Current models of centromere/kinetochore architecture are not sufficient to explain the number of molecules of histone H3 variant CENP-A observed in quantitative microscopy.
Abstract
The stoichiometries of kinetochores and their constituent proteins in yeast and vertebrate cells were determined using the histone H3 variant CENP-A, known as Cse4 in budding yeast, as a counting standard. One Cse4-containing nucleosome exists in the centromere (CEN) of each chromosome, so it has been assumed that each anaphase CEN/kinetochore cluster contains 32 Cse4 molecules. We report that anaphase CEN clusters instead contained approximately fourfold more Cse4 in Saccharomyces cerevisiae and ∼40-fold more CENP-A (Cnp1) in Schizosaccharomyces pombe than predicted. These results suggest that the number of CENP-A molecules exceeds the number of kinetochore-microtubule (MT) attachment sites on each chromosome and that CENP-A is not the sole determinant of kinetochore assembly sites in either yeast. In addition, we show that fission yeast has enough Dam1–DASH complex for ring formation around attached MTs. The results of this study suggest the need for significant revision of existing CEN/kinetochore architectural models.
Introduction
Accurate protein counts are crucial for building structural and mathematical models and reconstituting multiprotein complexes. Quantitative fluorescence microscopy is used to determine stoichiometries in live cells (Hirschberg et al., 2000; Dundr et al., 2002; Wu and Pollard, 2005; Joglekar et al., 2006, 2008). For example, Wu and Pollard (2005) used immunoblotting to calibrate fluorescence intensity measurements of 28 cytoskeletal and signaling proteins in fission yeast. Joglekar et al. (2006) used an internal protein standard to count molecules in budding yeast kinetochores.
Centromeres (CENs) specify the sites to assemble kinetochores that mediate attachment of chromosomes to the spindle microtubules (MTs). Centromeric nucleosomes contain the histone H3 variant CENP-A/CenH3/Cse4 (Palmer et al., 1987; Sullivan et al., 1994; Stoler et al., 1995). In budding yeast, one MT attaches to each kinetochore (Winey et al., 1995) assembled at the point CEN (∼120 bp). Cse4 is essential for kinetochore assembly (Collins et al., 2005). Chromatin immunoprecipitation (ChIP) shows that each CEN contains only one Cse4 nucleosome (Furuyama and Biggins, 2007), whereas another study suggests a wider range of Cse4 binding (Riedel et al., 2006).
The CENs and kinetochores of all 16 chromosomes are clustered throughout the cell cycle in budding yeast. Assuming a Cse4 dimer at each nucleosome, an anaphase cluster is presumed to contain 32 Cse4 molecules (Joglekar et al., 2006; Camahort et al., 2009), although the nucleosome structure at CENs is still disputed (Dalal et al., 2007; Mizuguchi et al., 2007; Furuyama and Henikoff, 2009; Black and Cleveland, 2011; Xiao et al., 2011). This value was used as a standard to count the kinetochore proteins and to build an architectural model of kinetochores (Joglekar et al., 2006, 2009). However, whether anaphase clusters contain only CEN-associated Cse4 has not been evaluated (Westermann et al., 2007).
In contrast to budding yeast, fission yeast and most other eukaryotes have regional CENs that span several kilobases to megabases and contain many CENP-A nucleosomes (Partridge et al., 2000; Takahashi et al., 2000; Irvine et al., 2004). A major question is whether the kinetochore assembled on point CENs represents a conserved subunit of each kinetochore-MT attachment site in regional CENs. To address this question, Joglekar et al. (2008) compared kinetochore protein intensities in Schizosaccharomyces pombe with Cse4-GFP. They reported that the S. pombe CENP-A/Cnp1 is present in approximately two to three nucleosomes per CEN and concluded that numbers of CENP-A nucleosomes do not scale with CEN lengths. They also found that the ratio of kinetochore proteins is conserved on each MT attachment site (two to four per CEN in S. pombe) except for too few Dam1–DASH complexes to form a ring (Joglekar et al., 2008). Numerous other studies have also used a standard based on Cse4 to determine stoichiometry of cellular structures (Gardner et al., 2008; Moore et al., 2008; Yeh et al., 2008; Anderson et al., 2009; Markus et al., 2009; Ribeiro et al., 2009; Shimogawa et al., 2009; Tang et al., 2009; Thorpe et al., 2009; Gao et al., 2010; Johnston et al., 2010; Lin et al., 2010).
Here, we used independent standards to count molecules in CEN/kinetochore clusters. We found that each anaphase cluster contains 680 Cnp1 and 122 Cse4 molecules, much higher than the earlier studies. Thus, CENP-A copy number scales to CEN lengths in fission yeast and exceeds MT attachments in both yeasts. Our data present several challenges to the current CEN/kinetochore models.
Results and discussion
Visible cytokinesis nodes contain less than one Cdc12 molecule?
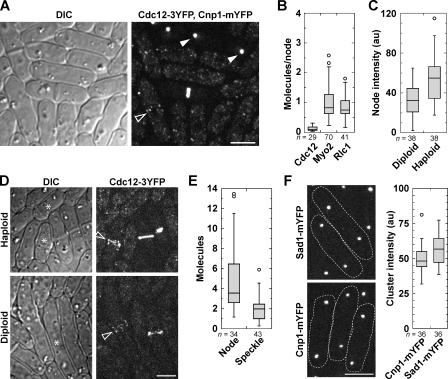
Cytokinesis nodes, discrete protein clusters at the equatorial plasma membrane during the G2/M transition (Fig. 1, A and D), are precursors of the contractile ring in fission yeast cytokinesis (Wu et al., 2006; Vavylonis et al., 2008; Coffman et al., 2009; Laporte et al., 2011). We used the Wu and Pollard (2005) sum intensity method (see Materials and methods) to compare fluorescence intensity of cytokinesis node proteins with Cnp1 anaphase clusters. Based on the prediction of ∼15 molecules of Cnp1 per cluster, each visible cytokinesis node contained <0.2 molecules of Cdc12 and ∼1 molecule of myosin-II heavy chain Myo2 or its light chain, Rlc1 (Fig. 1, A and B). These results contrast the reported levels of ∼4 molecules of Cdc12 and ∼40 molecules of Myo2 and Rlc1 per cytokinesis node (Wu and Pollard, 2005) and suggest either severe quenching of tagged node proteins or more Cnp1 at anaphase clusters.
Figure 1.
Relative quantification of Cnp1 and S. pombe cytokinesis proteins. (A) Differential interference contrast (DIC) and fluorescence image of S. pombe cells showing Cdc12-3YFP nodes (strain KV346; open arrowhead) or Cnp1-mYFP anaphase clusters (strain JW1469; closed arrowheads). (B) Box plots (see Materials and methods) of the number of molecules in individual 3YFP- or mYFP-tagged cytokinesis nodes using Cnp1 as a standard (set at 15 Cnp1 molecules/anaphase cluster, n = 60 clusters; strain JW1469). From left to right, strains KV346, JW1110, and JW949 are shown. (C) Relative intensity of individual Cdc12-3YFP nodes in a heterozygous diploid strain and a haploid strain (JW1404-1). au, arbitrary unit. (D) DIC and fluorescence images of cells quantified in C. Arrowheads mark cells with nodes. Asterisks on the DIC images mark interphase cells with speckles (haploid) or without speckles (diploid). (E) Quantification of Cdc12-3YFP in nodes and speckles in haploid cells. (F) Micrographs and quantification of spindle pole body protein Sad1 (JW1141) and Cnp1 (JW1469) during anaphase. Dashed lines on micrographs show cell boundaries. Bars, 5 µm.
We hypothesized that if quenching could make several Cdc12 molecules appear to be <0.2 molecules, nodes in diploid cells expressing one tagged and one untagged copy of Cdc12 should display an intensity similar to nodes in haploids. Instead, nodes in diploids were about half as intense as in haploids (Fig. 1, C and D), suggesting that quenching was not significant. Cytoplasmic speckles of Cdc12 are the dimmest visible structure and are assumed to contain at least a dimer because formins dimerize with strong affinity (Moseley et al., 2004; Xu et al., 2004; Coffman et al., 2009). Consistent with this assumption, we observed fewer cytoplasmic speckles in the diploid cells (Fig. 1 D). Cdc12 nodes appeared two to three times as bright as Cdc12 speckles (Fig. 1 E), indicating at least four to six Cdc12 molecules per node (Laporte et al., 2011). Moreover, the intensities of Cnp1 clusters and Sad1 at spindle pole bodies were similar (Fig. 1 F). These data suggest >450 Cnp1 per cluster (Wu and Pollard, 2005) and indicate that another standard is needed to resolve the discrepancy.
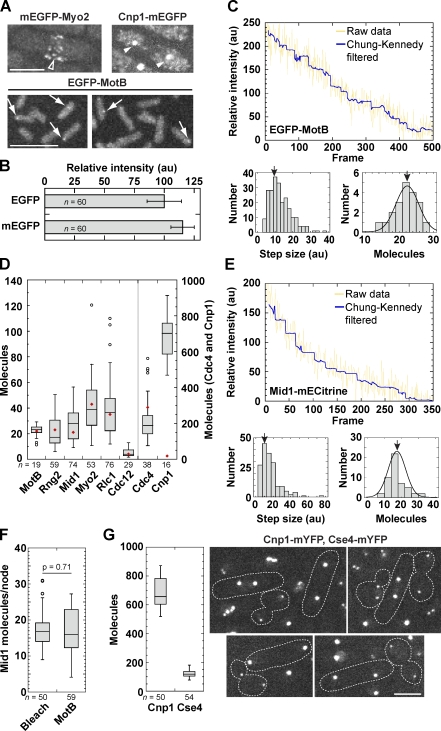
Anaphase CEN clusters harbor additional CENP-A
The intensity of EGFP-MotB in Escherichia coli flagellar motors is 22 times the intensity of a single EGFP molecule, determined by stepwise bleaching (Leake et al., 2006). Cytokinesis nodes of monomeric EGFP (mEGFP)–Myo2 were similar to EGFP-MotB in intensity, whereas Cnp1-mEGFP was much brighter (Fig. 2 A). The mEGFP was 14% brighter than EGFP, and we used this number to correct our data (see Materials and methods; Fig. 2 B). We repeated the bleaching experiments of Leake et al. (2006) to obtain the fluorescence intensity of each EGFP molecule. We calculated the average step size of intensity loss during bleaching and divided the initial intensity of the MotB motors by that value to find 22.5 ± 3.4 molecules per MotB motor (Fig. 2 C). Quantification of molecules in individual nodes of six cytokinesis proteins using MotB as a standard (Fig. 2 D) reproduced the previously estimated values (Wu and Pollard, 2005). However, each anaphase cluster had 680 ± 119 Cnp1-mEGFP molecules (Fig. 2 D), which is much higher than the reported ∼15 Cnp1 molecules (Joglekar et al., 2008). Counting Mid1 by stepwise bleaching in S. pombe yielded a similar result to the ratio measurements (Fig. 2, E and F). When S. pombe and Saccharomyces cerevisiae were observed together, each anaphase cluster contained 680 ± 100 Cnp1 and 122 ± 24 Cse4 molecules, respectively (Fig. 2 G).
Figure 2.
Quantification of node proteins and CENP-As using MotB as a reference. (A) Micrographs of Myo2 nodes (JW1109; open arrowhead), Cnp1 anaphase clusters (JW1470; closed arrowheads), and MotB motor of E. coli (JPA750; arrows) with the same imaging settings and contrast adjustment. (B) Comparison of EGFP (MLP198) and mEGFP (JW948) intensity (mean ± SD). au, arbitrary unit. (C) Quantification of the intensity of a single EGFP molecule and MotB molecules in each motor using stepwise bleaching experiments. (top) A representative bleaching trace of EGFP-MotB. Each plateau ranges from 10 to 50 frames. The yellow line shows raw data after background subtraction, whereas the blue line shows Chung-Kennedy filtered data. (bottom left) The histogram shows the step sizes from 20 bleaching traces. (bottom right) The number of EGFP-MotB molecules per motor calculated from the measured step size. The arrows indicate the peak (9.52 units [left] and 22.5 ± 3.4 molecules [right]). (D) Molecules measured in individual cytokinesis nodes and Cnp1 anaphase clusters (JW1470) compared with the MotB motor. Red diamonds indicate published mean values (Wu and Pollard, 2005; Leake et al., 2006; Joglekar et al., 2008). (E) Quantification of Mid1-mECitrine (JW1790) in interphase nodes using stepwise bleaching as in C from eight bleaching traces. The arrows indicate the peak. (F) Number of Mid1 molecules in interphase nodes using the bleaching method (left) or intensity ratio to the MotB motor (right). (G, left) Comparison of Cnp1 (JW1469) and Cse4 (JW2686-2) in anaphase clusters using MotB as a standard. (right) Cells expressing mYFP-tagged CENP-As in the same field for comparison. Dashed lines on micrographs show cell boundaries. Bars, 5 µm.
We explored the discrepancy with published ratios using molecular tools. PCR and sequencing revealed that the Cnp1-GFP(S65T) strain (YWY277) used in Joglekar et al. (2008) has cnp1-GFP(S65T) inserted at the Cnp1 locus upstream of the native gene that lacks the start codon (Fig. S1 A). We hypothesized that the native gene is translated from the ATG for amino acid 7. This untagged Cnp1 likely inserts at CENs in strain YWY277 based on the N-terminal truncation data (Takayama et al., 2008). In a strain (JW3523) with cnp1-GFP(S65T) completely replacing the native gene, anaphase clusters were 4.4-fold as bright as in YWY277 cells (Fig. S1, B and C). These data and the new Cse4 copy numbers explain the ∼40-fold underestimation of Cnp1 and show that Cnp1 occupancy is proportional to CEN sizes as observed in Candida albicans and vertebrates (Irvine et al., 2004; Joglekar et al., 2008; Sullivan et al., 2011).
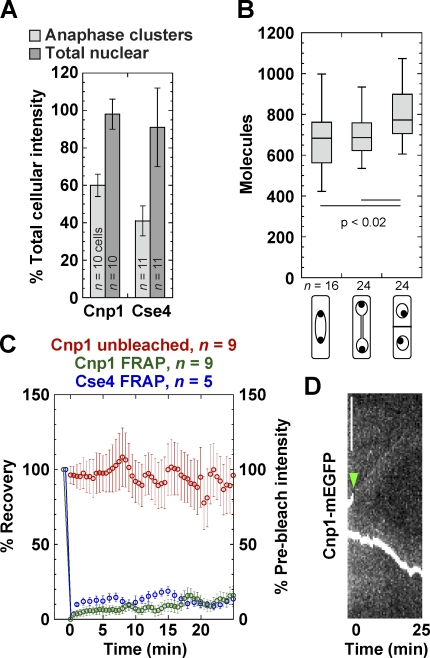
Cnp1 and Cse4 display similar dynamics at anaphase
Next, we investigated whether all CENP-A molecules at anaphase clusters are stably associated with chromosomes. Cse4 is incorporated into CEN nucleosomes during DNA replication and remains stably bound throughout most of the cell cycle in budding yeast (Pearson et al., 2004; Camahort et al., 2007). Cnp1 anaphase clusters (two per cell) and nuclei contained 61 ± 6 and 98 ± 8% of total Cnp1 intensity, respectively, whereas Cse4 anaphase clusters and nuclei contained 41 ± 8 and 91 ± 21% of total Cse4, respectively (Fig. 3 A). These data suggest that molecules could be available in the nucleus to exchange with those in the anaphase clusters. Cnp1 intensity increased during septum formation (Fig. 3 B; Takayama et al., 2008), which began ∼25 min after the start of anaphase (Wu et al., 2003). Cse4 recovered little in anaphase clusters, as reported (Pearson et al., 2004) using FRAP assays. Similarly, Cnp1 clusters bleached at early anaphase B did not recover much until ∼25 min later, nor did the unbleached clusters lose intensity (Fig. 3, C and D). This large immobile fraction (>80%) suggests that Cnp1 stably associates with nucleosomes and kinetochores.
Figure 3.
Cnp1 displays similar dynamics to Cse4 at anaphase clusters. (A) Percentage of total cellular CENP-A intensity (mean ± SD) contained in anaphase clusters or the nucleus of S. pombe (JW1469) and S. cerevisiae (JW2687) cells. (B) Quantification of Cnp1 at early anaphase B, late anaphase, and G1/S phase (JW1469) with cellular features diagrammed below the graph (see Materials and methods). (C) FRAP is plotted as the percentage of recovery (left axis), whereas the unbleached cluster’s intensity is plotted as the percentage of prebleach intensity (right axis; mean ± SEM). One CEN cluster in each cell was bleached at early anaphase B, and the recovery was monitored with a 30-s delay for Cnp1 (JW1470) and 1 min for Cse4 (KBY7006). The change in intensity of the unbleached cluster was also monitored. (D) A kymograph of a representative bleached cell expressing Cnp1-mEGFP. The arrowhead marks the bleach point. Bar, 5 µm.
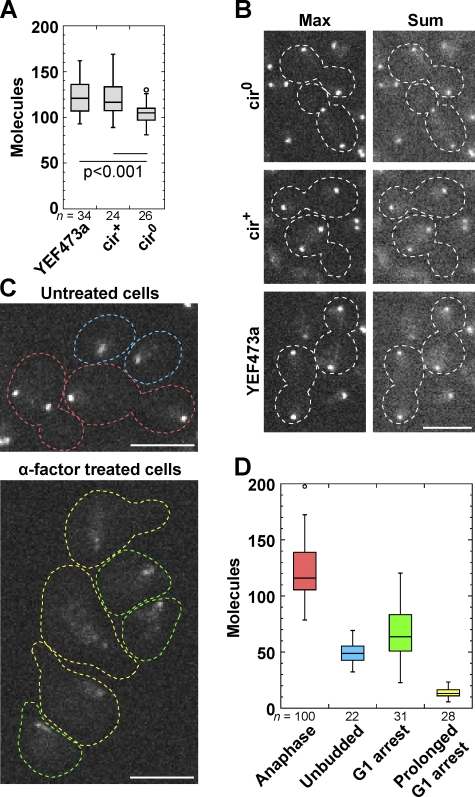
Some Cse4 bound to 2-µm circles and other locations outside CENs in budding yeast
Most laboratory budding yeast (but not fission yeast) strains contain ∼60–100 parasitic plasmids per cell, called 2-µm circles (2-µm), that partition using Cse4, cohesin, and other proteins (Velmurugan et al., 2000; Mehta et al., 2002, 2005; Hajra et al., 2006; Ghosh et al., 2007). The plasmids incorporate Cse4 nucleosomes and cluster near CENs (Velmurugan et al., 2000; Hajra et al., 2006), but it is unknown how many Cse4 molecules bind to 2-µm (Yeh and Bloom, 2006; Huang et al., 2011). Compared with 122 Cse4 molecules in YEF473a, the strain used above, each anaphase cluster contained 105 ± 13 Cse4 molecules in a cir0 strain (cured of 2-µm) and 121 ± 19 molecules in the isogenic cir+ strain (Fig. 4, A and B). Thus, 2-µm plasmids can bind ∼16 Cse4 molecules.
Figure 4.
Some Cse4 in S. cerevisiae binds to 2-µm, and most is lost from the CEN cluster during G1 phase. (A) Molecules in each anaphase cluster in Cse4-linker-mYFP strains with and without 2-µm. From left to right, strains JW2687, JW2679, and JW2683 are shown. (B) Maximum and sum intensity projections of representative anaphase clusters of the strains in A. Dashed lines on micrographs show cell boundaries. (C) Cells expressing Cse4-linker-mYFP (JW2687) imaged with the same settings and contrast adjustments. (D) Molecules in each CEN cluster for cells, color coded as in C. Prolonged G1 arrest is defined as cells with a long shmoo. Bars, 5 µm.
In budding yeast, MTs are attached to kinetochores throughout the cell cycle except briefly at S phase (Pearson et al., 2004; Westermann et al., 2007). Thus, Cse4 remains bound to CEN DNA during G1 phase. Cells in G1 phase from asynchronous cultures or after α-factor treatment had ∼40–50% Cse4 intensity in each CEN cluster compared with anaphase cells. The intensity was further reduced in prolonged G1 arrest (Fig. 4, C and D), suggesting that most Cse4 molecules are lost from the cluster in G1 phase.
Collectively, Cse4 molecules in each anaphase cluster are almost fourfold higher than the assumed 32 molecules (Joglekar et al., 2006). Some of the additional Cse4 associates with 2-µm plasmids (Fig. 4). However, the sizeable discrepancy suggests that the original conclusion of one Cse4 nucleosome per CEN may need further verification. This value derives from elegant ChIP experiments performed on micrococcal nuclease (MNase)–digested chromatin (Furuyama and Biggins, 2007). A critical assumption in these experiments is that terminal centromeric chromatin fragments generated by MNase digestion are mononucleosomes. The MNase-resistant fragment containing CEN DNA was >200 bp (Furuyama and Biggins, 2007), but recent biochemical analyses of nucleosomes constructed in vitro indicate that Cse4-containing nucleosomes only protect ∼115–130-bp DNA (Cole et al., 2011; Dechassa et al., 2011; Kingston et al., 2011). Thus, the size of the native MNase-resistant centromeric fragment is similar to a Cse4-containing dinucleosome. In addition, these experiments did not include a cross-linking step, so some less stably associated Cse4 at or near the CEN could have been lost. Alternatively, the additional Cse4 may bind to DNA surrounding the CENs and/or proteins at kinetochores (Akey and Luger, 2003; Riedel et al., 2006; Yong-Gonzalez et al., 2007), and these Cse4 may dissociate during G1 (Fig. 4, C and D).
Fission yeast kinetochores have sufficient Dam1–DASH complex to form a ring
Because of the underestimation of Cse4, the copy numbers of kinetochore components need to be adjusted accordingly and the architecture revisited to account for the additional proteins. The Ndc80 and Dam1–DASH complexes cooperate to processively maintain the kinetochore-MT attachment and produce force (Lampert et al., 2010; Tien et al., 2010). The Ndc80 complex binds the inner kinetochore at one end and MTs at the other (Wei et al., 2005; McIntosh et al., 2008; Powers et al., 2009). In vitro, Dam1 complexes can assemble into MT-binding rings with 16-fold symmetry (Miranda et al., 2005; Westermann et al., 2005), which harness the forces produced from depolymerizing MT ends and function as processivity factors for kinetochore-MT binding (Grishchuk et al., 2005; Asbury et al., 2006; Efremov et al., 2007; Burrack et al., 2011).
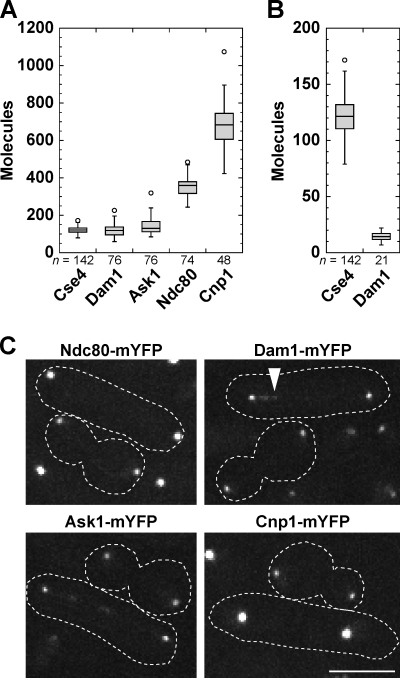
Previous data indicated that the Dam1 complexes at S. pombe kinetochores are insufficient to form rings around the MTs (Joglekar et al., 2008). Calculations using our data yield enough Dam1 complexes at each MT attachment site to form one ring in fission yeast and several in budding yeast. Indeed, occupancy of Dam1 complex proteins was 120 ± 33 Dam1 and 141 ± 40 Ask1, whereas Ndc80 was 351 ± 49 at each anaphase cluster (Fig. 5, A and C). Dam1 and Ask1 also formed foci on the spindle with 14 ± 4 Dam1 molecules each (Fig. 5 B). As a control, the ratio between Cse4 and Cnp1 was unchanged (Fig. 5, A and C). S. pombe has three chromosomes with two to four MT attachment sites each. Thus, enough Dam1 complexes exist at kinetochore-MT attachment sites to form rings. Single Dam1 complexes and small oligomers can interact with MTs with load-bearing capacity, but the cooperative binding of Dam1 complexes suggests a tendency toward oligomerization (Gestaut et al., 2008; Grishchuk et al., 2008; Gao et al., 2010; Lampert et al., 2010; Tien et al., 2010). Dam1 complex rings have not been observed in vivo (McIntosh, 2005), so the structure of Dam1 complexes requires further studies.
Figure 5.
Fission yeast kinetochores have sufficient Dam1–DASH complex to form a ring. (A) Molecules of Dam1 (JW3667), Ask1 (JW3666), Ndc80 (JW3668), and Cnp1 (JW1469) in anaphase clusters using Cse4-mYFP in the same field as a standard. (B) Quantification of Dam1 foci along the spindle. (C) Representative images of budding and fission yeast anaphase cells used for quantification in A and B. The arrowhead marks a Dam1 structure as quantified in B. Dashed lines on micrographs show cell boundaries. Bar, 5 µm.
How to specify the sites for kinetochore assembly?
Our data raise several challenging questions. Where are the additional Cse4 and Cnp1 molecules located? As each chromosome has one to a few MT attachment sites, how do cells select kinetochore assembly sites among the CENP-A nucleosomes? Could kinetochores assemble on several CENP-A nucleosomes instead of only one, as proposed in previous models (Joglekar et al., 2006, 2008)? What are the architectures of kinetochores from yeast to vertebrate cells based on the revised numbers of molecules?
In fission yeast, a temperature-sensitive allele of Mis6, which is required to recruit Cnp1 to CENs, displays nearly 100% minichromosome loss (Takahashi et al., 1994, 2000). In contrast, Fft3 deletion causes a 2.7× reduction in Cnp1 binding and only 11% minichromosome loss (Strålfors et al., 2011), suggesting that loss of some Cnp1 is tolerated. In human cells, 10% of the normal CENP-A level is sufficient to drive kinetochore assembly (Liu et al., 2006). Recent studies indicate that the constitutive CEN-associated network components CENP-C, -N, and -T are crucial for kinetochore assembly in fission yeast and other cells (Hori et al., 2008; Tanaka et al., 2009; Trazzi et al., 2009; Carroll et al., 2010; Gascoigne et al., 2011; Przewloka et al., 2011). In human and chicken cells, targeting CENP-C and -T to ectopic loci results in MT-attached kinetochores in the absence of CENP-A (Gascoigne et al., 2011). Drosophila melanogaster CENP-A and -C are interdependent for CEN localization (Erhardt et al., 2008). Based on these data, we hypothesize that a subset of CENP-A might recruit constitutive CEN-associated network components to assemble kinetochores in regional CENs. Distinguishing these assembly sites may be related to the mechanism of neo-CEN formation among ectopic CENP-A deposition sites (Heun et al., 2006; Zeitlin et al., 2009; Olszak et al., 2011). Thus, the CEN environment and interaction partners of CENP-As may both be necessary to direct kinetochore assembly at specific sites.
Collectively, our experiments challenge the idea that in yeast, each CENP-A nucleosome directs the assembly of one kinetochore-MT attachment. Instead, CENP-A occupancy exceeds the number of kinetochore-MT attachment sites in yeast as it does in higher eukaryotes. Our experiments also highlight the importance of understanding how kinetochore assembly sites are selected on CENP-A chromatin.
Materials and methods
Strains and growing conditions
Table S1 lists the E. coli, S. pombe, and S. cerevisiae strains used in this study. All tagged yeast genes were integrated at their native chromosomal loci under the control of endogenous promoters using pFA6a modules as previously described (Bähler et al., 1998; Longtine et al., 1998). The diploid strain expressing one copy of cdc12-3YFP was constructed using complementary ade6 mutant alleles by crossing strain JW81 to JW1404-1 and maintained in YE5S− adenine using standard genetic methods (Moreno et al., 1991). We found that all the tags for Cnp1, Cse4, and kinetochore proteins were free of mutations by sequencing genomic DNA after gene targeting. The cnp1 locus of strain YWY277 cnp1-GFP(S65T) was amplified and sequenced, which led to the discovery of the insertion and duplication at that locus (Fig. S1 A).
Because tagged histones are often less functional (Takayama et al., 2008), we tagged Cse4 in several strains using a flexible linker of 24 amino acids between the cse4 ORF and the monomeric YFP (mYFP) tag (Sandblad et al., 2006). The linker made the tagged Cse4 strains healthier than strains without a linker from 25 to 32°C, although we observed no obvious difference in growth rates at 25°C (Fig. S1, D and E). The linker strains also had no more than a 10% difference in anaphase cluster intensity from those without it, suggesting that the functionality of Cse4 makes little difference in its incorporation at 25°C. The strains did not show increased sensitivity to 20 µg/ml nocodazole (Fig. S1 E), suggesting that the tagged Cse4 is functional at 25°C but not fully functional at higher temperatures. Most of the tagged Cnp1 strains were functional, with the exceptions of the GFP(S65T)-tagged strains (Fig. S1 G), perhaps as a result of the thermosensitivity of GFP(S65T) itself. Cnp1 tagged at its N terminus under the endogenous promoter (Pcnp1 from −717 to +6 bp with respect to the ATG) affects its expression and level at CENs either with or without the kanMX6 marker (Fig. S1 F; Takayama et al., 2008).
Yeast cells were restreaked from −80°C stocks, grown 2–3 d on plates at 25°C, and then inoculated into 5–15 ml of liquid media. S. pombe cells were grown in YE5S and S. cerevisiae in yeast peptone dextrose (YPD) at 25°C. Cells were grown in liquid media at exponential phase for ∼48 h before microscopy (Wu et al., 2006). E. coli cells were streaked from −80°C stock, grown overnight on lysogeny broth plates, and then inoculated into 10 ml of liquid tryptone broth. Liquid cultures were grown overnight in tryptone broth at 30°C and washed into motility buffer at room temperature (10 mM potassium phosphate and 0.1 mM EDTA, pH 7.0) for imaging (Leake et al., 2006).
Microscopy
Yeast cells for microscopy were collected from liquid cultures. For some experiments, cells of different strains were mixed together just before collection to image under identical conditions. Cells were centrifuged at 5,000 rpm for 30 s and then washed into EMM5S or synthetic dextrose medium for imaging. Live-cell microscopy of S. pombe was performed using a thin layer of EMM5S liquid medium with 20% gelatin and 0.1 mM n-propyl-gallate and observed at 23–25°C as previously described (Wu et al., 2006). Live-cell microscopy of S. cerevisiae was performed either on synthetic dextrose medium plus 2% agar pads or on EMM5S with 20% gelatin with S. pombe cells to prevent clumping while immobilizing the cells. For synchronization, S. cerevisiae cells were treated with 8 µg/ml α-factor for 3 h (approximately one generation) at 25°C and imaged on agar pads containing 8 µg/ml α-factor. S. pombe cells for FRAP experiments were treated with 20 mM hydroxyurea for 4 h at 25°C to synchronize in S phase and were then released for 3 h before FRAP to ensure that all bleached cells and cells used for corrections were at similar cell cycle phases. E. coli cells were observed on bare slides under a coverslip sealed with Valap to observe rotational motility and to verify the functionality of EGFP-MotB motors (Leake et al., 2006) and were then were imaged immediately on motility buffer plus 20% gelatin to immobilize the cells for quantification. To calculate the full width half maximum (FWHM), we imaged 0.1-µm fluorescent beads (TetraSpeck microspheres; Invitrogen) at 0.05-µm spacing and calculated the FWHM of a Gaussian fit to the point spread function in the z axis.
We used a 100×/1.4 NA Plan-Apo objective lens (Nikon) on a spinning-disk confocal microscope (UltraView ERS [PerkinElmer] with a CSU22 confocal head on an Eclipse TE2000-U microscope [Nikon]) with 488- and 514-nm argon ion lasers and a cooled charge-coupled device camera (ORCA-AG; Hamamatsu Photonics). Image analyses were performed using ImageJ (National Institutes of Health). Images of yeast and E. coli cells in figures are maximum intensity projections of z sections spaced at 0.4 µm unless otherwise noted. We found no obvious signal variation as a result of focal plane depth, as previously reported (Joglekar et al., 2006), using our imaging system, perhaps because our imaging starts away from the coverslip (Fig. S2 A).
Comparison of the methods for measuring fluorescence intensity
A sum of intensity from multiple z sections spaced at the FWHM of the point spread function along the z axis was consistent with immunoblotting (Wu and Pollard, 2005), whereas a comparison of only the maximum intensity section was used to count molecules in kinetochores (Joglekar et al., 2006). We tested whether there is a difference in the quantification using these two methods.
We determined that the maximum intensity section does not represent the same fraction of the total intensity for structures with different apparent sizes by comparing S. pombe cells expressing fluorescent-tagged cytokinesis node proteins or Cnp1 and S. cerevisiae cells expressing Cse4-GFP (Fig. S2, B and C). Under our imaging conditions, 90% of the intensity in the maximum intensity plane of each CENP-A anaphase cluster was contained in a 0.5-µm2 (25 pixel) region of interest (ROI), whereas each node was contained in a 0.2-µm2 (9 pixel) ROI. The maximum z section of each node protein contained 51–65% of the sum intensity (Fig. S2 C), whereas anaphase clusters of Cnp1 and Cse4 had 46–48% in the maximum section. Comparing the maximum section of node and CEN cluster intensities yields a different ratio of fluorescence than the sum intensity ratio (Fig. S2 D). Because the maximum section contains a different fraction of the total intensity, the sum method is more accurate for comparing structures with different apparent sizes. Thus, throughout the manuscript, the data represented are the sum intensity of all 0.4-µm–spaced z sections that contain intensity above background for a given structure except where noted.
Counting proteins by ratio fluorescence imaging and bleaching
Uneven illumination was corrected using images of purified 6His-mYFP solution as previously described (Wu and Pollard, 2005; Wu et al., 2008). In brief, at least 10 images of purified YFP were averaged after offset subtraction and then divided by the maximum value in the field using the ImageJ Math process to get an image with intensity values ranging from ∼0.5 to 1. Offset-subtracted images of cells were divided by this correction image in ImageJ before measurements were taken. The offset is an image taken without laser lights. Background corrections were implemented as previously described (Wu and Pollard, 2005; Joglekar et al., 2006). In brief, an ROI size was chosen for each tagged protein that included at least 90% of the intensity (± two SDs of a Gaussian fit along two axes). The ROI sizes depended on the imaging conditions, but, in general, Cnp1 ROI was greater than Cse4 ROI, which was greater than cytokinesis node ROI. The same relationship exists between the numbers of z sections each protein occupied. Cnp1 and Cse4 were also compared with the same size ROI, and the ratio between their sum intensities was unchanged. Thus, as long as each ROI is chosen so that at least 90% of the intensity in the maximum plane is included, the ratios are consistent whether the ROIs are the same or different. The background intensity from a concentric ROI twice the size was calculated and subtracted from the measuring area in each imaging plane. All molecule number plots show the ratio of sum intensity except where noted. In box plots, the box contains the middle 50% of the data, the line in the box is the median, and circles above or below represent outliers. Outliers are defined by KaleidaGraph software based on the height of the box. Points >1.5× the box height above or below the box are defined as outliers.
The EGFP-tagged MotB structure in E. coli was verified to contain 22 molecules by imaging the cells in a single plane continuously with a 200-ms exposure for each frame and then quantifying the fluorescence intensity for each GFP molecule (Leake et al., 2006). Mid1-mECitrine nodes in interphase S. pombe cells were imaged continuously with a 100-ms exposure to verify the ratio comparison between fluorophores expressed in E. coli and S. pombe. The background-subtracted data were fit with a modified Chung-Kennedy filter that preserves the edges of steps (Leake et al., 2006; Engel et al., 2009). In brief, starting at the tenth frame, the mean and SD of two consecutive sets of 10 points (up to and just after the tenth frame) were determined, and the mean of the set with the lower SD was used at that frame. Some of the beginning and ending data are lost in this filter. In essence, the filter is a rolling mean that does not cross step boundaries. Thus, a lower SD suggests that the dataset lies on a plateau, whereas a higher SD suggests that the dataset crosses a boundary and is not reported. On the plot of filtered data, the discrete changes in fluorescence are more obvious and thus easier to measure. The fluorescence of a single molecule is determined by fitting the step sizes to a γ distribution, and the starting intensity is divided by the step size to get the number of molecules.
Molecules in cytokinesis nodes and anaphase CEN/kinetochore clusters were counted based on fluorescence intensity. Z sections spaced at 0.4 µm (except where noted) were collected for strains expressing mYFP- or mEGFP-tagged proteins in S. pombe and S. cerevisiae. Intensities of cytokinesis node proteins were obtained from the sum of two to three consecutive sections and compared with the sum intensity of anaphase clusters in five to six consecutive sections to obtain the mean number of each protein in individual structures. Intensities of Cnp1-mEGFP in anaphase clusters and mEGFP-tagged node proteins (Mid1 [strain JW1088], Rng2 [JW1112], and Myo2 [JW1109]) were compared with the intensity of EGFP-MotB motors, with a slight correction for the difference in intensity between EGFP and mEGFP. Ratios between Cnp1 and the three node proteins were the same using both mEGFP and mYFP tags (Mid1 [strain JW1089], Rng2 [JW1761], and Myo2 [JW1763]), verifying the consistency of the method. Additional node proteins were measured using mYFP (Rlc1 [strain JW1757-2], Cdc12 [JW1404-1], and Cdc4 [JW1797-2]). The 3YFP intensity of Cdc12-3YFP is corrected to the intensity of mYFP for comparison (Wu and Pollard, 2005). Then, Cse4- and Cnp1-mEGFP strains were imaged under the same conditions to extrapolate the MotB standard for Cse4 using the Cnp1 value. The ratios between Cnp1 and Cse4 were the same for mYFP- and mEGFP-tagged proteins or by comparing only the best focal plane.
In Fig. 3 B, the cell cycle stages of the cells were estimated as follows from the left to right: early anaphase B, with two CEN clusters 3–6.5 µm apart; late anaphase, with clusters >6.5 µm apart; and G1/S phase, with a septum that has begun to form. The first two stages (without a septum) were used to quantify anaphase clusters and were not significantly different by the t test. The Student’s two-tailed t test was used to calculate p-values.
FRAP analysis
FRAP was performed using the photokinesis unit on the (UltraView ERS) confocal system as previously described (Coffman et al., 2009; Laporte et al., 2011). In brief, the data were gathered using the Track-It function in the software (UltraView ERS) to find anaphase clusters in focus before bleaching. We collected at least three prebleach stacks (four z sections spaced at 0.3 µm) of anaphase clusters and 50 postbleach stacks at 30-s or 1-min intervals. For clusters that remained in focus during the movie, an ROI was selected at each site that was bleached >50% of the original signal. Unbleached cells at the same cell cycle stage as bleached cells were used for photobleaching corrections. Background corrections were performed using extracellular areas, and the data were normalized to the mean prebleach intensity set to 100% (Coffman et al., 2009). The intensity immediately after bleaching was set to 0% for the FRAP curves so that the graph shows the percentage of recovery. The plane with the maximum intensity from four sections was used for analysis at each time point, which is more accurate because the anaphase clusters do not always stay in the same focal plane. The unbleached Cnp1 cluster was also analyzed for loss of fluorescence because the S. pombe nuclei remained connected for most of the duration of our imaging. We attempted to fit the data to a single exponential curve equation given by y = m1 − m2 × exp(−m3 × x), in which m3 equals the off rate (Laporte et al., 2011), but the fit was not good as a result of the lack of recovery.
Online supplemental material
Fig. S1 shows that the abnormal cnp1 locus affects the intensity of Cnp1-GFP anaphase CEN clusters in strain YWY277 and effects of different tags on CENP-A intensity and function. Fig. S2 shows a comparison of ratio measurement methods. Table S1 lists the E. coli, S. pombe, and S. cerevisiae strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106078/DC1.
Acknowledgments
We thank Michael Chest, Huanyu Wang, and Mengzi Zhang for help with experiments and data analyses; Judith Armitage, Erfei Bi, Kerry Bloom, Makkuni Jayaram, and Matt Lord for strains; and members of the Wu laboratory for valuable discussion. Special thanks to Kerry Bloom and Edward Salmon for communication/discussion of unpublished results on Cse4.
V.C. Coffman is supported by an American Heart Association predoctoral fellowship. This work is supported by the National Institutes of Health grants R01GM062970 to M.R. Parthun and R01GM086546 to J.-Q. Wu.
Footnotes
Abbreviations used in this paper:
- CEN
- centromere
- ChIP
- chromatin immunoprecipitation
- DIC
- differential interference contrast
- FWHM
- full width half maximum
- mEGFP
- monomeric EGFP
- MNase
- micrococcal nuclease
- MT
- microtubule
- mYFP
- monomeric YFP
- ROI
- region of interest
- YPD
- yeast peptone dextrose
References
- Akey C.W., Luger K. 2003. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 13:6–14 10.1016/S0959-440X(03)00002-2 [DOI] [PubMed] [Google Scholar]
- Anderson M., Haase J., Yeh E., Bloom K. 2009. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol. Biol. Cell. 20:4131–4139 10.1091/mbc.E09-05-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury C.L., Gestaut D.R., Powers A.F., Franck A.D., Davis T.N. 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA. 103:9873–9878 10.1073/pnas.0602249103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.-Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951 [DOI] [PubMed] [Google Scholar]
- Black B.E., Cleveland D.W. 2011. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 144:471–479 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack L.S., Applen S.E., Berman J. 2011. The requirement for the Dam1 complex is dependent upon the number of kinetochore proteins and microtubules. Curr. Biol. 21:889–896 10.1016/j.cub.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R., Li B., Florens L., Swanson S.K., Washburn M.P., Gerton J.L. 2007. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 26:853–865 10.1016/j.molcel.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., Zhu D., Shilatifard A., Workman J.L., Gerton J.L. 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 35:794–805 10.1016/j.molcel.2009.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., Straight A.F. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Nile A.H., Lee I-J., Liu H., Wu J.-Q. 2009. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol. Biol. Cell. 20:5195–5210 10.1091/mbc.E09-05-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole H.A., Howard B.H., Clark D.J. 2011. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc. Natl. Acad. Sci. USA. 108:12687–12692 10.1073/pnas.1104978108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.A., Castillo A.R., Tatsutani S.Y., Biggins S. 2005. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol. Biol. Cell. 16:5649–5660 10.1091/mbc.E05-08-0771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y., Furuyama T., Vermaak D., Henikoff S. 2007. Structure, dynamics, and evolution of centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 104:15974–15981 10.1073/pnas.0707648104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa M.L., Wyns K., Li M., Hall M.A., Wang M.D., Luger K. 2011. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2:313 10.1038/ncomms1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., McNally J.G., Cohen J., Misteli T. 2002. Quantitation of GFP-fusion proteins in single living cells. J. Struct. Biol. 140:92–99 10.1016/S1047-8477(02)00521-X [DOI] [PubMed] [Google Scholar]
- Efremov A., Grishchuk E.L., McIntosh J.R., Ataullakhanov F.I. 2007. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc. Natl. Acad. Sci. USA. 104:19017–19022 10.1073/pnas.0709524104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B.D., Ludington W.B., Marshall W.F. 2009. Intraflagellar transport particle size scales inversely with flagellar length: Revisiting the balance-point length control model. J. Cell Biol. 187:81–89 10.1083/jcb.200812084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., Straight A.F. 2008. Genome-wide analysis reveals a cell cycle–dependent mechanism controlling centromere propagation. J. Cell Biol. 183:805–818 10.1083/jcb.200806038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S., Biggins S. 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA. 104:14706–14711 10.1073/pnas.0706985104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T., Henikoff S. 2009. Centromeric nucleosomes induce positive DNA supercoils. Cell. 138:104–113 10.1016/j.cell.2009.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Courtheoux T., Gachet Y., Tournier S., He X. 2010. A non-ring-like form of the Dam1 complex modulates microtubule dynamics in fission yeast. Proc. Natl. Acad. Sci. USA. 107:13330–13335 10.1073/pnas.1004887107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Bouck D.C., Paliulis L.V., Meehl J.B., O’Toole E.T., Haase J., Soubry A., Joglekar A.P., Winey M., Salmon E.D., et al. 2008. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 135:894–906 10.1016/j.cell.2008.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., Cheeseman I.M. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D.R., Graczyk B., Cooper J., Widlund P.O., Zelter A., Wordeman L., Asbury C.L., Davis T.N. 2008. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10:407–414 10.1038/ncb1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.K., Hajra S., Jayaram M. 2007. Faithful segregation of the multicopy yeast plasmid through cohesin-mediated recognition of sisters. Proc. Natl. Acad. Sci. USA. 104:13034–13039 10.1073/pnas.0702996104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Molodtsov M.I., Ataullakhanov F.I., McIntosh J.R. 2005. Force production by disassembling microtubules. Nature. 438:384–388 10.1038/nature04132 [DOI] [PubMed] [Google Scholar]
- Grishchuk E.L., Spiridonov I.S., Volkov V.A., Efremov A., Westermann S., Drubin D., Barnes G., Ataullakhanov F.I., McIntosh J.R. 2008. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc. Natl. Acad. Sci. USA. 105:6918–6923 10.1073/pnas.0801811105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajra S., Ghosh S.K., Jayaram M. 2006. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-µm circle partitioning locus and promotes equal plasmid segregation. J. Cell Biol. 174:779–790 10.1083/jcb.200603042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P., Erhardt S., Blower M.D., Weiss S., Skora A.D., Karpen G.H. 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 10:303–315 10.1016/j.devcel.2006.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg K., Phair R.D., Lippincott-Schwartz J. 2000. Kinetic analysis of intracellular trafficking in single living cells with vesicular stomatitis virus protein G-green fluorescent protein hybrids. Methods Enzymol. 327:69–89 10.1016/S0076-6879(00)27268-6 [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Huang C.C., Hajra S., Ghosh S.K., Jayaram M. 2011. Cse4 (CenH3) association with the Saccharomyces cerevisiae plasmid partitioning locus in its native and chromosomally integrated states: Implications in centromere evolution. Mol. Cell. Biol. 31:1030–1040 10.1128/MCB.01191-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine D.V., Amor D.J., Perry J., Sirvent N., Pedeutour F., Choo K.H., Saffery R. 2004. Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res. 12:805–815 10.1007/s10577-005-5377-4 [DOI] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D.C., Molk J.N., Bloom K.S., Salmon E.D. 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8:581–585 10.1038/ncb1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D., Finley K., Liu X., Wan Y., Berman J., He X., Salmon E.D., Bloom K.S. 2008. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 181:587–594 10.1083/jcb.200803027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K., Salmon E.D. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston K., Joglekar A., Hori T., Suzuki A., Fukagawa T., Salmon E.D. 2010. Vertebrate kinetochore protein architecture: Protein copy number. J. Cell Biol. 189:937–943 10.1083/jcb.200912022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston I.J., Yung J.S., Singleton M.R. 2011. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J. Biol. Chem. 286:4021–4026 10.1074/jbc.M110.189340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F., Hornung P., Westermann S. 2010. The Dam1 complex confers microtubule plus end–tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189:641–649 10.1083/jcb.200912021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Coffman V.C., Lee I-J., Wu J.-Q. 2011. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 192:1005–1021 10.1083/jcb.201008171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake M.C., Chandler J.H., Wadhams G.H., Bai F., Berry R.M., Armitage J.P. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 443:355–358 10.1038/nature05135 [DOI] [PubMed] [Google Scholar]
- Lin M.-C., Galletta B.J., Sept D., Cooper J.A. 2010. Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J. Cell Sci. 123:1329–1342 10.1242/jcs.065698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-T., Rattner J.B., Jablonski S.A., Yen T.J. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Markus S.M., Punch J.J., Lee W.-L. 2009. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr. Biol. 19:196–205 10.1016/j.cub.2008.12.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R. 2005. Rings around kinetochore microtubules in yeast. Nat. Struct. Mol. Biol. 12:210–212 10.1038/nsmb0305-210 [DOI] [PubMed] [Google Scholar]
- McIntosh J.R., Grishchuk E.L., Morphew M.K., Efremov A.K., Zhudenkov K., Volkov V.A., Cheeseman I.M., Desai A., Mastronarde D.N., Ataullakhanov F.I. 2008. Fibrils connect microtubule tips with kinetochores: A mechanism to couple tubulin dynamics to chromosome motion. Cell. 135:322–333 10.1016/j.cell.2008.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Yang X.-M., Chan C.S., Dobson M.J., Jayaram M., Velmurugan S. 2002. The 2 micron plasmid purloins the yeast cohesin complex: A mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 158:625–637 10.1083/jcb.200204136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Yang X.-M., Jayaram M., Velmurugan S. 2005. A novel role for the mitotic spindle during DNA segregation in yeast: Promoting 2 µm plasmid-cohesin association. Mol. Cell. Biol. 25:4283–4298 10.1128/MCB.25.10.4283-4298.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.J.L., De Wulf P., Sorger P.K., Harrison S.C. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143 10.1038/nsmb896 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Xiao H., Wisniewski J., Smith M.M., Wu C. 2007. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 129:1153–1164 10.1016/j.cell.2007.04.026 [DOI] [PubMed] [Google Scholar]
- Moore J.K., Li J., Cooper J.A. 2008. Dynactin function in mitotic spindle positioning. Traffic. 9:510–527 10.1111/j.1600-0854.2008.00710.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Moseley J.B., Sagot I., Manning A.L., Xu Y., Eck M.J., Pellman D., Goode B.L. 2004. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 15:896–907 10.1091/mbc.E03-08-0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak A.M., van Essen D., Pereira A.J., Diehl S., Manke T., Maiato H., Saccani S., Heun P. 2011. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat. Cell Biol. 13:799–808 10.1038/ncb2272 [DOI] [PubMed] [Google Scholar]
- Palmer D.K., O’Day K., Wener M.H., Andrews B.S., Margolis R.L. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805–815 10.1083/jcb.104.4.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge J.F., Borgstrøm B., Allshire R.C. 2000. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 14:783–791 [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Yeh E., Gardner M., Odde D., Salmon E.D., Bloom K. 2004. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 14:1962–1967 10.1016/j.cub.2004.09.086 [DOI] [PubMed] [Google Scholar]
- Powers A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., Asbury C.L. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., Glover D.M. 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21:399–405 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Ribeiro S.A., Gatlin J.C., Dong Y., Joglekar A., Cameron L., Hudson D.F., Farr C.J., McEwen B.F., Salmon E.D., Earnshaw W.C., Vagnarelli P. 2009. Condensin regulates the stiffness of vertebrate centromeres. Mol. Biol. Cell. 20:2371–2380 10.1091/mbc.E08-11-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C.G., Katis V.L., Katou Y., Mori S., Itoh T., Helmhart W., Gálová M., Petronczki M., Gregan J., Cetin B., et al. 2006. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 441:53–61 10.1038/nature04664 [DOI] [PubMed] [Google Scholar]
- Sandblad L., Busch K.E., Tittmann P., Gross H., Brunner D., Hoenger A. 2006. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 127:1415–1424 10.1016/j.cell.2006.11.025 [DOI] [PubMed] [Google Scholar]
- Shimogawa M.M., Widlund P.O., Riffle M., Ess M., Davis T.N. 2009. Bir1 is required for the tension checkpoint. Mol. Biol. Cell. 20:915–923 10.1091/mbc.E08-07-0723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K.C., Curnick K.E., Fitzgerald-Hayes M. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573–586 10.1101/gad.9.5.573 [DOI] [PubMed] [Google Scholar]
- Strålfors A., Walfridsson J., Bhuiyan H., Ekwall K. 2011. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 7:e1001334 10.1371/journal.pgen.1001334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K.F., Hechenberger M., Masri K. 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127:581–592 10.1083/jcb.127.3.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan L.L., Boivin C.D., Mravinac B., Song I.Y., Sullivan B.A. 2011. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 19:457–470 10.1007/s10577-011-9208-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamada H., Yanagida M. 1994. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell. 5:1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen E.S., Yanagida M. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 288:2215–2219 10.1126/science.288.5474.2215 [DOI] [PubMed] [Google Scholar]
- Takayama Y., Sato H., Saitoh S., Ogiyama Y., Masuda F., Takahashi K. 2008. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol. Biol. Cell. 19:682–690 10.1091/mbc.E07-05-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Chang H.L., Kagami A., Watanabe Y. 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell. 17:334–343 10.1016/j.devcel.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Tang X., Punch J.J., Lee W.-L. 2009. A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle. 8:3182–3190 10.4161/cc.8.19.9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe P.H., Bruno J., Rothstein R. 2009. Kinetochore asymmetry defines a single yeast lineage. Proc. Natl. Acad. Sci. USA. 106:6673–6678 10.1073/pnas.0811248106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien J.F., Umbreit N.T., Gestaut D.R., Franck A.D., Cooper J., Wordeman L., Gonen T., Asbury C.L., Davis T.N. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189:713–723 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S., Perini G., Bernardoni R., Zoli M., Reese J.C., Musacchio A., Della Valle G. 2009. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS ONE. 4:e5832 10.1371/journal.pone.0005832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.-Q., Hao S., O’Shaughnessy B., Pollard T.D. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Velmurugan S., Yang X.-M., Chan C.S.-M., Dobson M., Jayaram M. 2000. Partitioning of the 2-µm circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J. Cell Biol. 149:553–566 10.1083/jcb.149.3.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.R., Sorger P.K., Harrison S.C. 2005. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA. 102:5363–5367 10.1073/pnas.0501168102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H.-W., Niederstrasser H., Wong J., Drubin D.G., Nogales E., Barnes G. 2005. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 17:277–290 10.1016/j.molcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Westermann S., Drubin D.G., Barnes G. 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76:563–591 10.1146/annurev.biochem.76.052705.160607 [DOI] [PubMed] [Google Scholar]
- Winey M., Mamay C.L., O’Toole E.T., Mastronarde D.N., Giddings T.H., Jr, McDonald K.L., McIntosh J.R. 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129:1601–1615 10.1083/jcb.129.6.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., Pollard T.D. 2005. Counting cytokinesis proteins globally and locally in fission yeast. Science. 310:310–314 10.1126/science.1113230 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Kuhn J.R., Kovar D.R., Pollard T.D. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:723–734 10.1016/S1534-5807(03)00324-1 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Sirotkin V., Kovar D.R., Lord M., Beltzner C.C., Kuhn J.R., Pollard T.D. 2006. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 174:391–402 10.1083/jcb.200602032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., McCormick C.D., Pollard T.D. 2008. Chapter 9: Counting proteins in living cells by quantitative fluorescence microscopy with internal standards. Methods Cell Biol. 89:253–273 10.1016/S0091-679X(08)00609-2 [DOI] [PubMed] [Google Scholar]
- Xiao H., Mizuguchi G., Wisniewski J., Huang Y., Wei D., Wu C. 2011. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol. Cell. 43:369–380 10.1016/j.molcel.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Moseley J.B., Sagot I., Poy F., Pellman D., Goode B.L., Eck M.J. 2004. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 116:711–723 10.1016/S0092-8674(04)00210-7 [DOI] [PubMed] [Google Scholar]
- Yeh E., Bloom K. 2006. Hitching a ride. EMBO Rep. 7:985–987 10.1038/sj.embor.7400793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L.V., Joglekar A., Bond L., Bouck D., Salmon E.D., Bloom K.S. 2008. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 18:81–90 10.1016/j.cub.2007.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Gonzalez V., Wang B.-D., Butylin P., Ouspenski I., Strunnikov A. 2007. Condensin function at centromere chromatin facilitates proper kinetochore tension and ensures correct mitotic segregation of sister chromatids. Genes Cells. 12:1075–1090 10.1111/j.1365-2443.2007.01109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S.G., Baker N.M., Chapados B.R., Soutoglou E., Wang J.Y., Berns M.W., Cleveland D.W. 2009. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc. Natl. Acad. Sci. USA. 106:15762–15767 10.1073/pnas.0908233106 [DOI] [PMC free article] [PubMed] [Google Scholar]