Abstract
Using a screen for Wnt/β-catenin inhibitors, a family of 8-hydroxyquinolone derivatives with in vivo anti-cancer properties was identified. Analysis of microarray data for the lead compound N-((8-hydroxy-7-quinolinyl) (4-methylphenyl)methyl)benzamide (HQBA) using the Connectivity Map database suggested that it is an iron chelator that mimics the hypoxic response. HQBA chelates Fe2+ with a dissociation constant of ∼10−19 , with much weaker binding to Fe3+ and other transition metals. HQBA inhibited proliferation of multiple cell lines in culture, and blocked the progression of established spontaneous cancers in two distinct genetically engineered mouse models of mammary cancer, MMTV-Wnt1 and MMTV-PyMT mice, without overt toxicity. HQBA may inhibit an iron-dependent factor that regulates cell-type-specific β-catenin-driven transcription. It inhibits cancer cell proliferation independently of its effect on β-catenin signaling, as it works equally well in MMTV-PyMT tumors and diverse β-catenin-independent cell lines. HQBA is a promising specific intracellular Fe2+ chelator with activity against spontaneous mouse mammary cancers.
Keywords: Wnt/β-catenin, iron, iron chelator, drug therapy, mouse models of cancer
Introduction
Wnt signaling has diverse roles in development and during adult tissue homeostasis and repair. Dysregulated Wnt signaling is commonly found in cancer, often resulting in stabilization of β-catenin. Increased β-catenin protein abundance, followed by nuclear localization, leads to transcriptional regulation by interaction with members of the Lef/T-cell factor family and activation of gene expression. The ability of β-catenin to activate specific genes is determined by a number of cell- and tissue-specific factors, including co-expression of interacting transcriptional regulators, the epigenetic programming of the cells and the context of the T-cell-factor-response elements (Mosimann et al., 2009; Teo and Kahn, 2010). Because of this diversity of β-catenin-regulated genes, activation of Wnt/β-catenin signaling can stimulate proliferation or it can favor terminal differentiation (Henderson et al., 2010; Lucero et al., 2010). Although disruption of the signaling cascade leading to β-catenin stabilization is being pursued as a therapeutic option in cancer, in some settings, it may be useful to specifically inhibit subsets of Wnt/β-catenin signaling (Teo and Kahn, 2010).
Cell-based small molecule screens for the discovery of new compounds and novel signaling components offer the advantage of providing diverse and potentially novel targets for small molecules, with the disadvantage being the challenge of identifying those targets. The Wnt/β-catenin signaling pathway is an attractive system for drug development, as Wnt signaling participates in multiple processes (Coombs et al., 2008). Aberrant β-catenin signaling occurs in many cancers. The most prominent example is colon cancer, in which mutations in the adenomatous polyposis coli (APC) gene contribute to stabilization of β-catenin. However, targeting β-catenin alone may not be sufficient, as there is increasing evidence that β-catenin is not the critical target of APC mutations in colon cancers (Phelps et al., 2009; Rai et al., 2010). It might be useful, therefore, to find compounds that inhibit subsets of β-catenin targets and have additional antiproliferative effects that are independent of the Wnt/β-catenin pathway. Frequently, biochemical studies of small molecules can reveal effects reaching well beyond the originally targeted pathway. Although in cancer, the key indicator of therapeutic value for any small molecule is whether it can reverse tumors in relevant animal models and humans, a thorough understanding of its molecular, cellular and in vivo effects is invaluable to the design of preclinical and clinical studies.
In a cell-based screen for small molecule inhibitors of Wnt/β-catenin signaling, we identified a series of compounds that inhibited Wnt/β-catenin signaling with nanomolar inhibitory constant 50 values and acted downstream of β-catenin. The activities of one member of this family, which we term N-((8-hydroxy-7-quinolinyl) (4-methylphenyl)methyl)benzamide (HQBA), were investigated in depth. HQBA is a potent inhibitor of β-catenin signaling in a subset of cancer cell lines, all of which are derived from colon cancers. HQBA also inhibits cell proliferation in most cell lines. Most importantly, HQBA is able to reverse the growth of established cancers in two genetically engineered mouse models of mammary cancer, MMTV-Wnt1 and MMTV-PyMT mice. HQBA perturbs diverse signaling pathways, suggesting it functions by alterations in basic cellular physiology. Gene expression analysis suggested, and subsequent biophysical analysis confirmed that HQBA is a very high (Ka∼1020 −1) affinity ferrous iron chelator. Further experiments in cells and in embryos indicate that its cytotoxic effects and modulation of Wnt and survival signaling pathways can be attributed to its iron chelating ability. Its efficacy without overt toxicity in mouse models of cancer indicates it warrants additional study for use in man.
Results
Identification of a novel Wnt/β-catenin signaling inhibitor
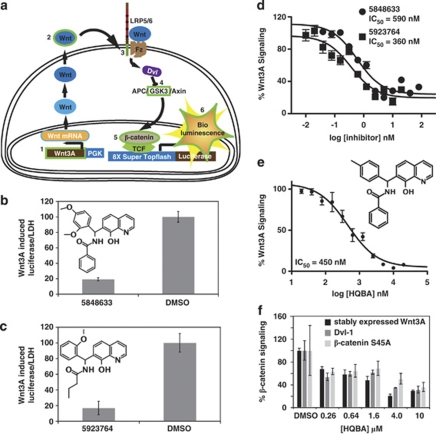
To identify therapeutic targets by which β-catenin-mediated Wnt signaling could be modulated in cancer, we constructed cell-based screens in which luciferase is expressed in response to initiation of signaling at various points in the pathway (Figure 1a; McCulloch et al., 2009a, 2009b; Coombs et al., 2010). In a screen of a 50 000 compound synthetic small molecule library, we identified compounds 5923764 and 5848633 (Figures 1b and c and Supplementary Table 2) that potently inhibited signaling. These compounds are >80% identical (as specified in search of Chembridge (San Diego, CA, USA) Hit2lead database), suggesting that they represent a single pharmacophore. Confirmatory dose ranging studies indicated that both compounds inhibited Wnt/β-catenin signaling with inhibitory constant 50 values between 300 and 600 n (Figure 1d).
Figure 1.
Identification of a novel pharmacophore that rapidly and specifically inhibits Wnt/β-catenin signaling. (a) Overview of the screens used to find Wnt/β-catenin inhibitors and identify their site of action. Reporter activity initiation points are outlined in green. They are (1) stable expression of WNT3A, (2) WNT3A-conditioned media, (3) stable expression of the constitutively active co-receptor mutant LRP6ΔN, (4) overexpression of Dvl-1, (5) stable expression of the non-degraded S45A mutant of β-catenin and (6) constitutive expression of luciferase from the SV40 promoter. (b, c) Chembridge compounds 5848633 and 5923764 at 10 μ inhibited autocrine WNT3A signaling by 81% and 83%, respectively, relative to vehicle-treated control. Bars represent luciferase activity normalized to endogenous lactate dehydrogenase (LDH). (d) Dose curves for each lead inhibitor indicate similar nanomolar inhibitory constant 50 (IC50) for inhibition of autocrine WNT3A signaling. (e) HQBA inhibits autocrine WNT3A-stimulated signaling with nanomolar efficiency. The structure of HQBA is shown in the inset. (f) HQBA acts downstream of stabilized β-catenin. Super Topflash reporter cells stably expressing WNT3A (black) or transfected with 500 ng of Dvl-1 plasmid (dark gray) or 50 ng of β-catenin S45A plasmid (light gray) were treated for 18 h with 260 n to 10 μ HQBA. The % β-catenin signaling is the ratio of luciferase activity over endogenous LDH activity normalized to % of vehicle controls.
To better define relevant components of the pharmacophore, we obtained additional compounds that were at least 70% structurally related to one of the lead compounds, and measured inhibitory constant 50 values for β-catenin signaling inhibition (for structure activity relationship data, see Supplementary Table 2). One variant, Chembridge compound 5839152 (HQBA) had the best ratio of Wnt inhibition to cytotoxicity after 24 h on HEK293-based reporter cell lines relative to the lead compounds. This compound was selected for scaled up synthesis (1 g) for mechanistic studies. The re-synthesized HQBA showed identical inhibition of Wnt/β-catenin signaling to that of the original library compound (Figure 1e).
HQBA specifically inhibits Wnt signaling downstream of β-catenin stabilization
To better define the site of action of HQBA, we tested doses ranging from 260 n to 10 μ on signaling in the HEK293 cells with an integrated Super Topflash reporter (Super Topflash cell line) initiated by stable expression of WNT3A or transient overexpression of Dvl-1 or S45A β-catenin (Figure 1f). We observed similar dose-dependent inhibition of signaling regardless of initiation point, suggesting that HQBA functions downstream of the stabilization of β-catenin.
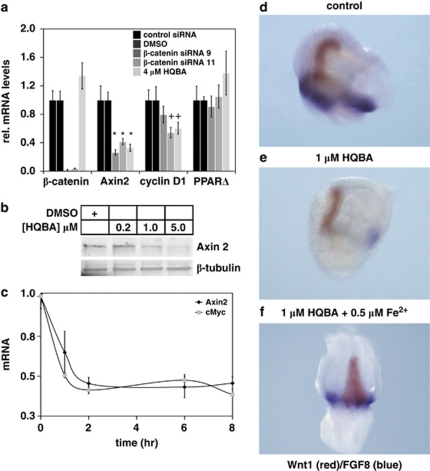
In colon cancers, β-catenin is stabilized due to mutations in the APC protein. We therefore tested whether HQBA functioned in APC-mutant cell lines, SW480 and DLD-1, with APC truncated at codons 1338 and 1416, respectively. SW480 and DLD-1 cells were treated overnight with 4 μ HQBA, and then relative transcript levels of several potential β-catenin-response genes were assessed. We first confirmed in SW480 cells that two distinct small interfering RNAs that knocked down β-catenin by >90% (Figure 2a) caused consistent reduction of axin2 and cyclin D1 gene expression, although they had no effect on peroxisome proliferator-activated receptor (PPAR)Δ expression. Treatment with HQBA paralleled the effect of β-catenin knockdown, with reduction of axin2 and cyclin D1 and no effect on PPARΔ gene expression. HQBA is not a general inhibitor of gene expression, as it had no effect on transcript level of β-catenin and a number of other tested transcripts (data not shown). In fact, HQBA treatment reproducibly leads to small increases in β-catenin mRNA abundance in a number of cell lines. Similar results were observed in DLD-1 cells (data not shown), and the dose-dependent knockdown of axin2 protein by HQBA was confirmed (Figure 2b).
Figure 2.
HQBA rapidly inhibits expression of endogenous Wnt/β-catenin-response genes. (a) Endogenous Wnt/β-catenin target genes axin2 and cyclin D1 are inhibited by both β-catenin knockdown and HQBA. DLD-1 cells were treated with either small interfering RNA (siRNA) against β-catenin siRNA (72 h) or 4 μ HQBA (18 h) as indicated, and then target gene expression was quantitated by quantitative PCR. HQBA phenocopies the effect of β-catenin knockdown. *P<0.002; +P<0.015 for difference from control by two-tailed t-test. (b) The 18-h treatment of DLD-1 colon cancer cells with HQBA decreases levels of endogenous axin2 protein as determined by SDS–polyacrylamide gel electrophoresis and immunoblot. (c) DLD-1 cells were treated for 0–8 h with 4 μ HQBA, and abundance of the Wnt-responsive genes axin2 and c-Myc were measured by quantitative PCR. (d) HQBA suppresses expression of Wnt target gene Fgf8. Control midbrain to rhombomere 2 explants positive for the expression of Wnt1 (red) and Fgf8 (blue). (e) HQBA (1 μ suppresses the expression of Fgf8 (blue). No change in the expression of Wnt1 was observed. (f) Ferrous ethylenediammonium sulfate (0.5 μ was sufficient to rescue the expression of Fgf8 in the presence of 1.0 μ HQBA.
To further investigate the mechanism of Wnt signaling inhibition by HQBA, we assessed the time-course of axin2 and c-Myc mRNA abundance in DLD-1 cells (Figure 2c). Treatment with HQBA reduces mRNA abundance of both genes quickly, with maximal effect within 1 h, suggesting that HQBA directly affects gene transcription or mRNA stability.
HQBA inhibits Wnt-mediated Fgf8 expression in vivo
To examine the specific effects of HQBA on Wnt target genes in vivo, we isolated chick embryo neural tissue that expresses Wnt1 and the Wnt-mediated target gene Fgf8. Canonical Wnt signals are required for the maintenance of expression of Fgf8 at the midbrain hindbrain boundary region in vertebrates (Canning et al., 2007; Coombs et al., 2010). Double in situ hybridization demonstrated the expression of both Wnt1 (red) and Fgf8 (blue) in tissue isolated from the midbrain to anterior hindbrain region (Figure 2d). Treatment of this tissue with HQBA results in the loss of Fgf8 mRNA, with no effect on the expression of endogenous Wnt1 mRNA. Hence, HQBA can block β-catenin target gene expression in cultured cells and in embryos (Figure 2e).
HQBA exhibits potent cytotoxicity in vitro
The initial signaling assays were performed with 12–24 h exposure to HQBA. We noted, however, that with longer incubation times, HQBA was increasingly cytotoxic toward a broad range of cancer cells (Table 1) derived from tumors of various origins including breast, cervical, colorectal, gastric and hepatocellular. The killing mechanism does not appear to require known Wnt/β-catenin pathway mutations, as diverse cell types without known β-catenin stabilization were also sensitive to the compound. Killing was slow in most cell lines. The lethal dose 50 values after 3 days of treatment averaged 20 μ with a range from 17 to 38 μ, with relatively few cell types displaying sub-μ sensitivity. By contrast, in a 7-day cytotoxicity assay, the average lethal dose 50 values dropped to 960 n, with a range of 48 n to 4 μ, with many cell lines showing a 10- to 100-fold increase in sensitivity.
Table 1. LD50s for HQBA toward cancer cell lines treated for 3 or 7 days.
| Cell line |
LD50s for HQBA (n) |
||
|---|---|---|---|
| Tissue of origin | 3-Day assay | 7-Day assay | |
| MCF7 | Breast cancer | Not effective | 420±160 |
| MDA-MB 231 | Breast cancer | 17 400±6900 | 370±130 |
| SK-BR3 | Breast cancer | 27 000±9000 | 97±35 |
| SUM159 | Breast cancer | 27 000±10 000 | 340±120 |
| 4T1 | Breast cancer (mouse) | 17±3 | 48±13 |
| HeLa | Cervical cancer | 420±170 | 570±160 |
| DLD1 | Colorectal cancer | 100±55 | 98±71 |
| HCT116 | Colorectal cancer | 1100±800 | 340±160 |
| RKO | Colorectal cancer | 1200±720 | 390±220 |
| HT29 | Colorectal cancer | 3700±1500 | 1700±500 |
| SW480 | Colorectal cancer | 27 000±14 000 | 340±150 |
| Rat2 | Fibroblast-rat embryo | 340±88 | 300±250 |
| AGS | Gastric cancer | 70±28 | 50±12 |
| HUH7 | Hepatocellular carcinoma | 38 000±60 000 | 470±460 |
| A204 | Rhabdomyosarcoma | Not effective | 140±70 |
| SJSA-1 | Osteosarcoma | Not effective | Not effective |
| TC-71 | Ewing's sarcoma | 49±15 | 140±80 |
| HT1080 | Fibrosarcoma | 37 000±33 000 | ND |
| PA-1 | Teratoma | 190±60 | 50±14 |
| hMEC 1 | hMEC (tert) | 42 000±17 000 | 3 300±1900 |
| hMEC 2 | hMEC (tert RAS ER) | 43 000±29 000 | 4000±1600 |
| hMEC 3 | hMEC (tert RAS ER ΔP53) | 43 000±28 000 | 4000±1600 |
| GMO1915C | Primary fibroblast | Not effective | Not effective |
| MMEC | Primary mouse mammary epithelial cells | 2500±1100 | 3100±1300 |
Abbreviations: ER, endoplasmic reticulum; hMEC, human mammary epithelial cell line; HQBA, N-((8-hydroxy-7-quinolinyl) (4-methylphenyl)methyl)benzamide; LD50s, lethal dose 50 values; ND, assay not done.
Cells were plated in clear 96-well plates at 5000 cells/well, allowed to adhere, and treated for 72 or 168 h with HQBA concentrations ranging from 20 μ to 9.8 n. Data were processed in Prism and errors are derived from 95% confidence intervals.
A number of cell types with known activation of the Wnt/β-catenin pathway (for example, HT29 and SW480, both with APC mutations) were less sensitive to HQBA than lines without known Wnt/β-catenin pathway mutations (for example, RKO, HeLa and TC-71). To determine whether HQBA might not affect Wnt/β-catenin signaling in all cell types, its ability to inhibit the endogenous β-catenin-response genes axin2 and c-Myc was assessed in a set of seven cancer cell lines (Table 2). HQBA inhibited expression of axin2 and c-Myc in some but not all cell types. Notably, HQBA did not inhibit axin2 in HT29 cells with mutant APC, nor did it inhibit axin2 in STF3A cells, in which it was able to inhibit signaling from the SuperTopFlash promoter. These findings suggest that HQBA does not inhibit all β-catenin-dependent transcription, and that it may therefore be acting on an as yet unidentified transcriptional regulator that is not critical in all tissues. We also noted that in some cell lines (for example, SW480 and DLD-1), HQBA increased β-catenin mRNA while still decreasing axin2 abundance. This is consistent with HQBA inhibition of β-catenin signaling downstream of β-catenin protein abundance. The cause of this increase in β-catenin mRNA is not yet understood.
Table 2. Inhibition of endogenous β-catenin-response genes in cancer cell lines.
| Cell line | Origin | Wnt pathway activated by: | Axin2 (% of control) | β-Catenin (% of control) | c-Myc (% of control) |
|---|---|---|---|---|---|
| SW480 | Colon cancer | APC | 24±13 | 134±5 | ND |
| HCT116 | Colon cancer | β-Catenin mutation | 24±9 | ND | 35±13 |
| DLD-1 | Colon cancer | APC | 43±4 | 205±6 | 59±11 |
| HT29 | Colon cancer | APC | 97±9 | 173±9 | 38±11 |
| STF3A | Neuroendocrine (HEK293) | High WNT3A | 122±10 | 173±6 | ND |
| MDA-MB-231 | Breast cancer | None | 103±16 | 74±12 | ND |
| HT1080 | Fibrosarcoma | None | 142±8 | ND | 150±3 |
Abbreviations: APC, adenomatous polyposis coli; ND, assay not done.
Cells were treated overnight with 10 μ N-((8-hydroxy-7-quinolinyl) (4-methylphenyl)methyl)benzamide, and then indicated gene expression was assessed by reverse transcriptase–quantitative PCR.
Not all cell types were sensitive to HQBA. Both non-transformed human mammary epithelial cells, and their isogenic RasV12 expressing, p53-knockdown-transformed counterparts were relatively insensitive to HQBA. Similarly to these human mammary epithelial cell lines, primary mouse mammary epithelial cells were 5.4- to 65-fold less sensitive than the majority of cancer cell lines to killing by HQBA after 7 days of treatment. The non-transformed fibroblast cell line GMO1915C was not killed at any drug concentration tested. This relative insensitivity of non-transformed cells suggests HQBA might be effective at preventing tumor progression without overt toxicity to normal tissues.
HQBA inhibits growth in mammary tumors of MMTV-Wnt1 and MMTV-PyMT mice
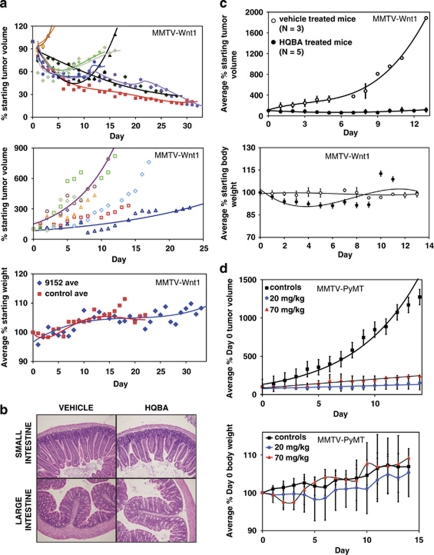
Many cancers are thought to require β-catenin signaling for continued proliferation. We therefore evaluated the efficacy of HQBA in vivo toward Wnt-initiated and Wnt/β-catenin-dependent spontaneously developing mammary tumors in MMTV-Wnt1-transgenic mice. About 15% of these mice develop mammary tumors between 6 weeks and 3 months of age. After the tumors reached at least 40 mm3, the affected mice were injected once daily with either vehicle (5% dimethyl sulfoxide in pure olive oil) or with 9 mg/kg HQBA for up to 32 days (Figure 3a). Sustained regression was observed to an average tumor volume of ∼20% of starting volume in 33% of treated mice. In the control group (n=8), all tumors grew steadily with an average doubling time of 5 days. The difference between the average tumor growth rates in the treated and untreated groups was highly significant (P<0.0001). HQBA did not cause obvious toxicity to normal tissues, as there was no significant difference in body weight between control and treated mice (P=0.37).
Figure 3.
HQBA inhibits growth of mammary cancer in MMTV-Wnt1 and MMTV-PyMT mice. (a) Upper panel: female virgin MMTV-Wnt1 mice with palpable tumors were treated with 9 mg/kg per day HQBA (N=9). Seven of the nine mice showed initial regression of tumor volume for ∼2 weeks. Three of the nine mice showed steady regression throughout the 5-week study with final tumor volumes ∼20% of starting volume. Center panel: growth of tumors in control, vehicle-treated MMTV-Wnt1 mice. The tumor volume doubled on average every 5 days. Lower panel: mice were weighed daily as an indicator of drug toxicity. Both control and treated mice gained weight at the same rate throughout the study. (b) Representative images of hematoxylin- and eosin-stained small and large intestine from mice treated for 2 weeks with vehicle or 9 mg/kg per day of HQBA. No deterioration of crypt/villi structure was seen in any part of the intestine. (c) Female virgin MMTV-Wnt1 mice with palpable tumors were treated with 200 μl of 25 μ HQBA daily for up to 13 days. Differences in mouse body weight translated this dosage into a range of 60 to 90 mg/kg per day. Upper panel: average tumor volume in the treated group (N=5) regressed to 60% of starting volume by day 6 and returned to starting volume by the end of the study. By contrast, average tumor volume in the control group (N=3) increased almost 20-fold. Lower panel: mouse body weight did not vary between arms by >10% throughout the course of the study. (d) Tumor bearing virgin female MMTV-PyMT mice were treated for 14 days with vehicle (N=4), 20 mg/kg per day HQBA (N=5), or 70 mg/kg per day HQBA (N=2). HQBA markedly reduced tumor growth rate (upper panel) without an effect on weight gain (lower panel).
One concern with drugs that might target Wnt/β-catenin signaling is toxicity in the intestine, as a number of studies have demonstrated loss of gut integrity when Wnt/β-catenin signaling is interrupted. We therefore specifically examined the gross and microscopic morphology of large and small intestines from HQBA-treated and control mice, and found no differences (Figure 3b). Thus, HQBA does not appear to be toxic to non-transformed intestinal cells.
To determine whether a higher HQBA dose would increase the response rate, a second study was subsequently conducted, in which five MMTV-Wnt1 mice with mammary tumors were treated with 75±15 mg/kg of HQBA and three mice were treated with vehicle as control (Figure 3c). HQBA treatment caused an average 40% decrease in tumor volume by day 6, and no net growth over the 13-day course of the study. By contrast, the tumors in the control mice grew rapidly, resulting in up to 20-fold increase in tumor volume in the same time frame. (P for both doses <0.0001.) At this higher dosage, treated mice initially lost body weight (10% by day 4), but caught up with control mice by the end of the study.
Our cell toxicity data suggested HQBA might be effective regardless of Wnt/β-catenin pathway dependence. To determine whether HQBA was effective in other tumor types in vivo, we tested it in a second genetically engineered mouse model. MMTV-PyMT-transgenic mice develop mammary tumors in response to oncogenic levels of Ras and phosphoinositide 3-kinase signaling induced by the expression of polyoma middle T antigen in mammary tissues (Guy et al., 1992). These tumors have no evidence of Wnt/β-catenin pathway activation (Herschkowitz et al., 2007). Female MMTV-PyMT mice that spontaneously developed mammary tumors were treated with HQBA at two doses, 20 mg/kg per day (n=5) and 70 mg/kg per day (n=2). HQBA treatment resulted in substantially decreased tumor growth, such that after 10 days tumors were on average only 1/5 the size of those in control mice, with no significant effect on body weight (Figure 3d). (P for both doses <0.0001.) This indicates HQBA is effective in genetically engineered mouse models of both Wnt-dependent and Wnt-independent tumors.
The molecular target of HQBA
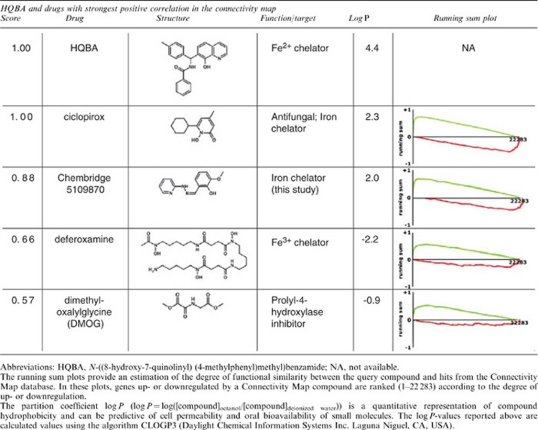
The ability of HQBA to inhibit β-catenin signaling in a subset of cell types with activating mutations, and its significant effects on growth of spontaneous murine mammary cancers, lead us to extensive efforts to identify the cellular target of the molecule. Both unmodified and photocrosslinker-containing biotinylated analogs of HQBA were synthesized, but despite exhaustive efforts, did not lead to identification of a functionally relevant protein target. As an independent approach, we queried the Connectivity Map (CMAP) (Lamb et al., 2006; Lamb, 2007) to determine whether HQBA altered gene expression in a pattern similar to compounds with a known mechanism of action. MCF7 cells were treated with HQBA or vehicle for 6 h and microarray signatures of upregulated and downregulated genes were generated (see Supplementary Table 3). Table 3 shows compounds in the CMAP database whose transcriptional signatures are most similar to those of HQBA. Strikingly, although none of the top matches appear chemically related to HQBA, two of the four compounds with the most similar transcriptome signatures are known iron chelators. The top hit, ciclopirox, is an iron chelator used clinically as a topical antifungal agent. Notably, ciclopirox was recently reported to have anti-cancer activity in vitro and in xenograft models (Eberhard et al., 2009; Zhou et al., 2010). The second best hit is a Chembridge compound (5109870) with no reported target. Of the more distant hits, deferoxamine is a clinically approved non-cell permeable high-affinity ferric iron chelator, whereas dimethyloxaloylglycine activates hypoxia-inducible factor-1α (HIF-1α) by inhibition of the iron-containing enzyme prolyl hydroxylase (Jaakkola et al., 2001). Thus, the CMAP data suggests HQBA functions through either iron chelation, or less likely, through induction of HIF-1α.
Table 3. Compounds from connectivity map analysis with significant positive correlation to the global effects on transcription caused by 4 μ HQBA (6 h on MCF7 cells).
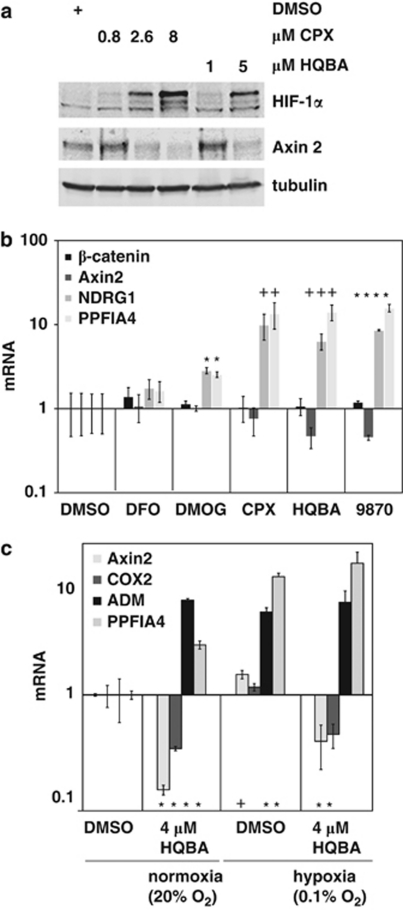
We confirmed that, as predicted from the CMAP profile, both ciclopirox and HQBA stabilize HIF-1α protein in SW480 cells. In addition, ciclopirox, similar to HQBA, causes a decrease in axin2 protein abundance (Figure 4a). As a further indication of overlap with the hypoxia response, HQBA, ciclopirox and 5109870 are more effective than dimethyloxaloylglycine and deferoxamine at inducing the hypoxia-inducible genes NDRG1 and PPFIA4 (Figure 4b). dimethyloxaloylglycine and deferoxamine differ from HQBA, as they do not decrease expression of the Wnt/β-catenin target gene AXIN2.
Figure 4.
HQBA induces a hypoxia response, but Wnt/β-catenin signaling is not affected by hypoxia. (a) Both HQBA and ciclopirox (CPX) induce HIF-1α and suppress axin2 protein. SW480 cells were treated for 18 h with dimethyl sulfoxide (DMSO) or the indicated amounts of HQBA or CPX, and then the lysates were analyzed by SDS–polyacrylamide gel electrophoresis and immunoblotting. (b) Diverse potential iron chelators including HQBA and 5109870 induce HIF-1α-responsive genes, but desferrioxamine (DFO) and dimethyloxaloylglycine (DMOG) do not repress Wnt/β-catenin-response genes. SW480 cells were treated with DMSO, or 10 μ HQBA, ciclopirox, deferoxamine, DMOG or 5109870 for 16 h. Relative levels of message for the hypoxia-inducible genes NDRG1 and PPFIA4 and β-catenin- and the Wnt-responsive gene axin2 were determined by quantitative PCR. *P<0.01; +P<0.05 by two-tailed t-test for difference from DMSO control. (c) Hypoxia does not repress expression of Wnt/β-catenin-response genes. SW480 cells grown at 20% or 0.1% O2 were each treated with vehicle or 4 μ HQBA for 16 h. Relative levels of message of the Wnt-response and hypoxia-induced genes shown were determined by quantitative PCR. +P<0.05; *P<0.01 by two-tailed t-test for difference from control.
To test whether the hypoxia response directly mediated the inhibition of β-catenin signaling, the effect of hypoxia on the β-catenin-response genes AXIN2 and COX2 was assessed. As Figure 4c shows, both HQBA and hypoxia robustly induced expression of ADM and PPFIA4. However, HQBA but not hypoxia represses expression of AXIN2 and COX2. We conclude that HQBA induces the hypoxia response but has additional hypoxia-independent activities that include inhibition of β-catenin signaling in selected cell types.
Metal-chelating activity of HQBA
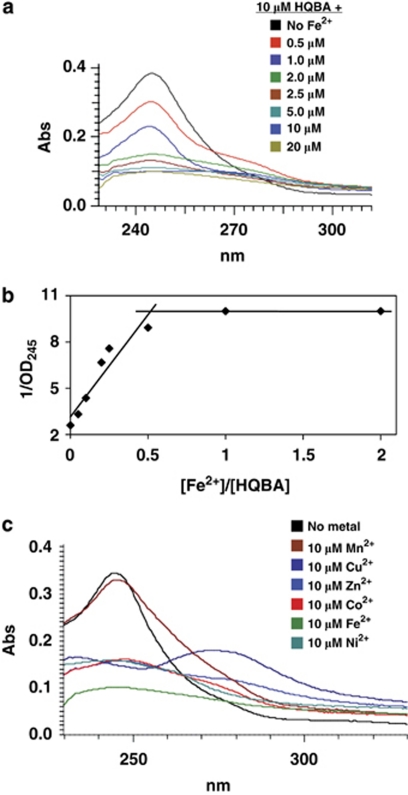
As an initial test of the metal chelating abilities of HQBA, we examined the ultraviolet absorbance spectrum of HQBA in the presence of increasing ferrous sulfate (Figure 5a). Increasing [Fe2+] caused a dose-dependent decrease in the A245, and a saturation plot indicated binding occurred with a stoichiometry of 2 mol HQBA to each mole of Fe2+ (Figure 5b). HQBA also chelates Co2+, Ni2+, Cu2+ and Zn2+ but not Mn2+ (Figure 5c). With the exception of Fe3+, which binds HQBA with 1:1 stoichiometry, all other metals bound with a one metal: two ligand ratio.
Figure 5.
HQBA is an iron chelator. (a) Mixture of 10 μ HQBA with varying concentrations of FeCl2 dose dependently reduced the absorbance of HQBA at 245 n. (b) Stoichiometry of Fe2+ binding is derived from plotting 1/OD245 versus [Fe2+]/[HQBA]. The intersection of slopes for Fe2+ saturated and unsaturated HQBA absorbance indicates that two molecules of HQBA bind one Fe2+ ion. (c) Absorbance (Abs) at 245 n of 10 μ HQBA with equimolar concentrations of various transition metals in the same series as Fe2+ indicates that it chelates Fe2+, Co2+, Ni2+, Cu2+ and Zn2+, but not Mn2+.
The clinically used iron chelator deferoxamine has a preference for binding ferric (Fe3+) iron, with a Kd of ∼10−31 compared with ∼10−10 for Fe2+ (Keberle, 1964). In assessing binding affinities and thermodynamic parameters by isothermal titration calorimetry (ITC), we found that HQBA bound most metals including ferric (Fe3+) iron with modest Kd values ranging from 100 n to 20 μ. However, in contrast to deferoxamine, HQBA bound ferrous iron (Fe2+) with an affinity ⩾1011 times higher than all other metals tested, with an estimated dissociation constant of 120 z (1.2 × 10−19 ). Stoichiometries determined by ITC agree with those obtained by spectroscopy (Table 4). We also verified that the compound 5109870 is an iron chelator, and displays identical two chelator: one Fe2+ stoichiometry (Supplementary Figure 1). Thus, HQBA is a novel high affinity, high selectivity ferrous iron chelator.
Table 4. Determination of stoichiometry (N), Ka and ΔH for chelation of transition metals by HQBA using isothermal calorimetry.
| Metal | n | Ka () | ΔH (kcal mol−1) |
|---|---|---|---|
| Fe (II) | 2.30±0.10 | (1.18±0.22) × 1019 | −37.4±2.1 |
| Fe (III) | 0.79±0.05 | (8.97±1.03) × 107 | −1.68±2.0 |
| Co (II) | 2.00±0.01 | (13.0±4.0) × 105 | −2.07±0.02 |
| Cu (II) (site 1) | 1.00±0.00 | (2.51±0.75) × 107 | −4.79±0.29 |
| Cu (II) (site 2) | 1.00±0.00 | (1.12±0.34) × 107 | −2.49±0.65 |
| Ni (II) | 2.28±0.04 | (1.92±0.18) × 105 | −2.23±0.6 |
| Zn (II) | 1.98±0.33 | (1.99±0.13) × 106 | −1.95±0.5 |
| Al (III) | 2.03±0.05 | (4.75±1.15) × 105 | −5.52±0.2 |
Abbreviation: HQBA, N-((8-hydroxy-7-quinolinyl) (4-methylphenyl)methyl)benzamide.
Binding affinities and relevant thermodynamic information for compound HQBA with Fe2+, Fe3+, Co2+, Cu2+, Ni2+, and Zn2+. Fe2+ displays ⩾1011-fold greater affinity than all other metals. Cu2+ was observed to fit best to a sequential binding sites model with two sites, as detailed above. All other metals with two ligand: one metal stoichiometry fit a one site binding model with a stoichiometry of 2.
The biological effects of HQBA are due to iron chelation
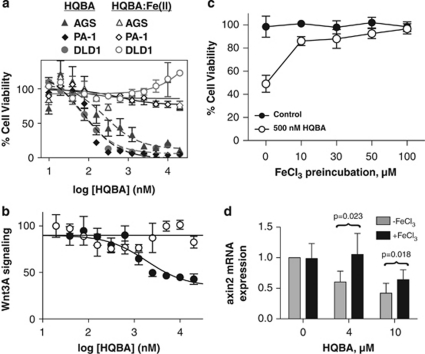
HQBA shares with the structurally unrelated small molecules ciclopirox and 9870 both the ability to chelate iron, and the ability to downregulate Wnt/β-catenin signaling. This strongly suggests that chelation of intracellular iron is responsible for the biological effects of HQBA. We investigated whether or not iron chelation was responsible for the observed cytotoxicity of HQBA. Pre-mixing of HQBA with 1 equivalent of Fe2+ completely abrogated its toxicity (Figure 6a) toward the highly sensitive cell lines AGS (gastric), DLD-1 (colorectal) and PA-1 (teratoma) and blocked its ability to inhibit Wnt/β-catenin signaling (Figure 6b). The effect of HQBA on Fgf8 expression in chick explants was similarly rescued by the addition of 0.5 μ ferrous ethylene diammonium sulfate (Figure 2f).
Figure 6.
The activity of HQBA is reversed by iron. (a) AGS, PA-1 and DLD-1 cells were incubated in the presence of 9.8 n to 20 μ HQBA±0.5 equivalents of ferrous ethylenediammonium sulfate tetrahydrate for 72 h in 96-well plates. Cells were then lysed and viability was assessed by measuring endogenous lactate dehydrogenase activity. (b) STF3A cells were plated in 96 well format and wells were treated with 9.8 n to 20 μ HQBA±0.5 equivalents of ferrous ethylenediammonium sulfate tetrahydrate for 18 h. Luciferase activity in lysates was measured and normalized to endogenous lactate dehydrogenase activity. Dose curves were plotted with Graphpad Prism (La Jolla, CA, USA). (c) DLD-1 cells were plated in 24-well plates in the presence of the indicated concentration of FeCl3, and allowed to grow for 24 h. Cells were washed three times with phosphate-buffered saline and then incubated with 500 n HQBA for an additional 72 h. Viability was measured as described. (d) DLD-1 cells were grown in the presence of 20 μ FeCl3 for 24 h, washed twice with phosphate-buffered saline, and then treated with the indicated concentration of HQBA in dimethyl sulfoxide for an additional 24 h before lysis and measurement of AXIN2 mRNA levels by quantitative reverse transcriptase–PCR. Data are shown as ±s.d. and significance calculated by paired t-test.
Confirming that the inhibition was due to the effect of intracellular iron, similar results were obtained when DLD-1 and HCT116 cells were preincubated with FeCl3 for up to 24 h followed by extensive washes, before the addition of HQBA (Figure 6c and data not shown). Likewise, pre-loading of DLD-1 cells with iron mitigated the effect of HQBA on axin2 expression (Figure 6d).
Discussion
In a cell-based screen for small molecules that inhibit Wnt/β-catenin signaling, we identified a closely related series of compounds with efficacy at high nanomolar concentrations. The selected lead, HQBA, is a previously unreported molecule that chelates Fe2+ with zeptomolar affinity, inhibits Wnt/β-catenin signaling in a subset of colon cancer cell lines and vertebrate embryos, has a broad spectrum of antiproliferative effects in cultured human cancer cell lines, and most importantly, inhibits tumor growth in two distinct genetically engineered mouse models of cancer without significant acute toxicity. The activity of HQBA in two genetically engineered mouse models of cancer without overt toxicity suggests the compound has good bioavailability and a reasonable half-life with once daily dosing. HQBA is a novel Fe2+ chelator with anti-cancer potential.
A number of small molecules that chelate iron have shown promise as cancer-cell-specific cytotoxic agents (Richardson et al., 2009). Diverse mechanisms have been proposed to explain how iron chelators kill cancer cells, possibly because of the fundamentally important role of iron in cells, and the ever-increasing number of proteins that have been found to require Fe (II) and/or iron–sulfur clusters for activity (Sheftel et al., 2010). Iron is required to prevent the hypoxic response, for DNA replication and repair, protein synthesis, and for mitochondrial energy generation. As HQBA inhibits transcription of β-catenin target genes in a subset of cells, our study additionally suggests a role for iron-containing proteins in β-catenin-dependent transcription in a tissue-specific manner. It is likely that different cell types and different tissues in animals have distinct iron requirements. Hence, iron can be a rate-limiting co-factor in a broad array of cellular activities. This may explain why we found HQBA induced a remarkable cell cycle arrest in G1 in some cell types, and induced an S phase arrest in others (data not shown). An additional consideration is that HQBA can chelate a number of other transition metals, although at lower affinity, which could also contribute to decreased cell proliferation.
Besides their ability to remove iron from key pathways, the formation of HQBA:Fe (II) complexes can have additional effects. Chelated iron can participate in generation of reactive oxygen species, an activity that contributes to the toxicity of the iron chelator Dp44mT (Jansson et al., 2010; Tian et al., 2010). In fact, we were able to detect a significant increase in both reactive oxygen species and reactive nitrogen species in several cell types treated with HQBA, but unlike with Dp44mT, the cytotoxicity of HQBA was not consistently rescued by pre-treatment with the reducing agent N-acetylcysteine nor with the nitric oxide synthase inhibitor nitro--arginine methyl ester (data not shown). Thus, although HQBA complexes may be redox active, reactive oxygen species and reactive nitrogen specie generation was not the cause of cytotoxicity in the colon cancer lines we tested.
The ability of HQBA to inhibit Wnt/β-catenin signaling downstream of β-catenin in several colon cancer cell lines and embryonic tissues suggests that it also functions at the level of transcription or mRNA stability. Although iron regulates some transcripts via message stability (via iron response elements and iron-responsive-element-binding proteins), here we suspect the effect is transcriptional, as it is cell type specific, and affects the artificial TOPFLASH promoter as well as several β-catenin-response genes. A number of iron-binding and iron-responsive transcription factors are known within the iron homeostasis pathways (Rutherford and Bird, 2004), and it is likely that iron and other metals control gene expression in other pathways.
The sensitivity of colonocytes and Wnt-β-catenin signaling to iron status has been noted recently. Brookes et al. (2008) found that several colon cancer cell lines, including Caco-2, SW480 and LS174 T, have increased β-catenin signaling in response to iron loading as demonstrated by both increase in TOPFLASH and increase in endogenous c-Myc and NKD gene expression. The increased baseline level of β-catenin-dependent signaling in cells with mutations in the APC or β-catenin gene may make this pathway particularly sensitive to changes in the abundance of an as yet-unidentified iron-dependent co-factor. We note that nuclear iron-dependent enzymes continue to be discovered; for example, the recently described mammalian cytosine hydroxymethylase, with strong tissue-specific and developmentally controlled expression, is an Fe (II)-containing enzyme (Tahiliani et al., 2009). Similarly, the iron-containing transcription factor, pirin, interacts with nuclear factor 1 and Bcl-3 (Pang et al., 2004). A β-catenin-dependent tissue-specific transcriptional regulator might be a particularly useful target for the treatment of APC-mutant cancers.
HQBA differs from the iron chelators, such as deferoxamine, used clinically to treat iron overload both in its cell permeability and its high selectivity for Fe2+ over Fe3+ (Table 3). Structure–activity studies indicate that the antiproliferative potential of iron chelators increases with increasing hydrophobicity (Hodges et al., 2004). This trend is thought to correlate with increasing ability of more hydrophobic chelators to enter the cell in which they have access to critical metalloenzymes and pools of stored iron. This notion is consistent with the high-calculated log P of HQBA, suggesting it is highly cell permeable. Antiproliferative capability also directly correlates with redox potential. Molecules such as deferoxamine, a hexadentate chelator of Fe3+, fully occupy the coordination sphere of iron, rendering it inaccessible to oxygen and reducing the formation of reactive oxygen species. Bidentate and tridentate chelators by contrast permit such interactions, and, therefore, may permit or even facilitate redox cycling of bound iron. Redox cycling and reactive oxygen species generation could therefore have a role in HQBA anti-cancer activity in some cell types.
The ultimate effectiveness of iron chelation as an anti-cancer strategy remains unclear. In part, this is because existing iron chelators are far from equivalent to each other in biological effects. In addition to differences in in vivo clearance rates, various iron chelators in clinical trials differ in their cell permeability, redox potential and potentially, ability to pull iron out of various iron-dependent enzymes and other intracellular pools of stored iron, as well as differences in side targets. The structure–activity relationship studies of desferrithiocin illustrate how dramatically iron chelators can differ (Bergeron et al., 1993, 1999). Desferrithiocin is an effective iron chelator with the adverse side effect of nephrotoxicity. Removal of a single methyl group abrogated the nephrotoxicity. Iron clearance in a rat model was unaffected, but was significantly reduced in a primate model. Other analogs, which replaced a pyridyl nitrogen with a carbon, were found to elicit severe gastrointestinal toxicity in rats (Bergeron et al., 1999).
Genetically engineered mouse models of cancer, while more difficult experimentally, are more predictive of whether a drug will have efficacy in humans than the easier xenograft models in immunocompromised mice. The ability of HQBA to block cancer progression without inducing weight loss supports the hypothesis that spontaneous cancers, even in chow-fed, iron-replete mice, are exquisitely sensitive to iron depletion. In addition, there may be additional properties of the intracellular HQBA:Fe (II) complexes that contribute to their anti-cancer activity.
Materials and methods
Reagents
A 50 000 compound library, as well as 5839152/HQBA, and related compounds were purchased from Chembridge. HQBA was re-synthesized, and derivatized versions were synthesized at the Duke University Small Molecule Synthesis Facility. Antibodies to axin2 were from Cell Signaling (Danvers, MA, USA). Antibodies to HIF-1α, β-catenin, β-actin and β-tubulin were from Abcam (Cambridge, MA, USA). The screen for Wnt3A inhibitors has been recently described (Coombs et al., 2010). For isothermal titration calorimetry experiments, most metals (Fe3+, Cu2+, Co2+, Al3+, Ni2+, Zn2+) used were obtained as atomic absorption standards in nitric acid from Aldrich (St Louis, MO, USA). Fe2+ was obtained as 2,2′-bipyridine ferrous perchlorate from GFS Chemicals (Powell, OH, USA). Glycine and acetohydroxamic acid were obtained from Aldrich.
Screen for Wnt3A inhibitors
HEK293 cells with stably integrated luciferase driven by 8X Super TopFlash promoter and stable expression of Wnt3A were incubated with 10 μ compound for 16–20 h.
Western blotting
Cells were treated with control or compound for times indicated and lysates were run on 10% SDS–polyacrylamide gel electrophoresis. Proteins were transferred to Immobilon-FL (Millipore, Billerica, MA, USA) polyvinylidene fluoride membranes and blocked with either 5% bovine serum albumin in phosphate-buffered saline+0.05% Tween 20 for detection of proteins by enhanced chemiluminescence, or with Sea Block (Pierce, Rockford, IL, USA) for fluorescent detection strategies. Primary antibody dilutions are 1:1000 for all antibodies except HIF-1α (1:2000), β-catenin (1:6000) and β-tubulin (1:15 000). Secondary antibodies are diluted to 1:5000 for detection by enhanced chemiluminescence, and 1:10 000 for fluorescence detection.
Determination of lethal dose 50 values in cancer cell lines
Cells were plated at 4000 cells/well in clear 96-well plates, and allowed to adhere overnight. Media was then exchanged for fresh and HQBA was added at concentrations ranging from 9 to 50 μ. After 72 h cells were washed in phosphate-buffered saline and lysed in phosphate-buffered saline containing 0.6% Igepal CA-630 (Sigma-Aldrich). Relative cell density/well was then determined by assaying endogenous lactate dehydrogenase activity. Lethal dose 50 values were obtained from nonlinear analysis using Prism. Assays were also conducted in which cells were plated at 2000 cells/well and incubated with HQBA for 7 days (168±4 h). In these assays, media and compound were refreshed every 72 h. In both long and short assays, assays were performed in triplicate.
Microarray and CMAP analysis
MCF7 cells were plated at 70–80% confluence in six-well plates and allowed to adhere overnight before incubation with dimethyl sulfoxide or 4 μ HQBA for 6 h. RNA was then isolated using RNeasy purification kit from Qiagen (Germantown, MD, USA). Labeled complementary RNA was prepared and hybridized to Affymetrix (Santa Clara, CA, USA) U133_Plus_2.0 microarrays according to the manufacturer's protocols. Gene expression signatures were analyzed using the Connectivity Map web tool (build 02), found at http://www.broad.mit.edu/cmap/. Full details of the Connectivity Map data set and analytics are provided elsewhere (Lamb et al., 2006; Lamb, 2007).
Small interfering RNA experiments
Cells were plated at 50% confluence in 6 or 12 well format, transfected next day with control (non-targeting) or small interfering RNAs targeting β-catenin and incubated for 48 (si β-catenin) or 72 h. Sequences of small interfering RNAs used are 5′-UAGCGACUAAACACAUCAA-3′ (control), 5′-GAUCCUAGCUAUCGUUCUU-3′ (β-catenin 09) and 5′-GCGUUUGGCUGAACCAUCA-3′ (β-catenin 11).
Quantitative PCR
For quantitation of mRNA, cells were treated as indicated. RNA was then isolated using an RNeasy purification kit from Qiagen. Generation of complementary DNA was performed according to manufacturer's instructions using the iscript complementary DNA synthesis kit from Biorad (Hercules, CA, USA). Quantitative PCR reactions were then built using IQ-Sybr Green kit or Ssofast Evagreen Supermix from BioRad. Primers are listed in Supplementary Table 1. Cycling was carried out in an iQ5 model icycler (BioRad).
Metal chelation analysis by spectrophotometry
Chelation of metals by HQBA and related compounds was evaluated by mixing 10–50 m compound with 0.5–40 m metal, and diluting 1000-fold in H2O in a quartz cuvette. Absorbance spectra over the range 210–550 n were obtained on a U-2810 spectrophotometer (Hitachi, Pleasanton, CA, USA).
Drug treatment in MMTV-Wnt1 and MMTV-PyMT mouse models of mammary carcinogenesis
MMTV-Wnt1 and MMTV-PyMT female virgin mice ranging from 6 to 20 weeks old were added to either control or treatment arms of the study, as they presented with palpable mammary tumors. Mice in the treatment arm were injected intraperitoneally every morning with 9 or 70 mg/kg drug prepared as follows: HQBA was dissolved to 100 m in dimethyl sulfoxide and 6–10 μl/mouse was added to 200 μl of vehicle. For studies of MMTV-Wnt1 mice, vehicle was 200 μl of pure olive oil. For the MMTV-PyMT mouse studies, vehicle was 5% cremophor, 22% (2-hydroxypropyl)-β-cyclodextrin in 0.9% sterile saline. Mice in the control arm were injected intraperitoneally daily with 200 μl of vehicle only. Mice were weighed daily before injection and their tumors measured post injection with calipers. Tumor volumes were calculated using the formula for volume of an ellipsoid: (½ (short dimension))2 × long dimension (Tomayko and Reynolds, 1989). Change in tumor volume is plotted relative to starting volume, which is normalized to 100% for each tumor.
Chick neural tube explant assays
Neural explants encompassing midbrain to rhombomere 2 tissue were isolated from Hamburger and Hamilton stage 10 embryos as previously described and cultured overnight until explants corresponded to Hamburger and Hamilton stage 16. Explants were embedded in collagen to maintain morphology and 1 μ HQBA and 0.5 μ Fe2+ were supplemented in the media as indicated. Double in situ hybridization for Wnt1 and Fgf8 were performed as described (Canning et al., 2007; Coombs et al., 2010)
Isothermal titration calorimetry
ITC data were collected on a VP-ITC titration microcalorimeter (Microcal Inc., Northampton, MA, USA); details of instruments and data reduction have been reported elsewhere (DiTusa et al., 2001). The cell volume of the calorimeter is 1.4346 ml. All protein and metal ligand solutions were degassed under vacuum before titrations. Samples consisted of HQBA (1–20 μ) in 10 m phosphate buffer, pH 6.80, with buffer as the reference. After cell equilibrium was reached, a 100–500 μ solution of metal (ZnNO3, CuNO3, NiNO3, Fe (bipy)3, AlNO3, FeNO3 or CoNO3) in the same buffer was injected using an injection schedule of 20–60 injections, 5–30 μl volumes, during 10–60 s and at 5–10 min intervals. All measurements were made at 25 °C. Data analysis was carried out using the ORIGIN software from Microcal. All binding enthalpies are reported after subtraction of the appropriate enthalpy of metal ion dilution.
Displacement titrations were conducted for Cu, Fe (II) and Fe (III) to ensure stability of the respective metal at the relevant oxidation state. Metals were pre-incubated with an appropriate chelator (Cu: glycine, Fe (II): bipy, Fe (III): acetohydroxamic acid) before titration. Displacement experiments were conducted as described above and data was analyzed according to the method of Sigurskjold (2000). During analysis of Fe (II) affinity, Fe (bipy)3 was added to the sample cell and a solution of HQBA was injected under the identical conditions described above.
Job plots: Co2+, Cu2+, Ni2+, Fe3+, Fe2+, Al3+
Solutions (500 μmol) containing varying ratios of 9152 and the appropriate metal in 1:1 MeOH/phosphate buffer (pH 6.80) were analyzed by ultraviolet/visible spectroscopy. The day to day transition was measured in absorbance units and plotted as mole fraction of metal versus absorbance at the appropriate wavelength after adjustment for residual metal absorbance. The job plot for zinc was measured by difference spectroscopy.
Acknowledgments
We thank Chuck Perou and Philipp Kaldis for assistance with the MMTV-Wnt1 and MMTV-PyMT mice, and Elizabeth Leibold for helpful discussions. The compound library was provided by the Huntsman Cancer Institute Drug Screening Resource at the University of Utah. This research was supported by the Duke-NUS Signature Research Program and the Singapore Translational Research (STaR) Investigator Award (to DMV) funded by the Agency for Science, Technology and Research, Singapore, and the Ministry of Health, Singapore. CAC and CMJ were funded by A*STAR, Singapore. AAS acknowledges support from the Duke Pharmacological Sciences Training Program, NIH T32-GM007105-37.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Bergeron RJ, Streiff RR, Creary EA, Daniels RD, Jr, King W, Luchetta G, et al. A comparative study of the iron-clearing properties of desferrithiocin analogues with desferrioxamine B in a Cebus monkey model. Blood. 1993;81:2166–2173. [PubMed] [Google Scholar]
- Bergeron RJ, Wiegand J, Weimar WR, Vinson JR, Bussenius J, Yao GW, et al. Desazadesmethyldesferrithiocin analogues as orally effective iron chelators. J Med Chem. 1999;42:95–108. doi: 10.1021/jm980340j. [DOI] [PubMed] [Google Scholar]
- Brookes MJ, Boult J, Roberts K, Cooper BT, Hotchin NA, Matthews G, et al. A role for iron in Wnt signalling. Oncogene. 2008;27:966–975. doi: 10.1038/sj.onc.1210711. [DOI] [PubMed] [Google Scholar]
- Canning CA, Lee L, Irving C, Mason I, Jones CM. Sustained interactive Wnt and FGF signaling is required to maintain isthmic identity. Dev Biol. 2007;305:276–286. doi: 10.1016/j.ydbio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Coombs GS, Covey TM, Virshup DM. Wnt signaling in development, disease and translational medicine. Curr Drug Targets. 2008;9:513–531. doi: 10.2174/138945008784911796. [DOI] [PubMed] [Google Scholar]
- Coombs GS, Yu J, Canning CA, Veltri CA, Covey TM, Cheong JK, et al. WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J Cell Sci. 2010;123:3357–3367. doi: 10.1242/jcs.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTusa CA, Christensen T, McCall KA, Fierke CA, Toone EJ. Thermodynamics of metal ion binding. 1. Metal ion binding by wild-type carbonic anhydrase. Biochemistry. 2001;40:5338–5344. doi: 10.1021/bi001731e. [DOI] [PubMed] [Google Scholar]
- Eberhard Y, McDermott SP, Wang X, Gronda M, Venugopal A, Wood TE, et al. Chelation of intracellular iron with the antifungal agent ciclopirox olamine induces cell death in leukemia and myeloma cells. Blood. 2009;114:3064–3073. doi: 10.1182/blood-2009-03-209965. [DOI] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson WR, Chi EY, Ye X, Nguyen C, Tien Y-T, Zhou B, et al. Inhibition of Wnt/beta-catenin/CREB binding protein (CBP) signaling reverses pulmonary fibrosis. Proc Natl Acad Sci USA. 2010;107:14309–14314. doi: 10.1073/pnas.1001520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges YK, Antholine WE, Horwitz LD. Effect on ribonucleotide reductase of novel lipophilic iron chelators: the desferri-exochelins. Biochem Biophys Res Commun. 2004;315:595–598. doi: 10.1016/j.bbrc.2004.01.101. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jansson PJ, Hawkins CL, Lovejoy DB, Richardson DR. The iron complex of Dp44mT is redox-active and induces hydroxyl radical formation: an EPR study. J Inorg Biochem. 2010;104:1224–1228. doi: 10.1016/j.jinorgbio.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Keberle H. The biochemistry of desferrioxamine and its relation to iron metabolism. Ann N Y Acad Sci. 1964;119:758–768. doi: 10.1111/j.1749-6632.1965.tb54077.x. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- Lucero OM, Dawson DW, Moon RT, Chien AJ. A re-evaluation of the ‘oncogenic' nature of Wnt/beta-catenin signaling in melanoma and other cancers. Curr Oncol Rep. 2010;12:314–318. doi: 10.1007/s11912-010-0114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch MW, Bugni TS, Concepcion GP, Coombs GS, Harper MK, Kaur S, et al. Carteriosulfonic acids A-C, GSK-3beta inhibitors from a Carteriospongia sp. J Nat Prod. 2009a;72:1651–1656. doi: 10.1021/np900336f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch MW, Coombs GS, Banerjee N, Bugni TS, Cannon KM, Harper MK, et al. Psammaplin A as a general activator of cell-based signaling assays via HDAC inhibition and studies on some bromotyrosine derivatives. Bioorg Med Chem. 2009b;17:2189–2198. doi: 10.1016/j.bmc.2008.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- Pang H, Bartlam M, Zeng Q, Miyatake H, Hisano T, Miki K, et al. Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J Biol Chem. 2004;279:1491–1498. doi: 10.1074/jbc.M310022200. [DOI] [PubMed] [Google Scholar]
- Phelps RA, Chidester S, Dehghanizadeh S, Phelps J, Sandoval IT, Rai K, et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–634. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Sarkar S, Broadbent TJ, Voas M, Grossmann KF, Nadauld LD, et al. DNA demethylase activity maintains intestinal cells in an undifferentiated state following loss of APC. Cell. 2010;142:930–942. doi: 10.1016/j.cell.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DR, Kalinowski DS, Lau S, Jansson PJ, Lovejoy DB. Cancer cell iron metabolism and the development of potent iron chelators as anti-tumour agents. Biochim Biophys Acta. 2009;1790:702–717. doi: 10.1016/j.bbagen.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Rutherford JC, Bird AJ. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot Cell. 2004;3:1–13. doi: 10.1128/EC.3.1.1-13.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheftel A, Stehling O, Lill R. Iron-sulfur proteins in health and disease. Trends Endocrinol Metab. 2010;21:302–314. doi: 10.1016/j.tem.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Sigurskjold BW. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal Biochem. 2000;277:260–266. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo JL, Kahn M. The Wnt signaling pathway in cellular proliferation and differentiation: a tale of two coactivators. Adv Drug Deliv Rev. 2010;62:1149–1155. doi: 10.1016/j.addr.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Tian J, Peehl DM, Zheng W, Knox SJ. Anti-tumor and radiosensitization activities of the iron chelator HDp44mT are mediated by effects on intracellular redox status. Cancer Lett. 2010;298:231–237. doi: 10.1016/j.canlet.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- Zhou H, Shen T, Luo Y, Liu L, Chen W, Xu B, et al. The antitumor activity of the fungicide ciclopirox. Int J Cancer. 2010;127:2467–2477. doi: 10.1002/ijc.25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.