Phosphorylation of the kinetochore component Nsk1 by Cdk1 antagonizes its localization to and function at the kinetochore and spindle during early mitosis.
Abstract
Cdk1 controls many aspects of mitotic chromosome behavior and spindle microtubule (MT) dynamics to ensure accurate chromosome segregation. In this paper, we characterize a new kinetochore substrate of fission yeast Cdk1, Nsk1, which promotes proper kinetochore–MT (k-MT) interactions and chromosome movements in a phosphoregulated manner. Cdk1 phosphorylation of Nsk1 antagonizes Nsk1 kinetochore and spindle localization during early mitosis. A nonphosphorylatable Nsk1 mutant binds prematurely to kinetochores and spindle, cementing improper k-MT attachments and leading to high rates of lagging chromosomes that missegregate. Accordingly, cells lacking nsk1 exhibit synthetic growth defects with mutations that disturb MT dynamics and/or kinetochore structure, and lack of proper phosphoregulation leads to even more severe defects. Intriguingly, Nsk1 is stabilized by binding directly to the dynein light chain Dlc1 independently of the dynein motor, and Nsk1–Dlc1 forms chainlike structures in vitro. Our findings establish new roles for Cdk1 and the Nsk1–Dlc1 complex in regulating the k-MT interface and chromosome segregation.
Introduction
The forces that drive chromosome motions during mitosis derive from the persistent interaction of dynamic microtubule (MT) plus-ends with kinetochores (Rieder and Salmon, 1998; Joglekar et al., 2010). Whereas stable kinetochore–MT (k-MT) attachments require the functions of the evolutionarily conserved Ndc80, KNL-1, and Mis12 complexes (Santaguida and Musacchio, 2009), other proteins act on k-MT plus-ends to regulate their stability and/or dynamics. For example, MT plus-end–tracking proteins, such as EB1 and CLASP, and members of the kinesin-8 family impact k-MT dynamics positively and negatively, respectively, and thereby influence the overall positioning of chromosomes within the mitotic spindle by defining their motile properties (Walczak et al., 2010).
Given the dynamic nature of k-MT interactions, it follows that k-MT regulators must be carefully controlled through posttranslational modification. In particular, Aurora B kinase, the catalytic subunit of the chromosomal passenger complex (CPC), is crucial for modulating the activity of kinetochore proteins to influence the stability of k-MT attachments (Lampson and Cheeseman, 2011). Other mitotic kinases, including Cdk1 and the Polo-like kinase 1, have also been implicated in regulating k-MT interactions, but few relevant kinetochore substrates are known (Petronczki et al., 2008; Ma and Poon, 2011).
Results and discussion
Nsk1 is a novel kinetochore and spindle protein
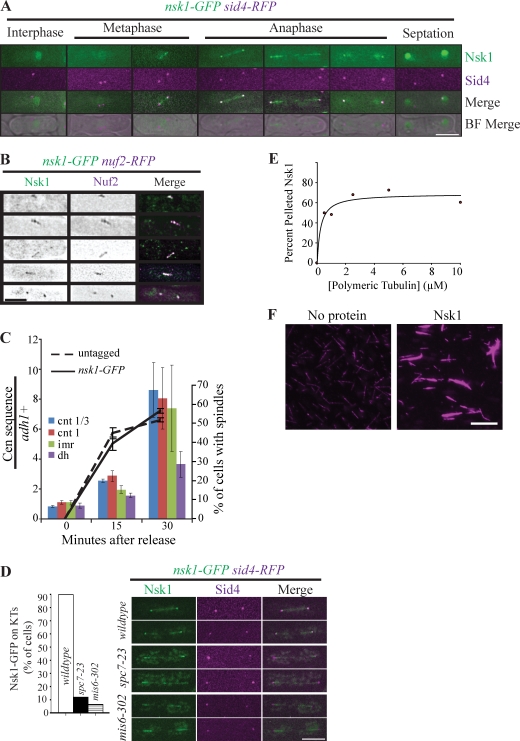
Cdc14 phosphatases antagonize Cdk1 in yeasts (Stegmeier and Amon, 2004; Mocciaro and Schiebel, 2010). To identify new Cdk1 substrates relevant to chromosome dynamics, tandem affinity purification (TAP) of a substrate-trapping mutant of the fission yeast Cdc14 phosphatase, Clp1, was performed from mitotic cells, and associated proteins were identified by 2D liquid chromatography mass spectrometry (MS). A top hit was an uncharacterized protein (Fig. S1 A) that we named Nsk1 (nucleolus spindle kinetochore 1) to reflect its localization pattern. This pattern was determined by imaging Nsk1 that had been tagged endogenously with GFP. During interphase, Nsk1-GFP was nucleolar (Fig. S1 B). During prometaphase/metaphase, Nsk1-GFP distributed throughout the nucleus but was also observed at puncta between spindle pole bodies (SPBs; Fig. 1 A) that colocalized with Nuf2-RFP, an outer kinetochore component (Fig. 1 B). During anaphase, Nsk1-GFP increased at kinetochores and also decorated the spindle, although it was excluded from the spindle midzone (Fig. 1, A and B).
Figure 1.
Nsk1 is a kinetochore-localized MT-binding protein. (A) Live-cell images of nsk1-GFP sid4-RFP. BF, brightfield. (B) Nsk1-GFP Nuf2-RFP localization in fixed cells. Fixation eliminated diffuse nucleoplasmic Nsk1, allowing kinetochore detection in early mitosis. The left two columns show inverted images, whereas the right column shows merged images. (C) Anti-GFP ChIPs were performed on nsk1-GFP and untagged cells synchronized by block and release of nda3-KM311. Real-time PCR was used to analyze association of anti-GFP immunoprecipitates with centromeric loci and with the euchromatic adh1+ control at the times indicated. Data represent the mean relative enrichment of centromeric sites over adh1+ in Nsk1-GFP immunoprecipitate samples, relative to the input material, and normalized to the immunoprecipitates of untagged strains. Error bars represent SEM (n = 4). Culture synchronicity was monitored by antitubulin staining of cells processed in parallel. The mean number of spindle-containing cells (±SEM) is shown. cnt, central core; dh, outer repeat; imr, innermost repeat. (D) Localization of Nsk1-GFP to kinetochores (KTs) was measured in ≥48 mitotic wild-type, spc7-23, or mis6-302 cells after shifting to 36°C for 3.5 h in a representative experiment of two. Representative images of fixed cells for each strain are shown. (E) Recombinant MBP-Nsk1 was incubated with paclitaxel-stabilized MTs and ultracentrifuged. The percentage of Nsk1 in pellet fractions is shown. A representative experiment of three is shown. (F) Nsk1 bundles MTs in vitro. Rhodamine-labeled GMP-CPP MTs were incubated with no protein or recombinant MBP-Nsk1 for 15 min and imaged by fluorescence microscopy. Bars: (A, B, and D) 5 µm; (F) 10 µm.
We verified the kinetochore localization of Nsk1 using chromatin immunoprecipitation (ChIP). Anti–Nsk1-GFP immunoprecipitates enriched centromeric sequences, with preference for the central core and innermost repeats over a segment of the outer repeats, relative to a euchromatic control (adh1+) as cells released from a prometaphase arrest imposed by the conditional β-tubulin mutant nda3-KM311 (Fig. 1 C). Nsk1 localization to kinetochores depended on the integrity of the outer kinetochore because it was lost from spindle poles, where kinetochores are located during anaphase, in mutants of spc7 and mis6 (Fig. 1 D; Santaguida and Musacchio, 2009). Because Nsk1 also localized to spindles, we tested whether it bound MTs using copelleting and microscopy assays. Nsk1 bound MTs with an apparent Kd of 0.23 µM (Fig. 1 E) and also bundled MTs (Fig. 1 F), suggesting that Nsk1 may localize to the spindle via a direct interaction with MTs.
Nsk1 promotes chromosome alignment and proper chromosome segregation
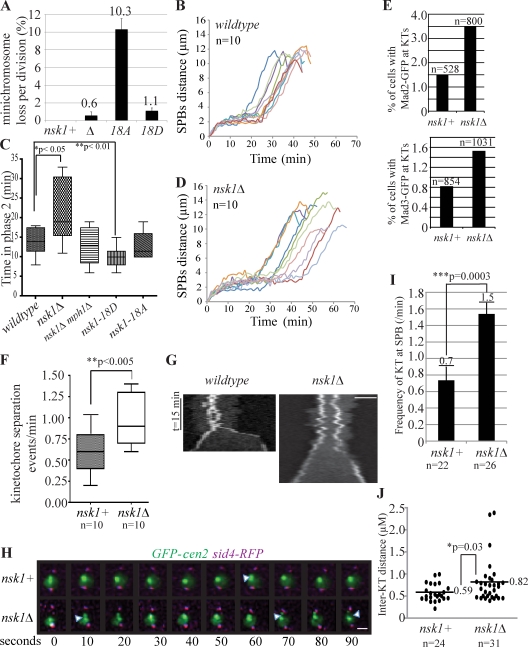
To determine the mitotic function of Nsk1, we examined cells lacking it. nsk1Δ cells were viable over a range of temperatures (Fig. S1 C), and their length at division was normal, indicating that nsk1Δ had generally unperturbed cell cycle control (Fig. S1 D). However, consistent with Nsk1 acting at kinetochores and spindle during mitosis, nsk1Δ cells exhibited mild sensitivity to thiabendazole (TBZ), an MT-destabilizing agent (Fig. S1 E) that was exacerbated by the deletion of Alp7/Mia1, a factor that promotes spindle integrity (Fig. S1 F; Oliferenko and Balasubramanian, 2002; Sato et al., 2004). Reciprocally, nsk1Δ suppressed the TBZ resistance of the klp5/kinesin-8 deletion, which hyperstabilizes MTs (Fig. S1 G; West et al., 2001). nsk1Δ also showed strong negative genetic interactions with a variety of mutations affecting kinetochore proteins (Fig. S1 H) and lost minichromosomes at an elevated rate (0.6 vs. 0% for wild type; Fig. 2 A), comparable with loss rates of mutants affecting centromeric gene silencing and DNA repair (Ekwall et al., 1996; Nakagawa et al., 2002; Greenall et al., 2006).
Figure 2.
Nsk1 is required for proper spindle dynamics and chromosome segregation. (A) Minichromosome loss rate was determined in the presence or absence of nsk1 mutations with SD from the mean of at least three experiments shown. (B–D) Spindle dynamics of the indicated strains were characterized by time-lapse microscopy. Spindle length (SPB to SPB, marked by Sid4-RFP) was measured at 1-min intervals. In B and D, each line represents data collected from an individual cell. In C, phase 2 duration data were extracted from spindle dynamics experiments as in B and D. (E) Asynchronous populations of the indicated strains were examined for the presence of Mad2-GFP (top)– or Mad3-GFP (bottom)–positive kinetochores (KTs) in the total number of cells indicated from more than two occasions of growth. (F–J) GFP-marked Cen2 and Sid4-RFP were imaged in mitotic nsk1+ and nsk1Δ cells at 10-s intervals. (F) In the 5 min before anaphase onset, the frequency of outward Cen2-GFP dot movements was quantified. (G) Kymographs of GFP-marked Cen2 in representative nsk1+ and nsk1Δ mitotic cells. Diffuse nucleoplasmic GFP signal shows the boundary of the nucleus. Bar, 2 µm. (H) Images from representative videos are shown. Arrowheads indicate kinetochore separation events. Bar, 1 µm. (I) Frequency at which kinetochores contacted an SPB during metaphase in the videos described above. Error bars represent SEM. (J) The interkinetochore distance before anaphase onset was also measured from the videos described above, and the maximum separation in each cell is provided. For all panels, n denotes the number of cells analyzed. Statistically significant differences were identified by two-tailed Student’s t tests with p-values indicated.
To understand why chromosomes missegregate in nsk1Δ, we used time-lapse microscopy to follow spindle dynamics, which in fission yeast occur in three phases (Hagan, 1998; Nabeshima et al., 1998). In phase 1, the spindle assembles. During phase 2, spindle length is constant, and, in phase 3, the pole-to-pole distance increases as anaphase B ensues. nsk1Δ cells spent significantly longer than wild-type cells in phase 2 (Fig. 2, B–D), which corresponds primarily to late prometaphase or metaphase.
A phase 2 delay can be caused by defects in kinetochore capture by spindle MTs that trigger activation of the spindle assembly checkpoint (SAC; Musacchio and Salmon, 2007). We did observe an elevated percentage of Mad2- and Mad3-GFP–positive kinetochores in nsk1Δ relative to wild type (Fig. 2 E), and phase 2 timing was similar to wild type in SAC-deficient nsk1Δ mphΔ cells (Figs. 2 C and S1 I). Thus, the phase 2 delay in nsk1Δ cells is a result of SAC activity. However, SAC mutants did not affect nsk1Δ cell growth significantly (Fig. S1 J), as would be expected if there were appreciable defects in kinetochore capture or biorientation. This suggested that nsk1Δ cells had defective k-MT interactions after biorientation. Interestingly, kinesin-8 klp5/6 mutants also display a SAC-dependent phase 2 delay and are viable with mad2Δ highlighting a commonality between Nsk1 and another factor that influences k-MTs after biorientation (Garcia et al., 2002).
Next, we tracked a single centromere (Cen2) at 10-s intervals to examine chromosome movements. In wild-type cells, sister kinetochores seldom separated, and their movements were coordinated. In contrast, nsk1Δ sister kinetochores moved apart from each other (Fig. 2, F–H) and also collided with SPBs (Fig. 2 I) more frequently. Accordingly, the mean interkinetochore distance in nsk1Δ cells was increased compared with wild type (Fig. 2 J), although spindle length was not (Fig. 2, B and D). These data suggest that k-MT plus-ends undergo catastrophe at higher frequencies in nsk1Δ cells, leading to defects in chromosome movements and segregation.
Nsk1 is a phosphoprotein regulated by Cdk1 and Clp1
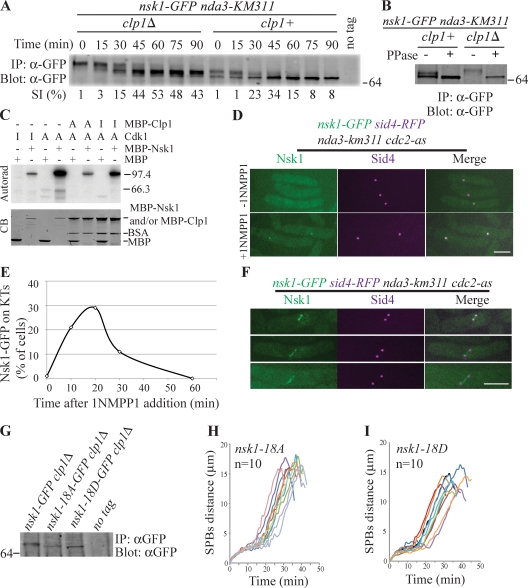
Given that Nsk1 was identified as a potential Cdk1 substrate, we tested whether Nsk1 was phosphorylated during mitosis to affect its function. Nsk1-GFP was phosphorylated at a prometaphase arrest and then dephosphorylated as cells progressed through mitosis (Fig. 3, A and B). In clp1Δ, Nsk1 was hyperphosphorylated in prometaphase-arrested cells, which is consistent with Nsk1 being a Clp1 substrate during early mitosis (Fig. 3, A and B). However, Nsk1 was partially dephosphorylated in clp1Δ cells progressing through mitosis (Fig. 3 A), suggesting that another phosphatase contributes to Nsk1 dephosphorylation at these stages. Next, we tested Nsk1 as a target for Cdk1 and Clp1 in vitro. Cdk1, but not catalytically inactive Cdk1, efficiently phosphorylated recombinant maltose-binding protein (MBP)–Nsk1 but not MBP (Figs. 3 C and S1 K). After washing out Cdk1, purified MBP-Clp1 but not catalytically inactive MBP-Clp1-C286S dephosphorylated Nsk1 (Fig. 3 C). Thus, Nsk1 is a substrate of both Cdk1 and Clp1.
Figure 3.
Nsk1 is a Cdk1 and Clp1 substrate. (A) Nsk1-GFP was immunoprecipitated (IP) from clp1+ or clp1Δ cells collected from an nda3-KM311 block and release at the indicated times and detected by immunoblotting. SI, septation index. (B) Nsk1-GFP was immunoprecipitated from clp1+ or clp1Δ cells arrested in prometaphase with nda3-KM311, and immunoprecipitates were treated (+) or were not treated (−) with λ-phosphatase (PPase) before immunoblotting. (C) Amylose resin–bound MBP or MBP-Nsk1 was incubated with active (A) or catalytically inactive (I) recombinant Cdk1-Cdc13 and γ-[32P]ATP. Four of the reactions were incubated further with active or catalytically inactive Clp1 after wash. 32P incorporation was detected by autoradiography (Autorad), and MBP proteins in the reactions were detected by Coomassie blue (CB) staining. (D–F) The indicated strain was arrested at prometaphase by a 6.5-h cold shift, and 1NMPP1 was added. (D) Representative images at 10 min are shown. (E) Fixed cells at the indicated time points were scored for the presence of Nsk1 at kinetochores (KTs). (F) Representative cells, not quantitated in E, which separated their SPBs and showed Nsk1-GFP between SPBs. Bars, 5 µm. (G) SDS gel mobilities of Nsk1-18A-GFP and Nsk1-18D-GFP were compared with Nsk1-GFP in clp1Δ strains. (A–C and G) Molecular mass is indicated in kilodaltons. (H and I) Spindle dynamics were characterized by time-lapse microscopy of the indicated strains. Spindle length (SPB to SPB, marked by Sid4-RFP) was measured at 1-min intervals. Each line represents an individual cell with the number of cells examined provided.
To determine whether Cdk1 phosphorylation affects Nsk1 function, we first tested the effect of Cdk1 inhibition on Nsk1 localization using a Cdk1 analogue–sensitive mutant (Dischinger et al., 2008). Nsk1 localized to kinetochores of nda3-arrested cells after Cdk1 inhibition but not before (Fig. 3, D and E). In nda3 cdc2-as cells that had progressed to metaphase, as judged by SPB separation, 1NMPP1 treatment led to Nsk1-GFP localization between SPBs, indicative of kinetochore and/or spindle localization (Fig. 3 F). These data suggest that Nsk1 may bind kinetochores independently of MTs and show that Cdk1 antagonizes Nsk1 kinetochore and spindle targeting.
Proper Nsk1 phosphoregulation is essential for accurate chromosome segregation
We used MS to identify 18 phosphorylated Cdk1 consensus sites on Nsk1 isolated from mitotic cells (see Materials and methods). When these 18 residues were mutated to alanines, Cdk1 phosphorylation of Nsk1 in vitro was abolished (Fig. S1 K). Strains were constructed in which the endogenous nsk1+ locus was replaced with nsk1-18A or a mutant in which the 18 serine/threonine residues had been converted to aspartic or glutamic acids, respectively, nsk1-18D. The mobility shift of Nsk1-GFP as a result of hyperphosphorylation in clp1Δ was abolished by the mutations (Fig. 3 G). Nsk1-18A isolated from mitotic cells comigrated with dephosphorylated Nsk1 from interphase cells (Fig. S1 L), indicating that the vast majority, if not all, Cdk1 phosphorylation sites in Nsk1 had been ablated.
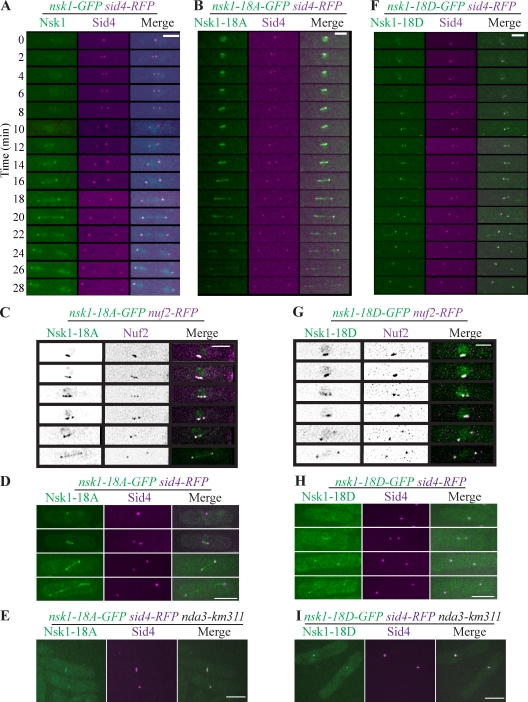
During mitosis, nsk1-18A cells had phase 2 kinetics similar to wild type (Figs. 2 C and 3 H). However, lagging chromosomes were observed in 53% of nsk1-18A cells (n = 41), whereas they were not detected in wild-type cells (n = 71; Fig. S2, A and B). Accordingly, minichromosomes were lost at a high rate (10.3%) in nsk1-18A cells (Fig. 2 A). In contrast to wild type (Fig. 4 A), Nsk1-18A-GFP localized strongly to kinetochores and the spindle at the onset of its assembly and remained there until the end of mitosis (Fig. 4, B–D). Also, in >50% of nda3-arrested cells, Nsk1-18A-GFP colocalized to kinetochores (Fig. S2 D) and SPBs (Fig. 4 E), to which kinetochores associate at this mitotic stage. These results are consistent with Cdk1 and Clp1 competitively controlling Nsk1 kinetochore localization, with Clp1 promoting kinetochore targeting (Fig. 3, A and B). We propose that kinetochore localization of Nsk1 in prometaphase is dynamic, a property that would allow destabilization of incorrect k-MT attachments. In this model, premature binding of Nsk1-18A would hamper correction mechanisms by inappropriately stabilizing k-MT interactions. Because merotelic attachments are frequent during prometaphase and unrecognized by the SAC (Gregan et al., 2011), their stabilization by Nsk1-18A would lead to a high incidence of lagging chromosomes and chromosome loss in the absence of a phase 2 delay, as we have seen (Figs. 2 [A and C] and 3 H). Consistent with merotelic attachments causing lagging chromosomes in nsk1-18A cells, we observed a high incidence of stretched and split kinetochores (Fig. S2 F). This differs from nsk1Δ cells, in which attachments are defective.
Figure 4.
Nsk1 localization is phosphoregulated. (A, B, and F) Time-lapse microscopy of the indicated strains. (C and G) Images from the indicated strains. The left panels show inverted images, whereas the right panels show merged images. (D, E, H, and I) Representative images of fixed cells from the indicated strains. Bars, 5 µm.
nsk1-18D had phase 2 timing similar to wild type (Figs. 2 C and 3 I), but its chromosome loss rate was higher than the nsk1Δ strain (Fig. 2 A). Like the Nsk1-18A mutant, Nsk1-18D-GFP was enriched on kinetochores relative to wild type during early stages of mitosis, but its localization to the spindle was attenuated (Fig. 4, F–H). Nsk1-18D-GFP also colocalized at SPBs (Fig. 4 I) and kinetochores (Fig. S2 E) in >50% of nda3-arrested cells, although the signals were typically less intense. The behavior of the Nsk1-18D mutant suggests that it is a weak phosphomimetic, not recapitulating the results of either full phosphorylation (that prevents kinetochore and spindle localization) or dephosphorylation (impelling both kinetochore and spindle association), although it might have defects unrelated to phosphoregulation. However, because there is no phase 2 delay in either nsk1-18A or nsk1-18D cells in contrast to nsk1Δ cells and both Nsk1-18A and Nsk1-18D are present on kinetochores, Nsk1 must impact spindle dynamics primarily at kinetochores.
Given the aforementioned data showing that Nsk1 promotes proper coupling between kinetochores and MT plus-ends, we tested whether Nsk1 opposes Aurora kinase (Ark1) function, which destabilizes incorrect k-MT attachments (Kelly and Funabiki, 2009; Liu and Lampson, 2009). As predicted, ark1-T7 nsk1Δ grew better at semipermissive temperatures (27 and 29°C) than ark1-T7 alone (Fig. S3 A). Furthermore, Nsk1-18D, which is enriched at kinetochores during early mitosis relative to Nsk1, exacerbated the ark1-T7 phenotype (Fig. S3 A). Reciprocally, two mutations in protein phosphatase 1, dis2cs and dis2Δ, which oppose Ark1, displayed negative genetic interactions with nsk1Δ but not nsk1-18D (Fig. S3 B). The high chromosome loss rate in nsk1-18A cells precluded meaningful genetic analysis with this strain. These genetic data support the proposed Nsk1 function in proper k-MT interactions.
Dynein light chain (DLC) promotes Nsk1 oligomerization and stability
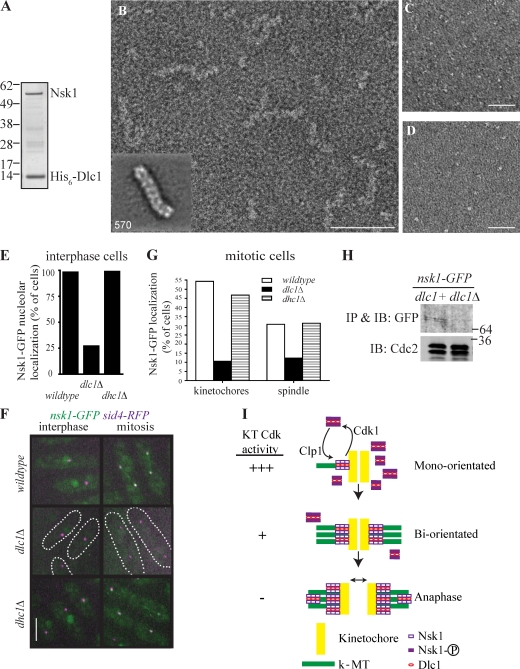
Interestingly, DLC 1 (Dlc1) was identified by 2D liquid chromatography MS as a top hit in both the Clp1-TAP (Fig. S1 A) and Nsk1 purifications from mitotic cells (Fig. S3 C). A structural study of the human DLCs TcTex1 and LC8/DYNLL1 in complex with a fragment of dynein intermediate chain shows that they are sandwiched in a configuration that is incompatible with binding other partners (Williams et al., 2007). These structures extended evidence that DLCs form dynein motor–independent complexes with, for example, Myosin Va and Nup159 (Hódi et al., 2006; Stelter et al., 2007; Barbar, 2008). Given the lack of Dhc1 in our purifications, we tested whether a discrete Nsk1–Dlc1 complex could exist; indeed, we were able to reconstitute it by coexpression in Escherichia coli (Fig. 5 A). By negative-stain EM, the Nsk1–Dlc1 complex was observed in chainlike structures of variable lengths and twists (Fig. 5 B) that are remarkably similar to structures formed by a fragment of budding yeast Nup159 in complex with Dyn2 (Stelter et al., 2007). Alignment and classification of relatively straight regions of the Nsk1–Dlc1 chains revealed discrete globular domains stacked into chains (Fig. 5 B, inset). Dlc1 or Nsk1 alone was globular (Fig. 5, C and D), indicating that the chainlike structures represent oligomerized Nsk1–Dlc1 complex. It is intriguing to note that fibrils, whose molecular composition are unknown, emanating from the kinetochore outer plate contact MT plus-ends in vertebrate cells (McIntosh et al., 2008). It will be interesting to learn whether DLCs, in complex with factors analogous to Nsk1, contribute to the formation of such fibrils and, in so doing, comprise functional units of the k-MT interface.
Figure 5.
Structural characterization of the Nsk1–Dlc1 complex. (A) Coomassie blue staining of reconstituted Nsk1–Dlc1 complex. (B–D) Typical micrograph of negatively stained recombinant and purified Nsk1–Dlc1 (B), Dlc1 (C), and Nsk1 (D). Bars: (B) 50 nm; (C and D) 100 nm. (B, inset) Representative class mean of Nsk1–Dlc1 oligomers. The number of particles is shown in corner. The side length of the panel is 415 Å. (E–G) Nsk1-GFP was visualized in dlc1+ dhc1+, dlc1Δ, and dhc1Δ cells containing Sid4-RFP. (E) The percentage of interphase cells with detectable nucleolar Nsk1-GFP signal was determined in >100 cells of each strain from three occasions of growth. (F) Representative live-cell images of the indicated strains. dlc1Δ cells are outlined. Bar, 5 µm. (G) The percentage of mitotic cells, identified by the position and number of Sid4-RFP–marked SPBs with Nsk1 at kinetochores or on spindle, was determined in >50 cells of each strain from five occasions of growth. (H) Western blot of Nsk1-GFP immunoprecipitates (IP) from dlc1+ and dlc1Δ cells. Cdk1 levels in lysates used for immunoprecipitates are shown. IB, immunoblot. (A and H) Molecular mass is indicated in kilodaltons. (I) A model of Nsk1 function and regulation in chromosome segregation. See Result and discussion for details. KT, kinetochore.
To determine how Dlc1 binding affects Nsk1, we examined Nsk1 localization in dlc1Δ cells. Nsk1 fluorescence was weak in both interphase and mitotic dlc1Δ cells (Fig. 5, E–G). This was also true for Nsk1-18A and Nsk1-18D, which in dlc1+ cells displayed stronger kinetochore and/or spindle targeting (Fig. S3, D and E). Consistent with diminished fluorescence intensity, Nsk1 levels were significantly reduced in dlc1Δ cells (Fig. 5 H), suggesting that complex formation with Dlc1 stabilizes Nsk1. In contrast, Nsk1 localization was unaffected by dhc1Δ (Fig. 5, E–G). Thus, we confirmed the prediction of a Dhc1-independent role for Dlc1 based on the finding that the double mutant dlc1Δ dhc1Δ has more severe meiotic defects than the dhc1Δ mutant alone (Miki et al., 2002). How many distinct complexes Dlc1 and other light chains participate in is not known, but the growing list of heavy chain–independent light chain interactions suggests that caution should be used in assigning dynein motor functions based on phenotypes caused by the loss of DLCs. In this regard, it is noteworthy that although it is not known whether it interacts directly with these proteins, LC8/DYNLL1 copurified with astrin and its binding partner kinastrin/small kinetochore-associated protein (Schmidt et al., 2010; Dunsch et al., 2011), human proteins with analogous functions to those of Nsk1 in binding kinetochores and spindle, stabilizing bioriented attachments, and facilitating proper chromosome movements (Yuan et al., 2009; Manning et al., 2010; Schmidt et al., 2010; Dunsch et al., 2011). Future studies will be useful in determining the extent of the functional and structural similarity between these proteins.
In conclusion, despite a principle role played by the CPC/PP1 system in regulating k-MT attachments (Lampson and Cheeseman, 2011), our results indicate the importance of fine tuning the phosphostatus of a Cdk1 substrate, Nsk1, that affects the proper coupling of kinetochores with MTs, ensuring that it interacts stably with the kinetochore and spindle only at the correct time during mitosis (Fig. 5 I). Nsk1 shares this property of carefully timed phosphoregulated redistribution with other Cdk1/Clp1 targets, namely monopolin (Choi et al., 2009; Rumpf et al., 2010), Klp9 (Fu et al., 2009), and the CPC (Tsukahara et al., 2010), although the specifics of timing and localization differ. Our evidence indicates that Cdk1 regulation of Nsk1–Dlc1 at the k-MT interface is crucial for protecting the fidelity of chromosome segregation.
Materials and methods
Strains and general yeast methods
The strains used in this study are listed in Table S1. Yeast strains were grown in yeast extract (YE) media or Edinburgh minimal media with supplements. nsk1, nsk1-18A, nsk1-18D, nuf2, and sid4 were tagged endogenously at the 3′ end with GFP::kanR, GFP::hygR, linker-GFP::kanR, TAP::kanR, RFP::kanR, and RFP::hygR cassettes as previously described (Bähler et al., 1998). A lithium acetate method (Keeney and Boeke, 1994) was used in Schizosaccharomyces pombe transformations for tag insertions, and integration of tags was verified using whole-cell PCR or fluorescence microscopy. Introduction of tagged loci into other strains was accomplished using standard S. pombe mating, sporulation, and tetrad dissection techniques. For spot assays, cells were grown to midlog phase at 25 or 32°C and resuspended in water to achieve an OD595 = 0.5. 10-fold serial dilutions were made. 5 µl of each dilution was plated on YE or YE plus different concentrations of TBZ in DMSO. Plates were incubated at 18, 25, 27, 29, or 32°C for 2–7 d.
Construction of nsk1Δ, nsk1-18A, and nsk1-18D strains
Deletion of nsk1+ was achieved by PCR-based homologous recombination. In brief, the DNA sequence of 600 bp upstream of the nsk1+ start codon was cloned at the 5′ end of ura4+ in pSK-ura4 (pKG700) with BamHI–PstI. The 600-bp sequence downstream of the nsk1+ stop codon was cloned at the 3′ end of ura4+ in the same plasmid with XhoI–KpnI. The entire insert was then amplified by PCR and transformed into diploid strain ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 h−/h+. Stable integrants were selected, and the deletion was confirmed by whole-cell PCR with primers corresponding to 800 bp upstream or downstream of the nsk1+ ORF and inside the ura4+ gene. After sporulation, tetrad analysis yielded a haploid nsk1::ura4+ strain (nsk1Δ). nsk1::kanR was subsequently obtained by a marker switch approach (Sato et al., 2005).
Using pREP81GFP-nsk1-18A or -18D as a template, the PCR product containing nsk1-18A or -18D with 100 bp of genomic flanking sequences was transformed into nsk1::ura4+ cells, and the successful integration and replacement of ura4+ by nsk1-18A or nsk1-18D were selected by plating on YE containing 1 mg/ml 5-FOA. Correct replacements were verified by whole-cell PCR. The DNA sequences of endogenous nsk1-18A and -18D were confirmed by whole-cell PCR followed by DNA sequencing. Introduction of nsk1-18A or -18D into other strains was verified by whole-cell PCR followed by restriction digestion. NaeI was used to differentiate wild type (1.4 kb) from nsk1-18A (582 and 807 bp). AccI was used for wild type (582 and 807 bp) and nsk1-18D (240, 510, and 636 bp).
ChIP
Untagged nda3-KM311 and nsk1+-GFP nda3-KM311 strains were grown in YE media to exponential growth phase at 32°C and then arrested by growing for 6.5 h at 18°C. Cells were fixed with 3% formaldehyde for 18 min at room temperature at 0, 15, and 30 min after release of cells to 32°C. Aliquots were taken for spindle analysis by immunofluorescence. ChIP was performed as previously described (Petrie et al., 2005). In short, fixed cells were resuspended in 400 µl of ChIP lysis buffer and disrupted by bead beating (2 × 1 min). Chromatin was then sheared by probe sonication (3 × 20 s) and immunoprecipitated with 0.75 µg monoclonal anti-GFP antibody (A11120; Invitrogen) for every 1.5 × 108 cells. Cross-linking was reversed by overnight incubation with TES buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS) followed by proteinase K digestion. After DNA purification, samples were subjected to real-time quantitative PCR on a Mastercycler (Eppendorf International) using the following primers: JPO-2239 (5′-GCTTAAAATCGCCATTTGTTTT-3′) and JPO-2240 (5′-TGATTCATGGATTGAAGTTGACA-3′) for cnt1 and 3; JPO-1631 (5′-TGAGTGTTTTGCCACATTACCC-3′) and JPO-1632 (5′-TTTGTGGTGCAACGTTGTTTG-3′) for cnt1 (outer region); JPO-2241 (5′-TGAAGGCTGTTGATTTGTGG-3′) and JPO-2242 (5′-ACAAACTGTTTTTGTTCAACGATT-3′) for imr; and JPO-769 (5′-CCAGACCATTACAAGCACTACATACG-3′) and JPO-770 (5′-GAATCTTCTCTTGAATAAAACCGCC-3′) for dh. Signal was all normalized to euchromatic control adh1+ JPO-793 (5′-AACGTCAAGTTCGAGGAAGTCC-3′) and JPO-794 (5′-AGAGCGTGTAAATCGGTGTGG-3′). The linear range of amplification for each set of primers was verified, and experiments were performed within this range. The ΔΔCt method was used for analysis. Results are shown as the mean ± SEM of four separate experiments for enrichment of centromeric relative to adh1 sequences in immunoprecipitated samples relative to input material. These data were all normalized to values obtained for the untagged strain at each time point (Partridge et al., 2007).
For spindle analysis accompanying the ChIP, fixed cells were permeabilized with 1 mg/ml zymolyase for 90 min in PEMS buffer (100 mM Pipes, pH 7.0, 1 mM EDTA, 1 mM MgCl2, and 1.2 M sorbitol). Standard immunofluorescence procedures followed (Hagan and Hyams, 1988), incubating the cells first with anti-TAT1 (provided by K. Gull, University of Oxford, Oxford, England, UK; Woods et al., 1989) followed by Texas red–conjugated secondary antibody and DAPI. Cells were then applied to polylysine-coated slides and scored for the presence of spindle MTs using a microscope (Axioskop II; Carl Zeiss) fitted with a Ludl filter wheel and chroma filters.
Vector construction
The nsk1 ORF was amplified from S. pombe genomic DNA using appropriate oligonucleotides and cloned into pREP81GFP and pREP81 N-terminal TAP vectors with NdeI–BamHI and into pMAL-c2X with EcoRI–BamHI. The phosphorylation site mutants nsk1-18A and nsk1-18D were created in pREP81-GFP-nsk1 using the QuikChange multisite-directed mutagenesis kit (Agilent Technologies). The amino acids mutated in Nsk1 are S71, S103, S113, S140, S171, S195, S206, S221, S236, T240, T255, S257, S289, S344, S379, S399, T411, and T437. These residues were mutated to alanine in Nsk1-18A. In Nsk1-18D, T240 and T411 were mutated to glutamic acid, and all other sites were mutated to aspartic acid.
Microscopy
Cells were grown in YE medium at 25°C and visualized live or after fixation in 70% ethanol using a spinning-disk confocal microscope (Ultraview LCI; PerkinElmer) with a 100× NA 1.40 Plan-Apochromat oil immersion objective and a 488-nm argon ion laser (GFP) or a 594-nm helium neon laser (mCherry and RFP). Images were captured on a charge-coupled device camera (Orca-ER; Hamamatsu Photonics) and processed using MetaMorph software (version 7.1; Molecular Devices). For temperature-sensitive strains, cells were first grown at 25°C followed by shifting to 36°C for 2–4 h. For cold-sensitive strains, cells were first grown at 32°C followed by shifting to 18°C for 6.5 h before being fixed in 70% ethanol. Z series optical sections were taken at 0.5-µm spacing. For time-lapse imaging, cells on YE agar pads were sealed using Valap (a vaseline, lanolin, and paraffin mixture). Images were captured at 60-s intervals.
Time-lapse imaging of GFP-marked cen2 sid4-RFP cells was performed at 25°C using a personal microscope system (DeltaVision; Applied Precision). This system includes a microscope (IX71; Olympus), 60× NA 1.42 PlanApo and 100× NA 1.40 UPlanSApo objectives, fixed- and live-cell filter wheels, a camera (CoolSnap HQ2; Photometrics), and softWoRx imaging software. Z stacks corresponding to the entire cell volume were captured at 10-s intervals using the Optical Axis Integration acquisition option. Images were deconvolved and combined into maximum projections. Kymographs of GFP-marked Cen2 were extracted from the images with ImageJ software (National Institutes of Health).
Minichromosome loss assay
The minichromosome loss assay was performed as previously described (Allshire et al., 1995; Trautmann et al., 2004). In brief, an Ade+ single colony (white) was picked from a YE-lacking adenine plate, resuspended in 3 ml YE medium, and grown at 32°C for ∼7 h. Approximately 500 cells were then plated on YE containing 12 mg/L adenine and incubated at 32°C for 3–5 d followed by 2–3 d at 4°C to allow the red color to deepen. The rate of minichromosome loss per division was calculated by dividing the number of at least half red-sectored colonies by the number of total colonies (red colonies were excluded from the count).
Copurification of Nsk1 and Dlc1
nsk1+ and dlc1+ cDNAs were cloned into the bicistronic expression vector pETDuet-1 (EMD) with NdeI–BglII and EcoRI–BamHI, respectively, to produce Dlc1 tagged with His6 and untagged Nsk1. The construct was verified by sequencing and transformed into BL21-RIL bacteria, and expression was induced by 0.4 mM IPTG for 4 h at 36°C. The bacteria pellet was frozen at −80°C followed by resuspension in buffer A (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 15 mM imidazole, 5 mM 2-Mercaptoethanol, and 1% NP-40). After incubation on ice for 5 min, the pellet was dispersed with a pipette and sonicated four times (for 30 s, each interrupted with a 30-s break). Cell lysates were centrifuged at 13,000 rpm for 30 min, and supernatants were incubated with Ni-NTA agarose (QIAGEN) for 30 min at 4°C. The agarose beads were then transferred to a 0.8 × 4 Poly-Prep Chromatography Column (Bio-Rad Laboratories) and washed with 10 ml of buffer B (same with buffer A, except that the imidazole concentration was increased to 60 mM). The beads were then transferred to an Eppendorf tube, and proteins were eluted by incubation with 750 µl elution buffer (same with buffer A, except that the imidazole concentration was increased to 300 mM) for 5 min.
Protein purification and MS
TAPs were conducted as previously described (Gould et al., 2004) from 8-L cell cultures with OD595 around 0.7. The nda3-KM311 strains were arrested at 19°C for 6 h. TAPs were also conducted from clp-C286S-TAP nda3-KM311 cells that were first blocked at 19°C for 6 h and then released to 32°C for 30 min. In a second approach to obtain Nsk1-interacting proteins, protein lysate was made from 8-liter nda3-KM311 cells arrested at prometaphase and incubated with 0.5 mg of recombinant MBP-Nsk1 and MBP-Nsk1-18D on amylose beads for 1 h at 4°C. After washing with 10 ml NP-40 buffer, proteins were eluted with 200 mM NaCl in NP-40 buffer (10 × 100 µl) followed by 500 mM NaCl (15 × 100 µl). The final elution was made with 10 mM of maltose (4 × 150 µl). All fractions were collected and precipitated with TCA. The identification of proteins from the samples by MS was performed as previously described (Kouranti et al., 2010).
Immunoprecipitations and immunoblotting
Whole-cell S. pombe lysates were prepared in NP-40 buffer (McDonald et al., 1999), and immunoprecipitations were performed using anti-GFP antibodies as previously described (McDonald et al., 1999), with minor modifications in that only 2 µg of antibody was used in each immunoprecipitation. Immunoblotting was performed with 0.4 µg/ml anti-GFP antibodies. Immunoblotting was performed using the infrared imaging system and protocol (Odyssey; LI-COR Biosciences). Goat anti–mouse Alexa Fluor 680 (LI-COR Biosciences) was used at a 1:5,000 dilution. Images were exported from Odyssey software in TIFF format.
In vitro kinase and phosphatase assays
In vitro kinase assays were conducted as previously described (Yoon et al., 2006), except that the kinase buffer consists of 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, and 2 mM DTT supplemented with 10 µM cold ATP and 5 µCi γ-[32P]ATP. When used, 100–500 ng of recombinant MBP-Nsk1 served as a substrate. For phosphatase assays, recombinant MBP-Nsk1 was phosphorylated by kinase-active Cdk1 as described above on amylose beads. Beads were then washed in 1 ml of phosphatase assay buffer (50 mM imidazole, pH 6.9, 1 mM EDTA, and 1 mM DTT) three times before they were incubated in the presence of ∼100 ng of recombinant MBP-Clp1 or MBP-Clp1-C286S at 30°C for 30 min in phosphatase assay buffer. Reactions were terminated by the addition of sample buffer and boiling followed by separation on SDS-PAGE and visualization by Coomassie blue staining and autoradiography.
Phosphatase assays of immunoprecipitates were conducted as previously described (Yoon et al., 2006), with the following modifications. After immunoprecipitation from denatured cell lysates, sepharose-bound proteins were washed three times with 1 ml NP-40 buffer and two times with 1 ml of phosphatase buffer (25 mM Hepes-NaOH, pH 7.4, 150 mM NaCl, and 0.1 mg/ml BSA), divided in half, and pulsed down, and the supernatant was aspirated off. 20-µl reactions composed of 1× phosphatase buffer, 2 mM MnCl2, and 1 µl of λ-protein phosphatase (New England Biolabs, Inc.) or 1 µl H2O were then incubated at 30°C for 30 min with gentle mixing. The reactions were terminated by adding 5 µl of 5× SDS sample buffer. When the phosphatase was Clp1, ∼100 ng MBP-Clp1 or MBP-Clp1-C286S was used, and the phosphatase buffer for wash and assay was changed to 50 mM imidazole, pH 6.9, 1 mM EDTA, and 1 mM DTT.
MT methods
For cosedimentation experiments, varying concentrations of monomeric tubulin or paclitaxel-stabilized MTs were incubated with 50 ng of preclarified wild-type MBP-Nsk1 in BRB80 + 100 mM KCl. After 10 min at room temperature, the reactions were pipetted onto 100 µl of BRB80 + 40% sucrose and pelleted for 20 min at 60,000 rpm in a TLA100 rotor at 22°C. Cushions for polymeric tubulin reactions also contained 20 µM paclitaxel. For the supernatant, 40 µl was withdrawn from the top of the tube and mixed with 40 µl of sample buffer. The pellet was resuspended in 80 µl of sample buffer. Equal amounts of supernatant and pellet fractions were subjected to SDS-PAGE and processed for immunoblotting using anti-MBP to detect Nsk1 or Coomassie blue staining to visualize tubulin. Quantitation of Nsk1 in supernatant and pellet fractions was performed using the Odyssey imaging system.
MT bundling by Nsk1 proteins was analyzed using a microscopy assay. Rhodamine-labeled GMP-CPP MTs (500 nM tubulin) were incubated with 1 µg MBP-Nsk1 in BRB80 for 15 min at room temperature. Reactions were visualized by widefield fluorescence microscopy using a 100× NA 1.4 objective (Nikon).
EM and image processing
Uranyl formate (0.7% wt/vol) was used for conventional negative staining as previously described (Ohi, 2004). Images of Dlc1 and Nsk1 alone were recorded using a transmission electron microscope (Morgagni; FEI Company) run at 100 kV at a 36,000× magnification. Images were collected on 1K × 1K charge-coupled device camera (Advanced Microscopy Techniques). Images of the Nsk1–Dlc1 oligomers were collected on an electron microscope (Tecnai F20; FEI company) equipped with a field emission electron source and operated at an acceleration voltage of 120 kV under low-dose conditions at a magnification of 62,000× and a defocus value of −1.5 mm. Images were collected using a 4K × 4K charge-coupled device camera (Gatan, Inc). Charge-coupled device images were converted to mixed raster content format and binned by a factor of two, resulting in final images with 3.46 Å/pixel. Particle images of the Nsk1–Dlc1 complex were selected with Boxer software (Ludtke et al., 1999) and windowed with 120-pixel side lengths. Only Nsk1–Dlc1 chains that contained relatively straight regions were chosen. Image analysis was performed with SPIDER (System for Processing Image Data from Electron microscopy and Related fields) and the associated display program WEB (Frank et al., 1996). 4,005 images of Nsk1–Dlc1 oligomers were rotationally and translationally aligned and subjected to 10 cycles of multireference alignment and K-means classification. Particles were grouped into 40 classes. From the 40 class means, 10 representative projections were chosen and used as references for another cycle of multireference alignment.
Online supplemental material
Fig. S1 presents additional characterization of nsk1Δ, nsk1Δ genetic interactions, and Nsk1 phosphoregulation. Fig. S2 presents images of chromosome segregation in nsk1Δ, nsk1-18A, and nsk1-18D cells. Fig. S3 presents additional characterization of nsk1 phosphomutants and MS results. Table S1 presents the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201105074/DC1.
Acknowledgments
We thank Drs. Richard Vallee and Tom Walz for valuable discussions, Drs. Dan McCollum and Janel McLean for helpful comments on the manuscript, and Dr. Keith Gull for the TAT-1 antibody.
This work was supported by the National Cancer Institute Cancer support grant P30CA021765 and the American Lebanese Syrian Associated Charities to J.F. Partridge, the National Institutes of Health operating grant 1DP2OD004483 to M.D. Ohi, the National Institutes of Health grant R01GM086610 to R. Ohi, the Howard Hughes Medical Institute, of which K.L. Gould is an Investigator, and the National Institutes of Health grant GM068786 to K.L. Gould.
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- CPC
- chromosomal passenger complex
- DLC
- dynein light chain
- k-MT
- kinetochore-MT
- MBP
- maltose-binding protein
- MS
- mass spectrometry
- MT
- microtubule
- SAC
- spindle assembly checkpoint
- SPB
- spindle pole body
- TAP
- tandem affinity purification
- TBZ
- thiabendazole
- YE
- yeast extract
References
- Allshire R.C., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218–233 10.1101/gad.9.2.218 [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951 [DOI] [PubMed] [Google Scholar]
- Barbar E. 2008. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry. 47:503–508 10.1021/bi701995m [DOI] [PubMed] [Google Scholar]
- Choi S.H., Péli-Gulli M.P., Mcleod I., Sarkeshik A., Yates J.R., III, Simanis V., McCollum D. 2009. Phosphorylation state defines discrete roles for monopolin in chromosome attachment and spindle elongation. Curr. Biol. 19:985–995 10.1016/j.cub.2009.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dischinger S., Krapp A., Xie L., Paulson J.R., Simanis V. 2008. Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network. J. Cell Sci. 121:843–853 10.1242/jcs.021584 [DOI] [PubMed] [Google Scholar]
- Dunsch A.K., Linnane E., Barr F.A., Gruneberg U. 2011. The astrin–kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J. Cell Biol. 192:959–968 10.1083/jcb.201008023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K., Nimmo E.R., Javerzat J.P., Borgstrøm B., Egel R., Cranston G., Allshire R. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109:2637–2648 [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. 1996. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199 10.1006/jsbi.1996.0030 [DOI] [PubMed] [Google Scholar]
- Fu C., Ward J.J., Loiodice I., Velve-Casquillas G., Nedelec F.J., Tran P.T. 2009. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev. Cell. 17:257–267 10.1016/j.devcel.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Koonrugsa N., Toda T. 2002. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 12:610–621 10.1016/S0960-9822(02)00761-3 [DOI] [PubMed] [Google Scholar]
- Gould K.L., Ren L., Feoktistova A.S., Jennings J.L., Link A.J. 2004. Tandem affinity purification and identification of protein complex components. Methods. 33:239–244 10.1016/j.ymeth.2003.11.019 [DOI] [PubMed] [Google Scholar]
- Greenall A., Williams E.S., Martin K.A., Palmer J.M., Gray J., Liu C., Whitehall S.K. 2006. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J. Biol. Chem. 281:8732–8739 10.1074/jbc.M512170200 [DOI] [PubMed] [Google Scholar]
- Gregan J., Polakova S., Zhang L., Tolić-Nørrelykke I.M., Cimini D. 2011. Merotelic kinetochore attachment: Causes and effects. Trends Cell Biol. 21:374–381 10.1016/j.tcb.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I.M. 1998. The fission yeast microtubule cytoskeleton. J. Cell Sci. 111:1603–1612 [DOI] [PubMed] [Google Scholar]
- Hagan I.M., Hyams J.S. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89:343–357 [DOI] [PubMed] [Google Scholar]
- Hódi Z., Németh A.L., Radnai L., Hetényi C., Schlett K., Bodor A., Perczel A., Nyitray L. 2006. Alternatively spliced exon B of myosin Va is essential for binding the tail-associated light chain shared by dynein. Biochemistry. 45:12582–12595 10.1021/bi060991e [DOI] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K.S., Salmon E.D. 2010. Mechanisms of force generation by end-on kinetochore-microtubule attachments. Curr. Opin. Cell Biol. 22:57–67 10.1016/j.ceb.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney J.B., Boeke J.D. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 136:849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Funabiki H. 2009. Correcting aberrant kinetochore microtubule attachments: An Aurora B-centric view. Curr. Opin. Cell Biol. 21:51–58 10.1016/j.ceb.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranti I., McLean J.R., Feoktistova A., Liang P., Johnson A.E., Roberts-Galbraith R.H., Gould K.L. 2010. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 8:e1000471 10.1371/journal.pbio.1000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Cheeseman I.M. 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21:133–140 10.1016/j.tcb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Lampson M.A. 2009. Regulation of kinetochore-microtubule attachments by Aurora B kinase. Biochem. Soc. Trans. 37:976–980 10.1042/BST0370976 [DOI] [PubMed] [Google Scholar]
- Ludtke S.J., Baldwin P.R., Chiu W. 1999. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- Ma H.T., Poon R.Y. 2011. How protein kinases co-ordinate mitosis in animal cells. Biochem. J. 435:17–31 10.1042/BJ20100284 [DOI] [PubMed] [Google Scholar]
- Manning A.L., Bakhoum S.F., Maffini S., Correia-Melo C., Maiato H., Compton D.A. 2010. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 29:3531–3543 10.1038/emboj.2010.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald W.H., Ohi R., Smelkova N., Frendewey D., Gould K.L. 1999. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 19:5352–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., Grishchuk E.L., Morphew M.K., Efremov A.K., Zhudenkov K., Volkov V.A., Cheeseman I.M., Desai A., Mastronarde D.N., Ataullakhanov F.I. 2008. Fibrils connect microtubule tips with kinetochores: A mechanism to couple tubulin dynamics to chromosome motion. Cell. 135:322–333 10.1016/j.cell.2008.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F., Okazaki K., Shimanuki M., Yamamoto A., Hiraoka Y., Niwa O. 2002. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol. Biol. Cell. 13:930–946 10.1091/mbc.01-11-0543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocciaro A., Schiebel E. 2010. Cdc14: A highly conserved family of phosphatases with non-conserved functions? J. Cell Sci. 123:2867–2876 10.1242/jcs.074815 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A.F., Murray A., Chikashige Y., Yamashita Y.M., Hiraoka Y., Yanagida M. 1998. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell. 9:3211–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H., Lee J.K., Hurwitz J., Allshire R.C., Nakayama J., Grewal S.I., Tanaka K., Murakami Y. 2002. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 16:1766–1778 10.1101/gad.997702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M.D., Li Y., Cheng Y., Walz T. 2004. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online. 6:23–34 10.1251/bpo70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliferenko S., Balasubramanian M.K. 2002. Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat. Cell Biol. 4:816–820 10.1038/ncb861 [DOI] [PubMed] [Google Scholar]
- Partridge J.F., DeBeauchamp J.L., Kosinski A.M., Ulrich D.L., Hadler M.J., Noffsinger V.J. 2007. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol. Cell. 26:593–602 10.1016/j.molcel.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Petrie V.J., Wuitschick J.D., Givens C.D., Kosinski A.M., Partridge J.F. 2005. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol. Cell. Biol. 25:2331–2346 10.1128/MCB.25.6.2331-2346.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Lénárt P., Peters J.M. 2008. Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev. Cell. 14:646–659 10.1016/j.devcel.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8:310–318 10.1016/S0962-8924(98)01299-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpf C., Cipak L., Schleiffer A., Pidoux A., Mechtler K., Tolić-Nørrelykke I.M., Gregan J. 2010. Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle. 9:3997–4004 10.4161/cc.9.19.13233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Vardy L., Angel Garcia M., Koonrugsa N., Toda T. 2004. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol. Biol. Cell. 15:1609–1622 10.1091/mbc.E03-11-0837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Dhut S., Toda T. 2005. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 22:583–591 10.1002/yea.1233 [DOI] [PubMed] [Google Scholar]
- Schmidt J.C., Kiyomitsu T., Hori T., Backer C.B., Fukagawa T., Cheeseman I.M. 2010. Aurora B kinase controls the targeting of the Astrin–SKAP complex to bioriented kinetochores. J. Cell Biol. 191:269–280 10.1083/jcb.201006129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. 2004. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Stelter P., Kunze R., Flemming D., Höpfner D., Diepholz M., Philippsen P., Böttcher B., Hurt E. 2007. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat. Cell Biol. 9:788–796 10.1038/ncb1604 [DOI] [PubMed] [Google Scholar]
- Trautmann S., Rajagopalan S., McCollum D. 2004. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev. Cell. 7:755–762 10.1016/j.devcel.2004.10.006 [DOI] [PubMed] [Google Scholar]
- Tsukahara T., Tanno Y., Watanabe Y. 2010. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 467:719–723 10.1038/nature09390 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Cai S., Khodjakov A. 2010. Mechanisms of chromosome behaviour during mitosis. Nat. Rev. Mol. Cell Biol. 11:91–102 10.1038/nrg2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Malmstrom T., Troxell C.L., McIntosh J.R. 2001. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell. 12:3919–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.C., Roulhac P.L., Roy A.G., Vallee R.B., Fitzgerald M.C., Hendrickson W.A. 2007. Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc. Natl. Acad. Sci. USA. 104:10028–10033 10.1073/pnas.0703614104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500 [DOI] [PubMed] [Google Scholar]
- Yoon H.J., Feoktistova A., Chen J.S., Jennings J.L., Link A.J., Gould K.L. 2006. Role of Hcn1 and its phosphorylation in fission yeast anaphase-promoting complex/cyclosome function. J. Biol. Chem. 281:32284–32293 10.1074/jbc.M603867200 [DOI] [PubMed] [Google Scholar]
- Yuan J., Li M., Wei L., Yin S., Xiong B., Li S., Lin S.L., Schatten H., Sun Q.Y. 2009. Astrin regulates meiotic spindle organization, spindle pole tethering and cell cycle progression in mouse oocytes. Cell Cycle. 8:3384–3395 10.4161/cc.8.20.9885 [DOI] [PubMed] [Google Scholar]