Cryo-EM tomography of wild-type and mutant cilia and flagella from Tetrahymena and Chlamydomonas reveals new information on the substructure of radial spokes.
Abstract
Radial spokes (RSs) are ubiquitous components in the 9 + 2 axoneme thought to be mechanochemical transducers involved in local control of dynein-driven microtubule sliding. They are composed of >23 polypeptides, whose interactions and placement must be deciphered to understand RS function. In this paper, we show the detailed three-dimensional (3D) structure of RS in situ in Chlamydomonas reinhardtii flagella and Tetrahymena thermophila cilia that we obtained using cryoelectron tomography (cryo-ET). We clarify similarities and differences between the three spoke species, RS1, RS2, and RS3, in T. thermophila and in C. reinhardtii and show that part of RS3 is conserved in C. reinhardtii, which only has two species of complete RSs. By analyzing C. reinhardtii mutants, we identified the specific location of subsets of RS proteins (RSPs). Our 3D reconstructions show a twofold symmetry, suggesting that fully assembled RSs are produced by dimerization. Based on our cryo-ET data, we propose models of subdomain organization within the RS as well as interactions between RSPs and with other axonemal components.
Introduction
Cilia and flagella play important roles in motility and sensory functions of eukaryotic cells ranging from protists to humans (Davenport and Yoder, 2005; Badano et al., 2006; Fliegauf et al., 2007; Satir and Christensen, 2007; Ginger et al., 2008). The radial spoke (RS) is a T-shaped macromolecular complex that is present in the 9 + 2 axoneme of most motile cilia and flagella. The importance of the RS in ciliary and flagellar motility was revealed by Chlamydomonas reinhardtii mutants that lack the entire RS complex or a part of it (Piperno et al., 1977; Witman et al., 1978; Huang et al., 1981). In these mutants, flagella display abnormal motility or are completely paralyzed. Similarly, in humans, the lack of RSs cause motility defects in cilia, resulting in the pathologies of primary ciliary dyskinesia and Kartagener’s triad (Sturgess et al., 1979; Castleman et al., 2009; Olm et al., 2011). Thus, the RS plays an important part in human health through its role in ciliary motility.
Ultrastructural studies have shown that the RS consists of a stalk, which is anchored to the A-microtubule extending toward the center of the axoneme, and an orthogonal head. The head domain is thought to have transient contacts with the inner sheath and the central pair (CP) apparatus (Warner and Satir, 1974; Witman et al., 1978; Goodenough and Heuser, 1985). The entire structure (stalk + head) is described as a T shape, which, depending on the organism, repeats in pairs or triplets every 96 nm along the A-tubule of each outer doublet microtubule (Warner and Satir, 1974; Goodenough and Heuser, 1985). The significance of this variability in RS periodicity is not known.
Comparisons of wild-type (WT) C. reinhardtii and RS-defective axonemes using 2D gels (Piperno et al., 1981) and analysis of intact 20S RSs isolated from the axoneme (Yang et al., 2001) revealed that RSs are composed of 23 polypeptides, RS proteins (RSPs) 1–23 (Piperno et al., 1981; Yang et al., 2006). Five of these proteins, RSP1, 4, 6, 9, and 10, localize to the spoke head (Piperno et al., 1981), whereas the remaining RSPs compose the stalk. More recently, a CaM- and spoke-associated complex (CSC) composed of CaM and three other proteins, has also been identified as a component of the axonemal RS structure (Dymek and Smith, 2007).
Genetic, biochemical, and motility studies have shown that the RS–CP complex regulates dynein force generation (Warner and Satir, 1974; Piperno et al., 1977; Witman et al., 1978; Huang et al., 1981, 1982; Brokaw et al., 1982; Kamiya, 1982; Goodenough and Heuser, 1985; Smith and Sale, 1992; Omoto et al., 1999; Smith, 2002; Mitchell and Nakatsugawa, 2004; Smith and Yang, 2004; Yang et al., 2004, 2006). Interdoublet microtubule sliding assays revealed that the dynein control system is modulated by a network of kinases, phosphatases, and potential sensors of second messengers, such as CaM (Habermacher and Sale, 1995, 1996, 1997; Porter and Sale, 2000). All these components are found inside the RS complex, and there is evidence that the RS transmits signals from the CP to the dynein arms through mechanical and/or chemical interactions (Warner and Satir, 1974; Witman et al., 1978; Huang et al., 1981, 1982; Brokaw et al., 1982; Kamiya, 1982; Habermacher and Sale, 1996; Omoto et al., 1999; Mitchell and Nakatsugawa, 2004; Smith and Yang, 2004), but the molecular mechanism of this regulation process is unknown. Identification of interactions occurring between RS and dynein and RS and CP is fundamental for understanding how the RS regulates the dynein motility to generate microtubule sliding and axonemal bending. In this study, we clarify details of the 3D structure of the RS and visualize connections between specific RS components and inner dynein arms (IDAs).
Loss of a single RSP can result in the loss of a subset of RSPs or the entire spoke from the flagellum (Huang, 1986; Curry and Rosenbaum, 1993). Investigations of the effect of these mutations on RS assembly, the analysis of predicted structural and functional domains within RSPs (Yang et al., 2006), GST pull-down, chemical cross-linking (Kohno et al., 2011), and in vitro reconstitution of RS particles (Diener et al., 2011) lead to information on possible protein–protein interactions between RSPs and potential functions of individual RSPs. Observation of the motility phenotypes of RS mutants, correlated with the 3D structure of WT and mutants will lead to a more complete understanding of how RSs function in ciliary motility.
In the present study, the 3D structures of in situ RSs are reconstructed by cryoelectron tomography (cryo-ET) and 3D subtomogram averaging. By comparative structural analysis of C. reinhardtii RS pairs (RS1 and RS2) and Tetrahymena thermophila RS triplets (RS1, RS2, and RS3), similarities and differences between RS1, RS2, and RS3 were clarified, and additional components of the C. reinhardtii RS pair architecture were identified. Using C. reinhardtii RSP mutants pf1, pf14, and pf24, the specific location of subsets of RSPs was shown. The 3D reconstructions also reveal a twofold rotational symmetry in the structure of the RS head, suggesting that the fully assembled axonemal RSs are produced by dimerization of cytoplasmic RS precursors. Based on our cryo-ET data, we propose models for RS assembly, interactions between the 23 RSPs, and the interaction of RSs with other axonemal components. Our results provide a fundamental step toward understanding how RSs perform mechanochemical signal transduction to regulate dynein activity.
Results
Although the importance of the RS as a regulator of ciliary/flagellar motility is well documented, its structure and mode of action remain poorly understood. To investigate the 3D architecture of RSs as multiprotein complexes, we used cryo-ET to reconstruct 3D structures of T. thermophila cilia and of flagella from WT C. reinhardtii cells and mutants that lack specific subsets of RSPs.
Overall arrangement of RSs in C. reinhardtii WT
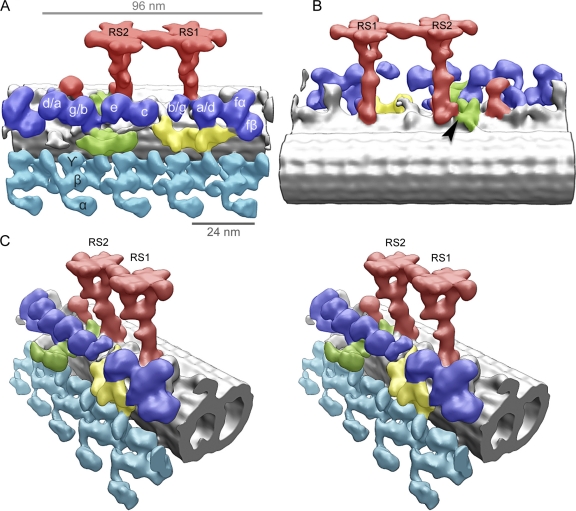
We extracted, aligned, and averaged 2,400 3D volumes containing RS pairs (96-nm periodicity) from electron tomograms of C. reinhardtii WT flagella embedded in amorphous ice. The extracted volumes contained the RSs, a portion of the microtubule doublet, outer dynein arms (ODAs), and IDAs as shown in Fig. 1 (A, B, and C). The averaged 3D reconstruction shows ODAs (Fig. 1, blue) and IDAs (Fig. 1, turquoise) as previously described (Ishikawa et al., 2007; Bui et al., 2008, 2009, 2011; Movassagh et al., 2010), the intermediate and light chains of IDAs (Fig. 1, yellow), the dynein regulatory complex (DRC; Fig. 1, green; Bui et al., 2008, 2009), and the relative position of the RSs (Fig. 1, red). The resolution of the WT model (Fig. 1 and Fig. 2) is ∼39 Å, as shown by the Fourier shell correlation in Fig. S1. This resolution allows us to visualize features of structure and assembly of the RS never shown before.
Figure 1.
Overall 3D architecture of the C. reinhardtii axonemal microtubule doublet (96-nm repeat). These surface renderings of 3D structure were averaged from 2,400 subtomograms and include the microtubule doublet (gray), ODAs (turquoise), IDAs (blue), the intermediate and light chains of IDAs (yellow), the DRC (green), and the RSs (red). RS1, radial spoke 1; RS2, radial spoke 2. (A and B) The models are oriented with the proximal end of the axoneme on the right (A) or left (B). The arrowhead indicates the interaction site between RS2 and DRC. (C) A stereo view of the reconstruction.
Figure 2.
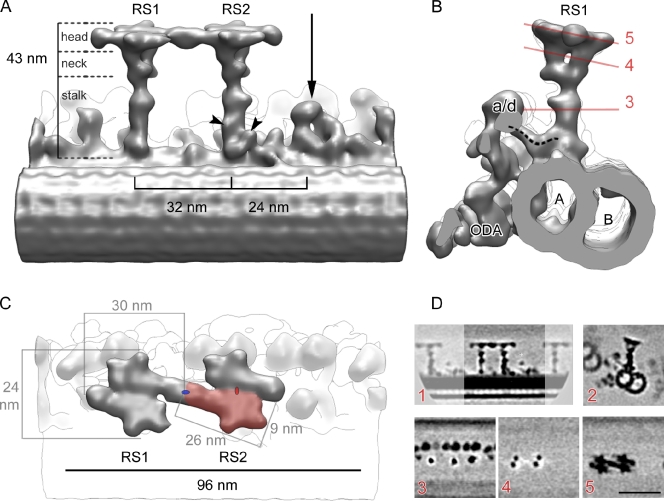
3D reconstruction of WT RSs in C. reinhardtii. (A–C) Surface renderings of tomographic reconstruction after 3D subtomogram averaging. (A) Longitudinal view showing the B-microtubule (foreground), radial spoke 1 (RS1), radial spoke 2 (RS2), the RS3 stump (arrow), and IDAs (fainted background). The proximal end of the axoneme points toward the left. Arrowheads indicate densities specific to RS2. The boundaries between the head, neck, and stalk domains are shown. (B) Side view seen from the proximal end showing RS1, IDA a or d (a/d), ODA, and the microtubule doublet. A, A-microtubule; B, B-microtubule. The dashed line indicates the dynein a/d tail connecting to the RS1 base. The red lines show the position of section planes through the original density map used to generate subfigures (shown in D, 3–5). (C) Top view showing the two RS heads. The proximal end points to the left as in A. The pale red area identifies one of the symmetrical subdomains composing the RS head. Two such subdomains build one RS head. The red ellipse indicates the twofold rotational symmetry between these subdomains. The two RS heads are also symmetrical, also following a twofold rotational symmetry, denoted by a blue ellipse. (D) Sections through the density map of the model shown in A–C. (1) Same orientation as in A; (2) Same orientation as in B; (3–5) Same orientation as in C. The proximal end is pointing toward the left in all sections, except for section 2, where the proximal end is oriented toward the reader. Bar, 50 nm.
In C. reinhardtii, WT flagella pairs of RSs repeat every 96 nm along the A-microtubule (Goodenough and Heuser, 1985). The stalks of RS1 and RS2 are centered 32 nm apart and appear perpendicular to the long axis of the microtubule (Fig. 2, A and B). RS1 and RS2 share the same overall structure, classically defined as T shaped (Fig. 2 and Video 1). Two additional densities are visible at the base of RS2 stalk (Fig. 2 A, arrowheads) that are not present in RS1. One of these occurs at the microtubule-anchoring area (Fig. 2 A) and connects RS2 to the DRC (Fig. 1 B, arrowhead). The DRC, together with RSs and the CP, regulates dynein activity (Huang et al., 1982) and also contains the nexin links, which connect adjacent microtubule doublets (Heuser et al., 2009). This density, at the base of the RS2 stalk, disappears in the spokeless mutant pf14 (see following paragraph), confirming that it is a component of the RS. The significance of the second density that is unique to the RS2 stalk (Fig. 2 A, arrowheads) has not been identified. The base of the RS2 stalk is connected to dynein c (Fig. S2, dotted line). A similar connection can be seen between the base of RS1 stalk and dynein a/d (Fig. 2 B, dotted line).
In both RS1 and RS2, the upper part of the stalk branches into a bifurcated “neck,” which connects the stalk to the head (Fig. 2, B and D, 4). At the base of this neck, a density protrudes toward heads of the IDAs (Fig. 2 B). In our reconstruction, the neck area appears identical in both RS1 and RS2. The head of each RS measures 30 nm long and 24 nm wide (Fig. 2 C) and is composed of two identical subdomains, ∼26 nm long and ∼9 nm wide (Fig. 2 C, red area), each connected to one of the two neck branches. Therefore, there are four identical head subdomains in the 3D reconstruction shown in Fig. 2. The two subdomains of each RS head are arranged with a twofold rotational symmetry (Fig. 2, C and D, 5), suggesting they have identical compositions but antiparallel orientations. The heads of R1 and R2 appear connected (Fig. 2, A and C), suggesting a tight interaction between adjacent RSs.
Our 3D reconstruction also shows a protrusion anchored to the A-microtubule 24 nm distant from RS2 (Fig. 2 A, arrow), as seen also by Bui et al. (2008). The nature of this bulbous-shaped structure has not been defined yet, but it is situated where RS3 attaches to the outer doublets in T. thermophila (see RS3 in T. thermophila and in C. reinhardtii).
Assembly of RSP subsets
Biochemical and genetic analyses in C. reinhardtii have demonstrated that the 20S axonemal RS complex is composed of ≥23 different proteins (Luck et al., 1977; Piperno et al., 1981; Yang et al., 2001, 2006). To identify the interactions between subsets of RSPs and their location in the RS, we reconstructed the structure of RS pairs from the following RS mutant C. reinhardtii strains (Table I): pf1, pf24, and pf14.
Table I.
C. reinhardtii mutants used in this study
| Mutant | Gene product | Motility phenotype | Morphological defect | Protein missing | Protein in reduced amount | References |
| pf1 | RSP4 | Paralyzed | Headless | RSP1, 4, 6, 9, and 10 | − | Huang et al., 1981 |
| pf14 | RSP3 | Paralyzed | Spokeless | RSP1–23 | − | Piperno et al., 1981 |
| pf24 | RSP2 | Paralyzed | Headless, missing part of the stalk | − | RSP1, 2, 4, 6, 9, 10, 16, and 23 | Huang et al., 1981; Patel-King et al., 2004; Yang et al., 2004 |
| ida4 | IDA4 | Reduced swimming velocity | Missing dynein a/d, c, and d/a | Dynein a, c, and d | − | Kamiya et al., 1991; Kagami and Kamiya, 1992 |
Minus signs indicate unavailable data.
The mutant pf1 is deficient in RSP4 (Huang et al., 1981), and biochemical data show that a subset of four other proteins, namely RSPs 1, 6, 9, and 10, is also missing in pf1 flagella (Luck et al., 1977; Piperno et al., 1977; Piperno et al., 1981). pf1 cells are paralyzed, suggesting that at least some of these five RSPs are structurally and/or mechanochemically necessary for the signal transduction from the CP to the dyneins (Warner and Satir, 1974; Piperno et al., 1977; Goodenough and Heuser, 1985; Yang et al., 2001). Early transmission EM investigations of embedded and sectioned pf1 axonemes indicated the absence of the top portion of the RS in this mutant (Piperno et al., 1977). Our 3D structural analysis shows that the stalk and the bifurcated neck are present in pf1 RSs, whereas the head is missing in both RS1 and RS2 (Fig. 3, yellow). No differences are visible between WT and pf1 stalks, whereas the neck looks slightly distorted in pf1 (Fig. 3, arrowhead). An increased flexibility of the two branches could account for the distortion of the neck domain and indicates that at least some of the head proteins are needed for the stabilization of the neck. Our analysis of pf1 RSs shows that RSPs 1, 4, 6, 9, and 10 build the head of both RS1 and RS2. The differential density map shown in gray in Fig. 4 and Video 2 represents the space occupied by RSPs 1, 4, 6, 9, and 10. A protein complex of ∼1,200 kD should fit in this volume, which corresponds to approximately four times that of the expected total mass of RSP1 (78.6 kD), RSP4 (49.8 kD), RSP6 (48.8 kD), RSP9 (29.5 kD), and RSP10 (23.5 kD; Table II; Piperno et al., 1981).
Figure 3.
Comparison of the 3D structure of RSs from three mutants and WT cells. (left) The proximal end is to the left. (right) The same model but oriented so that the distal end of the axoneme points toward the reader. For each mutant, the RSP that is not expressed genetically is indicated on the left and crossed out in red. On the right side are lists of the RSPs found in the flagella of the corresponding mutant. The arrows indicate the position of the adaptor protein complexes located at the base of RS1 and RS2.
Figure 4.
RS domains. Differential maps show the boundaries between the various domains of the RS structure (gray, heads; yellow, necks; blue, stalks; purple, adaptor protein complexes). (A) Longitudinal view with proximal end toward the left. (B) Side view (RS1 in the front) seen from the proximal end. RSPs localized in each domain are indicated on the left side (using the same color code). CaM and CaM-IP2, -IP3, and -IP4 are only present in RS2; the composition of the adaptor complex in RS1 is unknown. The volume of each domain (in cubic nanometers) is indicated on the right side of the figure.
Table II.
RSP molecular masses and estimated volumes in C. reinhardtii
| RS domain | RSP | MM I 2D-NEPHGEa | MM II theoreticala | Estimated number of RSP copies | Integrated MM I/integrated MM II | Estimated volume for integrated MM I/MM II | Volume from reconstruction |
| kD | kD | kD | nm3 | nm3 | |||
| Head | RSP1 | 123 | 78.6 | 4 | 1,264/920.8 | 1,500/1,100 | 1,470 |
| RSP4 | 76 | 49.8 | 4 | ||||
| RSP6 | 67 | 48.8 | 4 | ||||
| RSP9 | 26 | 29.5 | 4 | ||||
| RSP10 | 24 | 23.5 | 4 | ||||
| Neck | RSP2 | 118 | 77.4 | 2 | 508/354.8 | 600/420 | 600 |
| RSP16 | 34 | 39 | 2 | ||||
| RSP23 | 102 | 61 | 2 | ||||
| Stalk | RSP3 | 86 | 56.8 | 4 | 1,477/1,294.5 | 1,750/1,530 | 1,600 |
| RSP5 | 69 | 55.9 | 2 | ||||
| RSP7 | 58 | 55 | 2 | ||||
| RSP8 | 40 | 40.5 | 1 | ||||
| RSP11 | 22 | 21.5 | 2 | ||||
| RSP12 | 20 | 19.7 | 2 | ||||
| RSP13 | 98 | 98 | 1 | ||||
| RSP14 | 41 | 28.3 | 1 | ||||
| RSP15 | 38 | 38 | 1 | ||||
| RSP17 | 124 | 98.5 | 1 | ||||
| RSP18 | 210 | 210 | 1 | ||||
| RSP19 | 140 | 140 | 1 | ||||
| RSP20 | 18 | 18.3 | 4 | ||||
| RSP21 | 16 | 16 | 1 | ||||
| RSP22 | 8 | 10.3 | 2 |
MM, molecular mass; NEPHGE, 2D nonequilibrium pH-gradient gel electrophoresis.
RSP2 has been suggested as the link between the spoke stalk and head because in the mutant pf24, which is deficient in RSP2, the five spoke head proteins are missing from the flagella along with RSP2, RSP23, and RSP16 (Huang et al., 1981; Patel-King et al., 2004; Yang et al., 2004, 2005, 2008). To identify the location of these proteins in the RS, we investigated the 3D structure of pf24 RSs (Fig. 3, blue). Our reconstruction shows that pf24 (Fig. 3, blue) lacks the heads and necks of RS1 and RS2. RSP2, RSP16, and RSP23, therefore, form most of the branched RS neck (Fig. 4, yellow volume in segmented model) and are needed for the anchoring of the RS head to the rod-shaped stalk. The expected volume for the subset RSPs 2, 16, and 23 is ∼300 nm3, about one half of the volume calculated from our differential model (600 nm3; Fig. 4, yellow; and Table II). This suggests that one copy of RSP2, RSP16, and RSP23 is present in each of the two branches forming the RS neck. The remaining portion in both RSs of the pf24 model defines the stalk, a 28-nm elongated structure (Fig. 3 and Fig. 4, blue). We calculated the total volume of the stalk to be ∼1,600 nm3 (Table II).
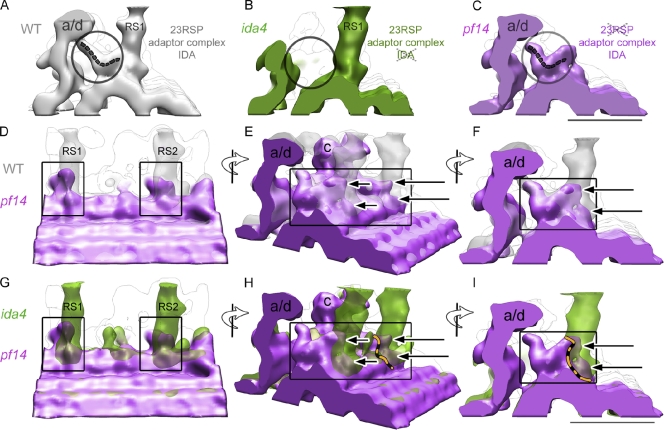
The last RS mutant we analyzed was pf14, usually defined as spokeless. In this mutant, the absence of the 23 RSPs is caused by a mutation in the single gene for RSP3 (Luck et al., 1977; Piperno et al., 1981; Williams et al., 1989). In the absence of RSP3, RSs are not assembled, and the flagella are immotile (Luck et al., 1977; Piperno et al., 1981; Williams et al., 1989; Diener et al., 1993; Wirschell et al., 2008). RSP3 is the stalk protein required for the assembly/anchoring of the T-shaped RS structure to the outer doublet microtubule. In vitro binding experiments (Diener et al., 1993) showed that RSP3 translated in vitro can bind to isolated spokeless pf14 axonemes. However, RSP3 does not bind directly to naked axonemal microtubules or to chick brain microtubules repolymerized in vitro (Diener et al., 1993), suggesting that an unidentified axonemal adaptor protein may link RSP3 to the microtubule. Our pf14 reconstruction (Fig. 3, purple) shows densities at which RS1 and RS2 are expected to bind to the A-microtubule (Fig. 3 and Fig. 5, C–I, arrows), indicating the presence of putative adaptor proteins (or adaptor complexes). These newly identified densities are in continuity with the tails of IDAs. The RS1 adaptor complex connects to the tail of dynein a/d (IDA assigned either dynein a or dynein d in Bui et al. [2008]; Fig. 5, A–I), whereas the RS2 adaptor complex connects to the dynein c tail (Fig. 5, D, E, G, and H; and see Fig. 8).
Figure 5.
Localization of adaptor complexes at the RS bases in C. reinhardtii. (A) Portion of the WT reconstruction showing the RS1 stalk, IDA a/d, and dynein a/d tail (encircled). The dashed gray line traces the backbone of the dynein a/d tail. (B) Portion of the ida4 mutant reconstruction. Ida4 lacks IDA a, c, and d. Note the absence of dynein a/d and the dynein a/d tail in the model reconstruction (circle). (C) Portion of the pf14 reconstruction showing IDA a/d and the dynein a/d tail (encircled). The dashed gray line traces the dynein a/d tail direction. (A–C) The proximal end of the axoneme points toward the reader. (D–F) Comparison of WT (gray) and pf14 (purple) reconstructions. pf14 lacks the 23 RSPs but has adaptor complexes (i.e., CSC) at the base of RS1 and RS2. Dynein a/d is also present in pf14 flagella (as in WT). (G–I) Comparison between ida4 (green) and pf14 (purple) reconstructions. ida4 lacks the tails of dynein a/d and c but shows RS1 and RS2. The portions of the pf14 model (purple) that penetrate RS1 and RS2 stalks (green, ida4) identify the adaptor protein complexes. (H) The orange and black dashed line indicates the boundary between the dynein c tail and the adaptor complex (CSC) at the base of RS2. (I) The dashed line (orange and black) indicates the boundary between the dynein a/d tail and the adaptor complex at the base of RS1 (the protein composition of this complex is unknown). (D and G) The proximal ends of the models are to the left; view from the adjacent doublet microtubules. (E and H) the same models as in D and G after 45° rotation. (F and I) the same models as in E and H after 45° rotation. The proximal end of the axoneme points toward the reader. (E, F, H, and I) Arrows point to densities of pf14 models located at the canonical RS binding sites. (D–I) Interface regions between RS and IDA are indicated by rectangles. (C and I, valid also for A and B and D–H) Bars, 25 nm.
Figure 8.
Model for RS structure. (A) The diagram summarizes current knowledge about the protein composition and the stoichiometry of one ideal RS. Assignment of the 23 RSPs to the four domains (head, neck, stalk, and CSC) is based on the literature and on our 3D reconstructions. The gray background marks the two 12S RS precursors and their protein composition according to Diener et al. (2011). The numbers highlighted in light blue indicate RS stalk proteins that might potentially break the twofold symmetry within one RS. The existing interactions between RSPs are also indicated. (B and C) Probable RSP locations within the RS reconstruction. (B) Side view; the proximal end of the axoneme points to the left. (C) End-on view; the distal end of the axoneme points toward the reader. (D) Top view showing the head of one RS. Possible molecular interactions based on A are indicated with dotted lines.
To accurately define the position of the IDA tails and RS adaptor complexes in pf14, we performed a comparative analysis of pf14, ida4, and WT (Fig. 5). Fig. 5 A shows the base of RS1 stalk, the RS1 adaptor complex, the inner dynein a/d, and the dynein a/d tail (Fig. 5 A, dashed line) in WT. Fig. 5 B shows the ida4 model as seen from the same viewpoint. ida4 is an IDA mutant that lacks dynein a, c, and d but retains RSPs and RS adaptor complexes. Fig. 5 C shows pf14, which lacks all 23 RSPs but retains the adaptor complexes and the IDAs (dynein a/d tail is indicated by the dashed line). The only components shown that are present in both ida4 and pf14 are the RS adaptor complexes. This allows us to look at the intersection of these two models to localize the shared complexes (Fig. 5, G–I, rectangles) and identify the adaptor complexes. In Fig. 5 (G–I), the adaptor complexes are represented by the portions of the pf14 model (Fig. 5, G–I, purple) that penetrate into the ida4 RS stalks (Fig. 5, G–I, green). In Fig. 5 (G–I), arrows indicate the adaptor complexes, whereas orange dashed lines show the IDA tail–adaptor complex contact surface. As shown in Fig. 5 (E, F, H, and I), adaptor complexes at the bases of RS1 and RS2 share similar morphology, suggesting two domains (Fig. 5, E, F, H, and I, two arrows) for each RS. Both domains are in contact with RSs. However, only one domain binds to IDA, whereas the other is on the A-tubule. This interpretation is in agreement with the recent finding of the CSC, which is part of the 20S RS complex and is thought to be responsible for the interaction between RSs and IDAs (Fig. 4 [purple], Fig. 5, and see Fig. 8; Dymek and Smith, 2007; Dymek et al., 2011).
Overall arrangement of RSs in T. thermophila
In T. thermophila, cilia RSs repeat in triplets, rather than pairs, every 96 nm along A-microtubules. Using cryo-ET and subtomogram averaging, we reconstructed the 3D structure of RS triplets of T. thermophila (Fig. 6, C and E). RS1 and RS2 are separated by 30 nm and are 43 nm tall, very similar in size and shape to the RSs observed in C. reinhardtii flagella. In both organisms, it is possible to distinguish three areas in the RS1 and RS2 structure: a rod-shaped stalk, a bifurcated neck (Fig. 6 F), and a head (Fig. 6, C, E, and F). As in C. reinhardtii, RS2 has two additional structures at the base of the stalk (Fig. 6 C). The shape of the RS1 and RS2 heads are similar to that observed in C. reinhardtii, although they have a slightly different shape and size (29 × 27 nm; Fig. 6 E). As with C. reinhardtii, these heads share a twofold symmetry, suggesting they are composed of identical head subdomains, and the heads are firmly connected to each other (Fig. 6 E).
Figure 6.
3D structure of RSs in T. thermophila and RS3 in C. reinhardtii. (A and B) Longitudinal (proximal end of the axoneme to the left; A) and side (as seen from the distal end; B) views of the WT C. reinhardtii extended RS pair. The RS3 stump is encircled. Bar, 50 nm. (C–F) Surface rendering of the RS triplet in T. thermophila. The base of RS3 is encircled, and a dashed line marks the approximate location corresponding to the upper side of RS3 stump found in C. reinhardtii. Note the striking similarity between the RS3 base in T. thermophila and the RS3 stump in C. reinhardtii (Fig. 2 and Fig. 3). (C) Longitudinal view with arrowheads pointing to RS2-specific densities. (D) Side view showing RS3. (E) Top view showing the three RS heads. The heads of RS1 and RS2 share the same structure (twofold rotational symmetry). Orange area shows one of the two subdomains that assemble in a single head and also has twofold rotational symmetry. The structure of the RS3 head differs from that of RS1 and RS2. (F) Side view showing RS1. a/d, IDA corresponding to C. reinhardtii dynein a/d; d/a, IDA corresponding to C. reinhardtii dynein d/a. See Fig. 1 A for IDA distribution.
RS3 in T. thermophila and in C. reinhardtii
The structure of the third RS (RS3) is probably the most surprising feature of our 3D reconstruction (Fig. 6, C–E). RS3 in T. thermophila shows a very different architecture from RS1 and RS2. Rather than extending perpendicular to the axonemal microtubule, RS3 leans toward RS2 and the B-microtubule (Fig. 6, C and D); a similar orientation of RS3 was observed in the sperm flagellum of Gallus domesticus (Burgess et al., 1991). The neck of RS3 doesn’t show any bifurcation (Fig. 6 D), and the head, which is smaller than the other RS heads, does not appear to be symmetrical (Fig. 6 E). A bridge connects the RS3 stalk to the head of the dynein corresponding to C. reinhardtii dynein g/b (Fig. S3), a feature that is not present in the C. reinhardtii reconstruction. The comparison between T. thermophila and C. reinhardtii WT 3D reconstructions, however, shows striking similarities between the base of the RS3 stalk in T. thermophila (Fig. 6, C and D, encircled) and the bulb-shaped structure of C. reinhardtii (Fig. 2 A, arrow; and Fig. 6, A and B, encircled). In T. thermophila, a bulb-headed structure forms the base of RS3 24 nm from RS2, perfectly matching the position of the bulbous structure of C. reinhardtii. In both cases, it is possible to distinguish an arclike density protruding from the bulbous structure (Fig. 6, A and C, right). This arc contacts the microtubule and the IDA (Fig. 6, B and D). Fig. 7 shows that the arclike structure is not actually part of RS3 but is a portion of the dynein d/a tail. This is best seen in the ida4 3D model (Fig. 7, green), in which the arclike density disappears together with dynein d/a (Fig. 7, arrows).
Figure 7.
RS3 and the tail of IDA d/a. 3D reconstruction of RS3 stumps from WT (gray; left) and ida4 (green; right) flagella. (A) View from the adjacent doublet microtubules. The proximal end of the axoneme is to the left. (B) The same two 3D reconstructions as in A after 80° rotation, as indicated by the open arrow. The IDA d/a is missing in ida4 flagella. The arrows and orange dashed lines show the approximate position of the dynein d/a tail in WT and its corresponding location in ida4. In ida4, no densities are visible at this location. d/a, dynein d/a; g/b, dynein g/b; arrowheads show the RS3 stump. Bar, 25 nm.
Nothing is known about the composition of RS3 in T. thermophila or its counterpart in C. reinhardtii. Our comparative analysis of the C. reinhardtii pf14 mutant (Fig. 3), whose flagella are devoid of all 23 RSPs, doesn’t show any striking alteration in the morphology of the RS3 basal portion, indicating that none of the known RSPs is a component of this RS3 stalk portion.
Discussion
We have used cryo-ET to reconstruct the 3D structure of RSs from C. reinhardtii and T. thermophila. Our reconstructions provide more structural details about RSs than the T-shape model obtained by freeze-fracture deep-etched replicas (Goodenough and Heuser, 1985), plastic embedded samples (Warner and Satir, 1974; Piperno et al., 1977; Huang et al., 1981; Diener et al., 1993; Dymek et al., 2011), and negatively stained samples (Yang et al., 2001, 2008; Qin et al., 2004). Based on our data, we can propose models for (a) subdomain organization of the RS, (b) interactions between the 23 RSPs, and (c) interactions between the RSs and other axonemal components.
RS pairs and triplets
At the current resolution (39 Å) in both C. reinhardtii and T. thermophila, RS1 and RS2 share a common overall structure, with two additional densities at the base of the RS2 stalk (Fig. 2 A and Fig. 6 C, arrowheads). The common structure suggests that RS1 and RS2 have the same, or at least a very similar, protein composition, with some minor differences at the base of the stalk. In contrast, RS3 in T. thermophila has a different orientation and structure, having a smaller asymmetric head, than RS1 and RS2. The RS3 stalk is directly connected to the dynein d/a tail and dynein g/b head, suggesting that RS3 is involved in the regulation of these dyneins. In C. reinhardtii flagella, only a stump is present at the position of RS3, appearing very similar to the basal half of the RS3 stalk of T. thermophila. This conserved portion of RS3 is also connected to dynein d/a (like in T. thermophila) but is too short to interact with the CP apparatus.
This similarity in RS3 structure in the two species suggests two possible pathways for the evolution of the C. reinhardtii RS3 stump: (1) it may have degenerated from the full-length RS3, or (2) it might represent an intermediate developmental stage before reaching full-length RS3. None of the known RSPs are components of the RS3 stump in C. reinhardtii, and nothing is known about the protein composition of RS3 in T. thermophila. It would be very interesting to perform a detailed analysis of the protein composition of RS3, which is a rather common structure (for avian species see Burgess et al. [1991]; for sea urchin see Nicastro et al. [2005]), and examine the effects of mutations of these proteins on RS activity and cilia/flagella motility.
The base of RS1 and RS2
The RS stalk is anchored to protofilaments A12 and A13 of the A-microtubule (Fig. S2). RSP3 is traditionally considered responsible for anchoring the complete RS structure to the microtubule because in the absence of RSP3, all 23 RSPs are missing from the axoneme (Piperno et al., 1977; Diener et al., 1993). Nevertheless, experimental evidence suggests that additional adaptor proteins are necessary to link RSP3 to the microtubule. In keeping with this hypothesis, Dymek and Smith (2007) discovered a complex of CaM and three CaM-binding proteins (CaM-IP2, -IP3, and -IP4), called CSC, that copurified with RSs yet was present in the spokeless axonemes of pf14. In C. reinhardtii, in which the expression of CaM-IP2 or CaM-IP3 was reduced using inhibitory micro-RNA, assembly of only RS2 was defective (Dymek et al., 2011), suggesting there are different adaptor complexes for RS1 and RS2.
Comparison of axonemes from pf14 and ida4 enabled us to visualize the presence of adaptor proteins as densities at the very base of the RSs. These densities are anchored to the A-microtubule and in direct contact with IDA tails. Our 3D reconstructions indicate that the adaptor density associated with RS1 interacts with the dynein a/d tail, whereas the adaptor density of RS2 is attached to the dynein c tail. These are plausible locations for enabling the CSC to act both as a stalk anchor and as a component of the dynein regulation pathway. Given the results of CSC artificial micro-RNA (Dymek et al., 2011), the adaptor density at the anchoring site of RS2 (Fig. 5 H, arrows and orange dashed line) in the pf14 reconstruction is apparently the CSC. The composition of the adaptor density at the base of RS1 (Fig. 5 I, arrows) remains mysterious, although it is clearly none of the known RSPs (Luck et al., 1977; Piperno et al., 1981, Williams et al., 1989) nor the CSC. Thus, RS1 and RSP2 have adaptor complexes of different morphology (Fig. 4 and Fig. 5) and protein composition.
The RS stalk
The stalk is composed of RSPs 3, 5, 7, 11–15, and 17–22, proteins that are rich in predicted functional motifs, including many motifs associated with signal transduction (Yang et al., 2006). This suggests that the spoke stalk probably serves as a scaffold for signaling molecules, having an important role in transducing chemical and mechanical signals (Yang et al., 2006).
Among these stalk proteins RSP3 contains an A-kinase–anchoring protein domain (Gaillard et al., 2001) and is involved in the anchoring of the RS complex to the microtubule and to the CSC (Diener et al., 1993; Dymek and Smith, 2007). Furthermore, RSP3 forms homodimers (Wirschell et al., 2008) that are central to the assembly of both 12S and 20S RSs (Wirschell et al., 2008; Diener et al., 2011). In building a model showing the positions of RSPs in the axonemal RS (Fig. 8), RSP3 must be located at the base of the stalk in contact with the adaptor complex (Diener et al., 1993). RSP3 is likely to bind directly to the neck (RSPs 2 and 23) or close to the neck, judging from the copurification of RSP3 and RSP2 in the 12S precursor, which is composed of RSPs 1–7, 9–12, and 23. Because RSPs 1, 4, 6, 9, and 10 are RSP head proteins, and homodimers of RSPs 7 and 11 (Colledge and Scott, 1999) are hypothesized to be localized at the bottom of the RS (Yang and Yang, 2006; Yang et al., 2006), RSP3 may bind RSP2 directly, or the two proteins could be linked only via RSPs 5 and 12 (detailed discussion in Diener et al. [2011]). This suggests that RSP3 dimers extend from the microtubule to the area where the stalk connects to the neck. Based on these previous biochemical studies, we provide the possible arrangement of RSPs in our 3D reconstruction as a diagram (Fig. 8, A–C).
RSP5 is thought to regulate coordination between the RSs and the outer arms (Yang et al., 2006). RSP5 has been proposed to localize at the spoke head/stalk boundary (Piperno et al., 1981). Our 3D reconstructions of pf24 spokes, however, indicate that RSP5 does not interact with the spoke head and that it is not located in the neck but somewhere in the stalk. GST pull-down and chemical cross-linking experiments (Kohno et al., 2011) also showed no interaction between RSP5 and head proteins. According to our results and to the literature (Huang et al., 1981; Patel-King et al., 2004; Yang et al., 2006; Kohno et al., 2011), two copies of RSP5 must be located in the stalk (Fig. 8, A and B; Diener et al., 2011).
RSP20 is a CaM (Yang et al., 2001). Because RSP2 and RSP23 contain CaM-binding domains and bind CaM in vitro (Yang et al., 2001, 2004; Patel-King et al., 2004), RSP20 is likely to be near, or in, the neck.
The position of the other RS stalk proteins remains difficult to predict at the current level of detail in our structural analysis. Based on the literature, we assume to find four copies of RSP20 (Yang et al., 2006) and RSP3 (Wirschell et al., 2008; Diener et al., 2011), two copies of RSP5, RSP7, RSP11, RSP12 (Diener et al., 2011), and RSP22 (Benashski et al., 1997), and one copy of RSP8, RSP21, RSP13–15, and RSP17–19 (Fig. 8; Yang et al., 2006) in the stalk of a WT RS and pf24 RS. Altogether these proteins would make up a complex of ∼1,400 kD, which matches the volume of the stalk, calculated on the basis of our 3D reconstructions. One, or several, of these single copy proteins may be specific to RS2, making up the two additional densities near the base of the stalk not seen in RS1.
The RS neck
Through comparative analysis of the 3D structure of RS in WT, pf1, and pf24 cells, we identified a bifurcated area located between the stalk and the head, which we called the neck. Based on the biochemistry of pf1 and pf24 flagella (Luck et al., 1977; Piperno et al., 1977; Huang et al., 1981; Patel-King et al., 2004; Yang et al., 2004, 2005), we conclude that the three RSPs, RSPs 2, 23, and 16, are part of the neck.
The two symmetric domains of the neck could arise from the dimerization of the 12S RS complex, each providing one domain. RSP2, but not RSP23, was identified as a component of this complex (Diener et al., 2011). In cytoplasmic extracts of pf1 and pf14, however, RSP2 was found in a 130–170-kD complex with an unidentified protein (Diener et al., 2011). This unidentified protein may have been RSP23, which was not probed for in these experiments. In keeping with this hypothesis, RSP2 and RSP23 synthesized in vitro bind to each other (Kohno et al., 2011). Both proteins also interact with the RS head: RSP2 binds to RSP4 and RSP10, and RSP23 binds to RSP1 (Kohno et al., 2011). Based on the neck map volume and the geometry of our RS reconstruction, the symmetrical part of the bifurcated neck can derive from the union of the two half-necks of two 12S RSs, each one containing one copy of RSP2 and RSP23 linking the RS stalk to the RS head.
The third component of the spoke neck is RSP16, which belongs to the HSP40 (DnaJ) family of molecular chaperones (Satouh et al., 2005; Yang et al., 2005, 2006, 2008). RSP16 is not part of the 12S RS precursors (Yang et al., 2008; Diener et al., 2011), rather it is transported into flagella separately, as a homodimer, to be subsequently incorporated into the fully assembled 20S spoke complex (Yang et al., 2005, 2006, 2008). Because RSP16 is not part of the 12S intermediate complex, which forms a major part of the 20S RS by dimerization, it could form an asymmetrical part of the neck (Fig. 8, B and C).
The RS head has a twofold rotational symmetry
In C. reinhardtii, two identical elongated domains build the spoke head (Fig. 2 C and Fig. 8 D). The two domains are positioned with twofold rotational symmetry. This is consistent with the hypothesis of dimerization of the 12S intermediate complex during 20S RS assembly (Diener et al., 2011). Each of the two identical elongated head domains binds to one of the two branches of the bifurcated neck as well (Fig. 2 B, Fig. 4, and Fig. 8, B and C), consistent with the 12S dimerization hypothesis.
A thought about this twofold symmetry might be of interest. The head of each RS is thought to transiently bind to the CP apparatus, which is unidirectional and not twofold symmetrical. A notion of direction, for allowing the RSs to interact with the microtubules in a specific orientation, would have to be introduced by a nonsymmetrical structure. However, a similar twofold symmetry was found in the intraflagellar transport of another microtubule-based flagellar protein complex (Pigino et al., 2009). Both structures might present a mechanism to enable motions in both directions along the axoneme.
Each symmetrical domain of the head shows five smaller subdomains, visible as bumps on the surface of the 3D reconstruction (Fig. 2 C, view from top; and Fig. 8), which are likely to correspond to the five component proteins, namely RSPs 1, 4, 6, 9, and 10 (Fig. 8 D; Yang et al., 2006). Actually, the expected amount of protein for each domain, as calculated from our 3D density map, indicates that two copies of RSPs 1, 4, 6, 9, and 10 are contained in each one of the two symmetrical head domains. This suggests that one bump contains two copies of the corresponding protein and that the spoke head contains, in total, four copies of each head protein, although we cannot exclude the possibility of heterodimer formation. Only RSP10, however, has been shown to form dimers (Kohno et al., 2011). Kohno et al. (2011) also detailed possible protein–protein interactions between the five head proteins RSP1, RSP4, RSP6, RSP9, and RSP10 in C. reinhardtii. Based on GST pull-down and chemical cross-linking experiments, they showed that RSP4 and RSP6 interact with RSP9 and RSP10 but not with each other. RSP1 interacts with RSP4 but not with the other RS head proteins (Kohno et al., 2011). In Fig. 8 D, the five head proteins have been arranged into the structure of the head according to the model proposed by Kohno et al. (2011).
The RS head is thought to interact with the projections of the CP and in some way transmit a signal down the spoke that ultimately regulates the activity of IDAs. The only motifs identified in the head proteins, however, were the membrane occupation and recognition nexus motifs present in RSP1 and RSP10, and these two proteins are close to each other in our dimerized model of the RS head (Fig. 8 D). It might be possible that the spoke head simply functions as a mechanical transducer of the signal from between the CP and the sensory area of the spoke neck, whereas the stalk propagates the signal from the neck to the CSC by such second messenger activity as phosphorylation, A-kinase, and Ca binding. The previous observation of tilted RSs (Warner and Satir, 1974) also suggests a role of mechanical stress for the function of RSs.
The symmetrical arrangement of the head is a conserved feature also in T. thermophila, although the shape of the head differed from the two organisms. Ueno et al. (2006) identified homologues of RSP4 and six in T. thermophila, and BLAST (basic local alignment search tool) analysis shows that homologues of RSP1, RSP9, and RSP10 are also present in T. thermophila. The difference in the shape between C. reinhardtii and T. thermophila RS heads could derive from a different organization of the five head proteins or from the presence of additional proteins.
In conclusion, our cryotomography investigation of RS1 and RS2 structure in C. reinhardtii strongly supports the hypothesis of 12S dimerization to form the mature RS. EM analysis of the negatively stained isolated 12S RS revealed a rod with a projection at one end forming a “7” or “L” (Diener et al., 2011). This putative 12S complex was ∼28 nm long, and the width of the head (the projection) was ∼20 nm (Diener et al., 2011). In view of their size and shape, one can easily imagine that two of these 7-shaped 12S intermediates bind together in a twofold symmetrical way to form the 3D structure of the completely assembled RS, as we present it in this paper (Fig. 2). Or, to be more precise, we propose that the symmetrical domains of the 20S RS (head and “backbone” of the neck and stalk) are generated by the dimerization of 12S particles. In Fig. 8 A, the RSPs highlighted in light blue are those that might potentially break the twofold symmetry in the RS stalk. Those RSPs are not components of the 12S and contribute, in a not yet defined fashion, to the final RS 20S axonemal complex.
Materials and methods
Cell culture and flagella isolation
The C. reinhardtii 137c WT and pf1, pf24, and pf14 mutant strains used in this study (Table I) were obtained from the Chlamydomonas Genetics Center. WT and mutant strains were cultured in TAP (Tris-acetate-phosphate) medium (Gorman and Levine, 1965). Data from the C. reinhardtii ida4 mutant were obtained from a previous study (Bui et al., 2008). T. thermophila WT cells obtained from the Tetrahymena Stock Center were used and cultured in proteose peptone medium (Orias et al., 2000).
Flagella and cilia from both organisms were isolated using the dibucaine method to induce deflagellation (Witman, 1986). The flagella were sedimented at 5,200 g for 20 min at 4°C, demembranated with 30 mM Hepes, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, 25 mM KCl, and 0.8% NP-40, and sedimented again at 5,200 g for 60 min at 4°C. The pellet of axonemes was resuspended in 30 mM Hepes, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, and 25 mM KCl and stored at 0°C before plunge freezing.
Quick plunge freezing and cryo-ET
The specimen was frozen with liquid ethane at liquid nitrogen temperature using a vitrification device (Vitrobot; FEI) and grids (Quantifoil Micro Tools GmbH) with holey carbon support film (200 mesh copper grid; R2/2; Quantifoil Micro Tools GmbH). 10-nm gold colloidal particles were applied on the grids during sample preparation and used later as fiducial markers for tomographic reconstruction. The grids with frozen-hydrated samples were transferred to a cryoholder (626; Gatan) cooled by liquid nitrogen. Images were collected as described previously (Ishikawa et al., 2007; Bui et al., 2008, 2009; Movassagh et al., 2010) using a transmission electron microscope (Tecnai F20; FEI) equipped with a field emission gun, an energy filter (GIF Tridiem; Gatan), and a 2,048 × 2,048 charge-coupled device camera (UltraScan 1000; Gatan) at the accelerating voltage of 200 kV, a magnification of 19,303×, and an under focus of 3–5 µm. Tomographic image series from −60 to 60°, with 2° tilt increments, were acquired using Explore3D software (FEI). Images were collected with a 0.1–0.2-s exposure time and energy filter width of 20 V. To select round-shaped flagella embedded in ice and to avoid deformation caused by compression (flattening), we carefully chose axonemes having a diameter of 220 nm (measured on single images) and later screened in 3D (Bui et al., 2009).
Image analysis
Tomograms were reconstructed by IMOD (Mastronarde, 1997) with fiducial marker alignment and R-weighted back projection. Subtomograms with pixel dimensions of 200 × 200 × 200 (∼137 nm in each dimension) were boxed out with a roughly 96-nm period from the original tomogram by Bsoft (Heymann, 2001) and aligned along the microtubule by SPIDER (Frank et al., 1996) followed by intermicrotubule alignment (Bui et al., 2008).
Averages were always deconvoluted by the total contribution of the missing wedge to keep data sampling isotropic as described in Bui et al. (2008). In brief, the deconvolution was performed by dividing the Fourier transformation of the averaged map by the weight from the contributions of missing wedges from all the original subtomograms, which have different orientations. Surface rendering was performed using the University of California, San Francisco Chimera software (Pettersen et al., 2004) after masking, denoising by a band-pass filter, and contrast inversion. The difference maps were generated using University of California, San Francisco Chimera (Pettersen et al., 2004).
The resolution of the reconstructions was measured using even/odd Fourier shell correlation between independent copies of the volume using the 0.5 criterion. Mass estimations of the head, neck, and stalk of the RS complex were calculated using the average density of 1.43 g/cm3 for proteins (Quillin and Matthews, 2000) and after normalizing the isosurface-rendering threshold to the mass of microtubules. The density maps of RS1 and RS2 were deposited in the EM Data Bank (accession no. EMD-1941).
Online supplemental material
Fig. S1 shows the Fourier shell correlation curve used to evaluate the resolution (in angstroms) of the 3D reconstruction shown in Fig. 1 and Fig. 2. Fig. S2 shows details of the interactions of the C. reinhardtii RS2 with the A-microtubule and the dynein c tail. Fig. S3 shows the connection of the T. thermophila RS3 with the head of dynein g/b. Video 1 shows the surface rendering of averaged axonemal 96-nm repeats from WT C. reinhardtii (the RSs are colored in yellow). Video 2 shows the surface rendering of the RS domains based on C. reinhardtii mutant analysis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106125/DC1.
Acknowledgments
We thank Prof. Joel Rosenbaum for the fruitful scientific discussions, Prof. Timothy J. Richmond for the biochemical facility, and Dr. Roger Wepf and Peter Tittmann (Electron Microscopy Center of Eidgenössische Technische Hochschule [ETH] Zürich).
This work was funded by grants from the Swiss National Science Foundation (NF31003A-125131/1), Swiss–Japan Cooperative Research Fund, and ETH Independent Investigators’ Research Awards (to T. Ishikawa) and by a European Molecular Biology Organization fellowship (to G. Pigino).
All commercial affiliations/conflicts of interest have been disclosed.
Footnotes
Abbreviations used in this paper:
- CP
- central pair
- cryo-ET
- cryoelectron tomography
- CSC
- CaM- and spoke-associated complex
- DRC
- dynein regulatory complex
- IDA
- inner dynein arm
- ODA
- outer dynein arm
- RS
- radial spoke
- RSP
- RS protein
- WT
- wild type
References
- Badano J.L., Mitsuma N., Beales P.L., Katsanis N. 2006. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7:125–148 10.1146/annurev.genom.7.080505.115610 [DOI] [PubMed] [Google Scholar]
- Benashski S.E., Harrison A., Patel-King R.S., King S.M. 1997. Dimerization of the highly conserved light chain shared by dynein and myosin V. J. Biol. Chem. 272:20929–20935 10.1074/jbc.272.33.20929 [DOI] [PubMed] [Google Scholar]
- Brokaw C.J., Luck D.J., Huang B. 1982. Analysis of the movement of Chlamydomonas flagella: the function of the radial-spoke system is revealed by comparison of wild-type and mutant flagella. J. Cell Biol. 92:722–732 10.1083/jcb.92.3.722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui K.H., Sakakibara H., Movassagh T., Oiwa K., Ishikawa T. 2008. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J. Cell Biol. 183:923–932 10.1083/jcb.200808050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui K.H., Sakakibara H., Movassagh T., Oiwa K., Ishikawa T. 2009. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J. Cell Biol. 186:437–446 10.1083/jcb.200903082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui K.H., Pigino G., Ishikawa T. 2011. Three-dimensional structural analysis of eukaryotic flagella/cilia by electron cryo-tomography. J. Synchrotron Radiat. 18:2–5 10.1107/S0909049510036812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S.A., Carter D.A., Dover S.D., Woolley D.M. 1991. The inner dynein arm complex: compatible images from freeze-etch and thin section methods of microscopy. J. Cell Sci. 100:319–328 [DOI] [PubMed] [Google Scholar]
- Castleman V.H., Romio L., Chodhari R., Hirst R.A., de Castro S.C.P., Parker K.A., Ybot-Gonzalez P., Emes R.D., Wilson S.W., Wallis C., et al. 2009. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 84:197–209 10.1016/j.ajhg.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge M., Scott J.D. 1999. AKAPs: from structure to function. Trends Cell Biol. 9:216–221 10.1016/S0962-8924(99)01558-5 [DOI] [PubMed] [Google Scholar]
- Curry A.M., Rosenbaum J.L. 1993. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil. Cytoskeleton. 24:224–232 10.1002/cm.970240403 [DOI] [PubMed] [Google Scholar]
- Davenport J.R., Yoder B.K. 2005. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am. J. Physiol. Renal Physiol. 289:F1159–F1169 10.1152/ajprenal.00118.2005 [DOI] [PubMed] [Google Scholar]
- Diener D.R., Ang L.H., Rosenbaum J.L. 1993. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J. Cell Biol. 123:183–190 10.1083/jcb.123.1.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener D.R., Yang P., Geimer S., Cole D.G., Sale W.S., Rosenbaum J.L. 2011. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken). 68:389–400 10.1002/cm.20520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek E.E., Smith E.F. 2007. A conserved CaM- and radial spoke–associated complex mediates regulation of flagellar dynein activity. J. Cell Biol. 179:515–526 10.1083/jcb.200703107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek E.E., Heuser T., Nicastro D., Smith E.F. 2011. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol. Biol. Cell. 22:2520–2531 10.1091/mbc.E11-03-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199 10.1006/jsbi.1996.0030 [DOI] [PubMed] [Google Scholar]
- Gaillard A.R., Diener D.R., Rosenbaum J.L., Sale W.S. 2001. Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP). J. Cell Biol. 153:443–448 10.1083/jcb.153.2.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger M.L., Portman N., McKean P.G. 2008. Swimming with protists: perception, motility and flagellum assembly. Nat. Rev. Microbiol. 6:838–850 10.1038/nrmicro2009 [DOI] [PubMed] [Google Scholar]
- Goodenough U.W., Heuser J.E. 1985. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100:2008–2018 10.1083/jcb.100.6.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman D.S., Levine R.P. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA. 54:1665–1669 10.1073/pnas.54.6.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermacher G., Sale W.S. 1995. Regulation of dynein-driven microtubule sliding by an axonemal kinase and phosphatase in Chlamydomonas flagella. Cell Motil. Cytoskeleton. 32:106–109 10.1002/cm.970320207 [DOI] [PubMed] [Google Scholar]
- Habermacher G., Sale W.S. 1996. Regulation of flagellar dynein by an axonemal type-1 phosphatase in Chlamydomonas. J. Cell Sci. 109:1899–1907 [DOI] [PubMed] [Google Scholar]
- Habermacher G., Sale W.S. 1997. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J. Cell Biol. 136:167–176 10.1083/jcb.136.1.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T., Raytchev M., Krell J., Porter M.E., Nicastro D. 2009. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell Biol. 187:921–933 10.1083/jcb.200908067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann J.B. 2001. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133:156–169 10.1006/jsbi.2001.4339 [DOI] [PubMed] [Google Scholar]
- Huang B.P.-H. 1986. Chlamydomonas reinhardtii: A model system for the genetic analysis of flagellar structure and motility. Int. Rev. Cytol. 99:181–215 10.1016/S0074-7696(08)61427-8 [DOI] [Google Scholar]
- Huang B., Piperno G., Ramanis Z., Luck D.J. 1981. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88:80–88 10.1083/jcb.88.1.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Ramanis Z., Luck D.J. 1982. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 28:115–124 10.1016/0092-8674(82)90381-6 [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Sakakibara H., Oiwa K. 2007. The architecture of outer dynein arms in situ. J. Mol. Biol. 368:1249–1258 10.1016/j.jmb.2007.02.072 [DOI] [PubMed] [Google Scholar]
- Kagami O., Kamiya R. 1992. Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J. Cell Sci. 103:653–664 [Google Scholar]
- Kamiya R. 1982. Extrusion and rotation of the central-pair microtubules in detergent-treated Chlamydomonas flagella. Prog. Clin. Biol. Res. 80:169–173 [DOI] [PubMed] [Google Scholar]
- Kamiya R., Kurimoto E., Muto E. 1991. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J. Cell Biol. 112:441–447 10.1083/jcb.112.3.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno T., Wakabayashi K., Diener D.R., Rosenbaum J.L., Kamiya R. 2011. Subunit interactions within the Chlamydomonas flagellar spokehead. Cytoskeleton (Hoboken). 68:237–246 10.1002/cm.20507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck D., Piperno G., Ramanis Z., Huang B. 1977. Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc. Natl. Acad. Sci. USA. 74:3456–3460 10.1073/pnas.74.8.3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. 1997. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120:343–352 10.1006/jsbi.1997.3919 [DOI] [PubMed] [Google Scholar]
- Mitchell D.R., Nakatsugawa M. 2004. Bend propagation drives central pair rotation in Chlamydomonas reinhardtii flagella. J. Cell Biol. 166:709–715 10.1083/jcb.200406148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movassagh T., Bui K.H., Sakakibara H., Oiwa K., Ishikawa T. 2010. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat. Struct. Mol. Biol. 17:761–767 10.1038/nsmb.1832 [DOI] [PubMed] [Google Scholar]
- Nicastro D., McIntosh J.R., Baumeister W. 2005. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc. Natl. Acad. Sci. USA. 102:15889–15894 10.1073/pnas.0508274102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olm M.A.K., Jr, Kögler J.E., Jr, Macchione M., Shoemark A., Saldiva P.H.N., Rodrigues J.C. 2011. Primary ciliary dyskinesia: evaluation using cilia beat frequency assessment via spectral analysis of digital microscopy images. J. Appl. Physiol. 111:295–302 10.1152/japplphysiol.00629.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto C.K., Gibbons I.R., Kamiya R., Shingyoji C., Takahashi K., Witman G.B. 1999. Rotation of the central pair microtubules in eukaryotic flagella. Mol. Biol. Cell. 10:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E., Hamilton E.P., Orias J.D. 2000. Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 62:189–211 10.1016/S0091-679X(08)61530-7 [DOI] [PubMed] [Google Scholar]
- Patel-King R.S., Gorbatyuk O., Takebe S., King S.M. 2004. Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol. Biol. Cell. 15:3891–3902 10.1091/mbc.E04-04-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pigino G., Geimer S., Lanzavecchia S., Paccagnini E., Cantele F., Diener D.R., Rosenbaum J.L., Lupetti P. 2009. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 187:135–148 10.1083/jcb.200905103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Huang B., Luck D.J. 1977. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 74:1600–1604 10.1073/pnas.74.4.1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Huang B., Ramanis Z., Luck D.J. 1981. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 88:73–79 10.1083/jcb.88.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M.E., Sale W.S. 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151:F37–F42 10.1083/jcb.151.5.F37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Diener D.R., Geimer S., Cole D.G., Rosenbaum J.L. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255–266 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin M.L., Matthews B.W. 2000. Accurate calculation of the density of proteins. Acta Crystallogr. D Biol. Crystallogr. 56:791–794 10.1107/S090744490000679X [DOI] [PubMed] [Google Scholar]
- Satir P., Christensen S.T. 2007. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69:377–400 10.1146/annurev.physiol.69.040705.141236 [DOI] [PubMed] [Google Scholar]
- Satouh Y., Padma P., Toda T., Satoh N., Ide H., Inaba K. 2005. Molecular characterization of radial spoke subcomplex containing radial spoke protein 3 and heat shock protein 40 in sperm flagella of the ascidian Ciona intestinalis. Mol. Biol. Cell. 16:626–636 10.1091/mbc.E04-09-0784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.F. 2002. Regulation of flagellar dynein by calcium and a role for an axonemal calmodulin and calmodulin-dependent kinase. Mol. Biol. Cell. 13:3303–3313 10.1091/mbc.E02-04-0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.F., Sale W.S. 1992. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 257:1557–1559 10.1126/science.1387971 [DOI] [PubMed] [Google Scholar]
- Smith E.F., Yang P. 2004. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton. 57:8–17 10.1002/cm.10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgess J.M., Chao J., Wong J., Aspin N., Turner J.A. 1979. Cilia with defective radial spokes: a cause of human respiratory disease. N. Engl. J. Med. 300:53–56 10.1056/NEJM197901113000201 [DOI] [PubMed] [Google Scholar]
- Ueno H., Iwataki Y., Numata O. 2006. Homologues of radial spoke head proteins interact with Ca2+/calmodulin in Tetrahymena cilia. J. Biochem. 140:525–533 10.1093/jb/mvj182 [DOI] [PubMed] [Google Scholar]
- Warner F.D., Satir P. 1974. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J. Cell Biol. 63:35–63 10.1083/jcb.63.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.D., Velleca M.A., Curry A.M., Rosenbaum J.L. 1989. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14 is an ochre allele. J. Cell Biol. 109:235–245 10.1083/jcb.109.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M., Zhao F., Yang C., Yang P., Diener D., Gaillard A., Rosenbaum J.L., Sale W.S. 2008. Building a radial spoke: flagellar radial spoke protein 3 (RSP3) is a dimer. Cell Motil. Cytoskeleton. 65:238–248 10.1002/cm.20257 [DOI] [PubMed] [Google Scholar]
- Witman G.B. 1986. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134:280–290 10.1016/0076-6879(86)34096-5 [DOI] [PubMed] [Google Scholar]
- Witman G.B., Plummer J., Sander G. 1978. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76:729–747 10.1083/jcb.76.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Yang P. 2006. The flagellar motility of Chlamydomonas pf25 mutant lacking an AKAP-binding protein is overtly sensitive to medium conditions. Mol. Biol. Cell. 17:227–238 10.1091/mbc.E05-07-0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Compton M.M., Yang P. 2005. Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Mol. Biol. Cell. 16:637–648 10.1091/mbc.E04-09-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Owen H.A., Yang P. 2008. Dimeric heat shock protein 40 binds radial spokes for generating coupled power strokes and recovery strokes of 9 + 2 flagella. J. Cell Biol. 180:403–415 10.1083/jcb.200705069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Diener D.R., Rosenbaum J.L., Sale W.S. 2001. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 153:1315–1326 10.1083/jcb.153.6.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Yang C., Sale W.S. 2004. Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot. Cell. 3:72–81 10.1128/EC.3.1.72-81.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Diener D.R., Yang C., Kohno T., Pazour G.J., Dienes J.M., Agrin N.S., King S.M., Sale W.S., Kamiya R., et al. 2006. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119:1165–1174 10.1242/jcs.02811 [DOI] [PMC free article] [PubMed] [Google Scholar]