug Screening Program, the Michael Hooker Distinguished Chair of Pharmacology, and grants from NIMH and NIDA. H-S.H. was supported by a NARSAD Young Investigator Award and NC TraCS (10KR20910). J.A.A. was supported by NIH T32HD040127-07, the UNC-Carolina Institute for Developmental Disabilities, and an Autism Concept Award AR093464 from the U.S. Department of Defense. A.M.M. was supported by a National Research Service Award from NINDS (5F32NS067712). I.F.K. was supported by a Joseph E. Wagstaff Postdoctoral Research Fellowship from the Angelman Syndrome Foundation. Assay development costs were partially supported by NINDS (5P30NS045892). Confocal and montage imaging was performed at the University of North Carolina at Chapel Hill Confocal and Multiphoton Imaging Facility, which is co-funded by grants from NINDS (5P30NS045892) and NICHD (P30HD03110).
Footnotes
Author Contributions
H-S.H., J.A.A., A.M.M., I.F.K., M.J.Z., B.L.R. and B.D.P. conceived and designed experiments, and wrote the manuscript. All authors reviewed and edited the manuscript. H-S.H., J.A.A., J.M., and H-M.L. performed drug screens and validations. H-S.H., B.T-B., J.M. and J.W.D. performed in vivo studies and immunofluorescence. H-S.H. and A.S.B. performed pharmacokinetic analyses. A.M.M. performed tests of UBE3A functionality. I.F.K. performed qRT-PCR and methylation analyses. N.S. oversaw high content screening instrumentation and implemented image processing algorithms. X.C. and J.J. synthesized the lactam E ring inactive camptothecin analog and the three indenoisoquinoline derivatives.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nature Genetics. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 2.Matsuura T, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nature Genetics. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 3.Sutcliffe JS, et al. The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome Research. 1997;7:368–377. doi: 10.1101/gr.7.4.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albrecht U, et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nature Genetics. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- 5.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nature Genetics. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 6.Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nature Genetics. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- 7.Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends in Neurosciences. 2011 doi: 10.1016/j.tins.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters SU, et al. Double-blind therapeutic trial in Angelman syndrome using betaine and folic acid. American Journal of Medical Genetics. 2010;152A:1994–2001. doi: 10.1002/ajmg.a.33509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- 10.Landers M, et al. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32:3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Numata K, Kohama C, Abe K, Kiyosawa H. Highly parallel SNP genotyping reveals high-resolution landscape of mono-allelic Ube3a expression associated with locus-wide antisense transcription. Nucleic Acids Res. 2011;39:2649–2657. doi: 10.1093/nar/gkq1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatani J, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang YH, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 14.Miura K, et al. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiology of Disease. 2002;9:149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- 15.Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum. Mol. Genet. 2008;17:111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- 16.Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nature Genetics. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 17.Runte M, et al. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe Y, et al. Genome-wide analysis of expression modes and DNA methylation status at sense-antisense transcript loci in mouse. Genomics. 2010;96:333–341. doi: 10.1016/j.ygeno.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nature Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 20.Hertzberg RP, et al. Modification of the hydroxy lactone ring of camptothecin: inhibition of mammalian topoisomerase I and biological activity. J. Med. Chem. 1989;32:715–720. doi: 10.1021/jm00123a038. [DOI] [PubMed] [Google Scholar]
- 21.Plaschkes I, Silverman FW, Priel E. DNA topoisomerase I in the mouse central nervous system: Age and sex dependence. J. Comp. Neurol. 2005;493:357–369. doi: 10.1002/cne.20793. [DOI] [PubMed] [Google Scholar]
- 22.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 23.Beaudenon S, Dastur A, Huibregtse JM. Expression and assay of HECT domain ligases. Methods Enzymol. 2005;398:112–125. doi: 10.1016/S0076-6879(05)98011-7. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Kao WH, Howley PM. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. Journal of Biological Chemistry. 1997;272:13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- 25.Bressler J, et al. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat Genet. 2001;28:232–240. doi: 10.1038/90067. [DOI] [PubMed] [Google Scholar]
- 26.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 27.Gammon DC, et al. Intrathecal topotecan in adult patients with neoplastic meningitis. Am J Health Syst Pharm. 2006;63:2083–2086. doi: 10.2146/ajhp060165. [DOI] [PubMed] [Google Scholar]
- 28.Lyu YL, et al. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol. Cell. Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greer PL, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bomgaars L, Berg SL, Blaney SM. The development of camptothecin analogs in childhood cancers. Oncologist. 2001;6:506–516. doi: 10.1634/theoncologist.6-6-506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.