Membrane organization by eisosomes is mediated by self-assembly of its main components into a membrane-bound protein scaffold with lipid-binding specificity.
Abstract
Spatial organization of membranes into domains of distinct protein and lipid composition is a fundamental feature of biological systems. The plasma membrane is organized in such domains to efficiently orchestrate the many reactions occurring there simultaneously. Despite the almost universal presence of membrane domains, mechanisms of their formation are often unclear. Yeast cells feature prominent plasma membrane domain organization, which is at least partially mediated by eisosomes. Eisosomes are large protein complexes that are primarily composed of many subunits of two Bin–Amphiphysin–Rvs domain–containing proteins, Pil1 and Lsp1. In this paper, we show that these proteins self-assemble into higher-order structures and bind preferentially to phosphoinositide-containing membranes. Using a combination of electron microscopy approaches, we generate structural models of Pil1 and Lsp1 assemblies, which resemble eisosomes in cells. Our data suggest that the mechanism of membrane organization by eisosomes is mediated by self-assembly of its core components into a membrane-bound protein scaffold with lipid-binding specificity.
Introduction
Cells are delimited by the plasma membrane, which mediates all communication and transport in and out of the cell. This necessitates the coordinated execution of many biochemical reactions simultaneously. To achieve this intricate task, the plasma membrane is highly organized in space and time. Despite the importance of membrane domains in cell biology, the mechanisms involved in domain formation are not well understood in many cases. The plasma membrane of Saccharomyces cerevisiae is patterned into at least three distinct nonoverlapping domains that are named after specific marker proteins, including membrane compartment containing either Pma1 (MCP), target of rapamycin complex 2 (MCT), or Can1 (MCC; Malínská et al., 2003; Berchtold and Walther, 2009). These domains differ in appearance, forming either a punctate pattern (MCC and MCT) or a network (MCP). In addition to harboring specific proteins, yeast plasma membrane domains also vary in lipid composition. In particular, the MCC is thought to be enriched in ergosterol, the major yeast sterol (Grossmann et al., 2007).
Ultrastructurally, MCCs appear as furrows in the plasma membrane (Strádalová et al., 2009). Their formation is mediated by large protein complexes underlying this domain, termed eisosomes (Walther et al., 2006). Eisosomes are remarkable cellular structures; they form a distributed pattern of complexes that are spaced at a minimal distance from each other. Moreover, they are extremely stable once formed and do not exchange subunits, nor do they move (Malínská et al., 2003; Walther et al., 2006). A typical yeast cell has ∼30 eisosomes, depending on its surface area, each of them consisting of many copies of two extremely abundant, highly similar proteins, Pil1 and Lsp1 (115,000 and 104,000 molecules per cell, respectively; Ghaemmaghami et al., 2003; de Godoy et al., 2008). Recently, we discovered that the molecular structure of Pil1 and Lsp1s core part consists of a BAR domain (for Bin1, amphiphysin, and Rvs proteins; Ziółkowska et al., 2011). These banana-shaped domains are found across species in a variety of proteins that commonly modulate membrane curvature in diverse processes, ranging from endocytosis to plasma membrane protrusion (Gallop and McMahon, 2005; Ren et al., 2006). In yeast, eisosome BAR domain proteins participate in membrane domain organization, as the normal plasma membrane domain pattern collapses and all fluorescently tagged MCC membrane proteins investigated so far mislocalize in pil1Δ cells, distributing uniformly over the membrane and forming one or a few large clusters, termed eisosome remnants (Walther et al., 2006; Grossmann et al., 2007; Fröhlich et al., 2009). In addition, pil1Δ cells have altered cellular signaling (Zhang et al., 2004) as well as endocytic rates of some, but not all, cargoes (Walther et al., 2006; Grossmann et al., 2008; Brach et al., 2011). Pil1 is not only required for normal plasma membrane distribution of proteins but also of lipids; in its absence, sterols distribute more evenly in the plasma membrane and accumulate at eisosome remnants (Grossmann et al., 2007). Thus, Pil1 provides an example of proteins that organize the plasma membrane in a highly tractable biological model system. In contrast, deletion of Lsp1 leads to only mild defects, but the molecular basis of the differences between these highly homologous proteins is unclear.
It is yet unknown how eisosomes are built, how they are targeted to the cell cortex, and how they organize the plasma membrane. To address these questions, we investigated the biochemical mechanisms of eisosome formation by Pil1 and Lsp1. Our study revealed a previously not recognized self-assembling scaffold that binds to and organizes the yeast plasma membrane.
Results
Recombinant Pil1 and Lsp1 assemble in vitro
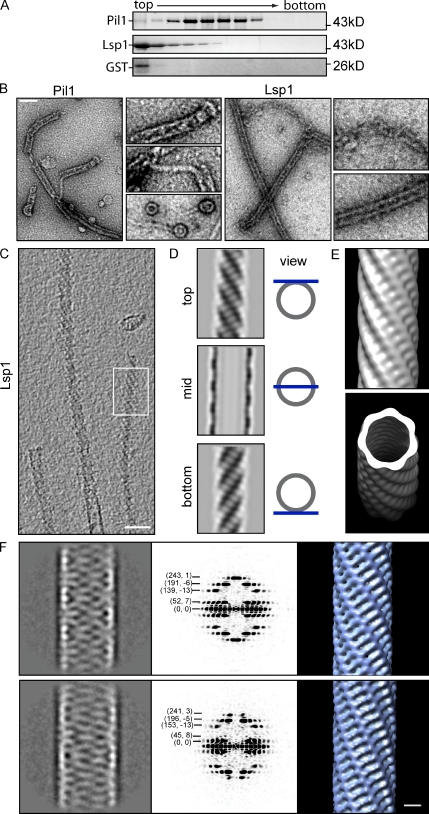
Eisosomes are very large protein complexes containing primarily Pil1 and Lsp1. To test whether complex formation is mediated by autonomous assembly of Pil1 and Lsp1, we assayed the properties of the purified recombinant proteins by velocity sedimentation in a sucrose gradient. Both Pil1 and Lsp1 migrated into the gradient, albeit at different speeds. In contrast, soluble control proteins, such as GST, remained at the top of the gradient (Fig. 1 A). As Pil1 and Lsp1 themselves are relatively small (∼38 kD), this result indicates that they assemble into large complexes.
Figure 1.
Pil1 and Lsp1 form filaments in vitro. (A) Pil1 and Lsp1 aggregate in vitro. SDS-PAGE of factions of a sedimentation velocity gradient analyzing recombinant Pil1 and Lsp1. Protein marker sizes are indicated on the right. (B) Recombinant Pil1 and Lsp1 form filaments visualized by negative staining and EM. Pil1 assembles into ringlike structures as well as thin and thick filaments. Lsp1 mostly forms thick filaments. Bar, 100 nm. (C) Cryo-EM and tomographic reconstructions of Lsp1 filaments have a distinct striation pattern. Bar, 50 nm. (D) Averaged tomographic top, mid, and bottom sections of a thick Lsp1 filament. (E) Surface rendering of the Lsp1 filament reconstruction. (F) Classification of Lsp1 segments reveal classes differing in diameter (left panels; the narrow class is shown on the top, whereas the wider class is shown on the bottom). Power spectra of both major classes are characteristic for filaments of helical symmetry (middle panels) and reveal differences in geometry, also visible in the resulting 3D maps (right panels). Bar, 10 nm.
To exclude the possibility that Pil1 and Lsp1 aggregate unspecifically, we investigated the structure of recombinant Pil1 and Lsp1 complexes by negative staining and EM. This analysis revealed intricate filamentous structures for both proteins, which are highly similar in sequence, but also some structural differences between their assemblies; Pil1 was present as a mixture of ringlike structures and two types of filaments, thin and thick ones (Fig. 1 B). Thick filaments appear to form by curling up thin filaments. In contrast, Lsp1 assemblies appeared structurally similar to thick Pil1 filaments but often ended into disordered chains. In Lsp1 samples, we rarely observed ringlike structures and never found thin filaments (Fig. 1 B, right).
Pil1 and Lsp1 assemble already during purification of the proteins, and the resulting filaments likely form when the concentration of subunits reaches a critical threshold. Once formed, the assemblies could remain in dynamic equilibrium with free subunits, or, alternatively, they could represent stable complexes. To distinguish between these possibilities, we monitored exchange of recombinant Pil1 subunits between preassembled complexes. Complexes of Pil1 labeled with different fluorescent dyes appeared as foci in light microscopy images. A dynamic equilibrium between free subunits and the assemblies predicts that subunits exchange between red- or green-labeled preassembled complexes over time. However, even if we monitored the complexes’ composition after >3 h, no exchange of labeled subunits occurred (Fig. S1 A). These data indicate that Pil1 assemblies are extremely stable in vitro, either as a result of high binding energy of the subunits to each other or as a result of a kinetic barrier for disassembly once a complex is formed.
We conclude that Pil1 and Lsp1 assemble into stable filamentous structures in vitro. Therefore, the formation of eisosomes is likely a consequence of intrinsic properties of Pil1 and Lsp1.
Lsp1 filaments have helical symmetry
To understand how Pil1 and Lsp1 filaments are built, we studied their structure in their native state by cryo-EM and 3D image reconstruction. We generated a structural model of Lsp1 filaments, as they are longer and much more ordered than analogous Pil1 structures, facilitating their analysis. Tomographic reconstructions of Lsp1 filaments display distinct striations (Fig. 1, C and D), and averaging of overlapping 3D segments along the axis of the filament shows grooves and ridges on the surface of a left-handed helix, corresponding to these surface features (Fig.1 E).
In a complementary approach, we used Fourier–Bessel analysis to reconstruct Lsp1 helices (DeRosier and Moore, 1970). Classification of segments revealed variation in the Lsp1 filaments, with at least two prominent classes, one being narrower and one being wider (Fig. 1 F and Table S3). Power spectra calculated for these two class averages show two very different helical symmetries, as manifested by differences in the position and Bessel order of the layer lines. For example, for the narrower filament (Fig. 1 F), the first layer line (52) has a Bessel order of 7, indicating the presence of a seven-start helix, whereas for the wider filament (Fig. 1 F), the first layer line (45) has a Bessel order of 8, indicating the presence of an eight-start helix. 3D reconstructions calculated from class averages revealed a similar helical ordering of subunits (Fig. 1 F) to that seen in the averaged structure from tomographic data (Fig. 1 E). As these two computational approaches were fully independent from each other, they cross-validate the derived structural models.
Pil1 and Lsp1 directly bind PI(4,5)P2-containing membranes
Pil1-GFP and Lsp1-GFP localization to the plasma membrane (Walther et al., 2006; Strádalová et al., 2009) could be mediated by direct binding to membranes, as observed for other BAR domain–containing proteins. To test this hypothesis, we incubated recombinant Pil1 or Lsp1 with liposomes mimicking the lipid composition of the plasma membrane and analyzed the resulting membrane structures by negative staining and EM. Fig. 2 A shows that both Pil1 and Lsp1 bind liposomes consisting of phosphatidylcholine (PC; 70 mol %)/phosphatidylserine (PS; 15 mol %) and phosphatidylethanolamine (PE; 15 mol %) only in the presence of 1.5% phosphatidylinositol (PI)-4,5, bisphosphate (PI(4,5)P2) and deform them into long tubules.
Figure 2.
Pil1 and Lsp1 directly bind PI(4,5)P2-containing membranes. (A) Pil1 and Lsp1 bind and tubulate PI(4,5)P2-containing liposomes. Negative staining and EM of recombinant Pil1 or Lsp1 incubated with liposomes containing PC/PS/PE (70%/15%/15%) or, in addition, 1.5% PI(4,5)P2. (B) Negative-stained samples of recombinant Pil1 or Lsp1 incubated with PC liposomes containing 1.5% PI or PI(4,5)P2. Insets show magnifications of Pil1 bound to liposomes. (A and B) Protein-covered membrane tubules are marked with yellow arrowheads. Bars, 100 nm. (C, top) Spin-down experiments of Lsp1 incubated with or without PC liposomes containing 1% PI or PI(4,5)P2 as indicated. Proteins bound to liposomes appear in the pellet (P). Lsp1 shows higher affinity to PI(4,5)P2 than to PI. S, supernatant. (bottom) Quantification of protein amounts in the pellet fractions from spin-down experiments represented in a box plot, consisting of the median (middle of the box), the upper and lower quartile (edges of the box), and whiskers at a 1.5–interquartile range distance from the upper and lower quartile. (D) Measurement of fluorescence from NBD-labeled pil1S45C (orange emission spectrum) alone as well as in the presence of PC/PS/PI liposomes (purple emission spectrum) or PC/PS/PI(4,5)P2 (green emission spectrum); the buffer control is shown in gray. n = 6.
To further test the lipid requirements for Pil1 and Lsp1 membrane binding, we incubated the proteins with liposomes made from PC, PC/PI, or PC/PI(4,5)P2. Even though we observed more abundant tubulation of PI(4,5)P2 containing liposomes than of liposomes containing 1.5% PI (Fig. 2 B), binding of Pil1 and Lsp1 is not strictly specific for PI(4,5)P2.
To independently confirm Pil1 and Lsp1 membrane binding, we used a biochemical copurification assay. In floatation assays, protein complexes never migrated to the top of density gradients under conditions in which we observed membrane binding by EM. Therefore, we used cosedimentation of protein and liposomes, as they copelleted with membranes under such conditions. The interpretation of these experiments was further complicated by the self-assembly of Pil1 and Lsp1. Pil1 assemblies pelleted alone in the absence of liposomes, preventing their further analysis. In contrast, Lsp1 had a larger soluble pool (>50% of the protein under the conditions we used), and inclusion of PI(4,5)P2-containing liposomes shifted all of Lsp1 to the liposome-containing bottom fraction, indicating membrane binding (86 ± 10%; Fig. 2 C). In agreement with our observations by EM, membrane binding of Lsp1 was not completely PI(4,5)P2 specific, but some binding also occurred in the presence of PI (Fig. 2 C) or PI(3P)-containing liposomes (not depicted).
As Pil1 and Lsp1 are highly homologous, we predict similar membrane-binding behavior for both proteins. As we could not analyze Pil1 by liposome cosedimentation, we used a different assay to test this hypothesis. To detect Pil1’s membrane association, we coupled an environmentally sensitive NBD (4-nitrobenzo-2-oxa-1,3-diazole) fluorophore to a version of Pil1 containing a single cysteine residue at a position likely facing the membrane (S45C; based on the crystal structure; see Fig. 6 B). The resulting pil1S45C mutant localized normally, as determined by fluorescence microscopy in yeast cells expressing its GFP-tagged derivative (unpublished data). A hydrophobic environment (e.g., caused by membrane binding) increases the fluorescence emission of NBD compared with its fluorescence in aqueous solution. Consistent with membrane binding, NBD-pil1S45C fluorescence increased 3.8-fold when incubated with liposomes containing PI(4,5)P2 (Fig. 2 D, green emission spectrum) compared with the signal of the protein alone (Fig. 2 D, yellow emission spectrum). In agreement with our observations by EM, binding was not completely specific for PI(4,5)P2, as we also observed a mild increase of NBD-pil1S45C fluorescence upon addition of PI-containing membranes (Fig. 2 D, purple emission spectrum). However, the increase in NBD-pil1S45C signal intensity induced by PI-containing liposomes was reduced in comparison with PI(4,5)P2-containing liposomes (2.1-fold vs. 3.8-fold, respectively; Fig. 2 D). From these data, we conclude that Pil1 and Lsp1 directly bind membranes, preferably those containing PI(4,5)P2.
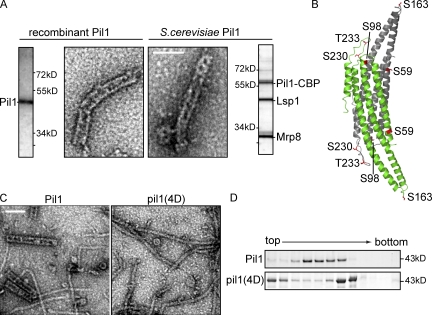
Figure 6.
Purified eisosome proteins from yeast structurally resemble recombinant Pil1 or Lsp1 protein assemblies. (A) Tandem affinity chromatography of tagged Pil1 enriches mainly Pil1, Lsp1, and Mrp8. Negative staining and EM reveal highly similar structures for purified eisosomes (right) as formed by recombinant Pil1 (left). Side panels show Coomassie blue–stained SDS-PAGE gels of the preparations used. (B) X-ray structure of dimeric Lsp1 BAR domain. Monomers are shown as a ribbon representation in green and gray. Residues that can be phosphorylated and that are represented in the structure of eisosome protein BAR domains are highlighted in red. (C) Purification of recombinant pil1(4D) and visualization by negative staining and EM show that pil1(4D) does not form thick helices but only long, thin filaments. Bars, 100 nm. (D) Precipitated fractions of sedimentation velocity gradients of recombinant Pil1 and pil1(4D) were analyzed by SDS-PAGE. They show different mobility of phosphorylation mutants compared with wild-type Pil1.
PI(4,5)P2 is required for the formation and organization of eisosomes in vivo
As Pil1 and Lsp1 preferentially bind PI(4,5)P2 in vitro, a reduction of the plasma membrane–specific PI(4,5)P2 pool may lead to impaired eisosome localization in vivo. To deplete PI(4,5)P2 from the plasma membrane and to test consequences on eisosomes, we used a yeast mutant containing a temperature-sensitive allele of MSS4 (mss4ts), encoding the kinase that converts PI-4-phosphate (PI4P) to PI(4,5)P2. Inactivation of Mss4 after a temperature shift results in the depletion of PI(4,5)P2 from the plasma membrane (Stefan et al., 2002). Under such conditions of reduced PI(4,5)P2 levels, no Pil1-GFP signal remained in an eisosome pattern in mss4ts cells, but, instead, all of the protein clustered into enlarged structures at the membrane or became cytosolic (Fig. 3 A). Time-lapse imaging of eisosomes, marked by Pil1-GFP, suggests that eisosomes progressively detach from the plasma membrane and aggregate under these conditions (Videos 1–3). This was specific for cells containing the mss4ts allele, as we did not observe a similar phenotype in control cells expressing Pil1-GFP. Depletion of other phosphoinositides, such as PI4P at the Golgi apparatus in temperature-sensitive pik1 mutants and PI3P in vps34Δ cells, did not lead to a comparable phenotype (Fig. S1, D and E). We also found that stt4 mutants impaired in generating PI4P at the plasma membrane and thus indirectly impaired in PI(4,5)P2 synthesis displayed abnormal Pil1 organization, albeit much weaker than mss4 cells (Fig. S1 D). Moreover, PI(4,5)P2 is important for normal plasma membrane domain organization, as PI(4,5)P2 depletion results in loss of the punctate Sur7 localization, an MCC domain marker, which was instead more evenly distributed over the plasma membrane of mss4ts cells after the temperature shift (Fig. 3 B).
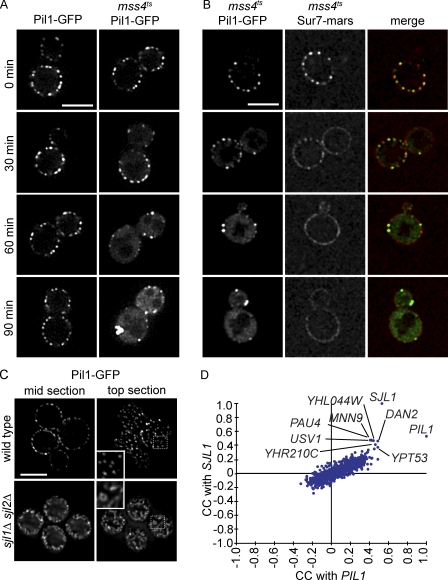
Figure 3.
PI(4,5)P2 is necessary for normal eisosomes in vivo, and PIL1 has a highly similar genetic profile to SJL1. (A) Fluorescence microscopy of Pil1-GFP in a yeast mutant strain containing a temperature-sensitive allele of MSS4 (mss4ts). Pil1-GFP loses its normal eisosome pattern but instead clusters to enlarged structures at the membrane after a 90-min (right column) temperature shift from 24 to 37°C. The control strain does not show this phenotype (left column). (B) Fluorescence microscopy of Sur7-mars and Pil1-GFP in mss4ts cells. After 30 min of temperature shift, Sur7-mars loses its localization to the MCC. After 60 min, it is evenly distributed in the plasma membrane. (C) Deletion of SJL1 and SJL2 results in increased Pil1-GFP assembly at the plasma membrane. Insets show magnified regions of cells in the boxed areas. Bars, 5 µm. (D) Comparison of correlation scores from an E-MAP focusing on lipid metabolism. SJL1, encoding the PI(4,5)P2 phosphatase, has the most similar genetic signature to PIL1, indicating similar gene function. CC, correlation of correlations.
In addition, increase of PI(4,5)P2 in the plasma membrane had the opposite effect as its decrease; deletion of two PI(4,5)P2 phosphatases encoded by the yeast synaptojanin-like proteins (SJL1 and SJL2) leads to increased PI(4,5)P2 levels (Stefan et al., 2002) and Pil1-GFP assembly into much larger structures that appear to protrude from the plasma membrane into the cytoplasm (Fig. 3 C). Thus, Pil1 interaction with PI(4,5)P2 is crucial for normal eisosome formation and plasma membrane domain organization.
To further test the physiological significance of eisosome protein interaction with PI(4,5)P2, we tested genetic interactions between PIL1 and SJL1 in epistatic miniarray profiles (E-MAPs; (Collins et al., 2006)). E-MAPs contain quantitative measurements of genetic interactions within a selected set of mutants. If two mutations have similar physiological consequences, they will share many suppressing or aggravating genetic interactions with other mutations, resulting in highly similar genetic interaction profiles. Strikingly, the genetic profiles of PIL1 and SJL1 are most similar to each other in two independently generated E-MAP datasets. For example, within an E-MAP containing 787 genes mainly functioning in lipid metabolism, PIL1 and SJL1 interactions with all other genes were more similar to each other than any other gene in the set (Fig. 3 D, correlation of genetic profiles of PIL1 and SJL1 = 0.8281 and 0.332 in an E-MAP focusing on plasma membrane function; Fig. S1 F; Aguilar et al., 2010), showing that deletions of PIL1 or SJL1 have very similar phenotypic consequences. These findings further argue that PI(4,5)P2 interaction of eisosome components is physiologically important.
Structure of membrane-bound Pil1 and Lsp1
Having established that eisosome proteins Pil1 and Lsp1 directly interact with PI(4,5)P2-containing membranes, we determined the structure of the resulting assemblies. To this end, we used a combination of cryo-EM, tomography, and 3D image reconstruction. Computational slices through 3D tomograms revealed that both Pil1 and Lsp1 decorate liposomal membranes exclusively from the outside, thereby constricting them to long tubes with a similar diameter as the helices formed by the protein alone (Fig. 4 A). Interestingly, the diameter of Pil1 or Lsp1-decorated tubules was always smaller than the diameter of the membrane tubule emerging from the end of the protein filaments, suggesting that the proteins bend membranes, thereby constricting them to a smaller diameter (Fig. 4 A). In addition, Pil1 especially formed much longer filaments on liposomes compared with the rather short and unordered filaments in the absence of membranes, indicating a possible role of protein–membrane interaction in assembly.
Figure 4.
Structure of membrane-bound Pil1 and Lsp1. (A) Structure of Lsp1 and Pil1 bound to PI(4,5)P2-containing liposomes. Tomographic midsections show that both proteins decorate liposomes and constrict them to a similar diameter. Bar, 50 nm. (B) Classification of Lsp1 filament segments in addition to membrane-bound Lsp1 and Pil1 reveals different diameters. N represents the number of segments used for the classification. (C) Helical reconstruction of prominent groups of Lsp1 filaments as well as membrane-bound Lsp1 and Pil1. Membrane-bound Lsp1 shows distinct density oriented toward the lipid bilayer. Bar, 10 nm.
Further classification analysis of filament segments from cryo-EM revealed variation in the diameter of the tubules, which was most pronounced for Pil1 bound to membranes (∼30–37 nm). Also, tubules formed by Lsp1 bound to membranes showed such diameter variation (∼32–36 nm), and the diameter of the protein alone was always smaller than in its membrane-bound state (∼29–32 nm; Fig. 4 B).
To gain more detailed information on the subunit arrangement in the thick filaments, we used a combination of two helical reconstruction approaches. 3D reconstructions from Fourier–Bessel analysis provided different low-resolution structures for supervised classification of all segments into respective symmetry classes and for refinement of the 3D reconstruction using iterative helical real-space reconstruction (IHRSR). This method is especially suited for flexible filaments with helical symmetry (Egelman, 2007). All structures showed a repeating unit with similar morphology and local twofold symmetry, consistent with their assembly from BAR domain dimers. However, the helical arrangement of subunits varied in the different structures (Fig. 4 C and Table S3). Notably, the repeating structural units in Lsp1 and Pil1 filaments closely resemble each other, consistent with the high primary sequence similarity between the two proteins (Fig. 4 C). In each of the filaments, subunits form a fenestrated protein coat around the liposome membrane, potentially allowing access of small molecules and proteins to the membrane. In the Lsp1 structure, additional density was visible in the model derived for membrane-bound protein compared with the one of the protein alone (Lsp1 + liposomes #1; Fig. S1 B). We believe that this density represents a part of Lsp1 that gains higher order upon membrane binding and thus becomes detectable.
Variation in the filament diameter and helical symmetry argues for high flexibility in these assemblies. Thus, we addressed whether the helical symmetry can change within a single filament. As only two main classes, narrow (30 nm) and wide (32 nm), were detected for Lsp1 filaments in the absence of the membrane (Fig. 4 C, Lsp1 #1 and #2), we used these data in our analysis for simplicity. The distribution of the two classes along the filament was not random. On the contrary, the probability for a narrow segment succeeding a narrow segment was 0.97. Conversely, the probability for a wide segment succeeding a wide segment was 0.89. This finding not only reflects filament architecture but also indicates that our analysis is sensitive enough to detect subtle differences. Continuous stretches of narrow or wide segments were often present in a single filament (Fig. S1 B). Extrapolation of these results for Lsp1 filaments in the absence of the membrane to Lsp1 and Pil1 filaments in the presence of the membrane indicates that the latter may have even more complicated mixtures of different helical symmetries.
Although variable, the diameter of the underlying membrane tubule correlated with the diameter of the outer protein coat. The consistent distance between visible membrane and proteins observed in different classes and samples argues for a specific protein–membrane interaction that imposes a specific curvature on the membrane tubule.
Pil1 and Lsp1 membrane binding requires an N-terminal protein segment and a patch of positively charged amino acids in their concave BAR domain surface
Recently, we discovered by crystallographic analysis of Lsp1 that eisosome proteins contain BAR domains (Ziółkowska et al., 2011). To investigate how Lsp1 BAR domains assemble into helical filaments and interact with the membrane, we computationally fitted models of Lsp1 dimers into our cryo-EM maps. Specifically, we used six different EM-derived density maps of assembled Lsp1 and Pil1 to fit three variations of the structural model of Lsp1 amino acids 51–264 varying in the conformation of the BAR domain tips (based on the comparison of AA, BB, and CC dimers, constructed by superimposition of A, B, and C chains present in the asymmetric unit on the crystallographic dimer AB; Ziółkowska et al., 2011). Fitting always placed the dimers at the same relative position in the cryo-EM map and always resulted in the same orientation of BAR domains with their concave side facing the membrane surface (Fig. 5 [A and B] and not depicted). For each map, the BB dimer gave slightly higher scores than AA and CC, most likely because its structure lacks a loop sequence and thus has a slightly smaller volume. To generate atomic models of the filaments, we imposed helical symmetry parameters of the cryo-EM density maps on the best-fitting dimers. This revealed clashes between the tips of the BAR domain dimer and its symmetry-related neighbors, in addition to densities, which were not occupied by the fitted chains (Fig. 5 B). We consider two explanations for these discrepancies. First, superimposition of AA, BB, and CC dimers suggests that the tips are the most flexible part of the Lsp1 BAR domain (Ziółkowska et al., 2011). Therefore, it is possible that the dimers change conformation in the assembled helices compared with the crystal structure and move their tips into the density that is located right next to the clashing region and which is not filled in our fitting (Fig. 5 B, green arrows). Second, some of the unaccounted density could be occupied by the Lsp1 N or C terminus, which are absent in the x-ray structure (Fig. 5 B, red arrows).
Figure 5.
Pil1 and Lsp1 membrane binding requires an N-terminal segment and a patch of positively charged amino acids on their BAR domain surface. (A) Computational rigid body fitting of the Lsp1 BAR domain dimer x-ray structure to cryo-EM density maps of Lsp1 tubules and Lsp1 bound to PC liposomes containing 1.5% PI(4,5)P2. The top view of tubules (top) shows Lsp1 BAR domain monomer chains colored blue to red from N terminus to C terminus. The side view of tubules (bottom) shows the Lsp1 helix colored blue to red from the bottom to the top. (B) A close-up of the side view and intersection of the tubules. Lsp1 BAR domain monomer chains are colored blue to red from the N terminus to the C terminus. Density that might be occupied by the flexible tips of the x-ray structure, adopting a slightly different conformation in the tubules than in the crystal, is indicated by green arrows. The density that could be filled by the C termini, which are missing in the x-ray structure, is indicated by red arrows. (C) Negative staining and EM of recombinant Pil1 or Lsp1 proteins incubated with PC liposomes containing either 1.5 or 3.5% PI(4,5)P2. Mutants with an N-terminal truncation (lsp1ΔN) or changes in the positively charged amino acid patch of the concave BAR domain surface of Pil1 or Lsp1 (lsp1KRE) retain the ability to bind and tubulate PI(4,5)P2-containing liposomes. Combination of both types of mutation (lsp1ΔNKRE) abolishes membrane binding. Protein-covered membrane tubules are marked with yellow arrowheads. Bar, 100 nm. (D) Spin-down experiments of Lsp1, lsp1KRE, lsp1ΔN, or lsp1ΔNKRE incubated with or without PC liposomes containing 0.1, 1, 1.5, or 3.5% PI(4,5)P2 as indicated. Panels showing different experimental conditions are separated by dotted lines for better visibility. P, pellet; S, supernatant. (E) Quantification of protein amounts in pellet fractions of experiments analogous to D. n = 3. Error bars represent SDs of three independent experiments.
These structural data show that Lsp1—and by extension, the almost identical Pil1 (Ziółkowska et al., 2011)—assembles by interactions at the end of the banana-shaped dimers with the concave surface facing the membrane, similar to other BAR domain proteins (Frost et al., 2008).
To test our model of Lsp1 assembly on membranes and to determine which regions of the protein are required for membrane interaction, we tested various mutants of Pil1 and Lsp1. Particularly, a patch of positively charged amino acids on the concave surface of the Lsp1 BAR domain is evolutionary conserved and required for normal localization of the proteins in vivo (Ziółkowska et al., 2011). However, mutation of two positively charged amino acids in this patch to glutamic acid (pil1KRE and lsp1KRE, carrying K130E and R133E mutations) did not completely abolish membrane binding, as determined by negative staining and EM with liposomes (Fig. 5 C). Similar results were obtained in spin-down experiments, which revealed reduced, but not abolished, membrane binding of lsp1KRE (Fig. 5, D and E). Intriguingly, in our structural model (Fig. 4 C), we observed one small protrusion per dimer of full-length Lsp1 proteins toward the membrane surface that were not filled by the crystal structure representing the BAR domain alone (missing the N terminus). Moreover, the most N-terminal segment of the crystal structure is oriented toward the membrane. Therefore, we hypothesized that, in analogy to membrane binding of other BAR domains, protein segments N-terminal of the BAR domain of two proteins together mediate membrane interaction. To test this model, we deleted the N-terminal 35 amino acids (pil1ΔN and lsp1ΔN). Intriguingly, when we analyzed these mutants in the absence of liposomes, we found only thin, but no thick, helices (Fig. S1 C). In the presence of liposomes, we observed thick helices and no defect in membrane binding by EM analysis or in spin-down experiments for pil1ΔN or lsp1ΔN (Fig. 5, C–E), indicating that the BAR domain alone is sufficient for membrane binding. Importantly, when we mutated the positive patch in the context of a mutant protein that lacks the N terminus (pil1/lsp1ΔNKRE), membrane binding of Pil1 or Lsp1 was completely abolished (Fig. 5, C–E). These data show that both the N terminus and a patch of positively charged residues on the concave surface of the BAR domains of Pil1 and Lsp1 are required for their interaction with membranes.
Purified eisosomes from yeast resemble recombinant Pil1 and Lsp1 helices
Next, we analyzed whether the structures of Pil1 and Lsp1 determined using in vitro methods reflect the arrangement of the proteins on eisosome membrane furrows observed in vivo (Strádalová et al., 2009). To this end, we first purified Pil1 fused to a tandem affinity purification tag by affinity chromatography together with associated eisosomes proteins from yeast cells. These experiments yielded a complex consisting of Pil1 (with a CaM-binding peptide remaining on the protein after elution by cleavage of the tag), Lsp1, and Mrp8 (Fig. 6 A; Walther et al., 2006; Wang et al., 2009). Mrp8 is a cytoplasmic protein, which associates with Pil1 and Lsp1 but does not play a structural role at eisosomes (Fig. S2). To test whether purified eisosome proteins from yeast assemble into structures similar to those formed by the recombinant proteins, we performed negative staining and EM of these preparations and found structures that resembled each other in overall shape and dimensions (Fig. 6 A). This indicates that the structural information derived from the recombinant proteins likely reflects the organization of eisosomes in cells.
Phosphomimicking mutations of Pil1 block formation of thick helices in vitro
To further test whether the arrangement of Pil1 and Lsp1 is physiologically relevant, we tested the prediction that their assembly should change in vitro under conditions that change eisosomes in vivo. Pil1 is phosphorylated on several residues in vivo, and alterations in its phosphorylation state affect eisosome assembly in yeast (Walther et al., 2007; Luo et al., 2008). In a mutated form of Pil1, denoted pil1(4D), four normally phosphorylated residues were changed to phosphomimicking aspartates (S45D, S59D, S230D, and T233D; Fig. 6 B). Pil1(4D) does not mimic all known phosphorylations of Pil1, but its four mutations are necessary and sufficient for mediating the Pil1 phosphorylation effect that results in just one or a few large clusters and a much stronger cytoplasmic signal compared with wild-type Pil1 (Walther et al., 2007). To investigate whether this effect is mediated by altering Pil1’s self-assembly, we first analyzed recombinant pil1(4D) by velocity gradient centrifugation and found that sedimentation of the mutant protein was significantly altered compared to the wild-type form (Fig. 6 D). Specifically, a large pool of the mutated protein remained at the top of the gradient, and, in addition, a faster-migrating species increased in abundance.
To visualize the effect of phosphomimicking mutations on Pil1 self-assembly, we analyzed the structure of pil1(4D) by negative staining and EM. These experiments showed that pil1(4D) is still able to assemble into thin filaments, which were abundantly present in the sample. However, we never observed thick helices that are common in wild-type Pil1 samples (Fig. 6 C). This indicates that the phosphomimicking mutations and, by extrapolation, phosphorylation of Pil1 lead to impairment of helical assembly. The analogous consequences of Pil1 phosphomimicking mutations observed in vivo and in vitro provide additional support for the interpretation that Pil1 and Lsp1 helices are equivalent to eisosomes.
Eisosomes form short helical lattices at the plasma membrane in yeast cells
To further test whether Pil1 and Lsp1 helices observed in vitro resemble eisosomes in vivo, we investigated their structure in yeast cells. Consistent with the data of Strádalová et al. (2009), freeze-fracture deep-etching EM (DEEM) analysis of the yeast plasma membrane showed abundant furrows, whose presence depended on PIL1 (not depicted; Strádalová et al., 2009). Previously, it was shown by immunogold labeling that these furrows contain Pil1 (Strádalová et al., 2009). Our images of the furrows revealed distinct striations resembling the surface pattern of Pil1 and Lsp1 assemblies formed in vitro (Fig. S3 A). However, as freeze-fracture DEEM images show the interface between the two lipid layers of the plasma membrane (P-face), eisosome proteins are not directly visible in such images, as they are still covered by a lipid monolayer. To overcome this limitation, we bound yeast cells to a grid as a solid support and removed most of the cells by a short burst of ultrasound waves (Fig. S3 D; Hanson et al., 2008). Notably, this technique requires spheroblasting of yeast cells, which we found can lead from mild to severe elongation of eisosomes, as visualized by fluorescent microscopy. Therefore, we developed a mild spheroplasting protocol, with only minor effects on the fluorescent signal of eisosomes (Fig. S3 C). This technique allowed us to image the cytoplasmic face of the plasma membrane that remains bound to the grid by DEEM, which exposed abundant filamentous structures that we suspected to be eisosomes (Fig. 7 A). To test this hypothesis, we labeled Pil1 or Lsp1 either with an affinity-purified polyclonal antibody against full-length Pil1 or against GFP in cells that express tagged versions of Pil1 or Lsp1 (Figs. 7 B and S3 B). Each of the antibodies specifically labeled the filamentous structures, thereby confirming the presence of Pil1 and Lsp1 within them. Importantly, the filaments showed striations characteristic for Pil1 and Lsp1 assemblies in vitro (Fig. 7 A). However, in contrast to the helical structures observed in vitro, eisosomes consist of half-cylinders that cover the cytoplasmic face of furrows, which can be best seen in 3D anaglyphs of DEEM images, representing the cytoplasmic side of the plasma membrane or the P-face (Fig. 7, C and D). In some, but not all, cases, these helical structures resided at the side of large membrane invaginations (Fig. 7, A and B, arrowheads). Collectively, these data show that eisosomes resemble the structure of Pil1 and Lsp1 assemblies determined in vitro.
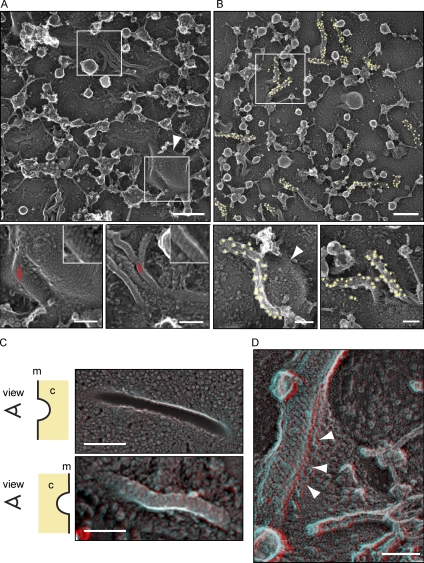
Figure 7.
Eisosomes in situ structurally resemble Pil1 and Lsp1 assemblies. (A) Representative image of the yeast plasma membrane from the cytosolic side (top). Bar, 300 nm. (insets) Magnifications of distinct areas (marked by white boxes) of the membrane show striated areas (red parallel lines) that resemble the pattern of recombinant Pil1 and Lsp1 structures. Bars, 100 nm. (B) Immunolabeling of plasma membranes of cells expressing Pil1-GFP using anti-GFP antibodies. Yellow circles highlight 18-nm gold particles for better visibility. Bars, 100 nm. (A and B) The structures are visible on the flat membrane as well as on the side of large invaginations (arrowheads). (C) DEEM images showing views on the plasma membrane from different perspectives. (top) View from the outside of a cell onto the inner leaflet of the plasma membrane. (bottom) View from the cytoplasm (marked as c) onto the plasma membrane (marked as m; red/cyan 3D glasses are recommended for 3D view, as well as for D). Bars, 300 nm. (D) View from the cytoplasm onto an eisosome at the plasma membrane. Arrowheads indicate how the plasma membrane protrudes underneath the eisosome protein coat to form a groove instead of a closed tube. Bar, 100 nm.
Discussion
Eisosome proteins play an important structural role in organizing the yeast plasma membrane. As expected from their architectural function, eisosome proteins localize very stably and are extremely high in abundance. For comparison, copy numbers of main eisosome components per cell (115,000 for Pil1 and 104,000 for Lsp1) are much larger than those of tubulin (5,590) or actin (60,000; Norbeck and Blomberg, 1997; Ghaemmaghami et al., 2003). As a result of these properties, Pil1 and Lsp1 could be classified as part of a membrane cytoskeleton. Consistent with this idea, we show that Pil1 and Lsp1 self-assemble into a protein scaffold that binds and deforms membranes, providing a mechanism for how these proteins organize the plasma membrane into domains.
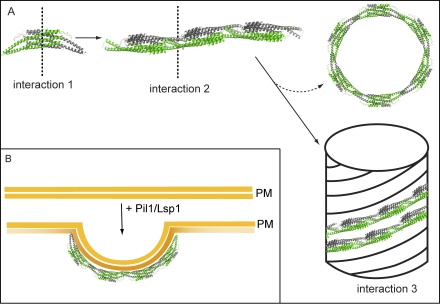
Similar to other BAR domain–containing proteins (such as F-BARs; Frost et al., 2008), Pil1 and Lsp1 form higher-order complexes on membranes. In contrast to other BAR domain proteins, however, the assemblies formed by eisosome proteins are extremely stable in the absence of membranes. Even though we cannot pinpoint exact contacts as a result of limited resolution, our current model of eisosome architecture posits three distinct interactions: one to form the BAR dimer (interaction 1), a second end-to-end contact of BAR domain dimers to form thin filaments (interaction 2), and a third lateral interaction to form helices (interaction 3; Fig. 8). The overall similarity and subtle differences of the assemblies formed by Pil1 and Lsp1 suggest that the strength of the different interactions may differ between the two proteins. For example, Lsp1 interaction 2 may have a lower affinity compared with Pil1, which would result in a requirement for combined binding energy from end-to-end interactions 2 and lateral interactions 3 to stabilize thick Lsp1 helices. Smaller assemblies, such as thin filaments, may be unstable and fall apart, explaining the absence of thin Lsp1 filaments and the increased pool of nonassembled Lsp1 in cells and biochemical assays. Immunolocalization of Pil1 and Lsp1 in EMs performed in this study (and Strádalová et al. [2009]) shows that both proteins are present in eisosomes covering membrane furrows, but how both proteins associate to form them and whether their different properties are used to modulate eisosome structure are yet unclear.
Figure 8.
Model for the assembly of eisosomes on the plasma membrane. (A) The assembly of eisosomes can be separated conceptually into three steps: interactions of the proteins to form dimers (interaction 1), association of dimers to form thin filaments (interaction 2), and assembly into helices (interaction 3). Rings observed for Pil1 are interpreted in this model as side products of the filament-to-helix assembly. (B) On the plasma membrane (PM), main eisosome components assemble into a scaffold similar to a half helix (see Discussion for details).
Interaction 3 is likely modulated by phosphorylation, as indicated by its sensitivity to phosphomimicking mutations of Pil1, which leads to formation of thin helices. Similarly, pil1ΔN formed only thin filaments, further suggesting that the N-terminal segment containing two of phosphorylation sites is required for interaction 3. These data may explain eisosome disassembly after overexpression of Pkh kinases, addition of myriocin, or other treatments that increase Pil1 phosphorylation (Walther et al., 2007; Luo et al., 2008; Fröhlich et al., 2009).
Pil1 and Lsp1 are most likely targeted to the plasma membrane by efficient membrane binding (Fig. 8 B). We predict that, initially, dimers or thin filament pieces interact with membranes and assemble in vitro into a stable helix with a membrane tubule inside or in vivo into a furrowlike lattice (Fig. 8 B). Several lines of evidence suggest that Pil1 and Lsp1 interact with PI(4,5)P2: (a) Pil1 and Lsp1 tubulate liposomes containing low amounts of PI(4,5)P2; (b) fluorescence spectroscopy of NBD-labeled Pil1 yields a strong signal consistent with membrane binding when PI(4,5)P2-containing liposomes are present; (c) in sedimentation assays, Lsp1 interacts more strongly with PI(4,5)P2-containing liposomes than with those containing other types of charged lipids at the same concentration; (d) inactivation of Mss4, leading to PI(4,5)P2 depletion, has a strong effect on eisosome localization in vivo; (e) conversely, deletion of two PI(4,5)P2 phosphatases (SJL1 and SJL2) and increased PI(4,5)P2 levels lead to enlarged Pil1-GFP assemblies; and (f) PIL1 and SJL1 show highly similar genetic interaction profiles in independently generated E-MAP datasets.
How can self-assembly of Pil1 and Lsp1 promote formation or stabilization of curved membranes, such as tubules and furrows? Two mechanisms for BAR domain–induced membrane bending are currently considered (Kozlov et al., 2010): protein scaffolding of the membrane and insertion of a wedge into one leaflet of the lipid bilayer. Both of these mechanisms are used by other BAR domain–containing proteins (Peter et al., 2004). Our reconstructions of Lsp1 with and without bound membranes show very similar structures, suggesting a scaffold mechanism. In addition, a part of the protein at the concave surface of the Pil1 or Lsp1 coat may be inserted as a wedge in one leaflet of the bilayer, for example, represented by the part of Lsp1 observed close to the membrane surface. Consistent with this notion, we found that a membrane-facing N-terminal segment of Pil1 or Lsp1 is required for efficient membrane binding of the proteins in addition to a positive patch of amino acids on the concave surface of the BAR domain. As a consequence of the insertion of a membrane wedge, the order of the outer membrane leaflet could be disordered, leading to absence of resolved density in this region and thus potentially explaining the gap apparent in our reconstructions between the lipid layer to the protein scaffold.
Many of these considerations are based on similarity between the models of Pil1 and Lsp1 assemblies in vitro and the structure of eisosomes, forming membrane furrows in yeast (Strádalová et al., 2009). This interpretation is supported by (a) a very similar structure for recombinant Pil1 and Lsp1 assemblies as for purified eisosomes isolated from yeast cells, (b) alterations of the in vitro structure caused by phosphomimicking mutations in Pil1, consistent with the phenotype of these mutations in yeast, and (c) the striated pattern of eisosomes on plasma membrane furrows or the cytoplasmic side of the plasma membrane, which resembles the pattern of thick helices formed by the recombinant proteins. Despite the overall close resemblance of the structures, there are at least two important differences. First, eisosomes contain both Pil1 and Lsp1 proteins. Thus, in vivo, the building blocks of the lattice could be Pil1 and Lsp1 heterodimers or a mixture of both types of homodimers, rather than a single species of homodimers present in vitro, and the different properties of the two proteins could be used to modulate the assembly. Second, whereas the in vitro filaments are closed cylinders coating a membrane tubule, eisosomes in vivo coat a membrane furrow, which likely resembles a half-cylinder. Attachment of the membrane to the cell wall and the large turgor pressure could prevent the closure of the lattice to a helix similar to the ones seen in vitro. Alternatively, a transition phase of specific lipid or protein composition at the eisosome boundary could prevent the closure of the tubules. It remains possible that the furrows are closed to a tube or otherwise remodeled as a result of the rearrangement of the proteins under some conditions. Such remodeling may be supported by flexibility of Lsp1 BAR domain tips and arrangement of subunits, reflected in tube diameter variability, observed in vitro. Interestingly, during uptake of the membrane dye FM4-64, some but not all eisosomes are labeled by bright dye-containing foci, indicating that the plasma membrane has a different structure at those sites.
From our work, several intriguing similarities between eisosomes and endophilin/amphyphysin BAR domain proteins emerge. Both protein families consist of BAR domains, can assemble into a scaffold on membranes, are connected to PI(4,5)P2-rich membranes, and function with synaptojanin proteins (Itoh et al., 2005). Additionally, both sets of proteins were linked to endocytosis, but their deletions have mild defects on protein uptake in most systems (Schuske et al., 2003; Verstreken et al., 2003; Walther et al., 2006; Grossmann et al., 2008; Brach et al., 2011). Endophilin recruits synaptojanin to endocytic sites through an SH3 domain (Schuske et al., 2003). Neither Pil1 nor Lsp1 contains such a domain. However, it was recently reported that the membrane-bending activity of endophilin particularly is important for many functions of the protein in Caenorhabditis elegans (Bai et al., 2010), and we now find that eisosome proteins also bend membranes. Based on these considerations and the similarity of interaction profiles between pil1Δ and sjl1Δ, it is possible that both genes participate in the same process, e.g., in PI(4,5)P2 turnover. Interestingly, membrane curvature, for example, caused by interaction with endophilin, aids synaptojanin activity (Chang-Ileto et al., 2011).
In summary, formation of an eisosome protein scaffold can mechanistically explain how the yeast plasma membrane is organized in domains of distinct composition, in particular for the MCC. We posit that membrane binding and assembly by Pil1 and Lsp1 will create a specific environment in the overlaying MCC, which is locally curved and may have increased PI(4,5)P2 concentration as a result of the presence of many binding sites for this lipid. This special environment then drives formation of the MCC domain.
Materials and methods
Yeast strains
All yeast strains were derived from the W303 or S288C strain background using PCR-based modification (Janke et al., 2004) and are listed in Table S1. Cells were grown for normal strains at 30°C or at 24°C for temperature-sensitive mutants, with shaking in standard rich medium (yeast peptone dextrose [YPD]) or in synthetic medium (Synthetic complete) supplemented with adenine, uracil, histidine, leucine, and tryptophane.
Protein expression and purification
Recombinant Pil1 and Lsp1 were expressed in Escherichia coli BL21(DE3) cells using pGEX-6P-1 vector (GE Healthcare) and IPTG induction for 3.5 h at 25°C. Pellets were frozen in liquid nitrogen and stored at −80°C. Cell pellets were thawed and broken up in lysis buffer (500 mM KoAc, 2 mM MgAc, 100 mM Hepes, pH 7.4, and 0.2 mM PMSF) by several passages through a microfluidizer (Microfluidics). Proteins were affinity purified with glutathione Sepharose (GE Healthcare), and the GST tag was cleaved off using PreScission protease. Eluted proteins were further purified by anion exchange chromatography on a HiTrap Q FF column (GE Healthcare) using an ÄKTA purifier system (GE Healthcare). Fractions were dialyzed overnight (150 mM KoAc, 2 mM MgAc, 20 mM Hepes, pH 7.4, and 5% glycerol) and concentrated to 30 µM in centrifugal filter devices (Amicon Ultra; Millipore). Aliquots were frozen in liquid nitrogen and stored at −80°C.
Sedimentation velocity gradients
100 µg of recombinant protein was loaded in 150 µl of buffer (150 mM KoAc, 2 mM MgAc, and 20 mM Hepes, pH7.4) on 12 ml of 10–40% sucrose gradient. Gradients were generated using a gradient master (BioComp Systems, Inc.). The gradients were spun in an ultracentrifuge (WX Ultra Series; Thermo Fisher Scientific) equipped with a TH-641 rotor at 40,000 rpm at 4°C for 3.5 h. 1-ml fractions were manually pipetted, and proteins were precipitated using TCA and analyzed by SDS-PAGE and Coomassie blue staining.
Vesicle preparation
Lipids in desired ratios (mole/mole) were mixed and dried in glass vials under a nitrogen stream. Before use, mixtures were desiccated under a vacuum for 2 h and hydrated in buffer. To obtain unilamellar vesicles, lipids were subjected to 5 cycles of freeze-thaw and extruded through a 200-nm pore-size polycarbonate filter (GE Healthcare) using a mini-extruder (Avanti Polar Lipids, Inc.).
Fluorescence microscopy
For fluorescence microscopy, cells were grown in synthetic media to OD600 = 0.5, mounted on coverslips covered with concanavalin A, and imaged either with a laser-based spinning-disk microscope (Andor Technology, TILL Photonics, and Agilent Technologies) using a 100× total internal reflection fluorescence microscopy objective (1.45 NA; Olympus) or using a DeltaVision system (Applied Precision) equipped with a microscope (IX-71; Olympus), a 1.35 NA 100× objective (Olympus), and a CoolSNAP HQ camera (Photometrics).
Spin-down assay
Liposome samples (4 mM) were incubated in the presence or absence of 3.75 µM recombinant protein in 40 µl of buffer (150 mM KoAc, 2 mM MgAc, and 20 mM Hepes, pH 7.4) for 20 min at room temperature. Samples were centrifuged in an ultracentrifuge (Discovery M120 SE; Thermo Fisher Scientific) using an S120-AT3 rotor at 85,000 rpm at 4°C for 30 min. Pellets and supernatants were separated and brought to 42 µl with SDS loading buffer, and both fractions were analyzed by SDS-PAGE and Coomassie blue staining.
EM
For negative staining, 3.5 µl of purified Pil1 or Lsp1 protein or protein liposome suspension was added on glow-discharged continuous carbon-coated Cu grids. After blotting with filter paper, the grid surface was stained with 1% uranyl acetate solution. Images were taken at magnifications of 23,000–31,000× using either a microscope (C120; Philips) equipped with a 1kx1k charge-coupled device camera (Gatan, Inc) and operated at 120 kiloelectron volts or on a microscope (Tecnai F20; FEI Company) equipped with a 4kx4k charge-coupled device camera (FEI Company) and operated at 200 kiloelectron volts. For cryo-EM, a 3-µl aliquot of purified Lsp1 or Pil1 protein or protein-liposome suspension was pipetted on a glow-discharged holey carbon–coated EM grid (C-Flat; Protochips, Incorporated). For cryoelectron tomography, 1 µl of diluted BSA-coated colloidal gold particles (10 nm in diameter) was added. Excess suspension was blotted with a filter paper, and the sample was vitrified by plunging it rapidly into liquid ethane. Cryo-EM was performed at liquid nitrogen temperature using a microscope (Tecnai F20) equipped with a 4kx4k charge-coupled device camera (FEI Company) operated at 200 kiloelectron volts. Single low-dose images (20e−/A2) or tilt series of 61 images from −60 to +60 degrees (80−130e−/A2) were collected with SerialEM (Mastronarde, 2005) at 1–3 µm of underfocus and at a nominal magnification of 50,000, giving a calibrated pixel size of 0.22 nm.
Tomography and subtomogram averaging
Tomographic reconstructions were calculated from the tilt series in IMOD (Mastronarde, 2005) and down-sampled by a factor of three, giving a final pixel size of 0.66 nm. Three filaments, oriented in the direction of the tilt axis, were traced in a tomographic reconstruction of Lsp1. To calculate an averaged structure of Lsp1 filament, 128 overlapping 3D filament segments (100 × 100 × 100 voxels, with a 90% overlap) were extracted. A cylindrical average of all segments was used as a reference in cross-correlation alignment, calculated using a custom Bsoft (Heymann and Belnap, 2007) program, Jsubtomo (available upon request; Huiskonen et al., 2010). The average of all aligned segments acted as a reference for a subsequent 20 rounds of alignment and averaging. The angle defining the orientation of the filament segment around the filament long axis was initially randomized, and changes of only 16 degrees were allowed during the alignment. This angular constraint and the orientation of the filament along the tilt axis ensured that the missing wedge artifact present in all tomographic data was nearly evenly averaged in the final structure, yielding an isotropic resolution in all directions.
Helical reconstruction
218 cryo-EM images were computationally down-sampled by a factor of two, giving a final pixel size of 0.44 nm. Contrast transfer function parameters were determined in the micrographs, and corresponding image distortions were corrected for in Bsoft. Filaments were traced in the images and cut into overlapping segments (90% overlap). Subimages (150 × 150 pixels), each containing a filament segment, were extracted (Table S2). Subimages were rotated so that the long axis of the segment was parallel to the image vertical axis. Rotated subimages were aligned horizontally to center the segments. The average of all segments was used as a template, and the process of averaging and alignment was iterated five times in SPIDER (Frank et al., 1996). Horizontally aligned images were subjected to multivariate data analysis and classification in SPIDER. The segments were first classified into 100 classes for each sample. The size of the Lsp1 dataset was reduced from 55,000 to 20,000 segments for classification. Class averages revealed variation in the datasets, most notably variation in the filament diameter. The initial horizontal alignment was improved by multireference alignment, using the first class means as references. These more accurately aligned segments were reclassified into 100 classes using the first 20 eigenimages. 3D reconstructions were generated by Fourier–Bessel analysis of the refined class averages using the Burnham-Brandeis Helical Package (designed by N. Volkmann, Sanford-Burnham Medical Research Institute, La Jolla, CA), an updated version of the original Brandeis Helical Package (Owen et al., 1996). In an attempt to detect all most prominent helical symmetries in the data, all Fourier transforms of class averages displaying strong layer lines consistent with either an even or an odd Bessel order were analyzed. Layer line indexes were assigned for 6 Fourier transforms of Lsp1 and Lsp-PI(4,5)P2 class averages and for 12 Fourier transforms of Pil1-PI(4,5)P2. In some cases, a few different assignments of Bessel orders were possible, and they were all considered in further analysis. 3D density maps were calculated from the Fourier transforms. The features of the repeating structural unit were similar between the density maps and also to the tomographic reconstruction of an Lsp1 filament, confirming that the layer line indexes and Bessel orders were assigned correctly. In the few ambiguous cases, the assignment, which resulted in a density map with consistent features to the other maps, was chosen to be the correct assignment.
Fourier–Bessel analysis provided multiple low-resolution template structures for the 3D reconstruction using IHRSR (Egelman, 2007). Several template structures with unique helical parameters were used: two for Lsp1, three for Lsp1-PI(4,5)P2, and four for Pil1-PI(4,5)P2 (Table S3). The filament segments that had been prealigned horizontally were classified into groups with different helical parameters and symmetries using projections from the template structures as references. Segment positions were adjusted horizontally to account for inaccuracies in the initial alignment against a common reference. IHRSR was run for five iterations for each group of segments. 80% of the best-correlating segments were chosen at each round to the reconstruction to count in possible inaccuracies in classification. The filament segments were reclassified against the refined models and five iterations of IHRSR were run. Because the starting models already had the correct helical symmetries present in the data, the helical symmetry parameters were kept constant during the iterations.
X-ray structure fitting to EM maps
Twofold symmetric Lsp1 dimers (AA, BB, and CC) were first generated from the three chains in the crystallographic asymmetric unit (A, B, and C) by superposition of the chains on the crystallographic dimer (AB) in the program LSQMAN (G.J. Kleywegt, Uppsala Software Factory, Sweden; Kleywegt, 1996). Only the core part of the dimer was used in the superposition. All three types of dimers—AA, BB and, CC—were fitted computationally on the six different cryo-EM density maps of Lsp1 and Pil1 filaments. Fitting was performed in CoLoRes (Situs package; Chacón and Wriggers, 2002) by calculating Laplacian-filtered correlation between the x-ray structure and the cryo-EM reconstruction to the resolution limit of each reconstruction. Helical symmetry of each of the reconstructions was applied on the best-fitting x-ray structure to generate atomic models of the filaments. For visualization in the University of California San Francisco Chimera program, the isosurface levels of cryo-EM reconstructions were set to correspond to the total mass of fitted Lsp1 structures, taking into account the presence of a membrane in some of the structures.
E-MAP
E-MAP datasets were either from Aguilar et al. (2010) or generated as previously described in Collins et al. (2006).
Spheroplasting and sample preparation for DEEM
For generation of yeast spheroplasts, cells were grown by shaking at 30°C to OD600 = 0.5 in YPD and treated with zymolyase for 10 min (modified from Ogg et al. [1992]). “Unroofing” of spheroplasts, as well as antibody decoration, freezing, replicating, and imaging of the samples, was performed as previously described (Ogg et al., 1992; Hanson et al., 2008). The primary antibodies used were rabbit anti-GFP (Invitrogen) or rabbit anti-Pil1 (Walther et al., 2007). For freeze-fracture DEEM, cells were grown to OD600 = 0.5 in YPD and quick-frozen by abrupt application of the sample on a coverslip onto a block of ultrapure copper cooled to liquid helium temperature (Heuser, 1989) before deep etching and platinum replication.
Data deposition
The cryo-EM reconstructions (Table S3) have been deposited in the Electron Microscopy Data Bank at the European Bioinformatics Institute under accession codes EMD-1865 (Lsp1 #1), EMD-1866 (Lsp1 #2), EMD-1867 (Lsp1-liposome #1), and EMD-1868 (Pil1-liposome #1).
Online supplemental material
Fig. S1 shows that Pil1 and Lsp1 form stable filaments of variable diameter in vitro and that their localization depends on plasma membrane PI(4,5)P2. Fig. S2 shows that Mrp8 does not localize in eisosomes and is not required for normal eisosomes at the plasma membrane. Fig. S3 shows that the yeast plasma membrane exhibits abundant furrows that depend on PIL1 and that immunogold-labeled Pil1 and Lsp1-GFP localize in elongated structures on plasma membranes from unroofed yeast cells. Videos 1, 2, and 3 show time lapse imaging of eisosomes marked by Pil1-GFP at different time points during Mss4 inactivation. Table S1 contains all yeast strains used in this study. Tables S2 and S3 present the data, collection, and reconstruction statistics for helical reconstruction. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201104040/DC1.
Acknowledgments
We thank Pietro DeCamilli, Robert Farese Jr., Shawn Ferguson, Boumediene Soufi, and Sebastian Schuck for critical reading of the manuscript and suggestions. We would like to especially thank Michael Shales for help in preparing the E-MAP figure and Judith Short and Edward Egelman for valuable suggestions concerning reconstruction of filament structures.
This work was supported by the Boehringer Ingelheim Fonds (L. Karotki), the Academy of Finland (grant 218080 to J.T. Huiskonen), the Max Planck Society (T.C. Walther), the German Research Foundation (T.C. Walther and K. Grünewald), and the Minna-James-Heineman Foundation (T.C. Walther).
Footnotes
Abbreviations used in this paper:
- DEEM
- deep-etching EM
- E-MAP
- epistatic miniarray profile
- IHRSR
- iterative helical real-space reconstruction
- PC
- phosphatidylcholine
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- YPD
- yeast peptone dextrose
References
- Aguilar P.S., Fröhlich F., Rehman M., Shales M., Ulitsky I., Olivera-Couto A., Braberg H., Shamir R., Walter P., Mann M., et al. 2010. A plasma-membrane E-MAP reveals links of the eisosome with sphingolipid metabolism and endosomal trafficking. Nat. Struct. Mol. Biol. 17:901–908 10.1038/nsmb.1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J., Hu Z., Dittman J.S., Pym E.C., Kaplan J.M. 2010. Endophilin functions as a membrane-bending molecule and is delivered to endocytic zones by exocytosis. Cell. 143:430–441 10.1016/j.cell.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold D., Walther T.C. 2009. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell. 20:1565–1575 10.1091/mbc.E08-10-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brach T., Specht T., Kaksonen M. 2011. Reassessment of the role of plasma membrane domains in the regulation of vesicular traffic in yeast. J. Cell Sci. 124:328–337 10.1242/jcs.078519 [DOI] [PubMed] [Google Scholar]
- Chacón P., Wriggers W. 2002. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 317:375–384 10.1006/jmbi.2002.5438 [DOI] [PubMed] [Google Scholar]
- Chang-Ileto B., Frere S.G., Chan R.B., Voronov S.V., Roux A., Di Paolo G. 2011. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev. Cell. 20:206–218 10.1016/j.devcel.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S.R., Schuldiner M., Krogan N.J., Weissman J.S. 2006. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 7:R63 10.1186/gb-2006-7-7-r63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy L.M., Olsen J.V., Cox J., Nielsen M.L., Hubner N.C., Fröhlich F., Walther T.C., Mann M. 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 455:1251–1254 10.1038/nature07341 [DOI] [PubMed] [Google Scholar]
- DeRosier D.J., Moore P.B. 1970. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J. Mol. Biol. 52:355–369 10.1016/0022-2836(70)90036-7 [DOI] [PubMed] [Google Scholar]
- Egelman E.H. 2007. The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. J. Struct. Biol. 157:83–94 10.1016/j.jsb.2006.05.015 [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. 1996. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199 10.1006/jsbi.1996.0030 [DOI] [PubMed] [Google Scholar]
- Fröhlich F., Moreira K., Aguilar P.S., Hubner N.C., Mann M., Walter P., Walther T.C. 2009. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J. Cell Biol. 185:1227–1242 10.1083/jcb.200811081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A., Perera R., Roux A., Spasov K., Destaing O., Egelman E.H., De Camilli P., Unger V.M. 2008. Structural basis of membrane invagination by F-BAR domains. Cell. 132:807–817 10.1016/j.cell.2007.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop J.L., McMahon H.T. 2005. BAR domains and membrane curvature: Bringing your curves to the BAR. Biochem. Soc. Symp. 72:223–231 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Grossmann G., Opekarová M., Malinsky J., Weig-Meckl I., Tanner W. 2007. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 26:1–8 10.1038/sj.emboj.7601466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G., Malinsky J., Stahlschmidt W., Loibl M., Weig-Meckl I., Frommer W.B., Opekarová M., Tanner W. 2008. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 183:1075–1088 10.1083/jcb.200806035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P.I., Roth R., Lin Y., Heuser J.E. 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180:389–402 10.1083/jcb.200707031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser J.E. 1989. Development of the quick-freeze, deep-etch, rotary-replication technique of sample preparation for 3-D electron microscopy. Prog. Clin. Biol. Res. 295:71–83 [PubMed] [Google Scholar]
- Heymann J.B., Belnap D.M. 2007. Bsoft: Image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157:3–18 10.1016/j.jsb.2006.06.006 [DOI] [PubMed] [Google Scholar]
- Hua W., Sheff D., Toomre D., Mellman I. 2006. Vectorial insertion of apical and basolateral membrane proteins in polarized epithelial cells revealed by quantitative 3D live cell imaging. J. Cell Biol. 172:1035–1044 10.1083/jcb.200512012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiskonen J.T., Hepojoki J., Laurinmäki P., Vaheri A., Lankinen H., Butcher S.J., Grünewald K. 2010. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 84:4889–4897 10.1128/JVI.00057-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Erdmann K.S., Roux A., Habermann B., Werner H., De Camilli P. 2005. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell. 9:791–804 10.1016/j.devcel.2005.11.005 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. 1996. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 52:842–857 10.1107/S0907444995016477 [DOI] [PubMed] [Google Scholar]
- Kozlov M.M., McMahon H.T., Chernomordik L.V. 2010. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 35:699–706 10.1016/j.tibs.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Gruhler A., Liu Y., Jensen O.N., Dickson R.C. 2008. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 283:10433–10444 10.1074/jbc.M709972200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malínská K., Malínský J., Opekarová M., Tanner W. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell. 14:4427–4436 10.1091/mbc.E03-04-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Norbeck J., Blomberg A. 1997. Metabolic and regulatory changes associated with growth of Saccharomyces cerevisiae in 1.4 M NaCl. Evidence for osmotic induction of glycerol dissimilation via the dihydroxyacetone pathway. J. Biol. Chem. 272:5544–5554 10.1074/jbc.272.9.5544 [DOI] [PubMed] [Google Scholar]
- Ogg S.C., Poritz M.A., Walter P. 1992. Signal recognition particle receptor is important for cell growth and protein secretion in Saccharomyces cerevisiae. Mol. Biol. Cell. 3:895–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera-Couto A., Graña M., Harispe L., Aguilar P.S. 2011. The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell. 22:2360–2372 10.1091/mbc.E10-12-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen C.H., Morgan D.G., DeRosier D.J. 1996. Image analysis of helical objects: The Brandeis Helical Package. J. Struct. Biol. 116:167–175 10.1006/jsbi.1996.0027 [DOI] [PubMed] [Google Scholar]
- Peter B.J., Kent H.M., Mills I.G., Vallis Y., Butler P.J., Evans P.R., McMahon H.T. 2004. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 303:495–499 10.1126/science.1092586 [DOI] [PubMed] [Google Scholar]
- Ren G., Vajjhala P., Lee J.S., Winsor B., Munn A.L. 2006. The BAR domain proteins: Molding membranes in fission, fusion, and phagy. Microbiol. Mol. Biol. Rev. 70:37–120 10.1128/MMBR.70.1.37-120.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuske K.R., Richmond J.E., Matthies D.S., Davis W.S., Runz S., Rube D.A., van der Bliek A.M., Jorgensen E.M. 2003. Endophilin is required for synaptic vesicle endocytosis by localizing synaptojanin. Neuron. 40:749–762 10.1016/S0896-6273(03)00667-6 [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Audhya A., Emr S.D. 2002. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell. 13:542–557 10.1091/mbc.01-10-0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strádalová V., Stahlschmidt W., Grossmann G., Blazíková M., Rachel R., Tanner W., Malinsky J. 2009. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J. Cell Sci. 122:2887–2894 10.1242/jcs.051227 [DOI] [PubMed] [Google Scholar]
- Verstreken P., Koh T.W., Schulze K.L., Zhai R.G., Hiesinger P.R., Zhou Y., Mehta S.Q., Cao Y., Roos J., Bellen H.J. 2003. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 40:733–748 10.1016/S0896-6273(03)00644-5 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Brickner J.H., Aguilar P.S., Bernales S., Pantoja C., Walter P. 2006. Eisosomes mark static sites of endocytosis. Nature. 439:998–1003 10.1038/nature04472 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Aguilar P.S., Fröhlich F., Chu F., Moreira K., Burlingame A.L., Walter P. 2007. Pkh-kinases control eisosome assembly and organization. EMBO J. 26:4946–4955 10.1038/sj.emboj.7601933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Kakaradov B., Collins S.R., Karotki L., Fiedler D., Shales M., Shokat K.M., Walther T.C., Krogan N.J., Koller D. 2009. A complex-based reconstruction of the Saccharomyces cerevisiae interactome. Mol. Cell. Proteomics. 8:1361–1381 10.1074/mcp.M800490-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lester R.L., Dickson R.C. 2004. Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein kinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p. J. Biol. Chem. 279:22030–22038 10.1074/jbc.M400299200 [DOI] [PubMed] [Google Scholar]
- Ziółkowska N.E., Karotki L., Rehman M., Huiskonen J.T., Walther T.C. 2011. Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat. Struct. Mol. Biol. 18:854–856 10.1038/nsmb.2080 [DOI] [PubMed] [Google Scholar]