In vitro, actin filament tension correlates with the binding and apparent activity of the filament-severing protein cofilin, suggesting a molecular mechanism by which cells respond to changes in mechanical force.
Abstract
Intracellular and extracellular mechanical forces affect the structure and dynamics of the actin cytoskeleton. However, the underlying molecular and biophysical mechanisms, including how mechanical forces are sensed, are largely unknown. Actin-depolymerizing factor/cofilin proteins are actin-modulating proteins that are ubiquitously distributed in eukaryotes, and they are the most likely candidate as proteins to drive stress fiber disassembly in response to changes in tension in the fiber. In this study, we propose a novel hypothesis that tension in an actin filament prevents the filament from being severed by cofilin. To test this, we placed single actin filaments under tension using optical tweezers. When a fiber was tensed, it was severed after the application of cofilin with a significantly larger delay in comparison with control filaments suspended in solution. The binding rate of cofilin to an actin bundle decreased when the bundle was tensed. These results suggest that tension in an actin filament reduces the cofilin binding, resulting in a decrease in its effective severing activity.
Introduction
Physical forces contribute to a wide range of biological processes, including survival (Chen et al., 1997), development (Maniotis et al., 1997; Colombo et al., 2003), wound healing (Timmenga et al., 1991), and growth (Damien et al., 2000). The molecular mechanisms by which cells sense and respond to mechanical signals are not fully understood. It is generally believed that force initiates signal transduction via stretch-activated ion channels in the cell membrane (Gillespie and Walker, 2001). However, cell mechanotransduction may involve numerous molecular mechanisms other than ion channels, such as force-initiated signal transduction via changes in cytoskeletal–matrix linkages (Sawada and Sheetz, 2002; Tamada et al., 2004). The assembly/disassembly of stress fibers is greatly affected by Rho-stimulated cytoskeletal contraction (Bershadsky et al., 2006; Pellegrin and Mellor, 2007) and extracellular mechanical force (applied to the fibers; Iba and Sumpio, 1991; Hayakawa et al., 2001; Kiyoshima et al., 2011). Actin-depolymerizing factor/cofilin proteins, actin filament-severing proteins, are ubiquitously distributed in eukaryotes (Bamburg et al., 1999) and are preferable candidates for the stress fiber disassembly induced by the loss of fiber tension (Ono et al., 1996). Theretofore, it was suggested that an unknown molecule senses the tension in stress fibers and inactivates cofilin by phosphorylation (Yang et al., 1998) or by phosphatidylinositol 4,5-bisphosphate (Yonezawa et al., 1990). We propose a novel hypothesis that tension changes are directly sensed by the actin filament by modulating its susceptibility to cofilin-mediated severing. To test this hypothesis, single actin filaments were tensed with optical tweezers, and the tension-dependent filament severing by cofilin was examined. Our results demonstrate that tension in the actin filament prevents its severing by cofilin and suggest that the actin filament itself is a tension sensor. This is the first demonstration that tension applied to a protein (e.g., actin filament) regulates its susceptibility to a modulating protein (e.g., cofilin).
Results and discussion
Single actin filaments function as a mechanosensor
An in vitro reconstituted system comprised of only actin filaments and recombinant (dephosphorylated) cofilin was used to directly test the aforementioned hypothesis. We prepared tetramethylrhodamine (TMR)-labeled single actin filaments (8–30 µm in length) with one end tethered to an N-ethylmaleimide (NEM)–myosin-coated bead (10 µm in diameter), whereas the other was trapped with a small (3 µm in diameter) NEM-myosin–coated bead that can be manipulated with optical tweezers. Tension was generated in the filament by displacing the trapping point (Fig. 1 A and Video 1). We measured the delay from the onset of the cofilin application to the severing of a single filament, with or without externally generated tension in the filament. In control experiments, cofilin was applied to a filament of which one end was suspended in solution. The actin filament was severed 17.0 ± 3.9 s (mean ± SEM; n = 27) after a cofilin (500 nM) application (Fig. 1 [B and C] and Video 2). This value agrees with that estimated by measuring the number of actin filament severed in vitro (Michelot et al., 2007) and actin filament turnover in vivo (Okreglak and Drubin, 2007) and direct kinetic measurements (Andrianantoandro and Pollard, 2006), showing that severing activity of cofilin is slower than cofilin binding. In contrast, when tension (∼30 pN) was generated in a filament, it was not severed or was severed by cofilin with a significantly larger (P < 0.01) delay (43.2 ± 5.1 s; n = 15; Fig. 1 E and Video 1). After the cessation of optical trapping, the filament was severed by cofilin within 15.1 ± 3.7 s (n = 10). These results demonstrate directly that tension in the actin filament prevents, or delays, the filament severing by cofilin (Fig. 1, F and G).
Figure 1.
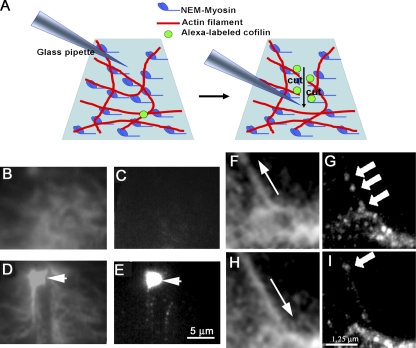
Tension in actin filament prevents or delays severing by cofilin. (A) A schematic drawing of the experimental setup. One end of a rhodamine-labeled actin filament was tethered to a NEM-myosin–coated bead (10 µm in diameter; left) fixed on a coverslip, and the other end of the filament was tethered to a small NEM-myosin–coated bead (3 µm in diameter; right) trapped by optical tweezers. (B) A fluorescence image of an actin filament attached to a bead before application of cofilin. The filament was suspended in the flow by perfusion (from left to right). (C–E) The filament was severed at 16 s (shown by an arrow) from the onset of cofilin application (see also Video 2). In contrast, when the actin filament (D) was stretched (E) by moving the microscope stage (a bead at the bottom middle of the panel was on the trapping point), severing of the filament by cofilin was prevented (E; see alsoVideo 1). (F) The distribution of the delay between cofilin application and the severing of the nontensed actin filaments (n = 27 from 24 independent experiments). (G) The duration of time observing actin filaments without being severed by cofilin (solid bars) was significantly prolonged (P < 0.01; n = 15 from 15 independent experiments) when the filament was tensed with optical tweezers (∼30 pN). Actin filaments were not severed (for >50 s) when control F-buffer solution was perfused (hatched bars in F for nontensed filaments and in G for tensed filaments). (H) A schematic drawing of the experimental setup used to apply magnetic force to actin filament. Actin filaments are pulled toward the electric magnet (indicated by a red arrow). (I) The plot shows the number of magnetic beads in a 1-mm2 area pulled toward the electromagnet during 2 min of observation after 250 nM cofilin application. The data (mean ± SEM; n = 7) were plotted with red squares against the force (from 3.4 to 0.17 pN). A similar plot in gelsolin (25 nM; mean ± SEM; n = 12) is shown with black circles. Vertical bars denote SEM. *, P < 0.05; **, P < 0.01, using the Mann-Whitney test.
Magnetic micromanipulation of micrometer-sized magnetic particles provides a means to probe force-dependent molecular interactions (Fig. 1 H; Wang et al., 1993). Tension-dependent severing of actin filaments by cofilin was examined by using magnetic beads (∼1 µm in diameter) conjugated with phalloidin and that were attached to the actin filaments tethered on the glass surface by NEM-myosin. Individual beads were pulled toward an electromagnet in a distance-dependent manner; the force was ∼3.4 pN near the tip of the electromagnet and quickly decreased with distance. Beads did not move to the tip of the magnet in control F buffer solution; however, in the presence of 250 nM cofilin, the beads in the area exposed to 0.2–0.7 pN of force were moved toward the tip of the magnet, suggesting that the actin filaments tethering the bead to the glass surface were severed by cofilin. In contrast, the beads exposed to a larger force (>3.4 pN) were rarely moved toward the tip during 2 min of observation (Fig. 1 I), showing the force-dependent inhibition of actin filaments severing by cofilin. The half-maximum inhibition was seen with ∼2 pN of force. For comparison, the effect of 25 nM gelsolin applied under the same conditions was examined, which also severs the actin filaments. Beads in the area exposed to the force >0.2 pN were moved toward the tip of the magnet (Fig. 1 I), and the rate of severing was higher where beads were exposed to larger forces.
Cofilin binds preferentially to relaxed, not tensed, actin filaments
The inhibitory effect of tension on the severing of actin filaments by cofilin may be accounted for by two possible mechanisms: tension in the actin filament affects the binding of cofilin to the filament, and/or the tension affects the severing activity of cofilin already bound to the filament. The effect of tension on the cofilin binding was examined using actin filaments under different mechanical conditions. Actin filaments were tethered at multiple sites on the coverslip by NEM-myosin, and 50 nM 5-iodoacetoamide fluorescein (IAF)–labeled cofilin was applied; brief fluorescence emission from IAF-cofilin was observed under total internal reflection fluorescence (TIRF) microscopy. The fluorescence was not seen in the absence of actin filaments, indicating that it takes place only during transient binding of cofilin to the actin filaments. Quantitative analysis showed that the cofilin-binding rate to actin filament meshwork was low (0.025 ± 0.006 events/s in 1 µm2 of meshwork; n = 3), and the mean duration of fluorescence was short (41 ± 26 ms; n = 147). However, the rate of binding and fluorescence duration increased (to 0.21 ± 0.12 events/s µm2 [n = 3] and 91 ± 102 ms [n = 415], respectively) when the meshwork of actin filaments was severed by scratching the mesh with a pipette tip (Fig. 2 A). Analysis was performed in a narrow (2-µm width) area facing the scratched region, and examples of the data analyzed are shown in Fig. 2 (B–E) and Video 3. Although most of the binding was brief, slow binding (1–3 s) was sometimes detected (a few percentages of binding events). The area free of F-actin gradually expanded during 60 s of observation, suggesting that cofilin had severed the actin filaments. The tethering of actin filaments at multiple sites on the glass surface decreases actin filament flexibility (Pavlov et al., 2007), whereas severing results in a freeing of the ends of the actin filaments, allowing their relaxation and tension reduction compared with the tethered filaments. These findings suggest that cofilin tends to bind to flexible, not tensed, actin filaments and severs them, as hypothesized (Michelot et al., 2007; Pavlov et al., 2007).
Figure 2.
Binding of cofilin to actin filaments. (A) A schematic drawing of the experimental setup. (B–E) Time-lapse imaging of rhodamine-labeled actin filament meshwork (B and D) and IAF-labeled cofilin (C and E). (B) A meshwork of F-actin observed with TIRF microscopy before severing. (C) An IAF-labeled cofilin image acquired during 1 s with TIRF illumination. (D) A meshwork of F-actin severed by a tip of a glass pipette (arrow). (E) Cofilin images acquired during 3 s after the meshwork was severed. The arrow shows the tip of the pipette. (F) The actin bundle was relieved to the original length by displacing the tip of pipette in the direction shown by the arrow. (G) A high accumulation level of cofilin-positive spots was observed along the actin bundle 4 s after relieving a strain (Video 4). (H) A bundle of actin filaments was stretched ∼20% by displacing the tip of the pipette attached to one end of the bundle in the direction shown by the arrow. (I) A few cases of binding of cofilin to the stretched bundle were observed (cofilin fluorescent spots are shown by the arrows in G and I). Images of IAF-cofilin were accumulated during 4 s (see also Video 5). Bars: (E) 5 µm; (I) 1.25 µm.
The dissociation time constant of cofilin from actin filaments was reported to be 0.18 s−1 (Cao et al., 2006). The long duration (1–3 s) of cofilin binding to the actin filaments detected in this study roughly agrees with the aforementioned estimation (i.e., the dissociation time constant 0.18 s−1 corresponds to a 5.6-s duration of binding).
Sliding a fine pipette along the surface, actin meshwork often formed a bundle of actin filaments from the actin meshwork, which consisted of F-actin and NEM-myosin; NEM-myosin binds to actin filaments and facilitates bundle formation by the zippering together of the actin filaments. The cofilin-binding rate to the bundle was 6.5 ± 4.5 events/s per 1 µm of bundles (n = 7; Fig. 2 [F and G] and Video 4). The rate decreased to 2.9 ± 2.7 events/s when the bundle was stretched 20–30% by moving the tip of the glass pipette attached to one end of the bundle (Fig. 2 [H and I] and Video 5). All these results suggest that tension in the actin filaments affects the binding rate of cofilin to actin filaments.
Tension-dependent cofilin binding to stress fibers
Tension-dependent cofilin binding to actin stress fibers was also examined in semi-intact human umbilical vein endothelial cells (HUVECs). Semi-intact cells on a prestretched (20%) elastic substrate were relaxed in ATP-free buffer (pH 6.5) with 500 nM purified cofilin, chemically fixed within 1 min, and stained with an anticofilin antibody. The experiment showed that cofilin distributed along the actin stress fibers in parallel to the axis of relaxation (Fig. 3, D–F). However, this characteristic staining pattern was not observed when the cells were 10% stretched (Fig. 3, A–C), demonstrating that tension prevents the cofilin binding to the tensed stress fibers in semi-intact cells. Quantitative fluorescence image analyses confirmed the tension-dependent cofilin distribution along the actin stress fibers (Fig. 3, G–K). Furthermore, in ATP-free DK buffer (Mackay et al., 1997), which reduces tension in stress fibers by attenuating actomyosin activity, the stress fibers in semi-intact cells were disassembled by 250 nM cofilin in relaxed cells (Fig. 3 M) but not in stretched cells (Fig. 3 L).
Figure 3.
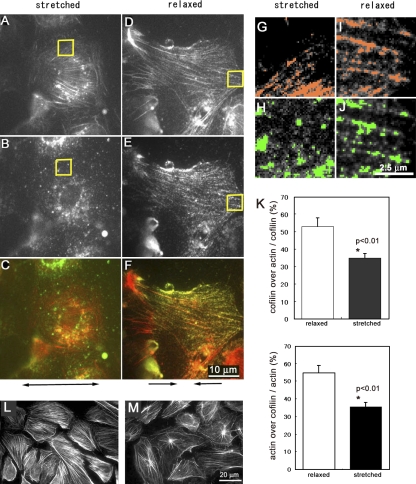
Cofilin binding to stress fibers is modulated by stretching of semi-intact cells. (A–F) Cofilin did not bind to stress fibers in semi-intact cells that were 10% stretched (A, B, and C) but did bind to stress fibers in the cells when they were 20% relaxed (D, E, and F). The specimen was fixed and then stained for actin (A and D) and cofilin (B and E). These images are merged in C (A and B) and F (D and E). The double-headed arrow below C shows the direction of stretching, and arrows below F show the direction of relaxation. The high magnification images of the areas enclosed by yellow squares in A–D are shown in G–J, respectively. (G–J) Colocalization of actin (colored red in G and I) and cofilin (colored green in H and J) was evaluated by MetaMorph software (measure colocalization). (K) The percentages of cofilin-positive pixels on actin stress fibers (top) and percentages of actin-positive pixels on cofilin-positive pixels (bottom) are shown. Data were obtained from five cells from two independent experiments. Vertical bars denote SEM. (L and M) Stress fibers in semi-intact cells bathed in ATP-free buffer with 250 nM cofilin were not disassembled when cells were kept stretched 10% (L) but were disassembled in the same solution when they were relaxed 20% (M).
To examine the tension-dependent cofilin binding to the actin stress fibers in living cells, a GFP-cofilin expression construct was introduced into HUVEC cells. In control cells, GFP-cofilin was distributed uniformly in the cytosol and in the lamellipodia (Fig. 4 A), as reported previously (Obinata et al., 1997). When the prestretched elastic substratum was relaxed (20%), GFP-cofilin was translocated to actin stress fibers within 1 min (Figs. 4 B and S2), suggesting that the binding of cofilin to stress fibers also depends on tension in the fiber in living cells. These stress fibers were disassembled within 30 min when the tension in the stress fibers was decreased; similar observations were described earlier (Ono et al., 1996; Katoh et al., 2001). These results account for the compressive stress-induced severing of actin bundles (Medeiros et al., 2006) and disassembly of stress fibers (Ono et al., 1996) in intact cells.
Figure 4.
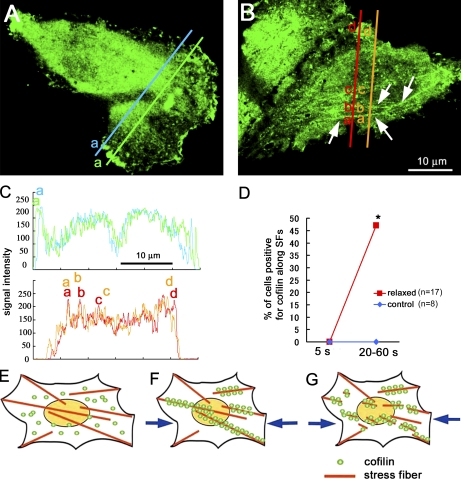
Cofilin binding to actin stress fibers in intact cells. (A) GFP-cofilin was distributed uniformly in the perinuclear cytosol but was often condensed in the ruffling membrane; GFP-cofilin did not associate with the stress fibers in control cells. (B) When the substratum was relaxed 20%, GFP-cofilin was translocated to actin stress fibers (shown by the arrows: see also Fig. S2) within 1 min. (C) Intensity profiles of the GFP-cofilin along the two lines are shown (the top profile for control cell and the bottom profile for relaxed cell). The broad peak (a) in the top profile corresponds to the cofilin condensed in the ruffling membrane (corresponding with letters in A). The narrow peaks (a, b, and c) in the bottom profile correspond to the stress fibers (corresponding with letters in B). (D) Percentage of cells positive for GFP-cofilin translocated to actin stress fibers increases from 0/17 to 8/17, with relaxing of the substrate (the number of cells examined was 17 from seven independent experiments). Translocation of GFP-cofilin to actin stress fibers (SFs) was not detected in control cells in the same period of observations (five independent experiments). *, P < 0.05, using Fisher’s exact test. (E–G) Schematic drawings of the mechanosensing by the actin stress fibers and the regulation of cofilin binding to actin stress fibers in cells. (E) Actin stress fibers generate contractile force in adherent cells, resulting in generation of tension in stress fibers, which prevents the binding of cofilin to the fibers. (F) When the tension declines (e.g., by relaxing the cell substratum or by decreasing the contractile force in the actin filaments), cofilin binds to and disassembles the fibers (G).
How does tension prevent cofilin binding to actin filaments?
Detailed examination by EM (McGough et al., 1997; Galkin et al., 2001) revealed a unique property of cofilin; it induces an ∼25% reduction in the pitch of the actin helix while keeping the original length of the filament (i.e., the binding of cofilin increases the degree of filament twisting). Such a conformational change in the filament is postulated to induce cooperative binding of cofilin to the filament (McGough et al., 1997; Galkin et al., 2001). Fluctuation analysis of actin filaments (Egelman and DeRosier, 1992) showed that the amplitude of spontaneous fluctuations would twist actin filaments to a degree comparable with the twist when filaments are decorated with cofilin (McGough et al., 1997). These results suggest that spontaneous structural fluctuations of actin filaments enable cofilin binding to the filaments and are consistent with the slow-association kinetics (Cao et al., 2006). Conceivably, cofilin prefers to associate with the twisted actin filament, which would induce twisting of the neighboring region of the filament, resulting in a cooperative form of cofilin binding.
Actin filaments behave like a twin strand of beads that can be easily twisted (Huxley and Brown, 1967). Precise x-ray diffraction studies indicate that stretching the actin filament is associated with changes in its helical structure (Huxley and Brown, 1967; Wakabayashi et al., 1994). Based on geometrical considerations of the helical structure of the actin filament, it can be postulated that stretching the filament causes untwisting of its right-handed genetic helix. Thus, when the filament is stretched, the amplitude of torsional fluctuations of the filament will be reduced, resulting in an inhibition of the cofilin binding to actin filaments. A tension-dependent reduction in the torsional fluctuations in the single actin filament was demonstrated very recently by molecular dynamics simulations (Matsushita et al., 2011). Besides, an evaluation of the effect of long axis tension on phalloidin fluorescence (Shimozawa and Ishiwata, 2009) suggests that external forces distort the filament structure. In this study, a direct measurement of the torsional fluctuations of a single actin filament under different stresses was performed by using a method of Tsuda et al. (1996). Experiments demonstrated that the SD of the fluctuations was reduced by 55–72% (P < 0.05) by an applied force of ∼5 pN (n = 5; Fig. S3). Cofilin binding increases the bending (McCullough et al., 2008; Pfaendtner et al., 2010) and twisting (Prochniewicz et al., 2005) elasticity of actin filaments, suggesting that any changes in elasticity will affect cofilin binding and severing. Therefore, reduction in the torsional fluctuation could be a potential molecular mechanism by which actin filament tension prevents cofilin binding.
This study demonstrates that a low level of tension (>2 pN) prevents (or delays) the severing of actin filaments by cofilin. This effect accounts for the frequent observations in cells that actin filaments with less tension, i.e., filaments not in use, are severed easily, whereas those in use, and thus generating larger tension, are not severed. The regulation of cofilin binding to the actin filament by tension may be behind these observations, at least in part, as schematically illustrated in Fig. 4 in addition to biochemical regulation of cofilin activity (Yang et al., 1998; Bamburg, 1999). This study raises the intriguing possibility that actin cytoskeletons are endowed with an ability to sense and respond to mechanical stress, as in the case of focal contact–associated proteins, e.g., p130Cas (Sawada et al., 2006).
Materials and methods
Direct observation and manipulation of single actin filaments
Rhodamine-labeled cytoplasmic β-actin (Cytoskeleton) was polymerized in F-buffer at a 1-mg/ml concentration for 12 h at 4°C. The F-actin solution was diluted at a final concentration of 1 µg/ml in F-buffer with beads (ϕ3 µm in diameter) coated with NEM-treated rabbit skeletal muscle myosin and placed on an observation chamber (0.5 × 2 × 18 mm). Phalloidin was not added to the solution because it inhibits the binding of cofilin to F-actin. During the experiment, an actin filament with one end attached to a ϕ10-µm bead was selected, whereas the other end of the filament was trapped with a ϕ3-µm bead that was manipulated by optical tweezers. The actin filaments were tensed by the optical tweezers. 500 nM mouse recombinant muscle cofilin (a gift from T. Obinata, Teikyo University, Tokyo, Japan) was applied by perfusion, allowing the measurement of the delay between the cofilin application and the severing of the actin filament, although 500 nM of cofilin severs actin filaments less effectively than 10 nM (Andrianantoandro and Pollard, 2006). Severing of actin filaments by mouse cofilin at the concentration 500, 250, 100, or 50 nM was examined. Rapid severing of the filaments was observed with 500 and 250 nM cofilin, and it was slower with 100 nM (or 50 nM) cofilin. We chose 500 nM in optical trapping experiments and 250 nM for magnetic force experiments to examine the effect of tension to the severing activity of cofilin. The severing of the actin filaments by cofilin was observed with an epifluorescence microscope (TE2000-U; Nikon) equipped with a high-NA lens (Plan Apo TIRF, 60×, 1.45 NA; Nikon) and a charge-coupled device camera (Cascade512B; Roper Scientific) or another epifluorescence microscope with image intensifiers (IIs; C8600; Hamamatsu Photonics) and charge-coupled device cameras (WAT-902H3-ULTIMATE; Watec Co., Ltd.). No severing was seen in the control perfusate (F-buffer with 10 mM of dithiothreitol, pH 8.0) without cofilin in both tensed or not tensed actin filaments up to 120 s in our experimental condition. The speed of solution flow was ∼100 µm/s, which was not different between control and cofilin-containing solutions. Therefore, the filament fluctuations were not changed obviously during perfusion. All imaging experiments were performed at room temperature (25°C–27°C).
Recombinant mouse muscle cofilin was provided by T. Obinata (Teikyo University, Tokyo, Japan). It should be noticed that vertebrate (mouse and chick) cofilin and vertebrate (rabbit and human) actin were used in this study, which may provide useful information of interactions between these molecules in cells (De La Cruz, 2005).
The optical trapping system was based on a previous study (Ashkin et al., 1987). The maximum retention force of the 1,064-nm laser (32 mW) was calibrated using the viscous dragging method (Tatsumi and Katayama, 1999). The mean retention force was estimated at ∼30 pN when a bead (ϕ3.0 µm; PolyScience) was trapped. All data in this study are presented as mean ± SEM.
Imaging cofilin bound to actin filaments
The F-actin solution (one nonlabeled/one rhodamine-labeled actin) at 10–100 µg/ml in F-buffer was put on a glass coverslip coated with 50 µg/ml NEM-treated muscle myosin, blocked by 1% casein in F-buffer, and washed with F-buffer (pH 6.5) before use. An equal volume of 100 nM IAF-labeled chicken cys-cofilin (Nagaoka et al., 1995) in F-buffer (pH 6.5) was applied (final concentration of 50 nM). IAF-labeled cofilin and rhodamine-labeled F-actin were observed under TIRF microscopy. The mean time constant of the photobleaching of IAF-labeled cofilin was 5.0 ± 0.8 s (n = 4) under these recording conditions. The rate of the association of cofilin (50 nM) with the glass surface that had been blocked by casein was very low (0.0001 events/s in 1 µm2 of glass surface and 50 nM cofilin), suggesting that the nonspecific binding of cofilin with the glass surface coated by NEM-myosin was negligible. The images were recorded 10–15 s after scratching the meshwork with a pipette tip. When 1 µg/ml of fluorescent Alexa Fluor 488 BSA was used as a control, no apparent binding of fluorescent BSA to the actin meshwork was detected under our experimental conditions.
We examined the actin-binding activity of the IAF-cofilin by cosedimentation assay (Nagaoka et al., 1995). As Fig. S1 shows, IAF-cofilin was cosedimented with F-actin at pH 8.4 and 7.0. As this property is comparable with those of the recombinant human cofilin (Fig. S1, lanes a and b), we conclude that the IAF-cofilin possesses basically the same binding activity as the nonlabeled native cofilin. In addition, the depolymerizing activity of IAF-cofilin was also detected but was reduced compared with the native cofilin; actin was present in the supernatant together with IAF-cofilin after ultracentrifugation, but the Coomassie brilliant blue staining was fainter than the staining of recombinant human cofilin.
Colocalization of actin and cofilin was made by using the measure colocalization feature of MetaMorph software (Molecular Devices). The fluorescence intensity of each pixel was measured at first, and the pixels with high fluorescence intensities (10% of pixels) were chosen for actin and for cofilin and were processed by measure colocalization.
Distribution of GFP-cofilin (expression vector for the GFP-cofilin fusion protein was a gift from T. Obinata) was imaged with a standard epifluorescence microscope equipped with Cascade512 charge-coupled device camera for time-lapse imaging. The number of cells with GFP-cofilin translocated to actin stress fibers was counted in each microscopic field between 20 and 90 s from the onset of relaxing the elastic substratum (20%), which was prestretched before plating cells. The cells endowed with more than two filamentous GFP-cofilin (5–20 µm in length) were considered positive for translocation of GFP-cofilin to stress fibers (as shown in Fig. 4). One to three cells were in the field of the microscope, and images were taken every 1 s. We sometimes used the FFT filtering function of ImageJ (National Institutes of Health) to remove interference patterns if necessary.
Magnetic force application to beads attached to the actin filaments
Avidin-labeled magnetic beads (SeraMag; Thermo Fisher Scientific) were conjugated with biotin-phalloidin (Invitrogen) and were then attached to the meshwork of actin filaments that adhered on the glass surface. Magnetic beads were pulled by an electrical magnet as previously mentioned (Wang et al., 1993; Ueki et al., 2010). A Permalloy bar (ϕ0.5 mm; Nireco Corporation) was used as the core of the electrical magnet. The procedure of electrochemical polishing of the Permalloy bar, mentioned elsewhere (Matthews et al., 2004), sharpens the tip of the bar and enables applying strong magnetic force to the beads. An electrical magnet was made of ∼5,000 turns/cm of copper wire (ϕ0.1 mm) around the Permalloy bar.
Preparation of proteins
Rabbit skeletal muscle actin was prepared using the method of Spudich and Watt (1971). Myosin was prepared from rabbit skeletal muscle, according to Perry (1955), and treated with NEM as follows: 5 mg/ml myosin in 0.6 M KCl and 20 mM Hepes, pH 7.0, was incubated with 100 µM NEM for 1 h at 4°C. The reaction was stopped by adding dithiothreitol at a final concentration of 10 mM. The solution was diluted 20 times with cold water and centrifuged at 6,000 g. The pellet was dissolved in 0.6 M KCl and 20 mM Hepes, pH 7.0, and dialyzed against 0.6 M KCl and 20 mM Hepes, pH 7.0. An equal volume of glycerol was added and stored at −30°C just before use.
Measurement of rotational angular fluctuation of a bead attached to an actin filament while mechanically stretching the filament
A streptavidin-conjugated polystyrene bead (2 µm in diameter) was attached to a biotin-labeled actin (Cytoskeleton) containing actin filament (∼1 µm) that was tethered to a gelsolin molecule on the glass surface. The rotational angular fluctuation of the bead was observed by an inverted microscope (TE2000-E; Nikon). Biotin-conjugated small fluorescent beads (ϕ50 nm) were attached on the streptavidin-conjugated polystyrene bead to detect the changes in the rotational angle. The motion of the fluorescent bead was analyzed by a particle-tracking application of MetaMorph image-analyzing software. A schematic drawing of the experimental setup is shown in Fig. S3. The streptavidin-conjugated polystyrene bead was optically trapped, and the actin filament was stretched by moving the trapping point in the downward direction (∼5 pN).
Online supplemental material
Fig. S1 shows the interaction of IAF-cofilin with actin. Fig. S2 shows the colocalization of GFP-cofilin with actin stress fibers in cells upon relaxation. Fig. S3 shows a decrease in the rotational angular fluctuation of a bead attached to an actin filament by mechanical stretching of the filament. Video 1 shows that tension in the actin filament prevents the filament from being severed by cofilin. Video 2 shows time-lapse images of an actin filament severed by cofilin. Video 3 shows the binding of cofilin to a severed actin meshwork. Video 4 shows the binding of IAF-cofilin to the relaxed actin bundle. Video 5 shows the binding of IAF-cofilin to the tensed actin bundle. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201102039/DC1.
Acknowledgments
We thank Mr. H. Makita (University of California, Los Angeles, Los Angeles, CA) and F. Funato (Nagoya University, Nagoya, Japan) for preparation of samples and data analyses. We thank Dr. T. Obinata for providing native and IAF-cofilin proteins and plasmids and for helpful discussion. We thank Dr. H. Abe (Chiba University, Chiba, Japan) for helpful discussion. We thank Dr. H. Machiyama for technical advice regarding single–actin filament manipulation.
This work was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology (to H. Tatsumi and M. Sokabe) and a grant from the Japan Space Forum (to H. Tatsumi and M. Sokabe).
Footnotes
Abbreviations used in this paper:
- HUVEC
- human umbilical vein endothelial cell
- IAF
- 5-iodoacetoamide fluorescein
- II
- image intensifier
- NEM
- N-ethylmaleimide
- TIRF
- total internal reflection fluorescence
- TMR
- tetramethylrhodamine
References
- Andrianantoandro E., Pollard T.D. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 24:13–23 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Ashkin A., Dziedzic J.M., Yamane T. 1987. Optical trapping and manipulation of single cells using infrared laser beams. Nature. 330:769–771 10.1038/330769a0 [DOI] [PubMed] [Google Scholar]
- Bamburg J.R. 1999. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15:185–230 10.1146/annurev.cellbio.15.1.185 [DOI] [PubMed] [Google Scholar]
- Bamburg J.R., McGough A., Ono S. 1999. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 9:364–370 10.1016/S0962-8924(99)01619-0 [DOI] [PubMed] [Google Scholar]
- Bershadsky A.D., Ballestrem C., Carramusa L., Zilberman Y., Gilquin B., Khochbin S., Alexandrova A.Y., Verkhovsky A.B., Shemesh T., Kozlov M.M. 2006. Assembly and mechanosensory function of focal adhesions: Experiments and models. Eur. J. Cell Biol. 85:165–173 10.1016/j.ejcb.2005.11.001 [DOI] [PubMed] [Google Scholar]
- Cao W., Goodarzi J.P., De La Cruz E.M. 2006. Energetics and kinetics of cooperative cofilin-actin filament interactions. J. Mol. Biol. 361:257–267 10.1016/j.jmb.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Chen C.S., Mrksich M., Huang S., Whitesides G.M., Ingber D.E. 1997. Geometric control of cell life and death. Science. 276:1425–1428 10.1126/science.276.5317.1425 [DOI] [PubMed] [Google Scholar]
- Colombo K., Grill S.W., Kimple R.J., Willard F.S., Siderovski D.P., Gönczy P. 2003. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 300:1957–1961 10.1126/science.1084146 [DOI] [PubMed] [Google Scholar]
- Damien E., Price J.S., Lanyon L.E. 2000. Mechanical strain stimulates osteoblast proliferation through the estrogen receptor in males as well as females. J. Bone Miner. Res. 15:2169–2177 10.1359/jbmr.2000.15.11.2169 [DOI] [PubMed] [Google Scholar]
- De La Cruz E.M. 2005. Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. J. Mol. Biol. 346:557–564 10.1016/j.jmb.2004.11.065 [DOI] [PubMed] [Google Scholar]
- Egelman E.H., DeRosier D.J. 1992. Image analysis shows that variations in actin crossover spacings are random, not compensatory. Biophys. J. 63:1299–1305 10.1016/S0006-3495(92)81716-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin V.E., Orlova A., Lukoyanova N., Wriggers W., Egelman E.H. 2001. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J. Cell Biol. 153:75–86 10.1083/jcb.153.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie P.G., Walker R.G. 2001. Molecular basis of mechanosensory transduction. Nature. 413:194–202 10.1038/35093011 [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Sato N., Obinata T. 2001. Dynamic reorientation of cultured cells and stress fibers under mechanical stress from periodic stretching. Exp. Cell Res. 268:104–114 10.1006/excr.2001.5270 [DOI] [PubMed] [Google Scholar]
- Huxley H.E., Brown W. 1967. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J. Mol. Biol. 30:383–434 [DOI] [PubMed] [Google Scholar]
- Iba T., Sumpio B.E. 1991. Morphological response of human endothelial cells subjected to cyclic strain in vitro. Microvasc. Res. 42:245–254 10.1016/0026-2862(91)90059-K [DOI] [PubMed] [Google Scholar]
- Katoh K., Kano Y., Amano M., Kaibuchi K., Fujiwara K. 2001. Stress fiber organization regulated by MLCK and Rho-kinase in cultured human fibroblasts. Am. J. Physiol. Cell Physiol. 280:C1669–C1679 [DOI] [PubMed] [Google Scholar]
- Kiyoshima D., Kawakami K., Hayakawa K., Tatsumi H., Sokabe M. 2011. Force- and Ca2+-dependent internalization of integrin in cultured endothelial cells. J. Cell Sci. In press [DOI] [PubMed] [Google Scholar]
- Mackay D.J., Esch F., Furthmayr H., Hall A. 1997. Rho- and rac-dependent assembly of focal adhesion complexes and actin filaments in permeabilized fibroblasts: An essential role for ezrin/radixin/moesin proteins. J. Cell Biol. 138:927–938 10.1083/jcb.138.4.927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis A.J., Chen C.S., Ingber D.E. 1997. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 94:849–854 10.1073/pnas.94.3.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita S., Inoue Y., Hojo M., Sokabe M., Adachi T. 2011. Effect of tensile force on the mechanical behavior of actin filaments. J. Biomech. 44:1776–1781 10.1016/j.jbiomech.2011.04.012 [DOI] [PubMed] [Google Scholar]
- Matthews B.D., Lavan D.A., Overby D.R., Karavitis J., Ingber D.E. 2004. Electromagnetic needles with submicron pole tip radii for nanomanipulation of biomolecules and living cells. Appl. Phys. Lett. 85:2968–2970 10.1063/1.1802383 [DOI] [Google Scholar]
- McCullough B.R., Blanchoin L., Martiel J.L., De la Cruz E.M. 2008. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. J. Mol. Biol. 381:550–558 10.1016/j.jmb.2008.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A., Pope B., Chiu W., Weeds A. 1997. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 138:771–781 10.1083/jcb.138.4.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros N.A., Burnette D.T., Forscher P. 2006. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8:215–226 10.1038/ncb1367 [DOI] [PubMed] [Google Scholar]
- Michelot A., Berro J., Guérin C., Boujemaa-Paterski R., Staiger C.J., Martiel J.L., Blanchoin L. 2007. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 17:825–833 10.1016/j.cub.2007.04.037 [DOI] [PubMed] [Google Scholar]
- Nagaoka R., Kusano K., Abe H., Obinata T. 1995. Effects of cofilin on actin filamentous structures in cultured muscle cells. Intracellular regulation of cofilin action. J. Cell Sci. 108:581–593 [DOI] [PubMed] [Google Scholar]
- Obinata T., Nagaoka-Yasuda R., Ono S., Kusano K., Mohri K., Ohtaka Y., Yamashiro S., Okada K., Abe H. 1997. Low molecular-weight G-actin binding proteins involved in the regulation of actin assembly during myofibrillogenesis. Cell Struct. Funct. 22:181–189 10.1247/csf.22.181 [DOI] [PubMed] [Google Scholar]
- Okreglak V., Drubin D.G. 2007. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178:1251–1264 10.1083/jcb.200703092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Abe H., Obinata T. 1996. Stimulus-dependent disorganization of actin filaments induced by overexpression of cofilin in C2 myoblasts. Cell Struct. Funct. 21:491–499 10.1247/csf.21.491 [DOI] [PubMed] [Google Scholar]
- Pavlov D., Muhlrad A., Cooper J., Wear M., Reisler E. 2007. Actin filament severing by cofilin. J. Mol. Biol. 365:1350–1358 10.1016/j.jmb.2006.10.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S., Mellor H. 2007. Actin stress fibres. J. Cell Sci. 120:3491–3499 10.1242/jcs.018473 [DOI] [PubMed] [Google Scholar]
- Perry S.V. 1955. Myosin Adenosinetriphosphatase. Methods in Enzymology. Colowick S.P., Kaplan N.O., editors. Vol. 2 Academic Press, New York: 582–588 [Google Scholar]
- Pfaendtner J., De La Cruz E.M., Voth G.A. 2010. Actin filament remodeling by actin depolymerization factor/cofilin. Proc. Natl. Acad. Sci. USA. 107:7299–7304 10.1073/pnas.0911675107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochniewicz E., Janson N., Thomas D.D., De la Cruz E.M. 2005. Cofilin increases the torsional flexibility and dynamics of actin filaments. J. Mol. Biol. 353:990–1000 10.1016/j.jmb.2005.09.021 [DOI] [PubMed] [Google Scholar]
- Sawada Y., Sheetz M.P. 2002. Force transduction by Triton cytoskeletons. J. Cell Biol. 156:609–615 10.1083/jcb.200110068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y., Tamada M., Dubin-Thaler B.J., Cherniavskaya O., Sakai R., Tanaka S., Sheetz M.P. 2006. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 127:1015–1026 10.1016/j.cell.2006.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozawa T., Ishiwata S. 2009. Mechanical distortion of single actin filaments induced by external force: Detection by fluorescence imaging. Biophys. J. 96:1036–1044 10.1016/j.bpj.2008.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871 [PubMed] [Google Scholar]
- Tamada M., Sheetz M.P., Sawada Y. 2004. Activation of a signaling cascade by cytoskeleton stretch. Dev. Cell. 7:709–718 10.1016/j.devcel.2004.08.021 [DOI] [PubMed] [Google Scholar]
- Tatsumi H., Katayama Y. 1999. Growth cones exhibit enhanced cell-cell adhesion after neurotransmitter release. Neuroscience. 92:855–865 10.1016/S0306-4522(99)00055-X [DOI] [PubMed] [Google Scholar]
- Timmenga E.J., Andreassen T.T., Houthoff H.J., Klopper P.J. 1991. The effect of mechanical stress on healing skin wounds: An experimental study in rabbits using tissue expansion. Br. J. Plast. Surg. 44:514–519 10.1016/0007-1226(91)90008-8 [DOI] [PubMed] [Google Scholar]
- Tsuda Y., Yasutake H., Ishijima A., Yanagida T. 1996. Torsional rigidity of single actin filaments and actin-actin bond breaking force under torsion measured directly by in vitro micromanipulation. Proc. Natl. Acad. Sci. USA. 93:12937–12942 10.1073/pnas.93.23.12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y., Sakamoto N., Sato M. 2010. Cyclic force applied to FAs induces actin recruitment depending on the dynamic loading pattern. Open Biomed Eng J. 4:129–134 10.2174/1874120701004010129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K., Sugimoto Y., Tanaka H., Ueno Y., Takezawa Y., Amemiya Y. 1994. X-ray diffraction evidence for the extensibility of actin and myosin filaments during muscle contraction. Biophys. J. 67:2422–2435 10.1016/S0006-3495(94)80729-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Butler J.P., Ingber D.E. 1993. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 260:1124–1127 10.1126/science.7684161 [DOI] [PubMed] [Google Scholar]
- Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., Nishida E., Mizuno K. 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 393:809–812 10.1038/31735 [DOI] [PubMed] [Google Scholar]
- Yonezawa N., Nishida E., Iida K., Yahara I., Sakai H. 1990. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 265:8382–8386 [PubMed] [Google Scholar]