The Tim50 subunit of the mitochondrial TIM23 complex contains a presequence-binding domain that is essential for viability and precursor transport across the inner membrane.
Abstract
N-terminal targeting signals (presequences) direct proteins across the TOM complex in the outer mitochondrial membrane and the TIM23 complex in the inner mitochondrial membrane. Presequences provide directionality to the transport process and regulate the transport machineries during translocation. However, surprisingly little is known about how presequence receptors interact with the signals and what role these interactions play during preprotein transport. Here, we identify signal-binding sites of presequence receptors through photo-affinity labeling. Using engineered presequence probes, photo cross-linking sites on mitochondrial proteins were mapped mass spectrometrically, thereby defining a presequence-binding domain of Tim50, a core subunit of the TIM23 complex that is essential for mitochondrial protein import. Our results establish Tim50 as the primary presequence receptor at the inner membrane and show that targeting signals and Tim50 regulate the Tim23 channel in an antagonistic manner.
Introduction
Targeting signals in newly synthesized proteins provide directionality to intracellular transport processes and guide proteins to and across the limiting membranes of the endoplasmic reticulum, nucleus, peroxisomes, chloroplasts, or mitochondria (Schnell and Hebert, 2003; Wickner and Schekman, 2005; Rapoport, 2007). The signals are recognized by receptors, which initiate organelle-selective transport steps. Besides providing directionality for the transport process, targeting signals frequently initiate downstream events in the precursor translocation processes. Thus, signal recognition by cognate receptors may activate diverse processes essential for precursor transport such as translocase activation, receptor-membrane recruitment, nucleotide hydrolysis, or channel gating. In this regard, targeting-signal receptor complex formation can be considered as a molecular switch that transforms the translocase from its resting conformation into the transport active state.
In the case of mitochondria, the majority of proteins are imported from the cytoplasm post-translationally. Different types of targeting signals drive protein transport across the outer membrane and subsequently direct precursor proteins into one of the four mitochondrial subcompartments: outer membrane, intermembrane space (IMS), inner membrane, or matrix (Neupert and Herrmann, 2007; Chacinska et al., 2009; Endo and Yamano, 2010; Mick et al., 2011). The most prominent type of mitochondrial targeting signals are cleavable presequences. Presequences represent an N-terminal segment of a precursor with an average length of 15–55 amino acids, characterized by a net positive charge and the propensity to form amphipathic α-helices (von Heijne, 1986; Vögtle et al., 2009). The presequence directs the precursor protein across the outer and inner mitochondrial membranes. Membrane translocation of the precursor is mediated through aqueous pores formed by the translocase of the outer membrane (TOM complex) and the presequence translocase of the inner membrane (TIM23 complex). Both translocase complexes recognize presequences with specific receptor subunits.
A redundant set of receptors for presequence-containing precursors on the mitochondrial surface directs the precursor to the pore-forming subunit, Tom40. Upon exit from the Tom40 channel, presequence-containing precursors engage with the presequence translocase of the inner membrane. Tim50 is an essential subunit of the TIM23 complex that interacts with the channel-forming Tim23 (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2009; Tamura et al., 2009). Tim23 exposes a presequence receptor domain into the intermembrane space, to which Tim50 binds as well. While presequence recognition by Tim23 activates channel activity (Truscott et al., 2001; Meinecke et al., 2006; van der Laan et al., 2007), Tim50 binding to Tim23 antagonizes this activation and induces channel closure when no precursor is transported (Meinecke et al., 2006; Alder et al., 2008a). Thus, the activity of the inner membrane translocase is tightly regulated by targeting signals in a yet undefined manner.
Despite the obvious fundamental importance of receptor–presequence interactions for precursor transport, surprisingly little is known on how presequences are recognized by their receptors. The only available molecular information on a presequence-binding site is the structure of rat ALDH (aldehyde dehydrogenase) presequence in complex with Tom20 (Abe et al., 2000). This lack of knowledge significantly limits our experimental means to address how signal recognition regulates mitochondrial translocases and concomitantly the transport process.
To identify and characterize mitochondrial presequence receptors in structural and functional terms, we chose a biochemical approach that reveals presequence-binding proteins through preparative receptor-peptide photo cross-linking, allowing for mass spectrometric delineation of presequence-binding domains. Our analyses revealed Tim50 as a presequence receptor and identified its targeting signal receptor domain. We demonstrate that presequence and translocase binding by Tim50 can be uncoupled into two functionally distinct and largely independent domains. Presequence recognition by Tim50 is essential for protein transport across the inner membrane of mitochondria, but does not depend on an interaction between the Tim23 channel and Tim50. Our analyses reveal that presequence and Tim50 binding to the Tim23 channel occur mutually exclusive, and thus provide insight as to how channel activity is antagonistically regulated during precursor transport.
Results
Design and characterization of presequence probes
To identify presequence receptors and determine their presequence-binding domains, we devised an approach based on preparative scale presequence peptide photo cross-linking. The available structure of rat ALDH presequence in complex with Tom20 (Abe et al., 2000) was used to design multifunctional probes of presequence peptides. In the peptide probes, the photoreactive amino acid derivative para-benzoylphenylalanine (BPA) replaced either serine 16 (pS16B) or leucine 19 (pL19B). Upon activation by UV irradiation, the benzophenone moiety of the photo-probe preferentially reacts with C-H bonds of the target protein to form C-C bonds. The positioning of the BPA was designed such that it was exposed to the hydrophilic, in case of pS16B, or hydrophobic side of the helix, in case of pL19B. A trypsin cleavage site was introduced N-terminally of the affinity tags to enable their release before mass spectrometric analysis. For affinity purification and detection purposes, a biotinyl lysine and a hexa histidine tag were placed at the C terminus (Fig. 1 A). Because photo cross-linking of peptide probes via BPA to a receptor generates covalent bonds, we used mass spectrometry to localize these bonds at the amino acid level and define binding regions for presequences. To validate our strategy, we purified the cytoplasmic domain of yeast Tom20 and GST, as a control, and incubated these with or without presequence probes followed by UV irradiation. As expected, both presequence probes cross-linked to Tom20, but did not significantly react with GST (Fig. S1 A). Interestingly, pS16B formed significantly less photo-adducts than pL19B, in agreement with the fact that the BPA group was positioned on the charged, lateral side of the helix (Abe et al., 2000). We subjected tryptic digests of pL19B photo-adducts to LC MALDI MS/MS analyses and identified cross-links of the peptide probe to Met53 and Met105 of Tom20 (Fig. S1, B and C), in agreement with some preference of BPA to react with methionine residues (Wittelsberger et al., 2006). Modeling of yeast Tom20CD in complex with the pALDH based on a representative structure of the rat Tom20 (Abe et al., 2000) allowed positioning of pALDH within the binding groove of yeast Tom20 (Fig. S1 D). The distance of the BPA group to the Met105 was within the reactive radius of 3–15 Å. Met53 could not be modeled in the yeast structure as the available structures of rat Tom20 lack the first 50 amino acids. This finding supported our strategy and showed that the photo-affinity labeling approach is in fact able to proximate a receptor domain for presequences.
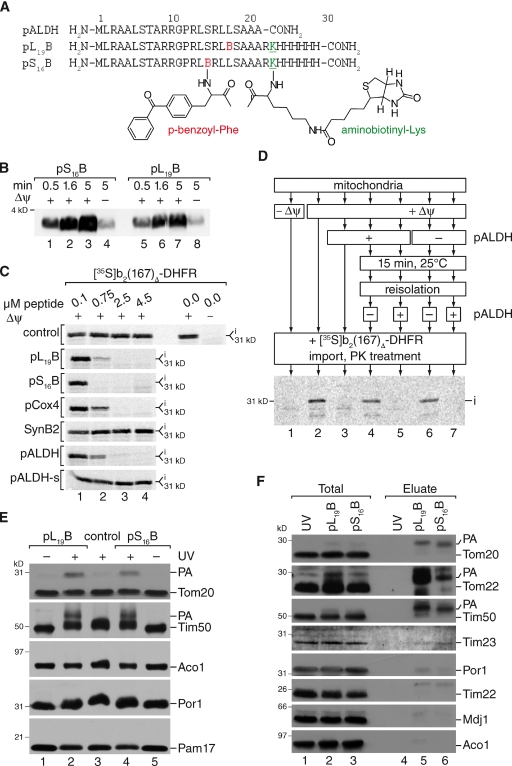
Figure 1.
Engineering presequence probes for receptor screening. (A) Primary structure of presequence probes and rat ALDH presequence (pALDH). (B) Presequence probes were imported into isolated mitochondria in the presence or absence of a Δψ for the indicated times at 25°C. After import, mitochondria were treated with proteinase K and samples analyzed by Western blotting using streptavidin-HRP (horseradish peroxidase). (C) Radiolabeled precursor was imported for 15 min at 25°C into isolated mitochondria in the presence of indicated amounts of peptides. After proteinase K treatment, import reactions were analyzed by SDS-PAGE and digital autoradiography. i, intermediate. (D) Mitochondria were incubated with and without presequence peptide in the presence or absence of Δψ. After re-isolation of mitochondria, protein import of a radiolabeled precursor protein was assessed in a second incubation. Therefore, radiolabeled precursor was added to treated or untreated mitochondria and import performed for 15 min. A control import was performed in the presence of peptide. Samples were analyzed by SDS-PAGE and digital autoradiography as in C. i, intermediate. (E) Isolated mitochondria were incubated with 2 µM of the respective photo-peptide, equilibrated for 10 min, and subjected to UV irradiation for 30 min. PA, photo-adduct. (F) After in organello photo cross-linking for 30 min, photo-adducts (PA) were purified from mitochondria by streptavidin agarose chromatography. Samples were analyzed by Western blotting with indicated antibodies. Total, 5%; Eluate, 100%. PA, photo-adduct.
To test if the presequence probes could be applied as tools to analyze other presequence receptors, peptides were initially tested for their ability to cross mitochondrial membranes in an in vitro import reaction. Both peptides were efficiently imported into mitochondria in a membrane potential (Δψ)–dependent manner, indicating that they had crossed the outer and inner membranes (Fig. 1 B). This was further supported by protease protection analyses (Fig. S2 A). To assess if presequence probes were transported along the authentic transport pathway into mitochondria, we performed import competition assays. Therefore, we imported the radiolabeled precursor of the model matrix-targeted preprotein b2(167)Δ-DHFR (comprising the N-terminal portion of cytochrome b2 fused to dihydrofolate reductase) into isolated mitochondria in the presence or absence of increasing amounts of pS16B or pL19B and compared their effect on precursor transport with that of different established functional presequence peptides and inactive versions thereof. Import of b2(167)Δ-DHFR was efficiently inhibited in the presence of Cox4 presequence peptides (pCox4) and rat ALDH presequence peptides (pALDH). In contrast, nonfunctional versions of the peptides, SynB2 (Truscott et al., 2001) or pALDH-s (similar amino acid composition but shuffled primary sequence), had no effect on matrix protein transport. The tested peptide probes pS16B and pL19B efficiently blocked import of the matrix-destined precursor at a similar concentration range as the authentic presequence peptides pCox4 and pALDH (Fig. 1 C).
To exclude that the block of precursor import by presequence peptides was due to a damaging effect of the amphipathic peptides on mitochondrial membranes, we first incubated mitochondria in the presence or absence of pALDH. After re-isolation of mitochondria, b2(167)Δ-DHFR was imported, resulting in efficient translocation of the matrix-destined precursor independent of peptide pretreatment (Fig. 1 D). For comparison, we tested pL19B and found that, similar to the authentic pALDH peptide, it did not affect mitochondrial integrity (Fig. S2 B). Moreover, transport of carrier precursors, which use internal signals, was not affected by the tested peptides (Fig. S2 C). We thus concluded that the peptides follow the authentic presequence transport pathway into the mitochondrial matrix.
Because the presequence probes were efficiently imported into mitochondria, we assessed if they could be used to identify presequence receptors in organello during the import process. Therefore, we incubated isolated mitochondria in the presence or absence of pS16B or pL19B peptide probes and subjected the reactions to UV irradiation for cross-linking. Photo-adducts were solely detected upon UV irradiation of the samples (Fig. 1 E) and the amount of adducts increased depending on the incubation time with mitochondria (Fig. S2 D). After photo cross-linking in isolated mitochondria under import conditions, photo-adducts were purified from mitochondria via the peptide’s biotin tag. Besides Tom20, we detected photo-adducts of presequence probes with Tom22 (Fig. 1 F). Moreover, adduct formation of Tim50 with presequence probes was observed (Fig. 1, E and F). Surprisingly, in mitochondria we could not detect photo-adducts between presequences and Tim23 (Fig. 1 F). We confirmed that the slower-migrating Tim50 bands represented presequence photo-adducts by decorating Tim50 immunoprecipitations with Tim50 antibodies as well as streptavidin-HRP conjugates. As expected, the slower-migrating bands were recognized in both cases (Fig. S2 E). Accordingly, upon UV irradiation presequence probes were specifically photo cross-linked to cognate receptors in organello and the resulting photo-adducts could be purified via the peptide probe, revealing Tim50 as a presequence receptor in the physiological context.
Tim50 is a presequence receptor at the inner membrane
Tim50 is an essential constituent of the TIM23 complex that can be cross-linked to precursors, which are accumulated in the TOM complex (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). The approximately 40-kD Tim50 IMS domain binds to Tim23 to regulate the channel’s activity. This interaction is considered to be critical for recruitment of Tim50 to the TIM23 complex (Geissler et al., 2002; Yamamoto et al., 2002; Meinecke et al., 2006; Gevorkyan-Airapetov et al., 2009). On the primary structure level, Tim50 contains a small segment exposed to the matrix, a transmembrane segment, an intermembrane space located phosphatase-like core segment (NIF) domain, and a C-terminal region of unknown function (Fig. 2 A). In organello photo cross-linking revealed Tim50 as a presequence-binding protein in the inner mitochondrial membrane (Fig. 1, E and F).
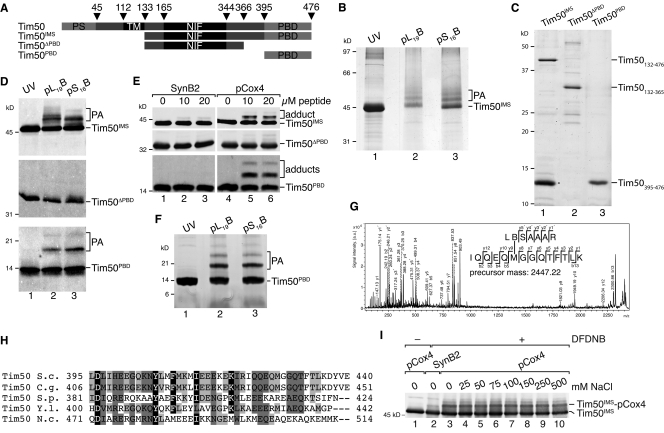
Figure 2.
Tim50 contains a C-terminal presequence-binding domain. (A) Schematic representation of Tim50 and truncation constructs used in this study. PS, presequence; TM, transmembrane domain; NIF, NIF domain; PBD, presequence-binding domain. (B) Purified Tim50IMS was incubated with presequence probes under UV irradiation for 30 min, samples analyzed by SDS-PAGE, and proteins subjected to in-gel digest before LC MALDI MS/MS analysis. PA, photo-adduct. (C) Indicated Tim50 constructs were purified from E. coli and analyzed by SDS-PAGE and stained with colloidal Coomassie. Asterisk denotes proteolytic Tim50PBD fragment. (D) Purified Tim50 variants were subjected to presequence photo cross-linking for 30 min and analyzed by Western blotting using anti-Tim50 antibodies. PA, photo-adduct. (E) Chemical cross-linking of pCox4 and SynB2 to isolated Tim50 variants using 100 µM DFDNB for 30 min on ice. Samples were analyzed as in D. (F) Photo cross-linking of presequences to Tim50PBD analyzed as in B. PA, photo-adduct. (G) Fragment ion mass spectrum of peptide Tim50422-435 cross-linked to pL19B18-24. Photo-adduct of pL19B with Tim50PBD (F) was digested with trypsin and subjected to LC-MALDI-MS/MS analysis. The indicated series of y- and b-ions revealed Met427 as the cross-link site. Signals of m/z 837.53 and 903.49 are frequently observed after fragmentation of cross-linked methionine side chains. (H) Alignment of Tim50 using ClustalW 2.0.11. Shown is the PBD domain; black, identical residues in four species, similar residues in at least four or three species are colored in dark or light gray, respectively. Similarity rules according to Erdmann et al. (1991). S.c., Saccharomyces cerevisiae; C.g., Candida glabrata; S.p., Schizosaccharomyces pombe; Y.l., Yarrowia lipolytica; N.c., Neurospora crassa. (I) Chemical cross-linking of 1 µM Tim50IMS to 5 µM pCox4 or SynB2 in the presence of increasing salt concentrations using 100 µM DFDNB for 30 min on ice. Samples were analyzed as in D.
To determine the presequence-binding site of Tim50, we purified Tim50IMS and performed in vitro photo cross-linking with presequence probes (Fig. 2 B). Both probes yielded photo-adducts that we mapped by LC MALDI MS/MS. In both cases, a photo-adduct was identified and located in the C-terminal portion of Tim50 (Fig. S3, A and B). To confirm this finding, we purified the C terminus of Tim50 (Tim50PBD) and the intermembrane space domain lacking the C terminus (Tim50ΔPBD) from Escherichia coli (Fig. 2 C). During Tim50IMS purification, a proteolytic fragment was generated that closely resembled the Tim50PBD construct as assessed by mass spectrometric analysis (unpublished data), suggesting that the C terminus of Tim50 forms a compact folded domain. Photo cross-linking lead to adduct formation of pL19B and pS16B with Tim50IMS and Tim50PBD, but not with Tim50ΔPBD (Fig. 2 D), supporting the idea that a binding site for presequences exists in the C terminus of Tim50. This finding was corroborated through an alternative chemical cross-linking approach in which cross-linking of the Cox4 presequence with a homo-bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene (DFDNB), was achieved to Tim50IMS and Tim50PBD, but not to a significant extend with Tim50ΔPBD. Only upon extended exposure of the blots was a faint adduct detected for Tim50ΔPBD, supporting the conclusion that the PBD is the primary binding site for presequences on Tim50IMS. The inactive variant of pCox4, SynB2, did not cross-link significantly to any of the Tim50 constructs (Fig. 2 E). We mapped photo-adducts of Tim50PBD from preparative scale analyses by LC MALDI MS/MS (Fig. 2 F). All photo cross-linking sites identified localized to a conserved element in the C terminus (Fig. 2, G and H; and Fig. S3, C–J).
Branched proteins generated by peptide cross-links between two polypeptide chains frequently display an aberrant migration pattern on SDS-PAGE with several adducts of appearingly different size (Junge et al., 2004; Alder et al., 2008b). Because cross-links of presequence peptides with Tim50 appeared as more than a single adduct on SDS-PAGE, we used mass spectrometry to analyze whether more than one peptide could be cross-linked to Tim50PBD. Only a minor fraction of photo-adducts between two peptides and Tim50PBD were detected (Fig. S4, A and B), indicating that 1:1 complexes between presequences and Tim50PBD predominate.
Because presequences are amphipathic in character, we addressed the biochemical properties that drive presequence binding to the PBD by salt titration. Our analyses show that adduct formation of presequences with Tim50IMS are insensitive to high salt concentrations, indicating that presequences interact with Tim50 through hydrophobic interactions (Fig. 2 I). In summary, these analyses provide evidence that Tim50 represents a presequence receptor at the inner membrane that recognizes the hydrophobic face of the presequence. Moreover, the analyses revealed the position of the receptor domain at the C terminus of the protein, a prerequisite for further functional analyses on the role of Tim50 in protein transport.
Presequence binding by Tim50 is essential for cell viability
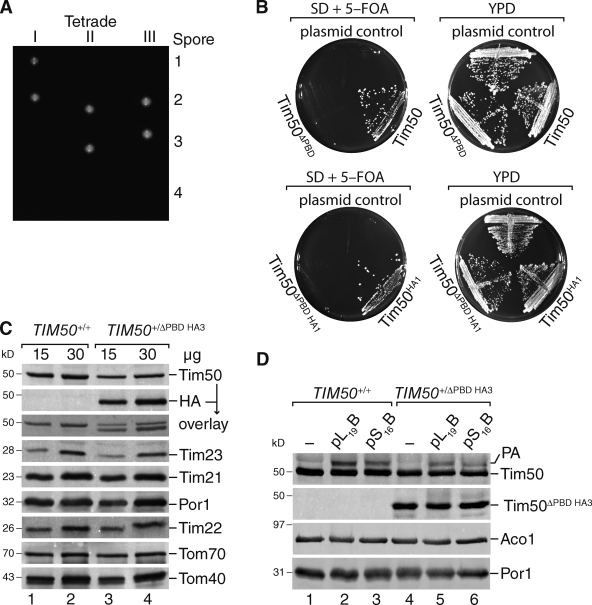
To assess the role of presequence binding in regards to Tim50’s function, we deleted the 3′ region of the ORF by homologous recombination in a diploid yeast strain, truncating the corresponding protein after amino acid 365. After sporulation and tetrad dissection, only two viable spores were recovered, indicating a 1:1 segregation of a lethal phenotype (Fig. 3 A). As an alternative approach, we applied a plasmid loss strategy using a strain that carried wild-type TIM50 on a plasmid with URA3 as selectable marker (Chacinska et al., 2005). The strain was transformed with an empty plasmid, a plasmid carrying TIM50 (or TIM50HA1), or a plasmid carrying TIM50 with 3′ deletion, encoding a protein lacking the C-terminal presequence-binding domain, TIM50ΔPBD (or TIM50ΔPBDHA1). 5-FOA selection yielded viable colonies only when full-length Tim50 was expressed from the second plasmid, but not upon expression of Tim50ΔPBD (irrespective of a C-terminal tag; Fig. 3 B). We purified mitochondria from diploid TIM50+/ΔPBD HA3 cells. Western blot analysis for selected mitochondrial Tim and Tom proteins did not reveal differences between the strains. Moreover, Tim50ΔPBD HA3 was readily detectable in mitochondria of the corresponding strain (Fig. 3 C). These mitochondria were subsequently used for UV-induced in organello photo cross-linking with presequence probes. In mitochondria, only full-length Tim50 but not Tim50ΔPBD HA3 was cross-linked to presequences (Fig. 3 D), indicating that the truncated Tim50 did not display presequence recognition in organello paralleling the results obtained for the in vitro cross-link. In summary, we find that presequence recognition by Tim50 is essential for cell viability.
Figure 3.
The presequence-binding domain of Tim50 is essential. (A) Diploid yeast cells carrying a single chromosomal deletion of TIM50366-476 were sporulated and subjected to tetrad dissection. (B) Yeast cells containing a chromosomal deletion of TIM50, complemented by a TIM50-containing plasmid carrying URA3, were transformed with plasmid constructs encoding the indicated Tim50 versions and subjected to plasmid loss on 5-FOA–containing medium. (C) Western blot analysis of mitochondria isolated from the cells described in A with the indicated antibodies. (D) In organello photo cross-linking of presequence probes using mitochondria from indicated strains for 30 min. Samples were analyzed as in C.
Presequence binding by Tim50 occurs independent of Tim23 interaction
A recent study suggested that the proximity of Tim50 to a matrix-destined precursor protein, as assessed by chemical cross-linking, depended on the presence of Tim23 (Mokranjac et al., 2009). Therefore, we analyzed binding of Tim50 and Tim50ΔPBD HA3 to the intermembrane space domain of Tim23 (Tim23IMS) to exclude that truncation of Tim50 abrogated binding to Tim23. Mitochondrial extracts from the diploid strain expressing Tim50 and Tim50ΔPBD HA3 were incubated with immobilized purified Tim23IMS. Both Tim50 and Tim50ΔPBD HA3 bound to Tim23IMS, indicating that Tim50ΔPBD retained its ability to bind to Tim23 in vitro (Fig. 4 A). However, in our assay, which addresses Tim50’s interaction with Tim23 in solution, Tim50ΔPBD displayed ∼30% binding compared with Tim50IMS. In mitochondria, Tim50 is a membrane protein associated with the TIM23 complex that can only diffuse in two dimensions and of which the local concentration at the translocase is unknown. Moreover, association between Tim50 and the first transmembrane span of Tim23 have been found (Alder et al., 2008b). Therefore, we addressed the association of Tim50 and Tim50ΔPBD with the TIM23 complex under more physiological conditions in mitochondria. We solubilized mitochondria expressing Tim50HA1 or Tim50ΔPBD HA1 in a background in which expression from the chromosomal TIM50 had been shut down and Tim50 levels, expressed from the chromosome, were drastically reduced (Fig. S5 A). Antibodies directed against HA efficiently immunoprecipitated Tim50HA1 or Tim50ΔPBD HA1 together with the core subunits of the TIM23 complex (Fig. 4 B). Thus, we find that in mitochondria presequence binding and Tim23 channel interaction of Tim50 are two topologically distinct functions, localized to different domains within Tim50IMS. This finding is further supported by the recent structural analysis of the Tim23-binding domain of Tim50 (Qian et al., 2011).
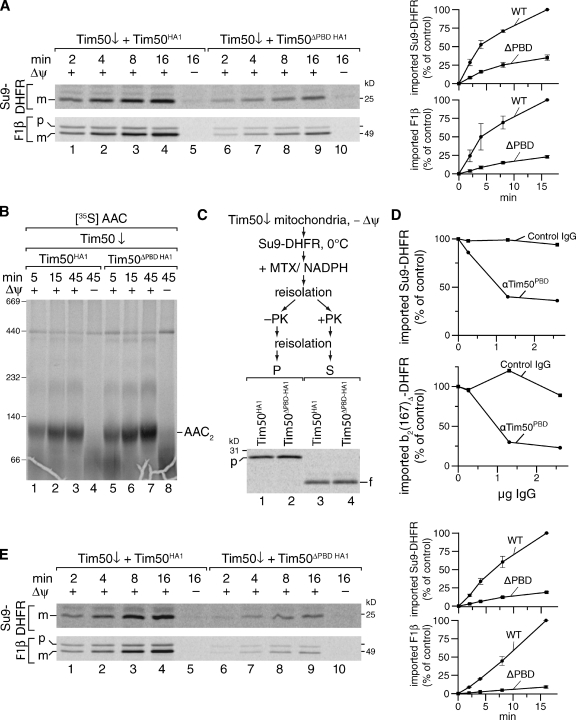
Figure 4.
Tim50 contains two separate functional domains for presequence and Tim23 binding. (A) Ni-NTA agarose with purified Tim23IMS or without (control) was incubated with mitochondrial detergent extracts, bound proteins eluted, and analyzed by Western blotting with the indicated antibodies. Total, 5%; Eluate, 100%. (B) Immunoprecipitation from solubilized mitochondria containing reduced levels of Tim50 and Tim50HA1 or Tim50ΔPBD HA1 using anti-HA or -6xHis (control) antibodies. Samples were analyzed as in A. Total, 5%; Eluate, 100%. (C) Isolated mitochondria from indicated strains were incubated with presequence probes and subjected to photo cross-linking for 30 min. Samples were analyzed as in A. PA, photo-adduct. Asterisk denotes cross-reactive protein. (D) Radiolabeled F1β was imported into wild-type and Tim50↑ mitochondria for the indicated times at 25°C. Subsequent to proteinase K digestion samples were analyzed by SDS-PAGE and digital autoradiography. Amounts of processed and protease-protected protein were quantified. The amount after import for 16 min in wild-type mitochondria was set to 100%. m, mature protein. (E) Steady-state protein analysis of isolated wild-type and Tim23 down-regulated mitochondria analyzed by Western blotting with the indicated antibodies. (F) Isolated mitochondria were incubated with 2 µM of the indicated presequence probes and subjected UV irradiation for 30 min. Samples were analyzed as in A. PA, photo-adduct.
Because presequence peptides were photo cross-linked to purified Tim50 and the Tim23–Tim50 interaction was independent of the presence of the PBD domain in mitochondria, we assessed if presequence binding by Tim50 was linked to its interaction with the TIM23 complex in organello. Thus, we performed in organello photo cross-linking of presequence probes in wild-type mitochondria and in mitochondria isolated from cells overexpressing Tim50. The amount of presequence Tim50 photo-adduct increased threefold when the level of Tim50 was elevated (Fig. 4 C), suggesting that Tim23 was not rate limiting for presequence recognition by Tim50. In agreement with this, we find that an increase of the Tim50 level in mitochondria stimulates in vitro import of a presequence-containing precursor protein (Fig. 4 D). To exclude that overexpression increased the probability of cross-linking due to dynamic association with Tim23, we analyzed presequence binding to Tim50 in mitochondria with a reduced level of Tim23 (Fig. 4 D). Tim23-depleted mitochondria displayed more than 95% reduction of Tim23 (Fig. 4 E) and a block in mitochondrial matrix import (Fig. S5 B). In these mitochondria, photo cross-linking of pL19B or pL16B to Tim50 (Fig. 4 F) and chemical cross-linking of Cox4 presequences to Tim50 (Fig. S5 C) were not significantly affected. Hence, presequence recognition by Tim50 under import conditions in organello as well as in vitro (Fig. 2) appears to occur independent of its association with Tim23.
Thus, binding of the presequence and Tim23 occur at different positions within Tim50’s IMS domain. Moreover, presequence binding to Tim50 does not depend on Tim23.
Presequence recognition by Tim50 is required for inner membrane translocation
To assess Tim50PBD’s function in matrix import, we used mitochondria with reduced levels of Tim50, but which contained Tim50ΔPBD HA1 for in vitro import studies. As expected, matrix import was severely affected in the mutant mitochondria (Fig. 5 A), whereas transport of AAC, a substrate of the carrier translocase, was unaffected (Fig. 5 B). Because Tim50 was suggested to play an undefined role during precursor import into the intermembrane space, which functionally preceded presequence association to Tom22IMS (Chacinska et al., 2005; Mokranjac et al., 2009), we addressed if presequence binding by Tim50 was required for the transport of matrix-targeted precursors across the outer mitochondrial membrane. Thus, we performed TOM translocation assays for the model matrix protein Su9-DHFR, consisting of the subunit 9 presequence of Neurospora crassa F1Fo-ATPase and mouse dihydrofolate reductase (Kanamori et al., 1999). Tim50 down-regulated mitochondria containing Tim50HA1 or Tim50ΔPBD HA1 were incubated with radiolabeled precursor in the absence of Δψ, blocking transport across the inner membrane, and in the presence of methotrexate, inducing a stable fold of the DHFR domain, thereby arresting the precursor in the TOM complex. To determine if the precursor was arrested in TOM, we treated mitochondria with proteinase K, which degrades unimported precursors, but clips the TOM-arrested precursors in a manner that retains the folded, protected DHFR fragment that is subsequently released into the supernatant (Kanamori et al., 1999). In comparison, Tim50HA1 and Tim50ΔPBD HA1 mitochondria displayed similar amounts of accumulated precursor arrested in the TOM complex (Fig. 5 C). Thus, presequence binding by the TIM23 complex constituent Tim50 is not required for early steps of precursor transport across the TOM complex.
Figure 5.
Presequence recognition by Tim50 is required for inner membrane transport. (A) Radiolabeled precursor was imported at 25°C into isolated mitochondria in the presence or absence of Δψ. Mitochondria were treated with proteinase K and analyzed by SDS-PAGE and digital autoradiography. Amounts of processed protein were quantified (right). The amount after import for 16 min in wild-type mitochondria was set to 100% (n = 3, SEM). p, precursor; m, mature. (B) Radiolabeled AAC was imported into indicated mitochondria at 25°C. After import and proteinase K treatment, mitochondria were solubilized and analyzed by BN-PAGE and digital autoradiography. (C) [35S]Su9-DHFR precursor was imported into mitochondria in the absence of Δψ, and subsequently diluted in buffer containing methotrexate and NADPH. After re-isolation, mitochondria were treated with proteinase K or left untreated as indicated. Samples were analyzed as in A. P, pellet; S, supernatant; p, precursor; f, protease-resistant DHFR fragment. (D) Radiolabeled precursor proteins were imported into mitochondria or mitoplasts for 15 min at 25°C in the presence of affinity-purified anti-Tim50PBD or control antibodies. Samples were analyzed as in A. 100%, import in the absence of antibody. Import into mitoplasts was calculated as percentage of the corresponding import into mitochondria. (E) Radiolabeled precursor was imported into mitoplasts in the presence or absence of Δψ. After import, mitoplasts were treated with proteinase K and analyzed and quantified as in A. p, precursor; m, mature.
Hence, presequence binding by Tim50 seemed to be required for precursor translocation across the inner mitochondrial membrane. We affinity purified antibodies directed against the presequence-binding domain of Tim50 (Tim50PBD) and performed in vitro import experiments into mitochondria or mitoplasts in the presence of anti-Tim50PBD or control antibodies. Tim50PBD antibodies specifically inhibited transport of presequence-containing precursors in mitoplasts (Fig. 5 D).
In a second, alternative approach, we used mitoplasts prepared from mitochondria with reduced levels of Tim50, containing Tim50HA1 or Tim50ΔPBD HA1, for in vitro import studies. Mitoplasting efficiency of Tim50ΔPBD HA1 mitochondria was close to that of Tim50HA1 mitochondria (Fig. S5 D). In Tim50ΔPBD mitochondria import of matrix proteins was severely affected (Fig. 5 E). Thus, we conclude that presequence binding by Tim50 is required for precursor transport across the inner mitochondrial membrane.
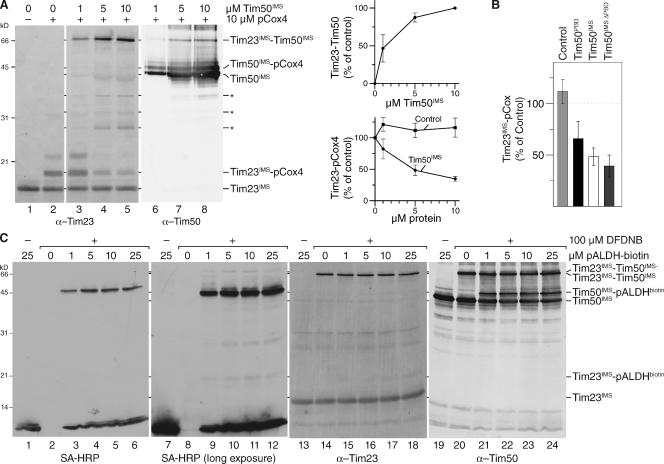
Presequence and Tim50 binding to Tim23 are mutually exclusive
Tim23 and Tim50 represent presequence receptors of the TIM23 complex. At the same time, they interact with each other through their IMS domains in a process that induces Tim23 channel closure in the resting state of the translocase (Meinecke et al., 2006; Alder et al., 2008b). Tim50 interacts with the second half of Tim23IMS (Geissler et al., 2002; Yamamoto et al., 2002; Tamura et al., 2009). Additionally, this region of Tim23 was suggested to be critical for presequence recognition (Bauer et al., 1996; de la Cruz et al., 2010). Thus, we investigated how Tim50IMS affected presequence binding to Tim23 and incubated purified Tim23IMS with an excess of pCox4 peptides in the presence or absence of Tim50IMS. While Cox4 presequence peptides were cross-linked to Tim23IMS in the absence of Tim50IMS, presequence cross-links were lost with increasing amounts of Tim50 while a Tim23–Tim50 adduct was formed (Fig. 6 A). Because Tim50IMS was expected to compete to a certain extent with Tim23 for presequences via its presequence binding domain, we used Tim50ΔPBD instead of Tim50IMS for the analyses. Despite a lack of presequence-binding ability by Tim50ΔPBD and the observed reduced interaction of the Tim50ΔPBD domain to Tim23 in vitro, it drastically reduced presequence binding to Tim23 (Fig. 6 B). Thus, in the absence of a competition effect for presequences, Tim50ΔPBD severely reduced the Tim23 presequence cross-link. As an additional control we used Tim50PBD alone, which does not interact with Tim23 and hence can be used to approximate the effect of competition on Tim23 presequence binding. The Tim50PBD displayed a high affinity to presequences compared with Tim23 and hence decreased the free pool of presequences, which in turn reduced the cross-linking efficiency of Tim23 to presequence peptides (Fig. 6 B). Hence, presequence and Tim50 binding to the receptor domain of Tim23 appear to occur in a mutually exclusive manner.
Figure 6.
Presequence and Tim50 binding to Tim23 are mutually exclusive. (A) 1 µM purified Tim23IMS was incubated with excess pCox4 in the absence or presence of the indicated amounts of Tim50IMS and subjected to chemical cross-linking by 100 µM DFDNB for 30 min. Samples were analyzed by Western blotting using the indicated antibodies (left). Quantification of Tim23–Tim50 adduct. 100%, adduct formed with maximal amount of Tim50 (SEM; n = 5; top right). Quantification of the Tim23–pCox4 adduct in response to increasing amounts of Tim50IMS or Tim21IMS (control). 100%, adduct formed without additional protein (SEM; n = 5; bottom right). Asterisks denote degradation products of Tim50IMS; Asterisk, Coomassie-stained degradation product of Tim50IMS detected as bleed-through using a fluorescence scanner at 685 nm. (B) 1 µM purified Tim23IMS was incubated with 10 µM pCox4 in the presence of 5 µM of indicated Tim50 constructs or Tim21IMS (control) and subjected to chemical cross-linking. The Tim23IMS–pCox4 adduct was quantified. 100%, adduct formed in the absence of additional protein (SEM; n = 5 for control and Tim50IMS, n = 3 for Tim50PBD, and n = 7 for Tim50ΔPBD). (C) Equimolar amounts (1 µM) of Tim50IMS and Tim23IMS were incubated with the indicated amounts of biotin-labeled pALDH. After chemical cross-linking using 100 µM DFDNB for 30 min the samples were analyzed by SDS-PAGE and Western blotting with the indicated antibodies.
To further support our finding, we asked if a ternary complex between Tim23, Tim50, and presequences existed. We performed chemical cross-linking between Tim23 and Tim50 in the presence of biotin-labeled pALDH peptide. Efficient Tim50 presequence adduct formation was detected in the presence of 1 µM peptide using streptavidin-HRP for detection. However, the cross-linking efficiency of presequences to Tim23 was much less and with the same detection method we were only able to show Tim23 presequence adducts upon long exposure. Using a Tim23 antibody, cross-links to presequences became apparent at 10–25 µM peptide (Fig. 6 C). Although we readily observed Tim23–Tim50 adduct formation, a presequence–Tim23–Tim50 adduct was not formed efficiently. We conclude that Tim50 displays a higher affinity to presequences than Tim23.
Discussion
Presequence interactions with their cognate receptors are essential to provide directionality to the transport process and affect protein dynamics in and between translocases in order to trigger precursor movement along the transport pathway (Neupert and Herrmann, 2007; Chacinska et al., 2009; Endo and Yamano, 2010; van der Laan et al., 2010). Hence, it is crucial to determine proteins that recognize targeting signals and to obtain information about the position where presequences are bound in order to address the consequences of presequence recognition at a given step of the import process. Despite ample insight into the composition of the mitochondrial protein import machinery, very little is known about how targeting signals are recognized and what effects presequences bring about at the level of translocase dynamics. At the analytical scale, radiolabeled precursors arrested in transit through translocases by folded C-terminal DHFR domains have been extensively used to probe precursor interactions along the import pathway; however, they did not provide specific presequence interaction sites (Kanamori et al., 1999; Yamamoto et al., 2002; Mokranjac et al., 2009; Chacinska et al., 2010). Moreover, structural analyses of mitochondrial-targeting signal receptor domains have only provided information on presequence recognition in the case of Tom20 (Abe et al., 2000; Perry et al., 2006).
Here, we applied photo-affinity labeling to identify presequence-binding domains by mass spectrometry. Using presequence peptides containing a photo-reactive amino acid, we identified the presequence receptor domain of Tim50 and concomitantly provide mechanistic insight into the role of presequence recognition for precursor transport.
Based on cross-links of Tim50 to radiolabeled precursors in transit through the TOM complex, Tim50 has been suggested to interact with precursors once they enter the IMS (Geissler et al., 2002; Mokranjac et al., 2003, 2009; Yamamoto et al., 2011). Moreover, the large Tim50IMS domain also interacts with Tim23IMS to regulate the Tim23 channel (Geissler et al., 2002; Yamamoto et al., 2002; Meinecke et al., 2006; Alder et al., 2008a,b; Gevorkyan-Airapetov et al., 2009; Mokranjac et al., 2009; Tamura et al., 2009). To assess how precursor and translocase interactions of Tim50 promote protein transport, it is essential to uncouple these processes. Thus, we used presequence probes to define a receptor domain in Tim50 and identified presequence cross-links in a conserved C-terminal element. This portion of the protein, termed presequence-binding domain (aa 395–476), is essential for viability and binds the presequence through hydrophobic interactions. Despite Tim50’s early interaction with precursors exiting TOM, which assist in their outer membrane translocation (Chacinska et al., 2005; Mokranjac et al., 2009), we show that presequence binding by Tim50 is required for transport across the inner membrane.
In a recent study Mokranjac et al. (2009) showed by using a chemical cross-linking approach that a precursor arrested in TOM is in proximity to Tim50 and that depletion of Tim23 affected this cross-link. This finding was interpreted such that precursor interaction of Tim50 depends on Tim23 and implies that Tim50 does not act as an independent receptor for presequences during protein transport. Here, we specifically addressed the receptor function of Tim50 and find that presequence recognition by Tim50 occurs independent of Tim23 binding, indicating that Tim50 is in fact an independent receptor. In agreement with this, not all Tim50 molecules are translocase-associated in mitochondria (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2009). Moreover, we show that Tim50IMS interacts with Tim23IMS in the absence of Tim50’s presequence-binding domain in mitochondria. This observation is further supported by the recent crystal structure of Tim50IMS lacking the presequence-binding domain (Qian et al., 2011). Thus, the intermembrane space domain of Tim50 can be dissected into two functionally distinct domains: (i) a Tim23-binding domain and (ii) a presequence-binding domain at the C terminus.
We suggest that the precursor-bound form of Tim50 recruits Tim23 and thus the translocase. This allows the presequence to be eventually transferred to Tim23. Thus, presequence recognition, which likely represents the initial step of Tim50 precursor contact, occurs independent of Tim23. However, subsequent transport steps of the precursor across the inner membrane depend on Tim23 binding to Tim50. Hence, the observed Tim23-dependent precursor cross-link to Tim50 (Mokranjac et al., 2009), which has not been mapped to a specific position of the precursor, appears to represent a stage after presequence binding during the transport process.
Interestingly, we find that in the Tim50-bound state Tim23 is unable to bind to presequences and that a ternary presequence–Tim23–Tim50 complex is formed inefficiently. Recent NMR studies indicate that the KD for the presequence–Tim23 complex is ∼0.47 mM (de la Cruz et al., 2010). Our analyses in mitochondria suggest that Tim50 displays a significantly higher affinity for presequences than Tim23. Thus, we propose that presequence binding by Tim50 precedes the receptor function of the Tim23 channel at the inner membrane. Based on these observations, we suggest that Tim50 acts as the primary presequence receptor of the TIM23 complex delivering the precursor to Tim23, which occurs most likely upon dissociation of Tim23 from Tim50. While these observations provide insight into the mechanism of precursor transport within the TIM23 complex, our findings also explain how antagonizing effects of Tim50 and presequences on Tim23 channel activity are mediated at the molecular level. Based on our results we speculate that once Tim50 binds to Tim23, the presequence is released from Tim50 to enter the Tim23 channel. As Tim23 is unable to bind presequences and Tim50 at the same time, due to overlapping binding sites and significantly reduced affinity (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2009; de la Cruz et al., 2010), it remains an open question as to how the handover of a presequence from Tim50 to Tim23 occurs, and one can be speculative on several scenarios of how this might occur. Moreover, the membrane potential across the inner membrane, which has been found to modulate the Tim23–Tim50 interaction in mitochondria, might play a yet undefined role in the process. Thus, further analyses will be required to critically test how presequence transfer within the translocase complex occurs.
Materials and methods
Import of precursor proteins and peptides
Precursor proteins were generated in the presence of [35S]methionine in reticulocyte lysate (Promega). Import into mitochondria was performed in import buffer (250 mM sucrose, 10 mM MOPS/KOH, pH 7.2, 80 mM KCl, 2 mM KH2PO4, 5 mM MgCl2, 5 mM methionine, and 1% fatty-acid free BSA) at 25°C in the presence of 2 mM NADH and 2 mM ATP. 1 µM valinomycin, 8 µM antimycin A, and 20 µM oligomycin were used to dissipate the membrane potential. Mitochondria were proteinase K treated, washed with SEM buffer (250 mM sucrose, 20 mM MOPS/KOH, pH 7.2, and 1 mM EDTA) and analyzed by BN-PAGE or SDS-PAGE in conjunction with autoradiography or Western blot.
For import competition experiments, mitochondria were preincubated with the indicated concentration of peptide for 2 min at 25°C.
For antibody inhibition, mitochondria were incubated for 20 min on ice in 10 mM MOPS, pH 7.4, to convert them to mitoplasts. After addition of the respective antibodies they were incubated for 15 min on ice, re-isolated, and resuspended in import buffer.
Arrest of radiolabeled Su9-DHFR was performed at 0°C, essentially as described in Kanamori et al. (1999). In brief, isolated mitochondria (0.5 mg/ml) were incubated in 10 mM MOPS/KOH, pH 7.2, 250 mM sucrose, 10 mM KCl, 5 mM MgCl2, 2 mM methionine, and 10 µM carbonyl cyanid 3-chlorophenylhydrazon (CCCP). After an incubation with radiolabeled Su9-DHFR precursor for 15 min on ice, mitochondria were diluted 1:5 using 10 mM MOPS/KOH, pH 7.2, 250 mM sucrose, 10 mM KCl, 20 µM CCCP, 0.5 µM methotrexate, and 1 mM NADPH, and re-isolated. After resuspension in the same buffer (0.5 mg/ml) the sample was split into two; one half was proteinase K treated, the other one not. The supernatant of the proteinase K–treated sample was precipitated using 15% TCA while the mitochondrial pellet of the untreated sample was analyzed.
Cross-linking analyses and purification of photo-adducts
For photo cross-linking, mitochondria were suspended in import buffer lacking BSA to 1 µg/µl, supplemented with 2 µM photo-peptide, and incubated 10 min on ice. UV irradiation was performed for 30 min on ice with a self-made device containing a halogen metal vapor lamp and a glass screen to filter protein-damaging wavelengths below 300 nm (Jahn et al., 2002). Subsequently, mitochondria were washed with SEM buffer and analyzed by Western blotting. For isolation of photo-adducts, mitochondria were resuspended (10 µg/µl) in lysis buffer (100 mM Tris/HCl, pH 8.0, 8 M urea, 1% SDS, 2% Triton X-100, 1 mM EDTA, 200 mM NaCl, and 1mM PMSF) and incubated for 10 min at room temperature. Samples were diluted to 1 µg/µl with dilution buffer (lysis buffer lacking SDS and containing 0.8 M urea, 2 µg/ml leupeptin, and 2 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride), and incubated for 10 min at 4°C. After removal of insoluble particles the sample was loaded onto streptavidin agarose (Thermo Fisher Scientific). Washing of the column material was performed using wash buffer A (as lysis buffer, but with 2% SDS), wash buffer B (as lysis buffer, but with 0.2% SDS and 1 M NaCl), and wash buffer C (100 mM Tris/HCl, pH 8.0, 0.2% Triton X-100, and 1 mM EDTA). Samples were eluted by incubation at 95°C for 15 min in protein loading buffer (2% SDS, 10% glycerol, 60 mM Tris/HCl, pH 6.8, 0.01% bromphenole blue, and 1% 2-mercaptoethanol).
Photo cross-linking of purified proteins was performed at a 0.5–1.5 molar ratio of protein to peptide for 30 min on ice.
Chemical cross-linking was performed using purified protein (1 µM) mixed with the indicated amount of peptide or additional protein for 15 min on ice in buffer containing 20 mM Hepes, pH 7.2, and 100 mM NaCl. 100 µM DFDNB was used to cross-link for 30 min on ice, after which the reaction was quenched for 15 min with 140 mM Tris/HCl, pH 7.5, and 5% β-mercaptoethanol.
For in organello DFDNB cross-linking 1 µg/µl mitochondria were suspended in 20 mM Hepes, pH 7.2, and 100 mM NaCl and incubated with 20 µM presequence peptide for 15 min at 25°C. Subsequent to cross-linking with 1 mM DFDNB for 30 min on ice the reaction was stopped by 250 mM Tris/HCl, pH 7.4, and washing of mitochondria with 20 mM Hepes, pH 7.2, 80 mM KCl, and 600 mM sorbitol.
Protein purifications, immunoprecipitation, and in vitro binding
Based on secondary structure prediction software included in Geneious Pro (Drummond et al., 2010) and the predicted transmembrane span, we generated Tim50IMS (aa 132–476), Tim50ΔPBD (aa 132–365), and Tim50PBD (aa 395–476), which were cloned into pPROEX HTc. These constructs, as well as Tom20CD (Brix et al., 1999), were purified via immobilized-metal affinity chromatography. GST was purified via glutathione–Sepharose 4B (GE Healthcare). For in vitro photo cross-linking, constructs were dialyzed against 10 mM MOPS/NaOH, pH 7.2, 5 mM MgCl2, 2 mM KH2PO4, and 20 mM KCl.
Tim23 pull-down was performed as described previously (Geissler et al., 2002). In brief, purified His-tagged Tim23IMS (Truscott et al., 2001) was immobilized on Ni-NTA agarose (QIAGEN). Mitochondria were solubilized (1 µg/µl) using 20 mM Hepes/KOH, pH 7.4, 100 mM KOAc, 10 mM Mg(OAc)2, 10% (vol/vol) glycerol, 20 mM imidazol, and 0.5% (vol/vol) Triton X-100 for 30 min at 4°C. Binding of the solubilized mitochondrial proteins to the immobilized protein was performed at 4°C for 1 h. Subsequent to washing with solubilization buffer (0.25% [vol/vol] Triton X-100), bound proteins were eluted with 8 M urea containing SDS-PAGE loading buffer.
Immunoprecipitation used HA and 6xHis antibodies, immobilized on Dynabeads ProtG. Mitochondria were solubilized (2 µg/µl) in 50 mM sodium phosphate, pH 7.4, 100 mM NaCl, 10% glycerol, and 1% digitonin before incubation with the antibody beads. Subsequent to washing, bound proteins were eluted by incubation in protein loading buffer (without β-mercaptoethanol) at 60°C for 10 min.
Peptide synthesis
Peptidic photo probes were synthesized with a peptide synthesizer (model 433A; Applied Biosystems) using standard fluorenylmethoxycarbonyl (Fmoc) chemistry in the 0.1 mmol scale. The photophore was directly introduced into the polypeptide chain by using the Fmoc derivative of the photoreactive amino acid para-benzoyl-Phe-OH (BPA; Bachem). The biotin moiety was introduced through biotinylated Lys. Due to the limited solubility of the corresponding amino acid derivative Fmoc-Lys(biotin)-OH (Novabiochem), its coupling was performed manually under visual inspection in a pear-shaped flask. For this purpose, the dried N-terminally deprotected resin was resuspended with 0.5 mmol Fmoc-Lys(biotin)-OH in N-methyl-2-pyrrolidon (5 ml), 0.9 mmol 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate in dimethylformamide (2 ml), 2 mmol diisopropylethylamine in N-methyl-2-pyrrolidon (1 ml), and rotated for 45 min at room temperature. The resin was washed with N-methyl-2-pyrrolidon (3x) and the coupling step was repeated as before. The resin was then transferred back into the synthesizer and automated synthesis was resumed with an Fmoc-deprotection cycle. The completed peptides were deprotected and cleaved from the resin via incubation in solution containing 95% trifluoroacetic acid, 2.5% triisopropylsilane, and 2.5% water for 4 h at room temperature. Purified products were characterized by reversed-phase HPLC and mass spectrometry. Biotin-labeled pALDH was synthesized in the same way as the photo-probes, but without the incorporation of the BPA.
pCox4 (MLSLRQSIRFFKPATRTLCSSRYLL), SynB2 (MLSRQQSQRQSRQQSQRQSRYLL), and scrambled pALDH-s (MLRGKQPTKSLLPQRSPKLSAAA) were purchased from JPT Peptide Technologies as an N-terminal amine and a C-terminal amide.
Genetic manipulation of yeast and growth conditions
Saccharomyces cerevisiae YPH499 or BY4743 were cultured in YP medium (1% yeast extract, 2% peptone) containing 2% glucose (YPD) or 3% glycerol (YPG) or 3% lactate (YPL) at 30°C. TIM50+/ΔPBD HA3 (in BY4743; yCS1) was generated through transformation of a C-terminal HA tagging cassette, resulting in the deletion of a region encoding amino acids 366–476 (Janke et al., 2004).
Sporulation of the BY4743 TIM50+/ΔPBD HA3 strain was achieved using a stationary culture from YPD, which had been washed in sporulation medium (2% KAc, 0.2% yeast extract, 0.1% glucose, 0.2% leucine, 0.04% histidine, lysine, and uracil) and cultivated at 24°C for 5 d in sporulation medium. Subsequently, tetrads were dissected on YPD plates and grown at 30°C.
For in vivo expression of different Tim50 constructs, full-length Tim50 (1–476) or Tim50ΔPBD (1–365) was cloned into pME2780 without (pCS22 and pCS20) or with a single HA tag (pCS23 and pCS21; Mumberg et al., 1994). Constructed plasmids were then used to replace the wild-type protein encoding plasmid within the gene deletion strain to yield strains yCS5-8 (Chacinska et al., 2005).
The strain containing TIM50 under the control of the GAL1 promoter (Geissler et al., 2002) was transformed with plasmids pCS27 and pCS26, encoding Tim50 (1–476) or Tim50ΔPBD (1–365), both of which contained a single C-terminal HA tag, under the control of the MET25 promoter (Mumberg et al., 1994) to give yCS2 and yCS3. Strains were precultured in selective lactate medium (0.67% YNB, 3% lactate, 0.003% lysine, 0.002% adenine, histidine, tryptophane, and uracil, pH 5.0) supplemented with 1% galactose and 1% raffinose. Subsequently, cells were grown in equivalent selective medium containing additional 0.01% glucose for 38 h at 30°C.
TIM23 was cloned into pME2804 under the control of the GALS promoter (Mumberg et al., 1994). The resulting plasmid (pCS28) was introduced into MB29 (Bömer et al., 1997) by plasmid shuffling (yCS4). Cultures were grown essentially as described previously (Geissler et al., 2002). In brief, cells were precultured in YPL containing 1% raffinose and 1% galactose for 28 h. The main culture was grown for 31 h in YPL with 0.1% glucose.
To isolate mitochondria, yeast cells were treated with 7 mg zymolase/g cell wet weight and opened in a Dounce Homogenizer. Mitochondria were isolated by differential centrifugation and were pelleted at 17,000 g as described previously (Stojanovski et al., 2007). Appropriate aliquots were stored in SEM buffer at −80°C.
Mass spectrometry
Photo-adducts purified by colloidal Coomassie-stained (Neuhoff et al., 1988) SDS-PAGE were subjected to an in-gel digest. In brief, gel pieces were cut out, washed with 25 mM NH4HCO3/water, 25 mM NH4HCO3/50% acetonitrile, and 100% acetonitrile, followed by reduction with 10 mM dithiothreitol, 25 mM NH4HCO3/water at 56°C for 1 h, washing (see above) and carbamidomethylation with 25 mM iodoacetamide in 25 mM NH4HCO3/water. After digestion with 120 ng trypsin at 37°C overnight, peptides were extracted from the gel with 0.1% trifluoroacetic acid (TFA) and separated by reverse-phase chromatography (EASY-nLC; Bruker Daltonics) using a PepMap100 C18 nano-column (Dionex) and an elution gradient from 9.5–90.5% acetonitrile in 0.1% TFA for 80 min. The eluate from the column was mixed with α-cyano-4-hydroxycinnamic acid (HCCA) as matrix (4.5% of saturated HCCA in 90% acetonitrile, 0.1% TFA, 1 mM NH4H2PO4) and spotted onto an anchorchip target using a robot (Proteineer fc II; Bruker Daltonics). Samples on the target were analyzed in a MALDI-TOF/TOF mass spectrometer (Ultraflextreme; Bruker Daltonics) recording MS and post-source decay MS/MS spectra using the software WARP-LC, AutoXecute, and FlexAnalysis (Bruker Daltonics). MS/MS spectra with precursor masses, which match any of the masses calculated for candidate cross-linked peptides, were selected for further manual evaluation supported by the software Biotools (Bruker Daltonics) to verify peptide identities and to identify the cross-linked amino acids.
MALDI-TOF-MS was also used to co-detect recombinant presequence receptor domains and their respective photo-adducts at the level of intact proteins. For this purpose, the photoreaction mixtures were desalted by RP C4 ZipTips (Millipore) and bound material was eluted with 80% acetonitrile/0.1% TFA. For dried droplet preparation, 1 µl eluate was mixed with 2 µl α-cyano-4-hydropxycinnamic acid (HCCA) matrix solution and spotted to a ground steel sample support. The matrix solution was prepared from a saturated HCCA solution (in 0.1% TFA/acetonitrile 2:1) by diluting the supernatant 1:6 with 70% acetonitrile/0.1% TFA. Mass spectra were acquired on a MALDI-TOF/TOF mass spectrometer (Ultraflex; Bruker Daltonics) operated in the linear mode as described previously (Dimova et al., 2009). In brief, positively charged ions in the mass-to-charge (m/z) range 5,000–25,000 were analyzed in the linear mode and a mixture of three standard proteins (Protein Calibration Mix 2; LaserBio Laboratories; m/z range 6181–23982) was used for external calibration with the post-processing software FlexAnalysis 3.3 (Bruker Daltonics).
Homology modeling and peptide docking
The cytoplasmic domain of the yeast Tom20 has been modeled using the homology modeling protocol as implemented in ROSETTA (Misura et al., 2006). Subsequent to screening the query sequence for regions that possess an experimentally characterized homologue, secondary structure prediction and preparation of the sequence alignments have been performed using HHpred server (Söding et al., 2005). The fragment libraries were obtained from the Rosetta Server. The solution NMR structure of rat Tom20 (Protein Data Bank accession no. 1OM2) was used as the template (for the modeled 74 residue–long fragment the compared sequences share 25 and 43% sequence identity and similarity, respectively; Abe et al., 2000). Generated models (2,500) have been sorted based on their energies (ROSETTA score) and compared with the template molecule by calculating the root mean square deviation (RMSD) on Cα positions in the LSQMAN program (Kleywegt and Jones, 1994). The best model was identified as the fourth on the list of top ROSETTA scores, exhibiting a calculated RMSD of 1.23 Å between 66 superposed Cα positions.
Docking of pALDH to the modeled yeast Tom20 fragment has been performed using the FlexPepDock protocol (Raveh et al., 2010) implemented in the ROSETTA package. The initial position of the peptide was obtained based on the superposition with the structure of rat Tom20 (Protein Data Bank accession no. 1OM2) in complex with the same peptide. Four different conformations of the peptide (models 5, 8, 11, and 12 out of 20 presented in the solution NMR structure) have been selected for the docking experiments and 2,500 decoys for each of them have been calculated. Docking of peptide model 11 gave the best ROSETTA scores (over 900 top scores out of 10,000) and the model with the lowest energy was selected.
Miscellaneous
BN-PAGE, SDS-PAGE, and Western blotting were performed by standard procedures. Proteins were detected using fluorescent dye coupled to secondary antibodies (LI-COR) using a fluorescence scanner (FLA-9000; Fujifilm) and the ImageReader FLA-9000 software or by enhanced chemiluminescence (GE Healthcare). Contrast adjustments were performed using Photoshop CS4 (Adobe). Quantifications were performed using ImageQuant TL (GE Healthcare).
Online supplemental material
Fig. S1 shows the validation of the photo cross-linking approach using the Tom20CD. Fig. S2 shows import, import block, and photo cross-linking experiments concerning the characterization of the photo-peptides. Fig. S3 shows the mass spectrometric identification of photo cross-linking sites. Fig. S4 shows the determination of the photo cross-linking stoichiometry. Fig. S5 presents further data on the characterization of the Tim23↓ and the Tim50ΔPBD-HA1 mutant mitochondria. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201105098/DC1.
Acknowledgments
We thank Drs. D. Görlich, K. Tittmann, M. Deckers, and D.U. Mick for helpful discussion and O. Bernhard, Dr. F. Mayer-Posner, K. Neifer, A. Pestov, and L. van Werven for excellent technical assistance.
This work has been supported by the Deutsche Forschungsgemeinschaft, SFB 860, the Göttingen Graduate School for Neurosciences und Molecular Biosciences, and the Max-Planck-Society (P. Rehling and O. Jahn). C. Schulz is a recipient of a doctoral fellowship from the Boehringer Ingelheim Fonds.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- PBD
- presequence-binding domain
References
- Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. 2000. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 100:551–560 10.1016/S0092-8674(00)80691-1 [DOI] [PubMed] [Google Scholar]
- Alder N.N., Jensen R.E., Johnson A.E. 2008a. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell. 134:439–450 10.1016/j.cell.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Alder N.N., Sutherland J., Buhring A.I., Jensen R.E., Johnson A.E. 2008b. Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. Mol. Biol. Cell. 19:159–170 10.1091/mbc.E07-07-0669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M.F., Sirrenberg C., Neupert W., Brunner M. 1996. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 87:33–41 10.1016/S0092-8674(00)81320-3 [DOI] [PubMed] [Google Scholar]
- Bömer U., Meijer M., Maarse A.C., Hönlinger A., Dekker P.J., Pfanner N., Rassow J. 1997. Multiple interactions of components mediating preprotein translocation across the inner mitochondrial membrane. EMBO J. 16:2205–2216 10.1093/emboj/16.9.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix J., Rüdiger S., Bukau B., Schneider-Mergener J., Pfanner N. 1999. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 274:16522–16530 10.1074/jbc.274.23.16522 [DOI] [PubMed] [Google Scholar]
- Chacinska A., Lind M., Frazier A.E., Dudek J., Meisinger C., Geissler A., Sickmann A., Meyer H.E., Truscott K.N., Guiard B., et al. 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 120:817–829 10.1016/j.cell.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell. 138:628–644 10.1016/j.cell.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., van der Laan M., Mehnert C.S., Guiard B., Mick D.U., Hutu D.P., Truscott K.N., Wiedemann N., Meisinger C., Pfanner N., Rehling P. 2010. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell. Biol. 30:307–318 10.1128/MCB.00749-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz L., Bajaj R., Becker S., Zweckstetter M. 2010. The intermembrane space domain of Tim23 is intrinsically disordered with a distinct binding region for presequences. Protein Sci. 19:2045–2054 10.1002/pro.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova K., Kalkhof S., Pottratz I., Ihling C., Rodriguez-Castaneda F., Liepold T., Griesinger C., Brose N., Sinz A., Jahn O. 2009. Structural insights into the calmodulin-Munc13 interaction obtained by cross-linking and mass spectrometry. Biochemistry. 48:5908–5921 10.1021/bi900300r [DOI] [PubMed] [Google Scholar]
- Drummond A.J., Ashton B., Buxton S., Cheung M., Cooper A., Heled J., Kearse M., Moir R., Stones-Havas S., Sturrock S., et al. 2010. Geneious v5.3, Available from http://www.geneious.com.
- Endo T., Yamano K. 2010. Transport of proteins across or into the mitochondrial outer membrane. Biochim. Biophys. Acta. 1803:706–714 10.1016/j.bbamcr.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Erdmann R., Wiebel F.F., Flessau A., Rytka J., Beyer A., Fröhlich K.U., Kunau W.H. 1991. PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell. 64:499–510 10.1016/0092-8674(91)90234-P [DOI] [PubMed] [Google Scholar]
- Geissler A., Chacinska A., Truscott K.N., Wiedemann N., Brandner K., Sickmann A., Meyer H.E., Meisinger C., Pfanner N., Rehling P. 2002. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell. 111:507–518 10.1016/S0092-8674(02)01073-5 [DOI] [PubMed] [Google Scholar]
- Gevorkyan-Airapetov L., Zohary K., Popov-Celeketic D., Mapa K., Hell K., Neupert W., Azem A., Mokranjac D. 2009. Interaction of Tim23 with Tim50 Is essential for protein translocation by the mitochondrial TIM23 complex. J. Biol. Chem. 284:4865–4872 10.1074/jbc.M807041200 [DOI] [PubMed] [Google Scholar]
- Jahn O., Eckart K., Brauns O., Tezval H., Spiess J. 2002. The binding protein of corticotropin-releasing factor: ligand-binding site and subunit structure. Proc. Natl. Acad. Sci. USA. 99:12055–12060 10.1073/pnas.192449299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Junge H.J., Rhee J.S., Jahn O., Varoqueaux F., Spiess J., Waxham M.N., Rosenmund C., Brose N. 2004. Calmodulin and Munc13 form a Ca2+ sensor/effector complex that controls short-term synaptic plasticity. Cell. 118:389–401 10.1016/j.cell.2004.06.029 [DOI] [PubMed] [Google Scholar]
- Kanamori T., Nishikawa S., Nakai M., Shin I., Schultz P.G., Endo T. 1999. Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl. Acad. Sci. USA. 96:3634–3639 10.1073/pnas.96.7.3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt G.J., Jones T.A. 1994. Halloween…Masks and Bones. From First Map to Final Model. Bailey S., Hubbard R., Waller D., Warrington, Great Britain, SERC Daresbury Laboratory; 59–66 [Google Scholar]
- Meinecke M., Wagner R., Kovermann P., Guiard B., Mick D.U., Hutu D.P., Voos W., Truscott K.N., Chacinska A., Pfanner N., Rehling P. 2006. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science. 312:1523–1526 10.1126/science.1127628 [DOI] [PubMed] [Google Scholar]
- Mick D.U., Fox T.D., Rehling P. 2011. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12:14–20 10.1038/nrm3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misura K.M.S., Chivian D., Rohl C.A., Kim D.E., Baker D. 2006. Physically realistic homology models built with ROSETTA can be more accurate than their templates. Proc. Natl. Acad. Sci. USA. 103:5361–5366 10.1073/pnas.0509355103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D., Paschen S.A., Kozany C., Prokisch H., Hoppins S.C., Nargang F.E., Neupert W., Hell K. 2003. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 22:816–825 10.1093/emboj/cdg090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D., Sichting M., Popov-Celeketić D., Mapa K., Gevorkyan-Airapetov L., Zohary K., Hell K., Azem A., Neupert W. 2009. Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol. Biol. Cell. 20:1400–1407 10.1091/mbc.E08-09-0934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Müller R., Funk M. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767–5768 10.1093/nar/22.25.5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D., Ehrhardt W. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 9:255–262 10.1002/elps.1150090603 [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J.M. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749 10.1146/annurev.biochem.76.052705.163409 [DOI] [PubMed] [Google Scholar]
- Perry A.J., Hulett J.M., Likić V.A., Lithgow T., Gooley P.R. 2006. Convergent evolution of receptors for protein import into mitochondria. Curr. Biol. 16:221–229 10.1016/j.cub.2005.12.034 [DOI] [PubMed] [Google Scholar]
- Qian X., Gebert M., Höpker J., Yan M., Li J., Wiedemann N., van der Laan M., Pfanner N., Sha B. 2011. Structural basis for the function of Tim50 in the mitochondrial presequence translocase. J. Mol. Biol. 411:513–519 10.1016/j.jmb.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T.A. 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 450:663–669 10.1038/nature06384 [DOI] [PubMed] [Google Scholar]
- Raveh B., London N., Schueler-Furman O. 2010. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins. 78:2029–2040 [DOI] [PubMed] [Google Scholar]
- Schnell D.J., Hebert D.N. 2003. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell. 112:491–505 10.1016/S0092-8674(03)00110-7 [DOI] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A.N. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(Web Server issue):W244–W248 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Pfanner N., Wiedemann N. 2007. Import of proteins into mitochondria. Methods Cell Biol. 80:783–806 10.1016/S0091-679X(06)80036-1 [DOI] [PubMed] [Google Scholar]
- Tamura Y., Harada Y., Shiota T., Yamano K., Watanabe K., Yokota M., Yamamoto H., Sesaki H., Endo T. 2009. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184:129–141 10.1083/jcb.200808068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott K.N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A.J., Rassow J., Pfanner N., Wagner R. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074–1082 10.1038/nsb726 [DOI] [PubMed] [Google Scholar]
- van der Laan M., Meinecke M., Dudek J., Hutu D.P., Lind M., Perschil I., Guiard B., Wagner R., Pfanner N., Rehling P. 2007. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 9:1152–1159 10.1038/ncb1635 [DOI] [PubMed] [Google Scholar]
- van der Laan M., Hutu D.P., Rehling P. 2010. On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim. Biophys. Acta. 1803:732–739 10.1016/j.bbamcr.2010.01.013 [DOI] [PubMed] [Google Scholar]
- Vögtle F.-N., Wortelkamp S., Zahedi R.P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., Meisinger C. 2009. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 139:428–439 10.1016/j.cell.2009.07.045 [DOI] [PubMed] [Google Scholar]
- von Heijne G. 1986. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 5:1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W., Schekman R. 2005. Protein translocation across biological membranes. Science. 310:1452–1456 10.1126/science.1113752 [DOI] [PubMed] [Google Scholar]
- Wittelsberger A., Thomas B.E., Mierke D.F., Rosenblatt M. 2006. Methionine acts as a “magnet” in photoaffinity crosslinking experiments. FEBS Lett. 580:1872–1876 10.1016/j.febslet.2006.02.050 [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Esaki M., Kanamori T., Tamura Y., Nishikawa S., Endo T. 2002. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell. 111:519–528 10.1016/S0092-8674(02)01053-X [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Itoh N., Kawano S., Yatsukawa Y.-i., Momose T., Makio T., Matsunaga M., Yokota M., Esaki M., Shodai T., et al. 2011. Dual role of the receptor Tom20 in specificity and efficiency of protein import into mitochondria. Proc. Natl. Acad. Sci. USA. 108:91–96 10.1073/pnas.1014918108 [DOI] [PMC free article] [PubMed] [Google Scholar]