Cortactin phosphorylation induces recruitment of the sodium-hydrogen exchanger NHE1 to invadopodia, resulting in pH changes that regulate cortactin-cofilin binding and invadopodium dynamics.
Abstract
Invadopodia are invasive protrusions with proteolytic activity uniquely found in tumor cells. Cortactin phosphorylation is a key step during invadopodia maturation, regulating Nck1 binding and cofilin activity. The precise mechanism of cortactin-dependent cofilin regulation and the roles of this pathway in invadopodia maturation and cell invasion are not fully understood. We provide evidence that cortactin–cofilin binding is regulated by local pH changes at invadopodia that are mediated by the sodium–hydrogen exchanger NHE1. Furthermore, cortactin tyrosine phosphorylation mediates the recruitment of NHE1 to the invadopodium compartment, where it locally increases the pH to cause the release of cofilin from cortactin. We show that this mechanism involving cortactin phosphorylation, local pH increase, and cofilin activation regulates the dynamic cycles of invadopodium protrusion and retraction and is essential for cell invasion in 3D. Together, these findings identify a novel pH-dependent regulation of cell invasion.
Introduction
Complications arising from cancer invasion and metastasis are the major cause of death in cancer patients. To escape from the primary tumor site, cancer cells must migrate through the ECM and intravasate through the basement membrane underlying blood vessels (Yamaguchi and Condeelis, 2007; Gimona et al., 2008). These complex and regulated processes are associated with the formation of specialized actin-rich membrane structures that degrade the ECM called invadopodia (Chen, 1989; Gimona et al., 2008). Several actin regulatory proteins, including N-WASP, cortactin, Arp2/3, and cofilin, are associated with invadopodium formation and function (Wang et al., 2007; Ayala et al., 2008), and each of these genes is up-regulated selectively in invasive breast carcinoma cells (Wang et al., 2004). Cortactin promotes invadopodium formation and maturation in many cancer cells (Artym et al., 2006; Ayala et al., 2008; Oser et al., 2009) and potentiates breast cancer metastasis in animal models (Li et al., 2001). Evidence suggests that invadopodia exhibit discrete stages of maturation, including the assembly of a cortactin-rich invadopodial precursor, cortactin phosphorylation and generation of barbed ends (actin polymerization), cortactin dephosphorylation (stabilization), and ECM degradation (Oser et al., 2009).
Cortactin phosphorylation is a key regulatory step of invadopodium maturation. Tyrosine phosphorylation of cortactin by a kinase cascade involving the Src and Arg nonreceptor tyrosine kinases (Tehrani et al., 2007; Mader et al., 2011) promotes recruitment of the Nck1 adaptor protein, N-WASP, and cofilin (DesMarais et al., 2009; Oser et al., 2009), leading to an increased Arp2/3 complex–mediated actin polymerization at invadopodia (Uruno et al., 2001; Weaver et al., 2001; Oser et al., 2010). Recent work has shown that phosphorylation of tyrosines 421 and 466, but not 482, is essential for both Nck1 binding and for barbed end formation at invadopodia (Oser et al., 2010). Together, these results suggest that cortactin-dependent regulation of N-WASP, Nck1, and cofilin is essential for actin polymerization in invadopodia. Cortactin is also known to regulate leading edge persistence (Bryce et al., 2005), matrix metalloproteinase (MMP) secretion (Clark and Weaver, 2008), and matrix degradation, contributing to cellular invasiveness (Clark et al., 2007; Weaver, 2008; Kirkbride et al., 2011). However, the mechanism linking cortactin phosphorylation to cofilin activation remains to be elucidated.
The F-actin–severing protein cofilin is essential for regulation of actin polymerization and remodeling during cell motility (Carlier et al., 1997; Ichetovkin et al., 2002; Ghosh et al., 2004; Cao et al., 2006; Sun et al., 2007). Cofilin promotes lead edge protrusion by increasing the number of free barbed ends (FBE) of actin available to initiate actin polymerization (DesMarais et al., 2004, 2005). In so doing, cofilin regulates cell migration behavior, cell directionality (Ghosh et al., 2004; Sidani et al., 2007), and, ultimately, cell invasion (Wang et al., 2007; Oser and Condeelis, 2009; van Rheenen et al., 2009). Cofilin activity is regulated via diverse mechanisms including phosphorylation/dephosphorylation (Moriyama et al., 1996), PIP2 binding (van Rheenen et al., 2007; Leyman et al., 2009), and local pH changes mediated by NHE1 (Frantz et al., 2008). NHE1 is a ubiquitously expressed transmembrane protein that regulates intracellular pH (pHi) by exchanging extracellular sodium for intracellular protons (Kemp et al., 2008).
We provide evidence here that cortactin phosphorylation promotes recruitment of NHE1 to regulate pH in invadopodia. We demonstrate that increased pH disrupts cortactin binding to cofilin, thus releasing cortactin’s inhibitory grip on cofilin. Finally, we demonstrate that these pH changes regulate invadopodial maturation and invadopodium-mediated invasion in 3D. Our findings reveal a novel mechanism by which cortactin phosphorylation and NHE1 control pH as a key step in cancer cell invasion.
Results
Cortactin phosphorylation and cofilin are required for invasion
We used a previously described 1-µm Transwell assay (Schoumacher et al., 2010) to characterize the role of cortactin phosphorylation in MDA-MB-231 invasion. Protrusive structures that crossed to the underside of the membrane (>12 µm; Fig. 1 A) were quantified as the relative number of invading structures (Fig. 1 B). Cells expressing wild-type (WT) cortactin extended cortactin-rich invasive structures that crossed to underside of the Matrigel-coated 1-µm-pore Transwell (Fig. 1 A). However, cells expressing only cortactin mutant lacking all three major tyrosine phosphorylation sites (3YF) or in which cofilin was knocked down (KD) exhibited severe defects in invasion (Fig. 1, A and B). Trans-membrane protrusion was inhibited by the protease inhibitor GM6001, which indicates a requirement for MMP activity in the process (Fig. 1 B).
Figure 1.
Cortactin tyrosine phosphorylation and cofilin are required for invadopodia elongation. (A) Representative 3D reconstructions of the 1-µm Transwell experiment. Data are based on three or more independent experiments. (B) The number of protrusive structures crossing to the bottom of the membrane (>12 µm) normalized to the cell area on top of the membrane is shown (>10 fields per group, n = 4; ***, P < 0.0001). (C) Quantification of cell protrusion through a 1-µm Transwell without Matrigel. The GM 6001 inhibitor was used to show that cell protrusion was independent of degradation (>10 fields/group, n = 4; ***, P < 0.0001). Cortactin tyrosine phosphorylation regulates Cofilin interaction in invadopodia. (D) Representative cofilin–cortactin FRET efficiency images of cells expressing either WT or 3YF cortactin. The white circle indicates the bleached spot. The top right insets show close-ups of the original images (indicated by the boxed regions; n = 5). (E) Quantification of cofilin–cortactin FRET/donor at mature invadopodia and precursors in MDA-MB-231 cell lines expressing WT or 3YF cortactin (endogenous cortactin KD). Light gray bars represent FRET on mature invadopodia and dark gray bars represent FRET on invadopodia precursors (*, P < 0.002; n = 5, >30 invadopodia/group). Error bars indicate SEM.
Cortactin 3YF and cofilin KD cells still exhibited severe defects in trans-membrane protrusion when surfaces were uncoated with Matrigel (Fig. 1 C). These data suggest that cortactin and cofilin regulate invadopodial elongation in MDA-MB-231 independently of degradation (Fig. 1 C). In agreement with these findings, cofilin KD cells (Fig. S2 C) exhibited impaired invasion through an 8-µm-pore Transwell (Fig. S1 A), even though cofilin KD cells exhibit only minor defects in matrix degradation (Fig. S1 B). Together, these results show that both cortactin phosphorylation and cofilin are required for invadopodia elongation and invasion.
Cortactin–cofilin interaction decreases during invadopodium maturation
Given their requirement for invadopodia elongation and invasion, we next determined whether cortactin–cofilin interactions change during invadopodium maturation. Cofilin is known to be the primary generator of actin FBE in invadopodia, whereas cortactin phosphorylation is a key regulator of cofilin activity (Oser et al., 2009). To test whether cortactin phosphorylation status changes during invadopodial maturation, we analyzed the presence of phosphorylated cortactin (P-cortactin) in both mature invadopodia and invadopodium precursors (Fig. S1 C). 95% of degrading invadopodia (mature invadopodia) have a clear accumulation of P-cortactin, whereas ∼25% of invadopodial precursors without degradation activity show an accumulation of P-cortactin (Fig. S1, C and D; Bowden et al., 2006). We found that EGF stimulation leads to a decrease in fluorescence resonance energy transfer (FRET) between cofilin and cortactin that coincides with the cortactin phosphorylation peak after EGF stimulation in MDA-MB-231 cells (Fig. S1 E), as we have described previously in MTLn3 cells (Oser et al., 2009). Using the degradation-based invadopodia maturation analysis, we also measured cofilin–cortactin interactions in precursor versus mature invadopodia using acceptor photobleaching FRET (AP-FRET) measurements. P-cortactin–rich mature invadopodia show a reduction in cofilin–cortactin FRET compared with invadopodium precursors (Fig. 1, D and E), which is in agreement with previous observations (Oser et al., 2009). These results suggest that a dynamic loss of cortactin–cofilin interaction correlates with invadopodium maturation. Elevated cortactin–cofilin FRET is observed in both precursors and mature invadopodia of 3YF cortactin–expressing cells (Fig. S2, A and B), which indicates that cortactin phosphorylation is associated with decreased cortactin–cofilin interaction and that both events correlate with invadopodial maturation.
Cofilin–cortactin interaction at invadopodia is regulated by local pH changes
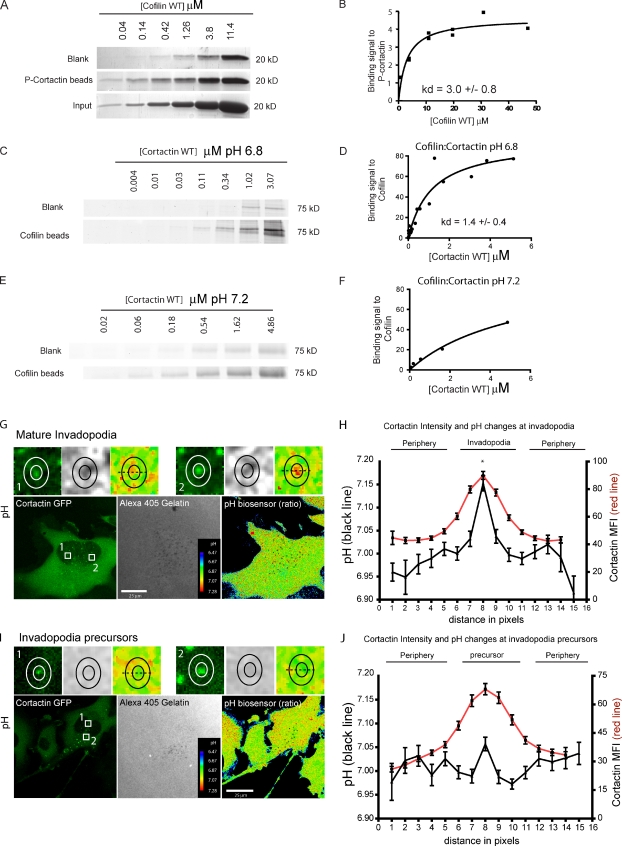
These results suggest that cortactin phosphorylation disrupts its interaction with cofilin. Thus, we conducted in vitro binding experiments to explore whether the cortactin phosphorylation was sufficient to disrupt cofilin–cortactin binding. As shown in Fig. 2 (A and B), cofilin binds efficiently to P-cortactin, with a dissociation constant (Kd = 3.0 ± 0.8 µM) similar to what was reported for the 3YF cortactin (2.8 ± 0.34 µM; Oser et al., 2009). These data indicate that cortactin phosphorylation does not disrupt its binding to cofilin directly, but must influence cofilin binding via an indirect mechanism.
Figure 2.
Cortactin–cofilin binding in vitro is not dependent on cortactin phosphorylation, but is pH sensitive. (A and B) Coomassie-stained gels (A) and quantification of the binding signal of cofilin to phosphorylated cortactin (B; P-cortactin; Kd = 3.0 ± 0.8 µM); data points to Kd calculation = 16. (C and D) Coomassie-stained gels (C) and quantification of the binding signal of cortactin to cofilin at pH 6.8 (D). Kd = 1.4 ± 0.4 µM; data points to Kd calculation = 22. (E and F) Coomassie-stained gels (E) and quantification of the binding signal of cortactin to cofilin at pH 7.2 (F). Kd > 4.1 ± 1.3 µM; data points =14. Invadopodia pH increases during maturation. MDA-MB-231 cells were plated on a 405-gelatin matrix for 4 h and pH changes were analyzed using the SNARF 5F pH biosensor. (G and H) Representative images of pH changes in mature invadopodia (G) and the line profile quantification of the pH changes (H). The inner ring represents the invadopodia core and the outer ring depicts the invadopodia periphery. Top panels in G show the local pH changes in detail. (I and J) Representative images of pH changes in invadopodia precursors (I) and the line profile quantification of the pH changes (J). The solid black line represents the pH fluctuations colocalizing with the cortactin accumulation (red line). n = 97 invadopodia and 106 precursors, three independent experiments, P < 0.001 between point 8 and all other points (Bonferroni test).
Frantz et al. (2008) demonstrated that cofilin activity is inhibited by phosphoinositide binding in a mechanism regulated by local pH changes. Cofilin undergoes pH-dependent structural changes, decreasing phosphoinositide binding at a higher pH. We tested whether pH regulates cofilin binding to cortactin. Cofilin binds to cortactin with a Kd of 1.4 ± 0.4 µM at pH 6.8 (Fig. 2, C and D). However, cortactin–cofilin binding affinity is reduced at pH 7.2 (Fig. 2, E and F). We could not reach saturation of binding at the tested concentrations in buffer at pH 7.2, and thus we were only able to place a lower limit on the Kd at pH 7.2 (limit Kd ≥ 4.1 ± 1.3 µM). The pH range used in these experiments is in agreement with the previously reported pH-dependent binding of cofilin to PIP2 (decreased binding at pH 7.5; Bailly and Jones, 2003; Frantz et al., 2008). This binding is specific, as several proteins do not bind cortactin under the conditions used here (Lapetina et al., 2009; Oser et al., 2010). Moreover, these binding affinities are within the expected range for interactions involving actin binding proteins (Pollard and Cooper, 1986; Cao et al., 2006).
The clear impact of pH changes on cofilin–cortactin binding led us to investigate pH fluctuations at invadopodia as a potential mechanism regulating cofilin–cortactin binding and cofilin activity. To characterize the local pH changes at invadopodial precursors (nondegrading) and mature invadopodia (degrading), MDA-MB-231 cells were plated on a fluorescent matrix and loaded with the pH indicator SNARF 5F. In mature invadopodia (Fig. 2 G), the peak of cortactin fluorescence (Fig. 2 H, red line) correlated with an increase in pH as determined by the SNARF 5F pH biosensor (Fig. 2 F, black line). On average, the pH in the invadopodium core was 7.15 ± 0.01 compared with a pH of 6.91 ± 0.03 in the surrounding cytoplasm. In contrast, analysis in invadopodium precursors showed that there is no significant pH change at the cortactin core of nondegrading precursors compared with the surrounding cytoplasm (pH range 6.97–7.05; Fig. 2, I and J). Together, these data suggest that cortactin–cofilin interaction in invadopodia is not directly affected by cortactin phosphorylation status but is pH dependent over the same range that pH varies in the invadopodium.
Barbed end formation in invadopodia is regulated by pH
Considering the observations that the cofilin–cortactin interaction is regulated by pH in vitro and that invadopodium maturation is accompanied by an increase in pH, we hypothesized that local pH changes regulate cofilin release from cortactin and the resulting increase in cofilin-dependent FBE in invadopodia. To test this hypothesis, we measured FBE when the pHi was stabilized to 6.8 or 7.2 using the ionophore nigericin and standard high K+ media (Chaillet and Boron, 1985; Kaplan and Boron, 1994). Upon EGF stimulation, control cells that were not treated with nigericin and maintained at pH 7.2 responded with an increase in FBE at invadopodia (Fig. 3, A and B; mean pHi of control cells was 7.08 ± 0.015). Cells that were equilibrated with a lower pH (6.8) failed to increase FBE in response to EGF, whereas the equilibration of the pHi to 7.2 alone induced a significant increase in FBE formation (Fig. 3, A and B). We show here that the pH increase from 6.8 to 7.2 observed during invadopodium maturation (Fig. 2, E and F) results in a significant increase in actin FBE. Moreover, NHE1KD cells failed to generate FBE after EGF stimulation (Fig. 3 C). These results support our hypothesis that invadopodium maturation and the generation of FBE within are regulated by pH.
Figure 3.
Actin barbed-end formation at the invadopodia is regulated by pH. (A) Representative images of a barbed-end assay in WT cortactin cells in different pH conditions. A preserved barbed end increase in response to EGF was observed in these conditions. Insets show enlarged views of the boxed regions. (B) Quantification of barbed ends in response to EGF. Results are normalized to the Starved control barbed intensity (>150 invadopodia/group, n = 3; ***, P < 0.0001). (C) Quantification of barbed ends in response to EGF in NHE1KD cells. Results are normalized to the Starved control barbed intensity (> 180 invadopodia/group, n = 3; ***, P < 0.0001). Error bars indicate SEM.
Tyrosine phosphorylation regulates cortactin–NHE1 interaction at invadopodia
The sodium hydrogen exchanger NHE1 is a key regulator of several pH-dependent cellular processes (Stock and Schwab, 2009). Recent studies suggest that NHE1 localizes to invadopodia (Busco et al., 2010) and has an important role in cancer invasion (Reshkin et al., 2000; Bourguignon et al., 2004; Busco et al., 2010). We found that endogenous NHE1 (Fig. 4 A) or ectopically expressed HA-tagged WT NHE1 (Fig. 4 B) colocalizes with cortactin in mature invadopodia in MDA-MB-231 cells. To confirm these results, we conducted a colocalization analysis using HA tag NHE1 constructs. An ERM binding–deficient NHE1 mutant (KRA-NHE1) with uniform membrane localization (Denker et al., 2000) also showed a reduced colocalization with cortactin (Fig. 4, C and E). We examined whether cortactin phosphorylation regulates recruitment of NHE1 to the invadopodia by measuring the colocalization of the 3YF cortactin mutant and NHE1. Cells expressing 3YF cortactin showed reduced cortactin–NHE1 colocalization at invadopodia (Fig. 4, D and E). Similarly, WT NHE1 showed increased colocalization with cortactin in mature invadopodia compared with invadopodia precursors (Fig. 4 F). We performed coimmunoprecipitation experiments to measure interactions between NHE1 and cortactin. NHE1 WT coimmunoprecipitated with WT cortactin, however; the nonphosphorylatable cortactin 3YF mutant exhibited a decreased interaction with NHE1 compared with WT cortactin (Fig. 4, G and H). These results suggest that cortactin phosphorylation increases NHE1 recruitment to invadopodia.
Figure 4.
Cortactin phosphorylation regulates NHE1 recruitment to the invadopodia. (A) Representative images of endogenous NHE1 and cortactin colocalization. (B) Representative image of MDA-MB-231 cells transiently expressing the WT NHE1-HA and WT cortactin. (C and D) The colocalization experiments were repeated in cells expressing KRA-NHE1–WT cortactin (C) and WT NHE1–3YF cortactin (D). Insets show enlarged views of the boxed regions. (E) Quantification of the cortactin–NHE1 colocalization. Results are based on the analysis of 15 cells/group; *, P < 0.01. (F) Quantification of NHE1-positive invadopodia precursors and mature invadopodia. The fraction of cortactin–NHE1–degradation colocalization was calculated in cells expressing WT NHE1-HA and WT cortactin. *, P < 0.05; **, P < 0.01. (G) Cells expressing WT cortactin or 3YF cortactin were lysed followed by coimmunoprecipitation of cortactin and NHE1. Although the NHE1 antibodies recognized cross-reacting bands, we used specific NHE1 siRNA to determine that the major identified bands were NHE1 (Fig. S2, D and E). (H) Quantification of NHE1 and cortactin coimmunoprecipitation. Results are based on three independent experiments. *, P < 0.03. (I) Quantification of cofilin–cortactin AP FRET at mature invadopodia and invadopodium precursors in MDA-MB-231 cells treated with NHE1 siRNA and rescued with either WT NHE1 or KRA NHE1. Mock cells are NHE1KD cells nucleoporated without a DNA construct (n = 2, >15 cells per group). Error bars indicate SEM. **, P < 0.01.
To test if NHE1 regulates cofilin–cortactin interaction at invadopodia, we analyzed the cortactin–cofilin AP FRET both in NHE1KD cells and also in rescue experiments with WT NHE1 and KRA NHE1. Considering that we observed a consistently higher FRET in invadopodia precursors compared with mature invadopodia, we used the precursor/mature invadopodia FRET ratio to report the results. As shown in Fig. 4 I, in control cells there was a twofold higher precursor/mature invadopodia FRET. The precursors/mature invadopodia FRET ratio was decreased in NHE1KD cells and fully rescued by the expression of WT NHE1 but not KRA NHE1 (Fig. S2 F). In agreement with these findings, NHE1 KD cells showed a reduced pHi, which was rescued by the expression of WT NHE1 (Fig. S3 A). Furthermore, cells expressing 3YF cortactin exhibited reduced average invadopodial pH, as NHE1 is not recruited to the invadopodia (Fig. S3 B) and NHE1 inhibition using 5-(N-ethyl-N-isopropyl)amiloride (EIPA) decreases pHi (Fig. S3 C). Together, these results show that NHE1 regulates the pH-dependent interaction between cortactin and cofilin in vivo.
Cortactin phosphorylation regulates the dynamic oscillatory behavior of invadopodia
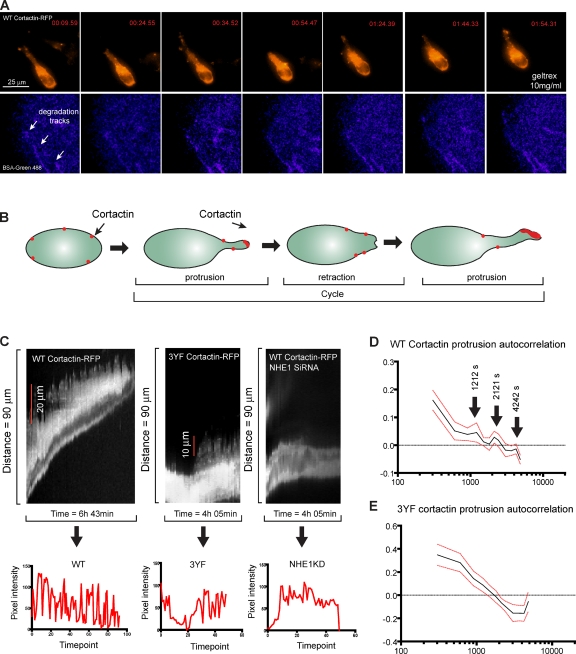
We propose that cortactin phosphorylation regulates NHE1 recruitment to invadopodia, resulting in release and activation of cofilin, leading to free barbed end generation within invadopodia. To quantify the dynamic recruitment and accumulation of cortactin and cofilin, we performed fluctuation analysis on time-lapse images of the cofilin and cortactin content in invadopodia. Both WT cortactin and cofilin exhibited periodic fluctuations in total intensity at invadopodia of steady-state MDA-MB-231 cells (Fig. 5, A and B). This oscillatory pattern was not caused by microscope focal changes because the analysis is done using 3D acquisition followed by the analysis of the maximum projection stack (Fig. 5 A, bottom inset). Correlation analysis showed that both WT cortactin and cofilin displayed an oscillation pattern of ∼800 s, as shown by the second peak on the curve (Fig. 5, C and D, arrows). 3YF cortactin expression inhibited the well-defined oscillatory pattern observed in WT cortactin-expressing cells (Fig. 5, E–G). In accordance with our model, we hypothesized that the oscillations in cortactin and cofilin at invadopodia were associated with pH fluctuations. We found that significant pH oscillations are observed at invadopodia (Fig. 5 H, bottom left) compared with the control areas in the cytoplasm (Fig. 5, H and I, bottom right). The coefficient of pH variation (standard deviation/mean, Fig. 5 I) shows that the pH oscillates more at invadopodia compared with other cytoplasmic areas of the same cell.
Figure 5.
Invadopodia display a dynamic oscillatory behavior in 2D. (A) Representative images of MDA-MB-231 cells expressing WT cortactin-RFP and cofilin-GFP. The cells were imaged for 4 h and the fluctuation in the relative fluorescence intensity of cofilin and cortactin was analyzed and plotted. The insets show the boxed region. (B) Representative traces of WT cortactin and cofilin pixel intensity representative of 1 invadopodia from 12 analyzed. (C and D) The fluctuations in WT cortactin and cofilin fluorescence intensity were analyzed using autocorrelation (see Material and methods). The autocorrelation for WT cortactin is shown in C and the corresponding cofilin-GFP autocorrelation is shown in D. Data are based on the analysis of 12 invadopodia in three independent experiments. (E–G) The experiment was repeated in cells expressing 3YF cortactin-RFP and cofilin-GFP. (E) Representative traces of cofilin and 3YF cortactin pixel intensity (1/10). Autocorrelation analysis of the fluorescence intensity of 3YF cortactin is shown in F and the corresponding cofilin-GFP signal is shown in G. Red traces represent the standard error of the mean. (H) Representative images of cells expressing WT cortactin and the SNARF-5F pH indicator. Inner white circles represent the invadopodial core and the outer circles represent the invadopodium periphery. (I) Quantification of the coefficient of pH variation. n = 18 invadopodia in three experiments; ***, P < 0.001. Error bars indicate SEM.
To test the role of NHE1 in the dynamic accumulation of cortactin and cofilin at the invadopodium, we treated MDA-MB-231 cells with the NHE1 inhibitor EIPA, which blocks NHE1-mediated pH regulation (Fig. S3 C), and analyzed cortactin and cofilin oscillations at invadopodia. Inhibition of NHE1 disrupts the stability of the invadopodium core, inducing its disassembly (Fig. S4, A and B). After addition of EIPA, the spatially stable fluorescent signals that characterize the invadopodial core disappear (Fig. S4, A and B). Together, these results define invadopodia as a very dynamic structure, characterized by cycles of cofilin and cortactin accumulation and pH increase. Furthermore, they provide evidence that cortactin phosphorylation determines the oscillatory behavior of invadopodia.
To corroborate these dynamic studies, we also analyzed the recovery of both cortactin and cofilin to invadopodia after photobleaching. As seen in Fig. S4 C, the expression of 3YF cortactin decreases the mobile fraction of both cofilin and cortactin, indicating that the invadopodium is less dynamic. These results indicate that cortactin phosphorylation regulates the dynamic recruitment of cofilin to invadopodia.
Cortactin phosphorylation, NHE1, and cofilin are essential for invadopodium elongation
The results described here identify a new pH-dependent mechanism that regulates invadopodia maturation and actin FBE generation. To analyze this mechanism in a more physiologically relevant environment, we investigated the role of cortactin tyrosine phosphorylation, cofilin, and NHE1 in cells moving in three dimensions, using a 3D invasion assay. This assay consisted of embedding cells in a thick Geltrex matrix and analyzing the transition of an early round cell to a polarized invasive cell over 72 h. After 24 h of 3D culture, MDA-MB-231 cells formed small, well-defined protrusive structures enriched in cortactin, Tks5, and MT1- MMPs (Fig. S5, A and B; and Video 1). By using the degradation marker DQ collagen IV or DQ-BSA-Green, we observed that these cortactin and Tks5-rich protrusive structures at the cell front had degradation activity and were therefore considered invadopodia (Fig. 6 A). Multiple well-defined invadopodia were observed in cells expressing WT cortactin 48 h after matrix embedding (Fig. 6 B), and these invadopodia elongated into long (>12 µm) structures, eventually leading to a polarized cell phenotype (∼72 h). A high-magnification transmission electron microscopy (TEM) image of an invadopodium (Fig. 6 B, lower right inset) shows a well-defined actin-rich protrusion surrounded by areas of matrix degradation (Fig. 6 B). Compared with WT cells, 3YF cortactin cells were rounder and had shorter invadopodia (Fig. 6, C and D). Closer examination revealed that these cells had less organized invadopodia, which was also confirmed by high-magnification TEM imaging (Fig. 6, C [bottom right inset] and D). Cofilin KD and NHE1 KD cells also showed shorter invadopodia as seen in the 3YF cortactin–expressing cells (not depicted). Thus, cortactin phosphorylation, cofilin, NHE1, and matrix proteolysis are all required for the formation of long (>12 µm) invadopodia (Fig. 6 D).
Figure 6.
Cofilin and cortactin phosphorylation are required for invadopodia protrusion. (A) Representative images of MDA-MB-231 cells expressing WT cortactin-RFP incubated in Geltrex + DQ Collagen IV (1:10) or Geltrex + BSA-Green 488 degradation marker. Images are representative of >50 cells analyzed in three independent experiments. (B) Representative images of WT cortactin cells embedded in the 3D matrix. The cells were imaged by confocal microscopy between 48 and 72 h after plating. The image is a 3D reconstruction of a representative field. Arrows and insets represent cells with invadopodia. The bottom right panel shows a representative TEM image of an invadopodium in a WT cortactin cell. (C) Representative images of 3YF cortactin cells embedded in the 3D matrix. The cells expressing 3YF cortactin-GFP were incubated as described (see Materials and methods). Arrows and bottom left insets represent high-magnification images of cells with invadopodia. The bottom right panel shows a representative TEM image of an invadopodium in a 3YF cortactin cell. Images are representative of 50 cells analyzed in three experiments. (D) Quantification of invadopodia length in 3D. Dominant protrusions enriched in cortactin were measured and the mean length analyzed. Approximately 20 cells per group were analyzed in four independent experiments. Where indicated, the cells were treated with specific SiRNA 24 h before seeding. The GM6001 inhibitor was used to show that the invadopodia protrusion is proteolysis dependent. (E) Quantification of cell protrusion through a 1-µm Transwell without Matrigel coating. Where indicated, cells were treated with NHE1 siRNA 72 h before the experiment and rescued with the described NHE1 constructs 24 h before the experiment (>8 fields/group, n = 3; **, P < 0.001). (F) Quantification of cell invasion through Matrigel-coated 1-µm Transwell. The number of protrusive structures crossing to the bottom of the membrane (>12 µm) was counted and normalized to the cell area on top of the membrane (>10 fields per group; **, P < 0.01).
We also tested the role of NHE1 in invadopodial elongation. Similarly to what we observed in the 3YF cortactin and cofilin KD cells (Fig. 1, B and C), NHE1 KD cells failed to elongate through a 1-µm Transwell filter in the absence of Matrigel (Fig. 6 E). Cell elongation was rescued by the expression of WT NHE1 but not the KRA NHE1 mutant (Fig. 6 E). Cell invasion in the presence of Matrigel was also inhibited in NHE1 KD cells (Fig. 6 F). These data suggest that the primary role of NHE1 at invadopodia is not associated with matrix degradation but rather linked to the regulation of focused actin polymerization.
Cortactin phosphorylation is required for the dynamic protrusion of invadopodia in 3D
Using the 3D invasion assay, we observed that proteolysis-dependent cell invasion is accomplished by a dynamic invadopodia protrusion/retraction cycle at the cell front (Fig. 7 A and Video 2). Interestingly, we observed a population of cells that actively degrade the matrix (Fig. 7 A) and a second population that would migrate through predegraded tracks (Fig. S5 C and Video 3; Takino et al., 2006). The mean migration speed of the degrading cells was 0.16 ± 0.02 µm/min, whereas the speed of cells migrating through the degraded areas was 0.85 ± 0.2 µm/min. Also, the protrusive structures in these cells did not resemble the structures observed on the leading edge of cells that were carving the tunnels (Fig. 7 A). Approximately 24 h after seeding, MDA-MB-231 cells extended long protrusions with a clear accumulation of cortactin at the tip (Fig. 7 A and Video 2). This invasive structure collapsed and elongated again, creating a well-defined cycle (Fig. 7 B). A few cortactin/Tks5-positive spots are also observed away from the leading invasive protrusion, and we propose that these are invadopodia precursors. Kymograph images of WT cortactin, 3YF cortactin, and NHE1KD cells (Fig. 7 C) showed that WT cells have a dynamic recycling of the invadopodia that are coupled to cell movements, whereas 3YF cortactin–expressing cells and NHE1KD cells have more stable structures with fewer cycling events. A similar phenotype was also observed for the cofilin KD cells (unpublished data). The kymograph images were analyzed in detail using the autocorrelation function of the intensity profiles for WT and 3YF cortactin. Fig. 7 D shows that WT cortactin cells had multiple peaks in the autocorrelation curve, indicating a recurrence of protrusion cycles. The correlation analysis of the 3YF cortactin cells (Fig. 7 E) and NHE1KD cells (Fig. S5 D) indicated slower and/or less defined protrusion events compared with WT. These results show that cortactin phosphorylation regulates cell invasion in 3D through the regulation of a dynamic invadopodia protrusion/retraction cycling at the cell front. Together, we provide evidence that cortactin phosphorylation regulates the dynamic cycles of cofilin and cortactin accumulation and pH increase at invadopodia, leading to an increased FBE generation. We propose that this mechanism explains the dynamic protrusion/retraction cycling of invadopodia at the cell front in 3D.
Figure 7.
Invadopodia are dynamic structures in 3D. (A) Representative images of MDA-MB-231 cells expressing WT cortactin-RFP incubated with Geltrex and BSA-Green 488 for 48 h. The cells were imaged for 8 h and the images represent snapshots of a time-lapse movie. The degradation marker channel was processed to highlight the degradation areas. (B) The cartoon shown represents the dynamics of invadopodia protrusion observed in these cells. Multiple cycles of invadopodia elongation and retraction are observed during cell invasion. Results are based on the analysis of 15 cells in three experiments. (C) Representative kymograph of a WT cortactin cell (left), a 3YF cortactin cell (middle), and a NHE1KD cell (right) migrating in 3D. The plots underneath represent the variations in pixel intensity of the protrusions over time. (D) Quantification of the WT cortactin protrusion autocorrelation. Cells expressing RFP-tagged WT cortactin were mixed with 10 mg/ml Geltrex and incubated for 24 h. Cells were imaged by between 24 and 48 h after plating. (E) Quantification of the 3YF cortactin protrusion autocorrelation. Cells expressing RFP-tagged 3YF cortactin were mixed with Geltrex and incubated for 24 h. Cells were imaged between 24 and 48 h after plating. n = 9 cells (WT) and 10 cells (3YF) in three independent experiments. Red traces represent the standard error of the mean.
Discussion
Both cortactin phosphorylation and cofilin activity are critical for invadopodium maturation and cancer cell invasion (Oser et al., 2009, 2010; Mader et al., 2011), but how cortactin phosphorylation regulates cofilin activity and especially how this mechanism regulates cell invasion in 3D was unclear. Here, we demonstrate that cortactin phosphorylation during invadopodia maturation recruits NHE1, thereby inducing a local increase in pH. The higher pH releases cofilin from cortactin and generates FBE required for invadopodium elongation. We also demonstrate that invadopodia are very dynamic structures, displaying a well-defined oscillation in pH and cortactin and cofilin content. We provide evidence that these events occur in a physiologically relevant 3D matrix system to control invasive migration. Our work demonstrates that cortactin phosphorylation–cofilin–NHE1 pathway regulates invadopodium elongation and dynamics at the cell front in 3D.
Cortactin phosphorylation regulates cofilin activity through a pH-dependent pathway
Cofilin is known to be the key generator of FBE in invadopodia, and its activation is known to be associated with cortactin phosphorylation at invadopodia. In particular, cortactin phosphorylation is correlated with dissociation of the cofilin–cortactin interaction, allowing cofilin to generate FBE (Oser and Condeelis, 2009; Oser et al., 2009). We have analyzed here the specific mechanism linking cortactin phosphorylation to the regulation of cofilin activity and found that the binding and release dynamics of cofilin from cortactin is not directly regulated by phosphorylation of cortactin. Rather, cortactin phosphorylation regulates cofilin binding indirectly, by controlling an NHE1-dependent pH increase that releases cofilin from cortactin’s inhibitory grip in invadopodia (Fig. 8).
Figure 8.
A model for how cortactin phosphorylation regulates invadopodium maturation and tumor cell invasion through a pH-dependent pathway. Cortactin tyrosine phosphorylation regulates the interaction between NHE1 and cortactin. NHE1 alkalinizes the local invadopodia pH and induces the release of cortactin-bound cofilin. Released cofilin activates barbed end generation at the invadopodia and is deactivated by either phosphorylation or by rebinding cortactin after a decrease in local pH.
The proposed inhibitory binding model is also supported by the following observations. First, we have found that cortactin concentration in MDA-MB-231 cells is in the micromolar range (1 µM, see Fig. S2 G). Additionally, the invadopodium compartment is unique and it contains on average approximately four times more cortactin than the mean cell levels, whereas cofilin shows only an approximately twofold increase over mean cell levels (Fig. S2 H). Although cofilin is found at a concentration of ∼6 µM in breast cancer cells (Chan et al., 2000), the majority of it is bound to actin or PIP2. Only an estimated 4% of cofilin is free in the cell (Tania et al., 2011), which suggests that there is enough cortactin available to sequester free cofilin in invadopodia. Altogether, these observations support our model of cofilin sequestration by cortactin at invadopodia.
Substantial evidence supports the finding that local pH increase is an evolutionarily conserved mechanism that regulates cytoskeleton dynamics and cell migration (King et al., 1994; Denker and Barber, 2002; Lovy-Wheeler et al., 2006). We describe here a novel pathway, where invasive cancer cells use a fine tuned pH regulation at invadopodia to regulate free barbed end generation. Cofilin is known to be a pH biosensor and its pH sensitivity is mediated by the protonation of the His133 (Frantz et al., 2008). Although we have strong evidence supporting an electrostatic interaction between cofilin and cortactin, the exact determinants that mediate this interaction and its disruption by elevated pH are unclear.
NHE1 is a key player in cancer invasion
NHE1 catalyzes an exchange of extracellular Na+ for intracellular H+, and its activity increases in response to growth factors (Putney et al., 2002; Frantz et al., 2007) and integrin engagement (Tominaga and Barber, 1998). NHE1 is necessary for directed migration in different cells, including fibroblasts (Denker and Barber, 2002) and leukocytes. Increasing evidence suggests that cancer cell migration and invasion rely on multiple pHi- and extracellular pH (pHe)-dependent pathways (Stüwe et al., 2007; Stock and Schwab, 2009) that regulate cell adhesion, cell volume, cytoskeleton dynamics, and ECM degradation (Stock et al., 2008; Stock and Schwab, 2009; Busco et al., 2010). More specifically, NHE1 is known to affect breast cancer invasion (Reshkin et al., 2000; Cardone et al., 2005; Cardone et al., 2007), melanoma migration (Stock et al., 2005), and ECM degradation through the regulation of pHe (Busco et al., 2010). We have used KD + rescue experiments to show that NHE1 regulates cortactin–cofilin interaction at invadopodia. NHE1 regulates the alkalization of the invadopodium, inducing cofilin activity and increasing actin filament free barbed end generation at mature invadopodia.
Upon receptor activation, cortactin is phosphorylated downstream of Src and Arg kinases (Mader et al., 2011). We propose that cortactin phosphorylation recruits NHE1 to the invadopodium, but the specific mechanism behind this step is unknown. The anchoring of NHE1 through the distal charged domain (550–564) has been demonstrated to determine the location of NHE1 (Denker et al., 2000). How this is linked to cortactin phosphorylation and then induction of the local pH changes that will drive cofilin off cortactin is unknown but may involve binding of both NHE1 and cortactin to F-actin (Weed et al., 2000; Weed and Parsons, 2001). Alternatively, NHE1 might interact with the cortactin-Nck1 invadopodium core through its binding to Nck-interacting kinase (NIK; Yan et al., 2001). Multiple mechanisms are known to regulate NHE1 exchanger activity, including phosphorylation by Rho-associated protein kinase (ROCK; Tominaga and Barber, 1998), p38 MAPK (Khaled et al., 2001), and NIK (Yan et al., 2001). Further analysis of the cortactin–NHE1 interaction is needed to elucidate this novel mechanism. The activation of NHE1 exchanger activity is also regulated downstream of growth factor receptors (Meima et al., 2009), which could represent a common EGF-dependent cortactin phosphorylation pathway. Recent work suggests that NHE1 localizes to invadopodia, where it acidifies the extracellular space, increasing ECM degradation (Busco et al., 2010). In addition to the proposed acidification of the peri-invadopodial extracellular space, we provide evidence here that the primary role of NHE1 may be to regulate barbed end formation and invadopodial elongation.
A model for invadopodial dynamics in 3D nonfibrillar matrices
The characterization of invadopodia in 3D models has been the interest of many research groups (Linder, 2007; Sabeh et al., 2009; Wolf et al., 2009; Li et al., 2010; Tolde et al., 2010). Invadopodial structure in 3D can differ significantly, depending on the cell type or the matrix model used. Using fibrillar collagen matrices or dermis slices, Wolf et al. (2009) have described cell invasion, proposing that invadopodia correspond to the lateral spikes associated with the sides but not the front of pseudopod extensions. However, this remains a speculation because no defined invadopodial markers were used in these studies. Similarly to what we describe here, Packard et al. (2009) showed that matrix degradation is localized at the leading front of migrating tumor cells. It is possible that as cells go through different ECM compositions, the mode of motility could change, making the organization of the cell front different in each specific situation (Wolf et al., 2003; Van Goethem et al., 2010).
In our investigation, we analyzed the invasion of MDA-MB-231 cells through a reconstituted basement membrane using a real 3D cell embedding technique and time-lapse imaging. We identified invadopodia using well-accepted invadopodium characteristics (Blouw et al., 2008; Crimaldi et al., 2009; Oser et al., 2010). First, invadopodia are actin-rich invasive protrusions. Second, the invadopodium is associated with matrix degradation. Finally, the scaffolding proteins cortactin and Tks5 are enriched in invadopodia. Considering these definitions and experimental conditions, we observed that the invadopodium compartment that is normally seen as a defined ventral surface structure in 2D forms a complex and dynamic invasive structure at the cell front in 3D. Also, a careful analysis of our movies indicates that invadopodial extension determines the direction of invasion. Although the crosstalk between directional sensing and the invadopodium has been suggested (DesMarais et al., 2009), the physical separation between these structures forced by 2D culturing methods always imposed a barrier to the detailed analysis of this mechanism.
Invadopodia are defined as actin-rich protrusions that localize matrix-degrading activity to cell–substratum contact points, converging sites of signaling, proteolytic, adhesive, cytoskeletal, and membrane trafficking pathways (Weaver, 2006). A comprehensive description of invadopodia in 3D awaits further work, but we propose that the distinct lamellipodium and invadopodium compartments observed in 2D are merged into one invasive structure at the cell front in 3D that preserves the characteristics of an invadopodium. Recently, Poincloux et al. (2011) found that MDA-MB-231 cells invade Matrigel based on cell rear contractility and a round phenotype (Poincloux et al., 2011). We observed a population of MDA-MB-231 cells that sustained a round phenotype but the majority of the cells in our experiments extended invadopodia and acquired a polarized phenotype after 48 h of 3D culturing. We believe that having the cells embedded in the matrix and the longer incubation times/analysis (>48 h) explain the differences observed in cell invasion.
Using our 3D model, we were able to observe the transition from a round cell to an invasive, polarized cell. This process is based on the formation of one leading edge invasive structure that goes through multiple cycles of protrusion and retraction. Overall, the defined frequency of cycles is dependent on cortactin phosphorylation as indicated by the random elongation frequency observed in the 3YF cortactin mutant. This evidence is the first to suggest that invadopodia are very dynamic structures in 3D, in agreement with what has been proposed in other 2D models (Oser et al., 2009; Artym et al., 2011). Our results suggest that cortactin phosphorylation, NHE1, and cofilin are not only required for the focal elongation of invadopodia but also the well-defined protrusion/retraction cycling observed in 3D. Interestingly, we also show that both cofilin and cortactin go through cycles of accumulation at invadopodia in 2D, resembling the frequency observed in 3D. The expression of the cortactin phosphorylation mutant 3YF induced a disorganized fluctuation of both cortactin and cofilin at invadopodia. Together, these results support that cortactin phosphorylation/dephosphorylation at invadopodia is the molecular mechanism behind the rhythmic protrusion/retraction necessary for cell invasion.
Materials and methods
Cell line
MDA-MB-231 cells were cultured in 10% FBS/DME with antibiotics. Where indicated, cells were starved in 0.5% FBS/0.8% BSA/DME for 12–16 h and then stimulated with 2.5 nM EGF (Invitrogen). All live cell experiments were imaged at 37°C in L15 media.
DNA constructs, RNAi, and transfection
KRA-NHE1-HA and WT-NHE1-HA were a gift from D. Barber (University of California, San Francisco, San Francisco, CA; Denker et al., 2000). KRA-NHE1-HA mutant contains K-A and R-A mutations in the distal charged domain, regulating ERM binding (Denker et al., 2000) and ion exchange activity (Aharonovitz et al., 2000). Mouse WT cortactin and cortactin 3YF mutants were cloned as described previously (Lapetina et al., 2009). Stable cell lines were generated using the retroviral expression vector pLXSN (BD; Oser et al., 2010). Where indicated, cofilin-GFP, Tks5-GFP (Stylli et al., 2009), and WT or 3YF cortactin-RFP (Oser et al., 2009) constructs were transiently transfected into MDA-MB-231 cells using the Nucleofection kit V protocol (Lonza) 24 h before each experiment. Control nonsilencing siRNA was from QIAGEN. For long time-lapse imaging and 3D experiments that required long incubations, cells with stable expression of WT or 3YF cortactin-GFP and stable KD of endogenous cortactin were used. To knock down the endogenous cortactin in these cells, the cortactin shRNA sequence 5′-GGACAAAGTGGATAAGAGC-3′ was subcloned into pSuper-Retro (Oligoengine) and transfected into Amphotropic Phoenix packaging cells. Retroviral sups were used to infect the GFP-cortactin WT or mutant cell lines, which were then selected with 10 µg/ml puromycin. Human-specific cortactin siGenome Smart Pool (M010508-00-0005) and SiGenome Smart pool NHE1 specific (SLC9A1: M005277-00-0005) were obtained from Thermo Fisher Scientific. 106 MDA-MB-231 cells were transfected with 2 µM siRNA using the Nucleofection kit V protocol 72 h before each experiment. For rescue experiments, the NHE1 constructs were transfected back into NHE1KD cells 24 h before the experiment.
Antibodies and inhibitors
Cortactin (ab-33333) and rabbit NHE1 (ab-67314) were obtained from Abcam; mouse anti-NHE1 (MAB3140) was obtained from Millipore. Chicken NHE11-A was obtained from Alpha Diagnostics. Arp2 (sc-H-84) and Tks5 (sc-30122) were obtained from Santa Cruz Biotechnology, Inc.; pY421 was obtained from Sigma-Aldrich; and anti-biotin FITC was obtained from Jackson ImmunoResearch Laboratories. Anti-cofilin (AE774) is a custom-made antibody (Yamaguchi et al., 2005). Alexa Fluor–conjugated secondary antibodies were from Invitrogen. For immunoprecipitation, cortactin 4F11 was obtained from Millipore. For immunoblots, cortactin (ab-33333) was obtained from Abcam; anti-β-actin (AC15) and pY421 were obtained from Sigma-Aldrich. Chicken NHE1 antibody was from D. Barber. All Western blot analyses were performed using the secondary antibodies from Li-Cor (mouse-680 and rabbit-800). EIPA was from Sigma-Aldrich and GM6001 was from Enzo Life Sciences.
3D cell imaging
25,000 cells were resuspended in cold 10 mg/ml Geltrex growth factor reduced basement membrane mix (Invitrogen). The mixture (80 µl) was plated in a 10-mm Mattek dish well for 20 min at 37°C. The dish was carefully filled with 2 ml 10% FBS/DME and incubated for 24–48 h. Before imaging, the media was replaced with 10% FBS/L15. Matrix degradation was analyzed in real time using the DQ BSA Green self-quenched BODIPY probe (Busco et al., 2010). In brief, the BSA green probe was mixed with Geltrex (30 µg/ml final concentration) before the cells were resuspended. Where indicated, DQ Collagen IV (Invitrogen) was also added to the Geltrex at a 1:10 dilution before the cells were resuspended. In motility/invasion experiments, the cells were imaged for 8 h using a microscope (DeltaVision; Applied Precision) and an EM charge-coupled device (CCD) camera (Photometrics). A 40-µm z stack (1-µm step) image was acquired every 5 min (60×, NA 1.42 oil objective lens). For quantification of invadopodia length and 3D reconstructions, the samples were imaged using a SP5 AOBS confocal microscope (Leica; 20×, NA 0.7 multi-immersion objective lens) 48 h after incubation. Mean invadopodia length was calculated using the 3D reconstructions and included invasive cell protrusions associated with degradation in any direction. Images were analyzed using Volocity 5.5 (PerkinElmer). Where indicated, a look up table filter was applied to the BSA-green channel to help visualize the degradation area.
Fluctuation analysis
All fluctuation analysis was done using custom software written in IDL (ITT Visual Information Solutions). Invadopodia fluctuation analysis can be divided into two steps: (1) generating the raw count rate trace from the invadopodium with a tracking algorithm and (2) autocorrelating the count rate trace offline with a multi-tau correlation algorithm (Schätzel, K. 1985. New concepts in correlator design. Institute of Physics Conference Series. Hilger, London. 77:175–184). The tracking algorithm consists of a Gaussian mask localization procedure and a method for assembling particle trajectories, both described previously (Crocker and Grier, 1996; Thompson et al., 2002; Larson et al., 2005). For fluctuation analysis based on the intensity of the invadopodium, the precise measure of integrated intensity in every frame is critical, and the preceding algorithms have been modified for fluctuation measurements in several ways. First, the intensity of the site is determined after a local background correction. The local background is determined by defining a border square with an edge size that is seven times the point spread function width, and the values at this position are fit to a plane of arbitrary orientation using a linear regression. The intensity of the site is then determined using a Gaussian mask fit on the background-subtracted spot. Second, the invadopodium trajectory must account for frames where the site disappears completely. If no site is found by the localization algorithm, the tracking procedure goes back to the last frame where a site was found and uses that position as the putative location of the site to calculate the intensity with the Gaussian mask (Thompson et al., 2002). This approach necessitates an approach for calculating intensity that does not depend explicitly on the existence of a visible site. In other words, using the Gaussian mask, one need only specify a position and a width of the point spread function to calculate intensity. Once the invadopodium is visible again, the tracking algorithm resumes based on this new position. This same procedure is used to generate the background trace from the cytosol. For the background trace, an arbitrary position in the cytosol, lacking an invadopodium, is chosen as a reference point, and intensity is calculated for each frame in the time stack.
After generation of a count rate trace using the tracking algorithm, the trace is autocorrelated using a multi-tau algorithm. This approach has been described in detail elsewhere (Schätzel, K. 1985. New concepts in correlator design. Institute of Physics Conference Series. Hilger, London. 77:175–184; Wohland et al., 2001) and is the basis of most commercial autocorrelator cards. In brief, the count rate, which is recorded in linearly spaced, invariantly sized frames (i.e., the intensity of a site as measured from a single frame of the time series), is rebinned to reduce noise at long autocorrelation delays. Rebinning consists of combining adjacent bins to increase the number of counts in the bin, and the bin size increases at longer delays. The change in bin size occurs at regularly spaced intervals and determines the signal-to-noise ratio and the resolution of the decay. For invadopodium autocorrelation, we used a doubling interval of four bins, meaning the size of the bin increases from one frame to two frames to four frames to eight frames, etc., at intervals of four times the fundamental imaging frequency. Therefore, if the imaging frequency is 0.125 Hz, the bin size at the 8-s delay is half the bin size at the 32-s delay. We refer the reader to Fig. 2 in Wohland et al. (2001).
Invadopodia protrusion/retraction analysis was also done using the autocorrelation analysis described. In brief, kymographs of cells invading through a 3D matrix were generated using ImageJ 1.44k. Measurements were made using a line tool crossing the protrusions on the kymograph image, and a pixel intensity profile plot was generated. The plots consisted of multiple peaks in intensity corresponding to the invadopodia protrusions. These plots were used for the autocorrelation analysis.
Cortactin–cofilin FRET experiments
MDA-MB-231 cells stably expressing WT cortactin-TagRFP or 3YF cortactin-TagRFP in which endogenous cortactin was transiently knocked down using siRNA were plated on Alexa Fluor 405–labeled gelatin for 4 h and fixed using 3.7% PFA. Immunofluorescence was performed using chicken Cofilin 774 + Alexa Fluor 488–goat anti–chicken (donor) and cortactin ab-33333 + Alexa Fluor 555–goat anti–mouse (acceptor). All the AP FRET was performed as described previously (Oser et al., 2010). In brief, invadopodia containing colocalized cortactin and cofilin were bleached (561 nm, 100% laser power, 10 iterations, bleach time ∼8.4s) and the changes in fluorescent signal in the 488 nm channel were measured and analyzed. Images were acquired using a laser scanning microscope (LSM 5 LIVE DuoScan; Carl Zeiss) with a CCD camera using LSM 5 Live Duoscan software. FRET efficiency was calculated as E = 1 − (Donor pre/Donor post). All images were corrected for laser fluctuations, sample bleaching, and background. For sensitized emission FRET (SE-FRET) experiments, MDA-MB-231 cells expressing Cofilin–GFP and WT cortactin-tagRFP were starved overnight as described previously and stimulated with 2.5 nM EGF. The live sensitized emission imaging was imaged on a microscope (IX70; Olympus) with 60× NA 1.4 objective lenses and a CCD (Sensicam QE; PCO), and the analysis was performed as described previously (Feige et al.,2005; Oser et al., 2010). In brief, control images were obtained to calculate the bleed-through: donor only, acceptor only, and donor excited acceptor. Images were analyzed using PixFRET 1.5.0. Results are shown as FRET/donor.
In vitro binding
Binding experiments were performed as described previously (Oser et al., 2009). Recombinant cortactin protein was covalently coupled to AminoLink Beads (1 mg/ml gel bed; 16.3 µM; Thermo Fisher Scientific) according to the manufacturer’s instructions. Cofilin was buffer exchanged into binding buffer (50 mM Hepes, pH 6.8, 20 mM NaCl, 0.01% NP-40, and 5% glycerol) and serially diluted 1:3; 20 µl was saved as input sample, and 490 µl of each dilution was added to 50 µl cortactin bead slurry or 50 µl blank bead slurry. The reaction was incubated for 30 min (4°C), the supernatant was removed, and beads were resuspended with binding buffer. Bound protein was recovered with 40 µl Laemmli sample buffer and analyzed on 12% SDS-PAGE gels, stained with Coomassie blue R-250, and scanned using a densitometer (Bio-Rad Laboratories). For the specific pH-binding experiments, cofilin was covalently coupled to the beads and cortactin was added to the binding buffer. Integrated density measurements of the band and equal background area were made using QuantityOne software (Bio-Rad Laboratories). The Kd was determined as described previously (Oser et al., 2009). Binding curves were generated using the standard one-site specific binding function of GraphPad Prism software (GraphPad Software).
pH measurements
Monitoring of pHi was performed using the cell-permeable SNARF-5F pH indicator (Invitrogen) according to the manufacturer’s instructions. In brief, 50,000 cells were plated on a thin Alexa Fluor 405–labeled gelatin matrix 24 h before the experiment. The pH indicator SNARF-5F (Invitrogen) was resuspended in DMSO/pluronic F-127 (Invitrogen) and mixed with the cells at a final concentration of 5 µM for 30 min, 37°C. Cells were washed twice in 10% FBS/L-15 and imaged using a confocal microscope (SP5 AOBS; Leica). The pH indicator was excited at 514 nm and emission was collected at 640 nm (625–655) and 580 nm (565–595). The 640/480 emission ratio was used to calculate the pH. In situ calibration was performed as described by the manufacturer. In brief, cells were incubated in a High K medium + 10 µM Nigericin (1 µM Nigericin, 105 mM KCl, 20 mM CaCl2, 1 mM MgSO4, 10 mM Hepes, 10 mM glucose, and TRIS for pH adjustment) with specific pH (6.6, 6.8, 7, 7.2, 7.4). All the image analysis was done using Volocity 5.5. Images were background corrected and a ratio image of 680 nm emission/580 nm (640/580) emission was generated. The mean cell 640/580 ratio of the calibration samples was used to generate a standard curve for pH calculations. Invadopodia were detected in the cortactin-GFP channel and matrix degradation was detected at using the Alexa Fluor 405–labeled matrix. At the end, a four-channel image was generated (405, matrix; 488, cortactin; 580, pH emission 1; and 640, pH emission 2). For the quantification of invadopodia pH, a linear measurement (∼15 pixels in length by 3 pixels wide) crossing the center of the cortactin spot was created for each invadopodia (Volocity measurements tool). The three pixels representing the peak intensity of cortactin were defined as the invadopodium core (pixel size xy = 0.39 µm, z step = 2.0 µm). The peak pH value in the 3 × 3 pixel matrix representing the invadopodium core was used to line up the structures for subsequent analysis. The same analysis was done in irrelevant areas of the cell and returned no changes in pH, confirming the accuracy of this technique. In time-lapse experiments, the ratio of invadopodium core pH and surrounding cytoplasm pH was used to compensate for photobleaching and laser fluctuations.
Colocalization experiments
For detection of endogenous NHE1, cells expressing WT cortactin or 3YF cortactin (100,000) were plated on thin 405-labeled gelatin matrix and fixed after 4 h in 3.7% PFA. Cells were immunostained for cortactin (ab-33333) NHE1 (ab-67314) followed by secondary antibodies (1:300 Alexa Fluor anti–mouse 555 and 1:300 Alexa Fluor anti–rabbit 488). Images were acquired using an inverted microscope (IX81; Olympus; 60×, NA 1.4 oil objective lens). For colocalization analysis of HA-tagged NHE1 mutants, cells were transfected with the indicated constructs and plated on a gelatin matrix and fixed as described earlier. Samples were imaged using a confocal microscope (SP5 AOBS; Leica), and colocalization was analyzed as described previously (Oser et al., 2010). In brief, images were processed using ImageJ Spot enhancing filter 2D (3.0 pixel Gaussian filter) and threshold levels were set to select only invadopodia (entropy threshold). The images were then analyzed for colocalization (Mander’s coefficient) using the Jacop colocalization plug-in for ImageJ.
Barbed end assay
The barbed end assay was performed as described previously (Oser et al., 2009). In brief, MDA-MB-231 cells were starved, stimulated with 2.5 nM EGF, and permeabilized with a permeabilization buffer (20 mM Hepes, pH 7.5, 138 mM KCl, 4 mM MgCl2, 3 mM EGTA, 0.2 mg/ml saponin, 1 mM ATP, and 1% BSA) containing 0.4 M biotin–actin (Cytoskeleton) for 1 min at 37°C. Cells were fixed in 3.7% formaldehyde for 5 min followed by blocking in PBS containing 1% FBS, 1% BSA, and 3 µM phalloidin. The samples were then stained with FITC anti-biotin to visualize barbed ends and Arp2 to identify invadopodium precursors. The actin–biotin intensity at invadopodium precursors was quantified as mean gray value (MGV) at invadopodium precursors minus MGV of the background. The data were normalized to the control condition for each experiment (Oser et al., 2009).
Co-immunoprecipitation
Immunoprecipitation experiments were performed as described previously (Oser et al., 2009). In brief, 106 MDA-MB-231 cells were plated on plastic tissue culture dishes 48 h before the experiment. The samples were stimulated with EGF (2.5 nM, 3 min), washed with cold PBS, and lysed in buffer containing 10 mM Hepes, 50 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 1% Triton X-100, and phosphatase (NaF and NaVO4) and protease inhibitors. The lysates were centrifuged at 18,000 g for 10 min at 4°C and precleaned with protein A/G agarose beads (sc-2003) for 30 min. The immunoprecipitate (IP) was mixed overnight with 2 µg cortactin antibody 4F11 or 2 µg control IgG1 antibody conjugated to protein A/G agarose beads. The IPs were washed three times in lysis buffer and analyzed by Western blotting. All Western blot analyses were done using QuantityOne (Bio-Rad Laboratories). Images were corrected for background levels and results are expressed as co-IP normalized to the IP optical density for each sample.
Electron microscopy
MDA-MB-231 cells expressing WT or 3YF cortactin-TagRFP where endogenous cortactin was transiently knocked down were plated on Geltrex (10 mg/ml)-coated Mattek dishes. Samples were fixed 24 h after incubation using 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, postfixed with 1% osmium tetroxide followed by 1% uranyl acetate then by dehydration through a graded series of ethanol, and embedded in LX112 resin (LADD Research Industries). Ultrathin (80-nm) sections were cut on an ultramicrotome (Ultracut UCT; Reichert), stained with uranyl acetate followed by lead citrate, and viewed on a transmission electron microscope (1200EX; JEOL) at 80 kv. To confirm that the matrix-free regions on the matrix were not artifacts, we compared areas around invadopodia and distant cell–matrix interface areas searching for matrix-free areas. The majority of matrix-free areas were associated with invadopodia. (Fig. S5 E)
Transwell invasion assay
Transwell assays were performed as described previously (Oser et al., 2010). In brief, 8.0-µm Transwell supports (Costar) were coated with 50 µl of reduced growth factor Matrigel (2.5 mg/ml) for 1 h at 37°C. The membranes were allowed to equilibrate in DME at 37°C for 1 h. 50,000 cells were resuspended in 200 µl 0.5% FBS/DME and plated in the upper chamber. The bottom chamber was filled with 1 ml 10% FBS/DMEM. Similarly, 1 µm Transwell assays were adapted from Schoumacher et al. (2010). In brief, 2.5 mg/ml Matrigel was mixed with 0.2 mg/ml FITC-gelatin (1/20 dilution) and applied to the upper surface of the membrane. Membranes were equilibrated as described and 50,000 cells were plated. Where indicated, cells were plated in Transwell membranes without Matrigel coating and allowed to invade for 24 h followed by fixation in 3.7% PFA. Intact membranes were imaged using an SP5 microscope (Leica; 3D stacks of the membrane section). Cell invasion in Matrigel-coated membranes was calculated as the number of invadopodia crossing to the underside of the membrane normalized to the cell area on the top of the membrane. Cell elongation on non–Matrigel-coated membranes was calculated using a ratio of cell area in the bottom and cell area on the top of the membrane to calculate invasion. In all Transwell experiments, the cells were stained with either Alexa Fluor 488 or Alexa Fluor 555 Phalloidin for 30 min before imaging. The MMP inhibitor GM6001 (25 µM) was added as indicated.
FRAP
FRAP curves were acquired on the Zeiss 5 Live Duoscan (Carl Zeiss) with dual bleaching using the 488 nm and 561 nm lasers, and dual fluorescence recovery monitoring in both red (550–615 nm) and green (495–525 nm) channels. Images were acquired with a time structure of 5 prebleach scans and 115 postbleach scans at time intervals of 250 ms, resulting in a total acquisition of ∼30 s. The integrated intensity of the invadopod at each time point was determined using a Gaussian mask tracking algorithm called Localize that has been described previously (Larson et al., 2005) to generate the raw FRAP curves. The curves were normalized to prebleach values, and the size of the bleach square was fixed at 10 × 10 pixels. Total cellular fluorescence was also measured to correct for possible cellular photobleaching. For the illumination intensities used to monitor fluorescence recovery over this time scale, we observed minimal photobleaching, obviating the need for additional normalization of the FRAP curve. The resulting normalized curves were fit using a nonlinear least squares fitting routine to the following function:
where F(t) is the integrated fluorescence of the invadopod at time t; k1 is the recovery time constant; A is the relative fractions of k1; and C is a constant offset to account for the bleach depth. All image processing was done with custom software implemented in IDL and curve fitting was done using GraphPad 5.03 (GraphPad Software).
Correlation coefficients between time constants determined for the same invadopodium but in different channels were calculated according to:
where kj are the time constants described above (j = 1,2) for a particular channel (red, green) and i is the summation variable that accounts for individual invadopodia. The mean value of the population is denoted by a superscript bar.
Statistical analysis
In experiments containing multiple groups, ANOVA was performed associated to Bonferroni tests. Where indicated, statistical analysis was calculated using the unpaired, two-tailed Student’s t test. Statistical significance was defined as P < 0.05. Error bars represent the SEM.
Online supplemental material
Fig. S1 complements the data shown in Fig. 1, including colocalization analysis, matrix degradation, and SE-FRET analyses. Fig. S2 shows the KD efficiency and expression levels of proteins tested. Fig. S3 shows the effects of NHE1KD and EIPA inhibitors on invadopodia and cell pH. Fig. S4 shows the effect of EIPA in invadopodia stability and the FRAP analysis of cortactin and cofilin at the invadopodia. Fig. S5 further describes the migration of MDA-MB-231 cells in 3D. Videos 1–3 show time-lapse images of MDA-MB-231 cells migrating in 3D. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201103045/DC1.
Acknowledgments
We thank Dr. Diane Barber for the NHE1 constructs, Dr. Bradley Webb for technical support and discussions, and Dr. Sergio Grinstein for helpful discussions. We also thank the Analytical Imaging Facility for technical help, as well as Ved Sharma and Minna Roh-Johnson for the discussions and Geoffrey Perumal for the EM preparations.
This work was funded by CA150344, CA100324 (to M. Oser, M.A.O. Magalhaes, and J. Condeelis), NIHCA133346, NIHNS39475; an Exceptional Project Grant from the Breast Cancer Alliance (to A.J. Koleske); and a National Science Foundation Graduate Research Fellowship (to C.C. Mader).
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- EIPA
- 5-(N-ethyl-N-isopropyl)amiloride
- FBE
- free barbed ends
- FRET
- fluorescence resonance energy transfer
- IP
- immunoprecipitate
- KD
- knockdown
- MMP
- matrix metalloproteinase
- pHi
- intracellular pH
- SE-FRET
- sensitized emission FRET
- TEM
- transmission electron microscopy
- WT
- wild type
References
- Aharonovitz O., Zaun H.C., Balla T., York J.D., Orlowski J., Grinstein S. 2000. Intracellular pH regulation by Na(+)/H(+) exchange requires phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150:213–224 10.1083/jcb.150.1.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym V.V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K.M., Mueller S.C. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66:3034–3043 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Artym V.V., Matsumoto K., Mueller S.C., Yamada K.M. 2011. Dynamic membrane remodeling at invadopodia differentiates invadopodia from podosomes. Eur. J. Cell Biol. 90:172–180 10.1016/j.ejcb.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala I., Baldassarre M., Giacchetti G., Caldieri G., Tetè S., Luini A., Buccione R. 2008. Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation. J. Cell Sci. 121:369–378 10.1242/jcs.008037 [DOI] [PubMed] [Google Scholar]
- Bailly M., Jones G.E. 2003. Polarised migration: cofilin holds the front. Curr. Biol. 13:R128–R130 10.1016/S0960-9822(03)00072-1 [DOI] [PubMed] [Google Scholar]
- Blouw B., Seals D.F., Pass I., Diaz B., Courtneidge S.A. 2008. A role for the podosome/invadopodia scaffold protein Tks5 in tumor growth in vivo. Eur. J. Cell Biol. 87:555–567 10.1016/j.ejcb.2008.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L.Y., Singleton P.A., Diedrich F., Stern R., Gilad E. 2004. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J. Biol. Chem. 279:26991–27007 10.1074/jbc.M311838200 [DOI] [PubMed] [Google Scholar]
- Bowden E.T., Onikoyi E., Slack R., Myoui A., Yoneda T., Yamada K.M., Mueller S.C. 2006. Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp. Cell Res. 312:1240–1253 10.1016/j.yexcr.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Bryce N.S., Clark E.S., Leysath J.L., Currie J.D., Webb D.J., Weaver A.M. 2005. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 15:1276–1285 10.1016/j.cub.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Busco G., Cardone R.A., Greco M.R., Bellizzi A., Colella M., Antelmi E., Mancini M.T., Dell’Aquila M.E., Casavola V., Paradiso A., Reshkin S.J. 2010. NHE1 promotes invadopodial ECM proteolysis through acidification of the peri-invadopodial space. FASEB J. 24:3903–3915 10.1096/fj.09-149518 [DOI] [PubMed] [Google Scholar]
- Cao W., Goodarzi J.P., De La Cruz E.M. 2006. Energetics and kinetics of cooperative cofilin-actin filament interactions. J. Mol. Biol. 361:257–267 10.1016/j.jmb.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Cardone R.A., Bagorda A., Bellizzi A., Busco G., Guerra L., Paradiso A., Casavola V., Zaccolo M., Reshkin S.J. 2005. Protein kinase A gating of a pseudopodial-located RhoA/ROCK/p38/NHE1 signal module regulates invasion in breast cancer cell lines. Mol. Biol. Cell. 16:3117–3127 10.1091/mbc.E04-10-0945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone R.A., Bellizzi A., Busco G., Weinman E.J., Dell’Aquila M.E., Casavola V., Azzariti A., Mangia A., Paradiso A., Reshkin S.J. 2007. The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol. Biol. Cell. 18:1768–1780 10.1091/mbc.E06-07-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.F., Laurent V., Santolini J., Melki R., Didry D., Xia G.X., Hong Y., Chua N.H., Pantaloni D. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136:1307–1322 10.1083/jcb.136.6.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillet J.R., Boron W.F. 1985. Intracellular calibration of a pH-sensitive dye in isolated, perfused salamander proximal tubules. J. Gen. Physiol. 86:765–794 10.1085/jgp.86.6.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.Y., Bailly M., Zebda N., Segall J.E., Condeelis J.S. 2000. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148:531–542 10.1083/jcb.148.3.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.T. 1989. Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells. J. Exp. Zool. 251:167–185 10.1002/jez.1402510206 [DOI] [PubMed] [Google Scholar]
- Clark E.S., Weaver A.M. 2008. A new role for cortactin in invadopodia: regulation of protease secretion. Eur. J. Cell Biol. 87:581–590 10.1016/j.ejcb.2008.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E.S., Whigham A.S., Yarbrough W.G., Weaver A.M. 2007. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67:4227–4235 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- Crimaldi L., Courtneidge S.A., Gimona M. 2009. Tks5 recruits AFAP-110, p190RhoGAP, and cortactin for podosome formation. Exp. Cell Res. 315:2581–2592 10.1016/j.yexcr.2009.06.012 [DOI] [PubMed] [Google Scholar]
- Crocker J.C., Grier D.G. 1996. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179:298–310 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
- Denker S.P., Barber D.L. 2002. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 159:1087–1096 10.1083/jcb.200208050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker S.P., Huang D.C., Orlowski J., Furthmayr H., Barber D.L. 2000. Direct binding of the Na—H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H(+) translocation. Mol. Cell. 6:1425–1436 10.1016/S1097-2765(00)00139-8 [DOI] [PubMed] [Google Scholar]
- DesMarais V., Macaluso F., Condeelis J., Bailly M. 2004. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J. Cell Sci. 117:3499–3510 10.1242/jcs.01211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V., Ghosh M., Eddy R., Condeelis J. 2005. Cofilin takes the lead. J. Cell Sci. 118:19–26 10.1242/jcs.01631 [DOI] [PubMed] [Google Scholar]
- DesMarais V., Yamaguchi H., Oser M., Soon L., Mouneimne G., Sarmiento C., Eddy R., Condeelis J. 2009. N-WASP and cortactin are involved in invadopodium-dependent chemotaxis to EGF in breast tumor cells. Cell Motil. Cytoskeleton. 66:303–316 10.1002/cm.20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige J.N., Sage D., Wahli W., Desvergne B., Gelman L. 2005. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc. Res. Tech. 68:51–58 10.1002/jemt.20215 [DOI] [PubMed] [Google Scholar]
- Frantz C., Karydis A., Nalbant P., Hahn K.M., Barber D.L. 2007. Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J. Cell Biol. 179:403–410 10.1083/jcb.200704169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C., Barreiro G., Dominguez L., Chen X., Eddy R., Condeelis J., Kelly M.J., Jacobson M.P., Barber D.L. 2008. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 183:865–879 10.1083/jcb.200804161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 304:743–746 10.1126/science.1094561 [DOI] [PubMed] [Google Scholar]
- Gimona M., Buccione R., Courtneidge S.A., Linder S. 2008. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 20:235–241 10.1016/j.ceb.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Ichetovkin I., Grant W., Condeelis J. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12:79–84 10.1016/S0960-9822(01)00629-7 [DOI] [PubMed] [Google Scholar]
- Kaplan D.L., Boron W.F. 1994. Long-term expression of c-H-ras stimulates Na-H and Na(+)-dependent Cl-HCO3 exchange in NIH-3T3 fibroblasts. J. Biol. Chem. 269:4116–4124 [PubMed] [Google Scholar]
- Kemp G., Young H., Fliegel L. 2008. Structure and function of the human Na+/H+ exchanger isoform 1. Channels (Austin). 2:329–336 10.4161/chan.2.5.6898 [DOI] [PubMed] [Google Scholar]
- Khaled A.R., Moor A.N., Li A., Kim K., Ferris D.K., Muegge K., Fisher R.J., Fliegel L., Durum S.K. 2001. Trophic factor withdrawal: p38 mitogen-activated protein kinase activates NHE1, which induces intracellular alkalinization. Mol. Cell. Biol. 21:7545–7557 10.1128/MCB.21.22.7545-7557.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K.L., Essig J., Roberts T.M., Moerland T.S. 1994. Regulation of the Ascaris major sperm protein (MSP) cytoskeleton by intracellular pH. Cell Motil. Cytoskeleton. 27:193–205 10.1002/cm.970270302 [DOI] [PubMed] [Google Scholar]
- Kirkbride K.C., Sung B.H., Sinha S., Weaver A.M. 2011. Cortactin: a multifunctional regulator of cellular invasiveness. Cell Adh Migr. 5:187–198 10.4161/cam.5.2.14773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapetina S., Mader C.C., Machida K., Mayer B.J., Koleske A.J. 2009. Arg interacts with cortactin to promote adhesion-dependent cell edge protrusion. J. Cell Biol. 185:503–519 10.1083/jcb.200809085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson D.R., Johnson M.C., Webb W.W., Vogt V.M. 2005. Visualization of retrovirus budding with correlated light and electron microscopy. Proc. Natl. Acad. Sci. USA. 102:15453–15458 10.1073/pnas.0504812102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman S., Sidani M., Ritsma L., Waterschoot D., Eddy R., Dewitte D., Debeir O., Decaestecker C., Vandekerckhove J., van Rheenen J., et al. 2009. Unbalancing the phosphatidylinositol-4,5-bisphosphate-cofilin interaction impairs cell steering. Mol. Biol. Cell. 20:4509–4523 10.1091/mbc.E09-02-0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tondravi M., Liu J., Smith E., Haudenschild C.C., Kaczmarek M., Zhan X. 2001. Cortactin potentiates bone metastasis of breast cancer cells. Cancer Res. 61:6906–6911 [PubMed] [Google Scholar]
- Li A., Dawson J.C., Forero-Vargas M., Spence H.J., Yu X., König I., Anderson K., Machesky L.M. 2010. The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr. Biol. 20:339–345 10.1016/j.cub.2009.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S. 2007. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 17:107–117 10.1016/j.tcb.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Lovy-Wheeler A., Kunkel J.G., Allwood E.G., Hussey P.J., Hepler P.K. 2006. Oscillatory increases in alkalinity anticipate growth and may regulate actin dynamics in pollen tubes of lily. Plant Cell. 18:2182–2193 10.1105/tpc.106.044867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader C.C., Oser M., Magalhaes M.A., Bravo-Cordero J.J., Condeelis J.S., Koleske A.J., Gil-Henn H. 2011. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 71:1730–1741 10.1158/0008-5472.CAN-10-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meima M.E., Webb B.A., Witkowska H.E., Barber D.L. 2009. The sodium-hydrogen exchanger NHE1 is an Akt substrate necessary for actin filament reorganization by growth factors. J. Biol. Chem. 284:26666–26675 10.1074/jbc.M109.019448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K., Iida K., Yahara I. 1996. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells. 1:73–86 10.1046/j.1365-2443.1996.05005.x [DOI] [PubMed] [Google Scholar]
- Oser M., Condeelis J. 2009. The cofilin activity cycle in lamellipodia and invadopodia. J. Cell. Biochem. 108:1252–1262 10.1002/jcb.22372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M., Yamaguchi H., Mader C.C., Bravo-Cordero J.J., Arias M., Chen X., DesMarais V., van Rheenen J., Koleske A.J., Condeelis J. 2009. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186:571–587 10.1083/jcb.200812176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M., Mader C.C., Gil-Henn H., Magalhaes M., Bravo-Cordero J.J., Koleske A.J., Condeelis J. 2010. Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J. Cell Sci. 123:3662–3673 10.1242/jcs.068163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard B.Z., Artym V.V., Komoriya A., Yamada K.M. 2009. Direct visualization of protease activity on cells migrating in three-dimensions. Matrix Biol. 28:3–10 10.1016/j.matbio.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poincloux R., Collin O., Lizárraga F., Romao M., Debray M., Piel M., Chavrier P. 2011. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc. Natl. Acad. Sci. USA. 108:1943–1948 10.1073/pnas.1010396108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. 1986. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55:987–1035 10.1146/annurev.bi.55.070186.005011 [DOI] [PubMed] [Google Scholar]
- Putney L.K., Denker S.P., Barber D.L. 2002. The changing face of the Na+/H+ exchanger, NHE1: structure, regulation, and cellular actions. Annu. Rev. Pharmacol. Toxicol. 42:527–552 10.1146/annurev.pharmtox.42.092001.143801 [DOI] [PubMed] [Google Scholar]
- Reshkin S.J., Bellizzi A., Albarani V., Guerra L., Tommasino M., Paradiso A., Casavola V. 2000. Phosphoinositide 3-kinase is involved in the tumor-specific activation of human breast cancer cell Na(+)/H(+) exchange, motility, and invasion induced by serum deprivation. J. Biol. Chem. 275:5361–5369 10.1074/jbc.275.8.5361 [DOI] [PubMed] [Google Scholar]
- Sabeh F., Shimizu-Hirota R., Weiss S.J. 2009. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 185:11–19 10.1083/jcb.200807195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R.D., Louvard D., Vignjevic D.M. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189:541–556 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidani M., Wessels D., Mouneimne G., Ghosh M., Goswami S., Sarmiento C., Wang W., Kuhl S., El-Sibai M., Backer J.M., et al. 2007. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 179:777–791 10.1083/jcb.200707009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock C., Schwab A. 2009. Protons make tumor cells move like clockwork. Pflugers Arch. 458:981–992 10.1007/s00424-009-0677-8 [DOI] [PubMed] [Google Scholar]
- Stock C., Gassner B., Hauck C.R., Arnold H., Mally S., Eble J.A., Dieterich P., Schwab A. 2005. Migration of human melanoma cells depends on extracellular pH and Na+/H+ exchange. J. Physiol. 567:225–238 10.1113/jphysiol.2005.088344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock C., Cardone R.A., Busco G., Krähling H., Schwab A., Reshkin S.J. 2008. Protons extruded by NHE1: digestive or glue? Eur. J. Cell Biol. 87:591–599 10.1016/j.ejcb.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Stüwe L., Müller M., Fabian A., Waning J., Mally S., Noël J., Schwab A., Stock C. 2007. pH dependence of melanoma cell migration: protons extruded by NHE1 dominate protons of the bulk solution. J. Physiol. 585:351–360 10.1113/jphysiol.2007.145185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylli S.S., Stacey T.T., Verhagen A.M., Xu S.S., Pass I., Courtneidge S.A., Lock P. 2009. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J. Cell Sci. 122:2727–2740 10.1242/jcs.046680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.X., Magalhães M.A., Glogauer M. 2007. Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J. Cell Biol. 179:239–245 10.1083/jcb.200705122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takino T., Watanabe Y., Matsui M., Miyamori H., Kudo T., Seiki M., Sato H. 2006. Membrane-type 1 matrix metalloproteinase modulates focal adhesion stability and cell migration. Exp. Cell Res. 312:1381–1389 10.1016/j.yexcr.2006.01.008 [DOI] [PubMed] [Google Scholar]
- Tania N., Prosk E., Condeelis J., Edelstein-Keshet L. 2011. A temporal model of cofilin regulation and the early peak of actin barbed ends in invasive tumor cells. Biophys. J. 100:1883–1892 10.1016/j.bpj.2011.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani S., Tomasevic N., Weed S., Sakowicz R., Cooper J.A. 2007. Src phosphorylation of cortactin enhances actin assembly. Proc. Natl. Acad. Sci. USA. 104:11933–11938 10.1073/pnas.0701077104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.E., Larson D.R., Webb W.W. 2002. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82:2775–2783 10.1016/S0006-3495(02)75618-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolde O., Rösel D., Veselý P., Folk P., Brábek J. 2010. The structure of invadopodia in a complex 3D environment. Eur. J. Cell Biol. 89:674–680 10.1016/j.ejcb.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Tominaga T., Barber D.L. 1998. Na-H exchange acts downstream of RhoA to regulate integrin-induced cell adhesion and spreading. Mol. Biol. Cell. 9:2287–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno T., Liu J., Zhang P., Fan Yx C., Egile C., Li R., Mueller S.C., Zhan X. 2001. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 3:259–266 10.1038/35060051 [DOI] [PubMed] [Google Scholar]
- Van Goethem E., Poincloux R., Gauffre F., Maridonneau-Parini I., Le Cabec V. 2010. Matrix architecture dictates three-dimensional migration modes of human macrophages: differential involvement of proteases and podosome-like structures. J. Immunol. 184:1049–1061 10.4049/jimmunol.0902223 [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Song X., van Roosmalen W., Cammer M., Chen X., DesMarais V., Yip S.C., Backer J.M., Eddy R.J., Condeelis J.S. 2007. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 179:1247–1259 10.1083/jcb.200706206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen J., Condeelis J., Glogauer M. 2009. A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J. Cell Sci. 122:305–311 10.1242/jcs.031146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Goswami S., Lapidus K., Wells A.L., Wyckoff J.B., Sahai E., Singer R.H., Segall J.E., Condeelis J.S. 2004. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 64:8585–8594 10.1158/0008-5472.CAN-04-1136 [DOI] [PubMed] [Google Scholar]
- Wang W., Eddy R., Condeelis J. 2007. The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer. 7:429–440 10.1038/nrc2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A.M. 2006. Invadopodia: specialized cell structures for cancer invasion. Clin. Exp. Metastasis. 23:97–105 10.1007/s10585-006-9014-1 [DOI] [PubMed] [Google Scholar]
- Weaver A.M. 2008. Cortactin in tumor invasiveness. Cancer Lett. 265:157–166 10.1016/j.canlet.2008.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A.M., Karginov A.V., Kinley A.W., Weed S.A., Li Y., Parsons J.T., Cooper J.A. 2001. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11:370–374 10.1016/S0960-9822(01)00098-7 [DOI] [PubMed] [Google Scholar]
- Weed S.A., Parsons J.T. 2001. Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene. 20:6418–6434 10.1038/sj.onc.1204783 [DOI] [PubMed] [Google Scholar]
- Weed S.A., Karginov A.V., Schafer D.A., Weaver A.M., Kinley A.W., Cooper J.A., Parsons J.T. 2000. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 151:29–40 10.1083/jcb.151.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohland T., Rigler R., Vogel H. 2001. The standard deviation in fluorescence correlation spectroscopy. Biophys. J. 80:2987–2999 10.1016/S0006-3495(01)76264-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Mazo I., Leung H., Engelke K., von Andrian U.H., Deryugina E.I., Strongin A.Y., Bröcker E.B., Friedl P. 2003. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160:267–277 10.1083/jcb.200209006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Alexander S., Schacht V., Coussens L.M., von Andrian U.H., van Rheenen J., Deryugina E., Friedl P. 2009. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 20:931–941 10.1016/j.semcdb.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Condeelis J. 2007. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta. 1773:642–652 10.1016/j.bbamcr.2006.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., Condeelis J. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 168:441–452 10.1083/jcb.200407076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Nehrke K., Choi J., Barber D.L. 2001. The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J. Biol. Chem. 276:31349–31356 10.1074/jbc.M102679200 [DOI] [PubMed] [Google Scholar]