Before ER tubule fusion, the atlastin GTPase undergoes a “prefusion” to “postfusion” conformational change that is mediated by an intramolecular salt bridge.
Abstract
Endoplasmic reticulum (ER) network branching requires homotypic tethering and fusion of tubules mediated by the atlastin (ATL) guanosine triphosphatase (GTPase). Recent structural studies on the ATL soluble domain reveal two dimeric conformers proposed to correspond to a tethered prefusion state and a postfusion state. How the prefusion conformer transitions to the postfusion conformer is unknown. In this paper, we identify an intramolecular salt bridge mediated by two residues outside the GTPase domain near the point of rotation that converts the prefusion dimer to the postfusion state. Charge reversal of either residue blocked ER network branching, whereas a compensatory charge reversal to reestablish electrostatic attraction restored function. In vitro assays using the soluble domain revealed that the salt bridge was dispensable for GTP binding and hydrolysis but was required for forming the postfusion dimer. Unexpectedly, the postfusion conformation of the soluble domain was achieved when bound to the nonhydrolyzable GTP analogue guanosine 5′-[β,γ-imido]triphosphate, suggesting that nucleotide hydrolysis might not be required for the prefusion to postfusion conformational change.
Introduction
The membrane-anchored atlastin (ATL) proteins belong to the dynamin superfamily of large GTPases (Zhu et al., 2003; Praefcke and McMahon, 2004). In humans, the neuron-specific isoform ATL1/SPG3A is enriched in the cis-Golgi apparatus, and mutations in it are linked to motor neurological deficits associated with hereditary spastic paraplegia (Zhu et al., 2003). ATL2 and ATL3, 62 and 60% identical to ATL1, respectively, are expressed in most, if not all, tissues and primarily ER localized (Rismanchi et al., 2008). siRNA-mediated depletion of isoforms 2 and 3 from HeLa cells, expressing little, if any, isoform 1 (Rismanchi et al., 2008), leads to an unbranched ER morphology, implying a function for ATL2/3 in ER network branching (Hu et al., 2009). This requirement could reflect a role for the molecule in tubule formation, extension, tethering, and/or fusion (Lee et al., 1989; Baumann and Walz, 2001). Though, based on the remarkable finding that the single Drosophila melanogaster orthologue of ATL, purified and incorporated into artificial liposomes, is sufficient to drive membrane tethering and fusion, ATLs have been proposed to mediate the homotypic tethering and fusion of membranes that underlies the branched ER network (Orso et al., 2009).
Recently, two groups have solved the x-ray crystal structure of the soluble domain of human ATL1 (Bian et al., 2011; Byrnes and Sondermann, 2011). The structures reveal a globular GTPase head connected through an eight–amino acid linker to a middle domain comprised of a three-helix bundle. As expected, the GTPase domain has an overall fold similar to that of GBP1 (Prakash et al., 2000), the closest relative of ATL1–3 in the dynamin superfamily (14, 14, and 16% identical to ATL1, ATL2, and ATL3, respectively). Dynamin superfamily members undergo conformational changes in a manner dependent on their nucleotide-bound state (Ghosh et al., 2006; Chappie et al., 2010). Accordingly, ATL1 crystallization by both groups was performed in the presence of a variety of GTP analogues. Both groups observed two strikingly distinct ATL1 conformers, indicating that, like GBP1 and dynamin, ATL1 indeed undergoes discrete conformational changes during its reaction cycle. Moreover, both structures showed ATL1 as a head to head dimer, reminiscent of the head to head dimers observed in crystal structures of the GTPase domains of dynamin bound to the transition state analogue GDP + AlFx (Chappie et al., 2010), and GBP1 bound to either guanosine 5′-[β,γ-imido]triphosphate (GMPPNP) or GDP + AlFx (Ghosh et al., 2006). Curiously, only GDP was observed in the nucleotide-binding pocket of the structures obtained, possibly because of either slow hydrolysis or increased mobility of the terminal phosphate of GMPPNP and other analogues (Bian et al., 2011; Byrnes and Sondermann, 2011). Therefore, how changes in the nucleotide-bound state of ATL1 relate to changes in its conformation remains to be clarified.
In the first of the two ATL1 dimer conformers (form 2), the monomers interact in a head to head fashion with an interfacial area of 756 Å2 because of contacts solely between the GTPase domains. The α-helical bundles of the respective middle domains point away from the dimer interface, and although not present in the crystal structures, the trans-membrane segments would be expected to anchor the interacting subunits in opposing membranes. In the second conformer (form 1), a similar head to head configuration is observed as in the form 2 dimer, though additional contacts increase the interfacial area between the GTPase domains to 1,226 Å2. In addition, the α-helical bundles of the middle domains are crossed over with respect to the head domains caused by a 90° rotation about a central conserved proline residue in the linker. In this crossed dimer configuration (form 1), a new set of contacts are made between the middle domains and the opposing heads. As a consequence of the crossover, the C termini of the two subunits of the form 1 dimer are within 14 Å of one another, necessarily placing the trans-membrane segments of the interacting subunits in the same membrane. Based on the orientation of the molecules relative to the presumed orientation of the lipid bilayer, the form 2 and form 1 dimers have been designated pre- and postfusion states, respectively (Bian et al., 2011; Byrnes and Sondermann, 2011; Daumke and Praefcke, 2011).
The dimer pairs suggest a compelling model for membrane tethering and fusion. First, head to head dimerization of ATL in trans (form 2) would initiate membrane tethering. Once tethered, crossover of the middle domains would catalyze membrane fusion, presumably by bringing opposing lipid bilayers into tight apposition and deforming them, consequently reducing the activation barrier for membrane fusion (Bian et al., 2011; Daumke and Praefcke, 2011). In part because dimerization of ATL1 in solution is nucleotide dependent (Bian et al., 2011; Byrnes and Sondermann, 2011), GTP binding has been suggested to form the prefusion dimer for the membrane-tethering step, whereas GTP hydrolysis and Pi release has been hypothesized to trigger the 90° rotation and crossover of the middle domains to achieve the fused state (Bian et al., 2011). According to this scenario, a cycle of GTP binding and hydrolysis would drive both membrane tethering and fusion, though how the postfusion complex is disassembled for further rounds of fusion remains to be clarified.
If the crossed dimer conformation indeed represents the postfusion state, contacts unique to this conformer should be important for driving membrane fusion. Conversely, inhibiting such contacts should block the conversion of prefusion dimers to the postfusion state. Here, we report a functional analysis of residues within the middle domain of ATL2 in search of ones that might be involved in the prefusion to postfusion transition. We then focus on a pair of residues that appear to mediate a postfusion conformer-specific salt bridge. We show that the salt bridge is not required for either GTP binding or hydrolysis but is necessary for transitioning to the postfusion dimer conformation. Furthermore, although GTP hydrolysis has been suggested to be required for formation of the postfusion conformation of the soluble domain, our results indicate that hydrolysis is not required, at least in the context of the soluble domain. The potential implications of this finding with regard to the ATL fusion mechanism are discussed.
Results
Loss of ER network branching upon ATL2/3 knockdown is rescued by wild-type ATL2 expression
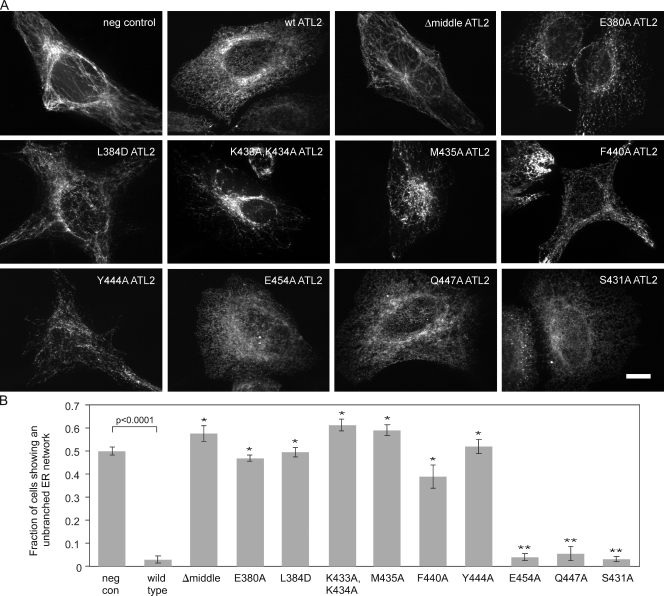
To identify ATL residues that participate in the interconversion between pre- and postfusion conformers, we used an RNAi knockdown replacement assay. The assay is based on the previously reported requirement for ATL2/3 in ER network branching in HeLa cells (Hu et al., 2009). As anticipated, treatment of HeLa cells with siRNAs identical to those previously shown to knock down ATL2 and ATL3 (Rismanchi et al., 2008) resulted in an abnormal ER morphology characterized by a notable reduction in network branch points (Fig. S1, A–C). In contrast to control knockdown cells with 200–400 ER network branch points per cell, ATL2/3 knockdown cells typically had <100 network branch points per cell (Fig. S1, C and D). Also consistent with a previous study, knockdown of both isoforms was required to elicit the unbranched phenotype (Fig. S1 A), indicating that either ATL2 or ATL3 is sufficient to maintain normal network morphology (Hu et al., 2009). To assess whether the unbranched ER phenotype is a specific consequence of ATL2/3 loss, the ATL2/3 siRNA was cotransfected with either a negative control DNA construct or an siRNA-immune HA-tagged ATL2 replacement construct. Whereas ∼50% of cells expressing the negative control construct displayed a largely unbranched network (<100 branch points per cell), few, if any, cells expressing wild-type HA-ATL2 displayed the phenotype (Fig. 1, A and B). Therefore, the unbranched ER phenotype can be attributed specifically to the loss of ATL2/3.
Figure 1.
Identification of ATL2 middle domain residues required for its ER network branching function. (A) Knockdown replacement assay. 48 h after transfection with a Myc-tagged DP1 negative control (neg con) construct, wild-type HA-ATL2, or each of the indicated HA-tagged ATL2 variants, cells were transfected with siRNAs targeting ATL2 and ATL3. 72 h after knockdown, cells were fixed and stained using an antibody against the Myc or HA epitope and viewed by confocal microscopy. Bar, 10 µm. (B) Quantification of the fraction of cells expressing the indicated proteins that had the unbranched ER phenotype. Values represent the means of three independent experiments ± SD. *, P < 0.0005 with respect to wild type; **, P < 0.0005 with respect to the Myc-DP1 negative control.
Specific middle domain residues are required for ATL2 activity
As our analysis was initiated before the recent determination of the ATL1 crystal structure, we started with a computationally derived ATL2 structure model based on its similarity to GBP1 (Prakash et al., 2000; Pieper et al., 2011). Domain boundaries defined by the model were applied to first test the importance of the middle domain. As anticipated, the HA-tagged variant ATL2Δ377–463 (or ATL2Δmiddle), lacking the entire middle domain, was stably expressed but failed to functionally replace endogenous ATL2/3 (Fig. 1, A and B). Then, several conserved middle domain residues were screened. RNAi knockdown replacement using ATL2 variants with either single or double amino acid substitutions to alanine revealed required residues within the middle domain (Fig. 1, A and B). Substitutions that blocked ATL2 function include E380A, L384D, K433A, K434A, M435A, F440A, and Y444A. Most of the residues identified by our analysis are located near the GTPase head (Fig. S2, A and B). Notably, M435 is equivalent to M408 in ATL1, which, when mutated, is associated with hereditary spastic paraplegia (Zhu et al., 2003), though the mutations have been reported to have only modest effects on the GTPase activity of ATL1 (Bian et al., 2011; Byrnes and Sondermann, 2011). Also, E380 is equivalent to residue E328 in the Drosophila homologue, whose charge reversal inhibits the in vitro liposome fusion reaction by 75% (Bian et al., 2011).
ER network morphology after replacement with the various ATL2 proteins appeared somewhat distinct, not only from one another but also from the morphology seen after knockdown. The exception was ATL2Δmiddle, whose network morphology was similar to the knockdown. The significance of these differences is unclear but may reflect a differing ability of each variant to engage in the ATL2 reaction cycle, with ATL2Δmiddle being the least functional. Many of the single alanine substitutions that blocked ER network branching were in highly conserved residues (Fig. S2 A), though substitution of at least one conserved residue, E454, had no apparent effect (Fig. 1, A and B). Less conserved or nonconserved surface residues, such as Q447 and S431, respectively, could also be substituted (Fig. 1, A and B).
Finally, many of the nonfunctional ATL2 variants, when expressed at high levels, exerted a dominant-negative effect on ER morphology that could be observed even in the presence of endogenous ATL2/3 (Fig. S2 C). To avoid the potentially complicating effects of such high level expression, only cells expressing each variant below a predetermined threshold level were included for quantification of functional replacement (see Materials and methods).
Middle domain mutations fall into three classes
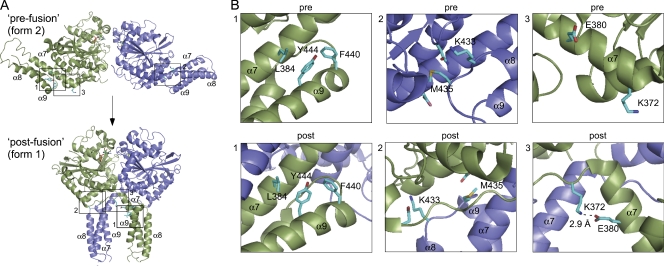
The two dimer crystal forms of ATL1 (Bian et al., 2011; Byrnes and Sondermann, 2011) allowed us to analyze the positions of our mutations. Because ATL2 is 73% identical to ATL1 in its cytoplasmic domain, its overall fold is likely very similar to ATL1 with only minor differences in the precise placement of the backbone and side chains. To aid in the analysis, computational models for ATL2 in both the pre- and postfusion conformation were derived based on the ATL1 structures (Pieper et al., 2011). Inspection revealed that the required residues fall into three categories (Fig. 2, A and B). The first category consists of L384, Y444, and F440 (Fig. 2, A and B, box 1). These residues pack together near the surface of the middle domain in the prefusion conformer, and the packing interactions remain relatively unchanged in the postfusion conformer. The second category consists of K433 and M435 (Fig. 2, A and B, box 2). These residues are in a loop connecting two helices (α8 and α9) of the middle domain. In the prefusion conformer, K433 and M435 contact residues within the head domain of the same monomer. In the postfusion conformer, K433 and M435 alter their contacts to residues in the head domain of the opposing monomer. The third and final category, consisting of E380 and K372, was of particular interest, as it pointed to residues appearing to make substantial contacts only in the postfusion conformer (Fig. 2, A and B, box 3). In the prefusion conformer, the nonpolar portion of the E380 side chain may be involved in a set of middle domain packing interactions, and K372 exhibits no obvious contacts. Significantly, K372 is immediately adjacent to the point of 90° rotation that converts the prefusion dimer to the postfusion dimer conformer, and in the postfusion conformer, it becomes involved in a salt bridge with E380.
Figure 2.
View of required ATL2 residues in the prefusion and postfusion conformer. Required residues identified by knockdown replacement (Fig. 1) are shown in cartoon and stick form. The Protein Data Bank coordinates for the prefusion (3QOF) and postfusion (3QNU) ATL1 conformers were downloaded from the RCSB Protein Data Bank database (Bian et al., 2011) and rendered in PyMOL (DeLano Scientific LLC). One subunit is green, and the other is blue. Bound GDP is highlighted in sticks. (A) The location of the three categories of required residues in both pre- and postfusion dimer conformers are boxed and numbered (1–3). (B) A close-up view of the required residues boxed (1–3) in A. Key residues are numbered by their position in the ATL2 sequence and shown in stick form. See the Results under Middle domain mutations fall into three classes for details.
A salt bridge specific to the postfusion dimer is required for ATL2 activity
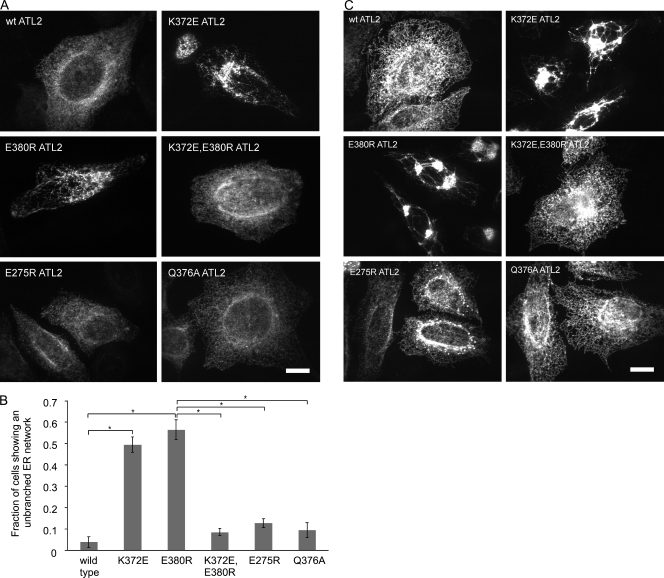
To test the functional significance of the K372-E380 salt bridge seen in the postfusion structure, we first examined the effect of charge reversal of either residue on the ER network branching function of ATL2. For this, two new variants, ATL2 K372E and ATL2 E380R, were generated. Each variant was stably expressed, but neither functioned in ER network branching (Fig. 3, A and B), confirming the importance of the respective charges at the two positions. Indeed, at high expression levels, both variants exerted a dominant-negative phenotype such that an abnormal network branching pattern was seen even in the presence of endogenous ATL2/3 (Fig. 3 C). Notably, although K372 and E380 are each seen to make an additional polar contact in the postfusion dimer structure with E275 and Q376, respectively, neither charge reversal of E275 nor alanine substitution of Q376 interfered with ATL2 function (Fig. 3, A–C). Thus, the latter contacts appear dispensable. Finally, to test whether the inability of ATL2 E380R and ATL2 K372E to function might indeed be caused by their inability to form a salt bridge, the double mutant variant ATL2 K372E,E380R was constructed. We reasoned that combining the mutations in the same molecule might serve a compensatory function, restoring electrostatic attraction. Remarkably, this variant functioned indistinguishably from wild-type ATL2 in ER network branching (Fig. 3, A and B) and exhibited no dominant-negative phenotype (Fig. 3 C). Thus, the salt bridge between K372 and E380, specific to the postfusion conformer, is required for ATL2 function.
Figure 3.
The K372-E380 salt bridge is required for ATL2 function. (A) Cells transfected with the indicated HA-ATL2 variants were treated 48 h later with siRNAs against ATL2 and ATL3. 72 h after knockdown, cells were fixed and stained with antibodies against the HA epitope and viewed by confocal microscopy. (B) Quantification of the fraction of cells expressing the indicated proteins that had the unbranched ER phenotype. Values represent the means of three independent experiments ± SD. *, P < 0.0005. (C) High level expression of the indicated nonfunctional HA-ATL2 variants also confers a dominant-negative ER phenotype seen here even without ATL2/3 knockdown. Bars, 10 µm.
The K372-E380 salt bridge is not required for either GTP binding or hydrolysis
Because the K372-E380 contact is specific to the postfusion dimer conformer, we anticipated that the most upstream steps of the proposed ATL reaction cycle, namely GTP binding, formation of the prefusion dimer, and nucleotide hydrolysis, would all be normal in the single charge reversal mutants. To assess the biochemical properties of the mutant proteins, the soluble cytoplasmic domain of the relevant ATL2 variants—wild type, K372E, E380R, and the double mutant as well as the nucleotide binding–deficient K107A—were expressed, purified, and subjected to GTP hydrolysis assays.
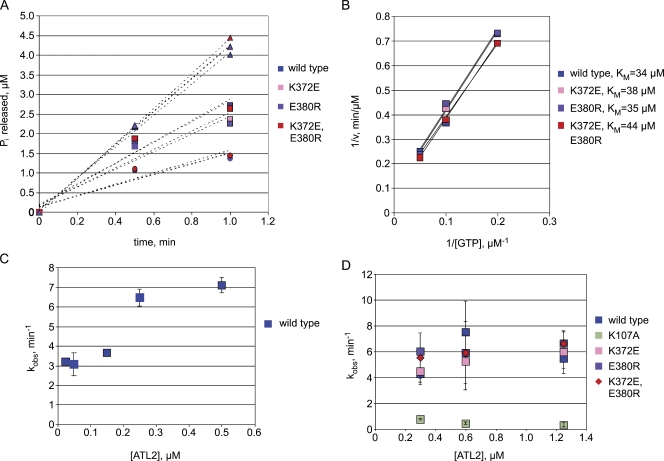
Members of the dynamin superfamily of large GTPases possess a core GTPase domain with a globular fold similar to that of Ras and other small GTPases (Prakash et al., 2000; Niemann et al., 2001), but their biochemical properties differ in significant ways. In contrast to Ras superfamily small GTPases that require guanine nucleotide exchange factors for GTP loading because of their high, subnanomolar nucleotide affinity (Neal et al., 1988), dynamin-related GTPases have a relatively low affinity for nucleotides (Song and Schmid, 2003). To assess nucleotide binding by ATL2 and how the K372-E380 salt bridge might influence it, the ability of each variant to bind and hydrolyze GTP was measured over a range of substrate concentrations (Fig. 4 A). When analyzed using a linearized form of the Michaelis–Menton equation (Fig. 4 B), wild-type ATL2 had an expected relatively high apparent Michaelis constant (KM) for GTP of 34 µM, well within the range of the 10–100-µM KM exhibited by other dynamin-related GTPases (Song and Schmid, 2003; Song et al., 2004). Under these conditions, K372E, E380R, and the double mutant ATL2 each exhibited a KM for GTP close to that of wild-type ATL2: 38, 35, and 44 µM for K372E, E380R, and the double mutant, respectively (Fig. 4, A and B). Therefore, the inability to form the K372-E380 contact appeared to have little impact on nucleotide binding.
Figure 4.
The K372-E380 salt bridge is not required for either nucleotide binding or hydrolysis. (A) 1 µM of the purified cytoplasmic domains (ATL2 1–467) of the indicated proteins were incubated with 5 µM (circles), 10 µM (squares), or 20 µM (triangles) GTP and assayed for phosphate release at the indicated times. Each point represents the means of two independent measurements. (B) A Lineweaver–Burk plot based on initial velocities from A was used to extract the KM of the indicated ATL2 proteins for GTP. (C) The indicated concentrations of wild-type ATL2 were incubated with 0.2 mM GTP followed by assaying for phosphate release. (D) 0.3, 0.6, or 1.25 µM of the indicated ATL2 variants was incubated with 0.2 mM GTP followed by an assay for phosphate release. Values represent the means of three independent measurements ± SD. v, initial velocity.
Dynamin-related GTPases are also distinguished from Ras superfamily GTPases by their relatively high intrinsic catalytic activity, which renders them independent of an external GTPase-activating protein (Song and Schmid, 2003). In the case of dynamin, self-assembly into higher order oligomers further stimulates hydrolysis by as much as 100-fold, in a manner dependent on its associated GTPase effector domain (Stowell et al., 1999; Song and Schmid, 2003). Other dynamin-related GTPases, such as GBP1, have intrinsic GTPase activity but do not undergo higher order assembly (Prakash et al., 2000). Consequently, these GTPases do not exhibit the dramatic assembly stimulated increase in activity that is observed for dynamin. Nevertheless, the basal GTPase activity of full-length GBP1 is stimulated three- to fivefold upon dimer formation, with a dimerization constant of 0.4 µM (Prakash et al., 2000; Ghosh et al., 2006). Notably, dimer-induced stimulation of GBP1 catalytic activity is observed even for truncated molecules retaining only the GTPase head domain, implying that stimulation by dimer formation requires only the head to head binding interface (Ghosh et al., 2006).
To assess the extent to which the ATL2 GTPase is stimulated by dimer formation, the ability of the wild-type ATL2 soluble domain to hydrolyze GTP was measured at saturated GTP concentrations under initial velocity conditions (Fig. S3 A) over a range of protein concentrations. As anticipated, the catalytic rate constant, kobs, was stimulated approximately twofold with an apparent dimerization constant of ∼0.2 µM (Fig. 4 C), presumably caused by enhancement of GTP binding by formation of the head to head dimer. kobs began to level off at 0.3–0.5 µM ATL2, suggesting that ATL2 was largely dimeric above this concentration. No further stimulation of the GTPase activity was observed, even at concentrations as high as 30 µM ATL2 (Fig. S3 B), consistent with a lack of higher order assembly under these conditions. Next, to determine the impact of the K372-E380 salt bridge on catalytic activity, the ability of both wild-type and mutant variants to hydrolyze GTP was measured over a range of protein concentrations at which the wild-type protein was expected to be dimeric. kobs for wild-type ATL2 was ∼6 min−1, consistent with previous measurements for ATL1 (Bian et al., 2011; Byrnes and Sondermann, 2011), and it did not vary significantly between 0.3 and 1.25 µM ATL2 (Fig. 4 D). As expected, the nucleotide binding–deficient K107A ATL2 exhibited only low activity, with a kobs of ∼0.6 min−1. Under these conditions, kobs for K372E and E380R ATL2, as well as the double mutant variant, was indistinguishable from that of the wild type. Therefore, the ability of ATL2 to bind GTP and form a head to head dimer as well as hydrolyze GTP does not appear to depend on the K372-E380 salt bridge.
The K372-E380 ionic contact is required for stable dimer formation
The undiminished ability of K372E ATL2 and E380R ATL2 to bind and hydrolyze GTP suggested that prefusion (head to head) dimer formation was normal. Formation of a GMPPNP-dependent dimer, as detected by size exclusion chromatography (SEC), has been suggested previously by others to reflect formation of the GTP-bound prefusion state (Bian et al., 2011; Byrnes and Sondermann, 2011). Therefore, we next subjected each variant to SEC analysis.
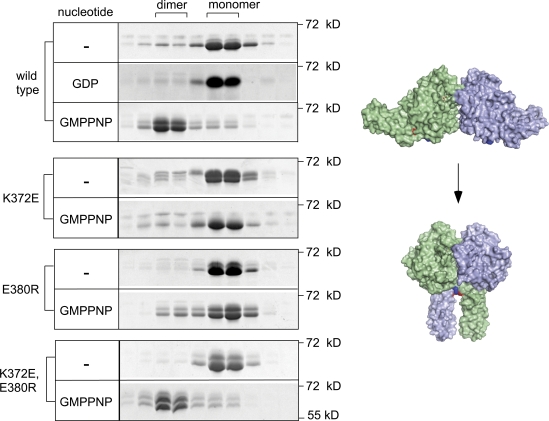
Consistent with previous studies (Bian et al., 2011; Byrnes and Sondermann, 2011), the soluble domain of wild-type ATL2 formed dimers in the presence, but not in the absence, of GMPPNP (Fig. 5). No ATL2 dimerization was observed with GDP, consistent with previous studies for ATL1 (Byrnes and Sondermann, 2011; Moss et al., 2011), though contrasting with another (Bian et al., 2011). Also as expected, stable dimer formation was diminished for the nucleotide binding–deficient ATL2 K107 (Fig. S4). Under these conditions, both ATL2 K372E and ATL2 E380R were expected to form stable dimers. To our surprise, the level of ATL2 K372E and ATL2 E380R in the dimer fractions was negligible, regardless of whether they had been incubated with GMPPNP (Fig. 5). Remarkably, however, the compensatory charge reversal mutation in ATL2 K372E,E380R fully restored GMPPNP-dependent dimer formation. Thus, in contrast to expectations, the K372-E380 salt bridge is required to form a stable GMPPNP dimer.
Figure 5.
The K372-E380 contact is required for GMPPNP-dependent stable dimer formation. The purified cytoplasmic domains of the indicated ATL2 variants (30 µM) were incubated with no nucleotide (−), 5 mM GDP, or 2 mM GMPPNP for 30 min at RT. Thereafter, samples were resolved on a Superdex 200 column. The Coomassie-stained proteins (∼60 kD) present in monomer (∼70 kD) and dimer peak (∼150 kD) positions from each column run are shown after SDS-PAGE. Also shown is a surface rendering of pre- and postfusion dimer conformers with K372 and E380 highlighted in blue and red, respectively. Models were drawn in PyMOL from Protein Data Bank no. 3QOF and 3QNU.
The soluble domain of ATL2 adopts the postfusion conformation in the GMPPNP-bound (GTP bound) state
The inability of ATL2 K372E and ATL2 E380R to form the GMPPNP-dependent solution dimer raised the possibility that the GMPPNP solution dimer actually corresponds to the postfusion, rather than prefusion conformer. The implications of this hypothesis are significant, as it would imply that the ATL2 soluble domain achieves the postfusion conformation in the GTP-bound state, not requiring nucleotide hydrolysis. To test this possibility, we used two independent means to probe the conformational state of ATL2 in the presence of various nucleotide analogues.
First, we attempted to visualize the GMPPNP-bound ATL2 solution dimer by single-particle EM of negatively stained samples. EM images of GMPPNP-bound ATL2 particles were processed, and a total of 571 individual particle images were boxed manually, band-pass filtered, and aligned with respect to their center of mass. To test whether GMPPNP-bound ATL2 adopts the prefusion or postfusion conformation, two 3D density maps from the same set of particle images were reconstructed using two different initial reference maps calculated from the atomic models of the prefusion (Protein Data Bank accession no. 3QOF) and postfusion (Protein Data Bank accession no. 3QNU) ATL1 conformers (shown in Fig. 2 A). The resulting 3D maps resembled the postfusion conformation more than the prefusion one (Fig. S5). Cross-correlation values between the final density maps and their respective initial references were 0.293 for the prefusion map and 0.425 for the postfusion map, indicating that the GMPPNP-bound ATL2 dimer more likely adopts the postfusion configuration.
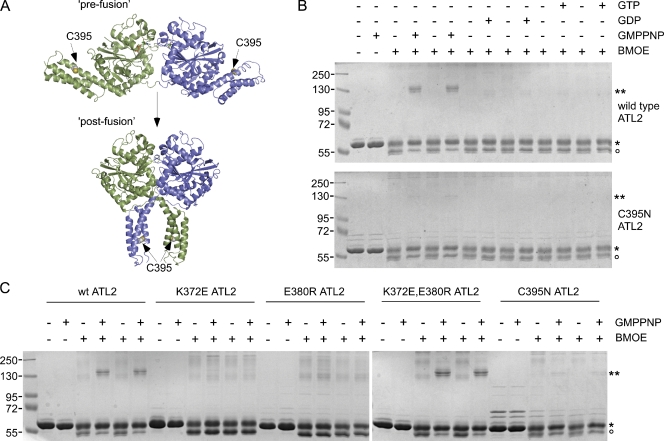
The second method was cross-linking. Bismaleimidoethane (BMOE) is a short bifunctional sulfhydryl cross-linker capable of conjugating two cysteine residues to one another if they lie in close enough (∼10 Å) proximity. ATL2 has three surface-exposed cysteines. Two are in the head (C59 and C144) and one is in the middle domain (C395). Of these, only the middle domain C395 residue is in a position to mediate cross-linking of one monomer to the other to form a covalently conjugated dimer. Importantly, C395-mediated dimer conjugation is predicted to occur in the postfusion state but not in the prefusion state (Fig. 6 A). This is because the C395 residues of the two monomers are <20 Å apart in the postfusion dimer, but they are >100 Å apart in the prefusion dimer. In contrast to C395, the other two cysteines in the head are too far apart, >50 Å, in either dimer configuration. Nevertheless, they may mediate cross-linking within the monomer, likely leading to the slightly more rapidly migrating species of the monomer seen even in the absence of nucleotide. Significantly, cross-linker–dependent dimer conjugation was observed only in the presence of GMPPNP (Fig. 6 B). Dimer formation was not observed in the absence of nucleotide or in the presence of GDP or GTP, indicating that the postfusion conformation is specific to the GMPPNP-bound (or GTP bound) state. Presumably, postfusion dimer formation also occurs transiently with bound GTP, but subsequent hydrolysis returns it to the monomer state. As a control to confirm that the GMPPNP dimer is cross-linked through the middle domain C395 residue, an ATL2 variant lacking the C395 sulfhydryl (ATL2 C395N) was tested. As predicted, it failed to form dimers either in the presence or absence of GMPPNP. To further validate our assay for postfusion dimer formation, ATL2 K372E and ATL2 E380R were each subjected to the same assay. As predicted by their behavior in SEC, neither of the single mutant variants formed the GMPPNP-dependent cross-linked dimer, whereas the compensatory double mutation restored dimer formation (Fig. 6 C). These results argue that the soluble domain of ATL2 adopts the postfusion conformation exclusively in the GMPPNP-bound (or GTP bound) state. Moreover, although nucleotide hydrolysis is dispensable for achieving the postfusion state, at least for the soluble domain, formation of the K372-E380 salt bridge is not.
Figure 6.
GTP hydrolysis is not required for the prefusion to postfusion conformational change. (A) Location of the C395 residue in ATL2 used to report on the postfusion conformation. The C395 side chain in ATL2 is highlighted as spheres in both pre- and postfusion ATL1 dimer structures rendered as detailed in Fig. 2. (B) The purified cytoplasmic domain of wild-type or C395N ATL2 (20 µM) was incubated in the presence or absence of the indicated nucleotides for 30 min at RT. Thereafter, samples were diluted, further incubated with or without BMOE for 1 h at RT, resolved by SDS-PAGE, and stained with Coomassie blue. The positions of non–cross-linked monomer and covalently cross-linked dimer ATL2 are indicated by single and double asterisks, respectively. The open circle indicates the position of ATL2 likely to have cysteine modifications not leading to dimer formation. (C) The purified cytoplasmic domains of the indicated ATL2 variants were subjected to the assay as described in B. Molecular masses are given in kilodaltons.
Discussion
Our findings reveal an intramolecular salt bridge required for the ER network branching function of ATL2. The importance of the K372-E380 ionic interaction for ATL2 function is likely caused by its stabilization of the postfusion dimer conformation. This might seem surprising given that the charge interaction occurs intramolecularly, within each monomer subunit of the dimer. Based on the position of the salt bridge in the context of the postfusion dimer, we speculate that the ionic contact constrains the linker in a kinked conformation relative to the head and middle domains. In so doing, it may serve to position M374 and L375, two intervening nonpolar residues that need to pack extensively against the opposing head, to form the postfusion conformation. In the absence of the salt bridge, the linker may be rendered too flexible, reducing the ability of M374 and L375 to pack effectively.
Previous observations are consistent with the K372-E380 salt bridge being required for ATL-catalyzed membrane fusion. A Drosophila ATL variant bearing a mutation equivalent to E380R is significantly reduced in its ability to catalyze liposome fusion (Bian et al., 2011), though neither its GTPase activity nor its ability to dimerize has been reported. In addition, a Drosophila variant with a mutation equivalent to K372E lacks fusion activity (Bian et al., 2011). Thus, the opposing charge carried by the two residues appears functionally important. Whether the salt bridge, per se, is required for fusion activity remains to be determined. But based on the cumulative evidence, it seems likely that the ability of the two residues to engage in a salt bridge will be required for stabilizing the postfusion dimer and hence for membrane fusion, even in the more distantly related organism.
Our finding that the ATL2 soluble domain can achieve the postfusion conformation in the GMPPNP (or GTP)-bound state without nucleotide hydrolysis contrasts with the conclusions from an earlier study. In that study, biochemical analyses performed on the ATL1 soluble domain were used to arrive at the opposite conclusion: that ATL1 adopts the prefusion conformation in the GTP-bound state and the postfusion conformation in the GDP-bound state (Bian et al., 2011). This has contributed, at least in part, to the current model for ATL proposing that the prefusion to postfusion conformational change is directly coupled to GTP hydrolysis and Pi release. The reasons for the conflicting results and conclusions remain unclear, and further work will be needed to resolve the discrepancy. Nevertheless, the results presented herein clearly indicate that GTP binding, in the absence of hydrolysis, is sufficient to induce the prefusion to postfusion conformational change, at least for the ATL2 soluble domain.
As shown previously for GBP1 (Prakash et al., 2000; Ghosh et al., 2006), we observed a modest concentration-dependent stimulation of the ATL2 soluble domain GTPase activity in the submicromolar range. By analogy to GBP1, this is likely caused by enhancement of GTP binding by formation of the head to head dimer. As the K372-E380 salt bridge is not present in the crystal structure of the initial nucleotide-bound prefusion dimer (Bian et al., 2011; Byrnes and Sondermann, 2011), mutations hindering formation of the salt bridge would not be predicted to impair the initial dimer-induced stimulation of GTP binding and hydrolysis. As expected, neither the K372E nor E380R mutant variants were diminished in their ability to bind or hydrolyze GTP. This result underscores the specificity of the defect in the K372E and E380R mutant ATL2 variants: they are likely able to form the initial head to head contact and bind GTP as well as the wild-type protein, yet they fail to transition to the postfusion conformation.
Our observation that GTP hydrolysis is neither required for, nor dependent on, formation of the postfusion conformation is somewhat surprising. It will be important to determine whether this behavior of the soluble domain reflects that of the full-length molecule in membranes. Finally, the interpretation of our findings with respect to the role of GTP hydrolysis in the ATL fusion mechanism also needs to be tempered by the uncertainty of whether the behavior of the ATL2 soluble domain, observed herein, reflects the behavior of the full-length, membrane-anchored protein. A possibility deserving of consideration is that membrane-anchored ATL is more conformationally constrained than its soluble counterpart. That is, whereas the soluble domain is sufficiently flexible to adopt the postfusion conformation when restricted to the GTP-bound state, membrane-anchored ATL may require an additional input of energy, provided perhaps by the hydrolysis of GTP within the prefusion dimer. A requirement for nucleotide hydrolysis for formation of the membrane-anchored postfusion dimer would serve to explain the requirement for GTP hydrolysis in ATL-catalyzed liposome fusion (Orso et al., 2009). On the other hand, this scenario depends on the membrane-anchored prefusion dimer being compatible with the GTP-bound state and the postfusion dimer being compatible with the GDP-bound state. This seems counterintuitive, given that the postfusion conformation of the soluble domain is clearly compatible with the GMPPNP (or GTP)-bound state but most likely not with the GDP-bound state (Fig. 6 B).
An alternative possibility is that the behavior of the soluble domain does indeed reflect the behavior of the membrane-anchored full-length protein. In the latter scenario, we would propose that nucleotide hydrolysis is not directly coupled to the prefusion to postfusion conformational change but rather that the energy released from hydrolysis is harnessed to drive another discrete step in the ATL reaction cycle. Whatever the identity of that hydrolysis-dependent step, it should explain the observed requirement for GTP hydrolysis in the in vitro fusion assay (Orso et al., 2009). Further work to determine the conformational states of GTP- and GDP-bound membrane-anchored ATL2, as well as the impact of the salt bridge identified herein on the catalytic properties of full-length and membrane-anchored ATL2, promises to more clearly delineate the role of nucleotide hydrolysis in the ATL2 mechanism.
Materials and methods
Cells, constructs, antibodies, and reagents
All experiments were conducted on HeLa cells maintained at 37°C in a 5% CO2 incubator in MEM (Sigma-Aldrich) and 10% FBS (Atlanta Biologicals) with 1% penicillin/streptomycin (Thermo Fisher Scientific). The N-terminally HA-tagged ATL2 isoform 2 construct was contributed by C. Blackstone (National Institutes of Health, Bethesda, MD). All ATL2 variant constructs were generated by PCR-mediated site-directed mutagenesis (QuikChange; Agilent Technologies). The siRNA-immune HA-ATL2 construct was generated using QuikChange to replace the 21 nucleotides targeted by the ATL2 siRNA 5′-GGAGCTATCCTTATGAACATTCATA-3′ with 5′-GGAGCTATCCGTACGAACACTCATA-3′. N-terminally Myc-tagged PRA2 and DP1 constructs were generated by PCR amplification of a HeLa cDNA library and cloning into the pCS2 Myc vector at the EcoRI and XbaI sites and the XbaI site, respectively. All constructs used herein were fully verified by sequencing (Genewiz). An mAb used to detect protein disulfide isomerase (Abcam) and the HA epitope (Sigma-Aldrich) were purchased, and the 9E10 mAb was used to detect the Myc epitope. The rhodamine anti–mouse secondary antibody was also purchased (Invitrogen). GTP, GDP, and GMPPNP were purchased (Sigma-Aldrich), reconstituted to 100 mM stocks in 10 mM Tris, pH 8.0, and 1 mM EDTA, and stored at −80°C.
Knockdown replacement assay
Cells plated on 60-mm culture dishes were transfected with ∼5 µg of the indicated HA-ATL2 replacement constructs using transfection reagent (jetPEI; VWR). Myc-tagged PRA2 and Myc-tagged DP1, both ER-localized proteins, served as negative controls. Neither affected either the percentage of cells showing the unbranched ER phenotype or the extent of loss of network branching relative to siRNA treatment alone. 24 h after DNA transfection, cells were trypsinized and replated onto 12-mm glass coverslips in a 24-well plate. siRNA treatment targeting both ATL2 and ATL3 was performed the next day using transfection reagent (Oligofectamine; Invitrogen) according to manufacturer’s recommendations. The ATL2 (#1) and ATL3 (#2) siRNAs were identical in sequence to those previously published (Rismanchi et al., 2008). 72 h after knockdown, cells were fixed in ice-cold methanol and processed for immunofluorescence. In brief, primary (1 h at RT) and secondary (30 min at RT) antibody incubations were performed in a blocking solution consisting of PBS + 2.5% calf serum + 0.1% Triton X-100, and washes were with 5× 1 ml PBS. All images were obtained using a spinning-disk confocal scanhead (Yokagawa; PerkinElmer) mounted on a microscope (Axiovert 200; Carl Zeiss) with a 100× 1.4 NA objective (Carl Zeiss) and acquired using a 12-bit camera (ORCA-ER; Hamamatsu Photonics). Maximal value projections of sections at 0.2-µm spacing (approximately six per cell) were acquired using Imaging Suite software (PerkinElmer) and imported as 8-bit images into Photoshop (Adobe). Quantification of functional replacement was performed manually on a wide-field fluorescence microscope (Axioplan; Carl Zeiss) with a 40× 1.4 NA objective. Images were acquired using a 12-bit camera (ORCA-ER) and QED software (Media Cybernetics). For quantification of functional replacement, the fraction of cells expressing the indicated HA-ATL2 that showed a loss of ER network branching (among ≥100 cells per experiment) was counted. Some of the HA-ATL2 variants, when expressed at high levels, exhibited a dominant-negative ER phenotype (even without ATL knockdown). For these variants, a threshold level of HA-ATL2 immunofluorescence below which expression of the replacement construct alone did not confer an ER phenotype was determined. Quantification of functional replacement was then restricted to those cells expressing the HA-ATL2 variant below the predetermined threshold.
Protein expression and purification
The 6His-tagged cytoplasmic domain of ATL2 was generated using PCR amplification of nucleotides encoding amino acids 1–467 from HA-ATL2 and cloned into the pRSETB vector at NheI and EcoRI sites. Variants were generated using QuikChange mutagenesis and sequence verified. Protein expression was induced with 0.5 mM IPTG in BL21(DE3)pLysS cells at 23°C for 16 h, and purification used standard protocols for purification of 6His-tagged proteins on Ni+2 agarose beads (QIAGEN). Proteins, eluted in 50 mM Tris, pH 8.0, 250 mM imidazole, 100 mM NaCl, 5 mM MgCl2, and 10% glycerol, were typically 8–24 mg/ml, >95% pure, flash frozen in liquid N2, and stored at −80°C.
GTPase assay
Purified 6His-ATL2 variant proteins, dialyzed into SEC and precleared by centrifugation in a rotor (TLA-100; Beckman Coulter) at 100,000 rpm for 15 min, were diluted to various concentrations in the same buffer. GTP at the indicated concentrations was added to the protein and incubated at 37°C for varying times. After quenching with 5 mM EDTA, 160 µl of each reaction was then added to 40 µl malachite green phosphate assay reagent (Accurate Chemical & Scientific Corp.) in a 96-well transparent flat-bottomed dish (Costar; Thermo Fisher Scientific) and developed for 10 min at 25°C before measuring the absorbance at 650 nm. Phosphate release was calculated using a standard curve according to the manufacturer’s instructions.
KM and kobs determinations
1 µM dialyzed and precleared 6His-ATL2 variant proteins were incubated with 5, 10, or 20 µM GTP at 37°C for varying times, quenched, and assayed for phosphate release (see previous paragraph). Initial velocities for each ATL2 variant were plotted against substrate concentration on a double reciprocal scatter plot, and the KM for each ATL2 variant was extracted from the x intercept of its best-fit line (R2 = 0.99 − 1; x intercept = −1/KM). For kobs determinations at differing protein concentrations, varying concentrations of 6His-ATL2 variants were incubated with 0.2 mM GTP (determined to be saturating for ≤2 µM ATL2) for 5 min at 37°C. During this time, product formation was predetermined to be linear with time. Samples were quenched and assayed for phosphate release (see previous paragraph). When assaying GTPase activity at high concentrations (3 and 30 µM ATL2), GTP was added at 1 mM in the reaction and incubated for 1 min at 37°C. Thereafter, samples were diluted 10-fold before assaying for phosphate release.
SEC
Purified 6His-ATL2 variant proteins, dialyzed into SEC buffer + 0.5 mM DTT and precleared by centrifugation in a rotor (TLA-100) at 100,000 rpm for 15 min, were diluted to 10 or 30 µM and incubated with or without 2 mM GMPPNP for 30 min at RT. 100 µl of each sample was then injected onto a fast protein liquid chromatography column (Superdex 200; GE Healthcare) preequilibrated in SEC buffer + 0.5 mM DTT and separated at a flow rate of 0.5 ml/min at 4°C. 0.5-ml fractions within the included volume of 24 ml were collected, precipitated with TCA using 0.5% Triton X-100 as a carrier, resuspended in reducing sample buffer, resolved by SDS-PAGE, and stained using Coomassie blue. Where indicated, wild-type ATL2 was incubated with 5 mM GDP for 30 min at RT and resolved on the same column, except that 1 mM GDP was also included in the column buffer.
Cross-linking
Purified 6His-ATL2 variant proteins were dialyzed into SEC buffer, pH 7.0, at 4°C and precleared by centrifugation in a rotor (TLA-100) at 100,000 rpm for 15 min. 20 µM of each protein was incubated at RT in SEC buffer, pH 7.0, in the absence or presence of 2 mM GMPPNP, GDP, or GTP. After 30 min at RT, the reaction was diluted fivefold into SEC (to 4 µM ATL2) in the absence or presence of 12 µM BMOE (Thermo Fisher Scientific) and incubated for 1 h at RT. Samples were then quenched with 20 mM DTT for 15 min, mixed with reducing sample buffer, and resolved by SDS-PAGE.
EM
20 µM purified 6His-ATL2 in buffer containing 25 mM Tris-HCl, pH 7.5. 100 mM NaCl, 5 mM MgCl2, 2 mM EGTA, 5% glycerol, and 0.5 mM DTT was diluted twofold in the same buffer without glycerol in the presence of 1 mM GMPPNP. After 30 min at RT, the reaction mixture was further diluted 30-fold into the same buffer and immediately applied onto glow-charged thin carbon foil grids, blotted with a filter paper, and stained with a 2% solution of uranyl acetate in water. The grids were examined at 120 kV with an electron microscope (Tecnai 12; FEI). Images were recorded with a 2,000 × 2,000 charge-coupled device camera (UltraScanT 1000; Gatan, Inc.) at a nominal magnification of 52,000.
Image processing and model docking
EM images were processed using the EMAN image analysis software (National Center for Macromolecular Imaging; Ludtke et al., 1999; Tang et al., 2007). Individual particles were boxed manually with 80 × 80 pixels (2.17 Å/pixel), normalized, and combined to yield one raw image stack file. A total of 571 individual particle images were collected, band-pass filtered, and aligned with respect to their center of mass. To test the likelihood of the conformations that the ATL GTPase could adopt, two simulated 3D density maps were computed with Chimera (version 3; University of California, San Francisco) from the atomic models of two conformers (shown in Fig.6 A), prefusion (Protein Data Bank accession no. 3QOF) and postfusion (Protein Data Bank accession no. 3QNU). These two density maps were then used as initial references for the reference-based projection matching of the particle images followed by the reconstruction of particle images in EMAN2. The iterative refinement cycles were ended when the calculated Fourier shell correlation between the 3D models generated in two consecutive iterations showed no further improvement. This indicated that the 3D reconstruction was converging to a stable optimum, and the final 3D density maps were calculated. For model docking, the atomic models of prefusion and postfusion were fitted into the reconstructed EM density map using the feature Fit model in map implemented in Chimera. Cross-correlation values between the final density maps and the simulated 3D density maps from two conformers were calculated. The value was 0.293 for the prefusion conformer and 0.425 for the postfusion conformer, with 55% volume included.
Online supplemental material
Fig. S1 shows that RNAi-mediated depletion of both ATL2 and ATL3 leads to a reduction of ER network branch points in HeLa cells. Fig. S2 shows that the middle domain ATL2 residues required for function lie near the GTPase head. Fig. S3 shows that the linearity of GTPase assay and ATL2 GTPase activity are not further stimulated at high ATL2 concentrations. Fig. S4 shows that stable formation of ATL2 soluble domain dimers depends on GTP binding. Fig. S5 shows an EM analysis of the GMPPNP-bound ATL2 solution dimer, indicating that the soluble domain of ATL2 is more likely to adopt the postfusion dimer conformation when it is bound to GMPPNP. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201105006/DC1.
Acknowledgments
The authors thank Craig Blackstone for his kind contribution of the HA-ATL2 isoform 2 construct, Mark Macbeth for help with PyMOL, and Adam Linstedt for helpful comments on the manuscript.
This work was supported by an American Cancer Society Research Scholar grant (07-041-01-CSM) and National Institutes of Health Exploratory/Development Research grant (1R21DK088208) to T.H. Lee and a Richard King Mellon Foundation fellowship to J. Morin-Leisk.
Footnotes
Abbreviations used in this paper:
- ATL
- atlastin
- BMOE
- bismaleimidoethane
- GMPPNP
- guanosine 5′-[β,γ-imido]triphosphate
- SEC
- size exclusion chromatography
References
- Baumann O., Walz B. 2001. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205:149–214 10.1016/S0074-7696(01)05004-5 [DOI] [PubMed] [Google Scholar]
- Bian X., Klemm R.W., Liu T.Y., Zhang M., Sun S., Sui X., Liu X., Rapoport T.A., Hu J. 2011. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 108:3976–3981 10.1073/pnas.1101643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes L.J., Sondermann H. 2011. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. USA. 108:2216–2221 10.1073/pnas.1012792108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie J.S., Acharya S., Leonard M., Schmid S.L., Dyda F. 2010. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 465:435–440 10.1038/nature09032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O., Praefcke G.J. 2011. Structural insights into membrane fusion at the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 108:2175–2176 10.1073/pnas.1019194108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Praefcke G.J., Renault L., Wittinghofer A., Herrmann C. 2006. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 440:101–104 10.1038/nature04510 [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., Blackstone C. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 138:549–561 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Ferguson M., Chen L.B. 1989. Construction of the endoplasmic reticulum. J. Cell Biol. 109:2045–2055 10.1083/jcb.109.5.2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S.J., Baldwin P.R., Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- Moss T.J., Andreazza C., Verma A., Daga A., McNew J.A. 2011. Membrane fusion by the GTPase atlastin requires a conserved C-terminal cytoplasmic tail and dimerization through the middle domain. Proc. Natl. Acad. Sci. USA. 108:11133–11138 10.1073/pnas.1105056108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal S.E., Eccleston J.F., Hall A., Webb M.R. 1988. Kinetic analysis of the hydrolysis of GTP by p21N-ras. The basal GTPase mechanism. J. Biol. Chem. 263:19718–19722 [PubMed] [Google Scholar]
- Niemann H.H., Knetsch M.L., Scherer A., Manstein D.J., Kull F.J. 2001. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 20:5813–5821 10.1093/emboj/20.21.5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G., Pendin D., Liu S., Tosetto J., Moss T.J., Faust J.E., Micaroni M., Egorova A., Martinuzzi A., McNew J.A., Daga A. 2009. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 460:978–983 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- Pieper U., Webb B.M., Barkan D.T., Schneidman-Duhovny D., Schlessinger A., Braberg H., Yang Z., Meng E.C., Pettersen E.F., Huang C.C., et al. 2011. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 39(Suppl. 1):D465–D474 10.1093/nar/gkq1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke G.J., McMahon H.T. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5:133–147 10.1038/nrm1313 [DOI] [PubMed] [Google Scholar]
- Prakash B., Praefcke G.J., Renault L., Wittinghofer A., Herrmann C. 2000. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 403:567–571 10.1038/35000617 [DOI] [PubMed] [Google Scholar]
- Rismanchi N., Soderblom C., Stadler J., Zhu P.P., Blackstone C. 2008. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 17:1591–1604 10.1093/hmg/ddn046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B.D., Schmid S.L. 2003. A molecular motor or a regulator? Dynamin’s in a class of its own. Biochemistry. 42:1369–1376 10.1021/bi027062h [DOI] [PubMed] [Google Scholar]
- Song B.D., Leonard M., Schmid S.L. 2004. Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin-mediated endocytosis. J. Biol. Chem. 279:40431–40436 10.1074/jbc.M407007200 [DOI] [PubMed] [Google Scholar]
- Stowell M.H., Marks B., Wigge P., McMahon H.T. 1999. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat. Cell Biol. 1:27–32 10.1038/8997 [DOI] [PubMed] [Google Scholar]
- Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Zhu P.P., Patterson A., Lavoie B., Stadler J., Shoeb M., Patel R., Blackstone C. 2003. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J. Biol. Chem. 278:49063–49071 10.1074/jbc.M306702200 [DOI] [PubMed] [Google Scholar]