Mammalian SEPT9 is positioned at the end of septin octamers and is necessary for octamer assembly into polymers necessary for abscission during cytokinesis.
Abstract
Septins are filamentous guanosine triphosphatase–binding proteins that are required for cytokinesis in a wide range of organisms from yeast to man. Several septins, including SEPT9, have been found to be altered in cancers, but their roles in malignancy and cytokinesis remain unclear. It is known that they assemble into rod-shaped oligomeric complexes that join end-on-end to form filaments, but whether SEPT9 incorporates into these complexes and how it does so are unanswered questions. We used tandem affinity purification of mammalian septin complexes to show that SEPT9 occupies a terminal position in an octameric septin complex. A mutant SEPT9, which cannot self-associate, disrupted septin filament formation and resulted in late abscission defects during cytokinesis but did not affect septin-dependent steps earlier in mitosis. These data suggest that mammalian SEPT9 holds a terminal position in the septin octamers, mediating abscission-specific polymerization during cytokinesis.
Introduction
Septins are a family of filament-forming GTP-binding proteins that have been implicated in diverse cellular processes (Hall, 2008). Much work has focused on the role of septins in cell division in which they mediate the physical division of the cytoplasm. This process, called cytokinesis, is necessary to ensure that each daughter cell inherits the correct complement of chromosomes. Because defects in cytokinesis have been linked to cancer (Fujiwara et al., 2005; Ganem et al., 2007; Sagona and Stenmark, 2010), it is not surprising that altered expression of individual septins is also associated with various human malignancies. For example, changes in the expression of SEPT9 or amplification of the SEPT9 locus has been linked to sporadic ovarian and breast cancers (Burrows et al., 2003; Montagna et al., 2003; Scott et al., 2006; Gonzalez et al., 2007). However, the precise role of septins in cytokinesis and malignant transformation remains unknown.
Septins associate with each other to form complexes, which in turn assemble in an end to end fashion to form long filaments. The best-characterized septin complex is that of the budding yeast Saccharomyces cerevisiae. This complex comprises near stoichiometric ratios of the four major yeast septins, Cdc3, Cdc10, Cdc11, and Cdc12, and electron microscopy revealed that these subunits had a rod-shaped appearance (Frazier et al., 1998). Moreover, by tagging individual septins, Bertin et al. (2008) demonstrated that these rods were nonpolar octamers composed of two tetramers with mirror symmetry and ordered Cdc11–Cdc12–Cdc3–Cdc10–Cdc10–Cdc3–Cdc12–Cdc11.
In mammals, 14 genes encode septin proteins (called SEPT1–SEPT14), and most of these genes undergo alternative splicing to produce multiple isoforms (Russell, 2008). In particular, SEPT9 has been shown to undergo alternative transcript initiation and splicing at the N and C termini, giving rise to 18 different transcripts that would encode 15 different isoforms (McIlhatton et al., 2001). The longer N-terminal variants (SEPT9_i1–3) have small unique sequences spliced to a common exon (exon 3), whereas SEPT9_i4 splices into exon 3, deleting most of it, and SEPT9_i5 lacks all of exon 3.
Only a subset of the 14 septin genes and isoforms appears to be expressed in a given cell, suggesting that mammalian septin complexes may differ in composition in different cell and tissue types. Based on sequence similarity, the 14 septin genes fall into the following four subgroups (Kinoshita, 2003; Peterson et al., 2007): the SEPT2 subgroup (septins 1, 2, 4, and 5), the SEPT3 subgroup (septins 3, 9, and 12), the SEPT6 subgroup (septins 6, 8, 10, 11, and 14), and the SEPT7 subgroup (septins 7 and 13). Although they do not have a simple orthologous relationship to the yeast septins (Pan et al., 2007), the four mammalian septin subgroups do have structural similarity to the four yeast septins (Versele and Thorner, 2005). For example, all yeast septins except Cdc10 have coiled-coil domains near their C termini. The same is true for three of the four mammalian septin subgroups, with SEPT3 subgroup members lacking a coiled coil. It has therefore been proposed that mammalian septins may assemble into complexes in a manner analogous to that observed in yeast. Recently, Sellin et al. (2011) demonstrated that mammalian septin complexes immunopurified from multiple cell types consist of a member of each of the four septin groups. Thus, mammalian septins likely assemble into complexes that might be analogous to budding yeast.
To date, the best description of a mammalian septin complex comes from crystallography analyses in which three mammalian septins coexpressed in bacteria were purified and crystallized (Sirajuddin et al., 2007). They were found to form an apolar hexameric complex that was ordered SEPT7–SEPT6–SEPT2–SEPT2–SEPT6–SEPT7. The crystal structure not only revealed the order of these three septins but also showed that the septin subunits interacted with each other at one of two interaction surfaces in an alternating manner. The first interaction surface involved the nucleotide-binding domains (G interface), whereas the other surface comprised the N- and C-terminal domains (NC interface). However, because the crystal structure was derived from a specific combination of recombinant septins, it remains unclear whether this organization holds true for native mammalian septin complexes. More importantly, biochemical and immunofluorescence studies on septin complexes from HeLa cells clearly show that SEPT9 is part of the septin complex (Surka et al., 2002; Estey et al., 2010), yet this septin was not included in the crystallography studies. How SEPT9 fits into the canonical 7–6–2–2–6–7 complex and whether septins might substitute for each other within the same subgroup were recently addressed by Sellin et al. (2011). They showed that native septin complexes immunoisolated from multiple mammalian cell types expressing different septin combinations were all capable of disassembling into protomeric octamers. This implied that cell lines expressing multiple members of one subgroup were likely generating many different types of octamers, with each member of a subgroup capable of substituting for another member of that subgroup. Furthermore, SEPT9 depletion resulted in a shift to a hexameric state, suggesting that SEPT9 might occupy a terminal position in the native octameric complex. An alternative, but not implausible, interpretation is that SEPT9 may occupy an internal position in the native octameric complex and that, after SEPT9 depletion, the trimeric 2–6–7 halves come together because of the plasticity of the septin complex. In support of this, the yeast septin complex displays remarkable plasticity. For instance, removal of the innermost yeast septin (Cdc10) results in a reordered hexameric complex with trimeric Cdc3–Cdc12–Cdc11 halves being joined together (McMurray et al., 2011). In addition, the innermost yeast septin (Cdc10) is structurally related to SEPT9 because both septins lack a coiled coil.
To definitively resolve the position of SEPT9 within the octamer, we generated mutations that would distinguish between SEPT9 occupying an innermost or outermost position. Using tandem affinity pull-downs and mutagenesis of individual septin interaction surfaces, we confirm the order of septins predicted by the crystal structure and demonstrate that SEPT9 occupies the ends of an octameric mammalian septin complex such that the order is 9–7–6–2–2–6–7–9. Moreover, by introducing mutations that prevent SEPT9 self-association at the NC interaction surface, we show that septin filament disruption causes defects in the late stages of cell division before abscission, similar to those observed upon SEPT9 depletion.
Results
SEPT9 associates with other septins on fibers and rings
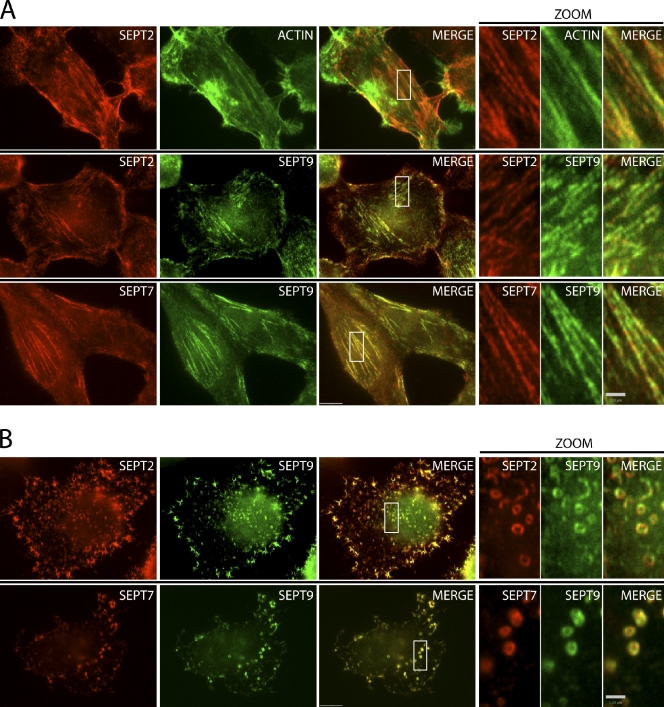
Crystallography of a complex of mammalian SEPT2, SEPT6, and SEPT7 expressed in bacterial cells revealed a nonpolar hexameric complex ordered SEPT7–SEPT6–SEPT2–SEPT2–SEPT6–SEPT7 (Sirajuddin et al., 2007). However, most cells characterized to date express additional septins, and we have demonstrated that HeLa cells express septins 2, 6, 7, 9, and 11 (Estey et al., 2010). Moreover, when SEPT9 was immunoprecipitated from HeLa cells, other septins coimmunoprecipitated, including SEPT2, SEPT6, SEPT7, and SEPT11 (Surka et al., 2002; Estey et al., 2010). We first asked whether SEPT9 associated with other septins in higher-ordered filamentous structures. Septins are known to adopt higher-order structures that resemble filaments and rings in vitro and in vivo. In fibroblast cells that contain central actin stress fibers, septins appear filamentous and have been shown to colocalize with subnuclear actin stress fibers (Kinoshita et al., 1997, 2002; Frazier et al., 1998; Xie et al., 1999; Nagata et al., 2004; Kremer et al., 2007). As shown in Fig. 1 A, in HeLa cells, SEPT2 and SEPT7 appear filamentous, colocalizing with actin stress fibers. Immunostaining with SEPT9 antibodies revealed strong colocalization with both SEPT2 and SEPT7 in these fibers.
Figure 1.
Colocalization of SEPT9 with septin fibers and rings. (A) Actin-templated septin fibers in interphase HeLa cells. (top) Immunofluorescent staining of SEPT2 fibers (red) and colocalization with phalloidin-stained actin stress fibers (green). Boxed areas of septin fibers are magnified on the right. (middle) Colocalization of SEPT9 with SEPT2 fibers. (bottom) Colocalization of SEPT9 with SEPT7 fibers. (B) Actin-independent septin self-assembly into rings upon treatment with cytochalasin D. (top) Immunofluorescent staining of SEPT2 rings (red) and colocalization with SEPT9. (bottom) SEPT9 colocalizes with SEPT7 rings upon cytochalasin D treatment. Bars: (main images) 5 µm; (zoom) 1 µm.
Latrunculin B or cytochalasin D disrupts F-actin and uncouples septins from actin, causing them to form short curvilinear structures that curl and ultimately form rings (Xie et al., 1999; Kinoshita et al., 2002). In vitro, when salt concentrations are lowered, septin complexes have been shown to form filaments that bundle laterally and self-assemble into rings (Kinoshita et al., 2002). Upon disruption of actin with cytochalasin D, SEPT2 and SEPT7 filaments reorganize into actin-independent rings with typical diameters of 0.45 µm (Fig. 1 B). SEPT9 also strongly colocalized with both SEPT2 and SEPT7 in these rings (Fig. 1 B). Thus, SEPT9, a member of the fourth septin subgroup, is associated with other septins on actin-templated septin fibers and actin-independent septin rings.
SEPT9 depletion disrupts septin fibers and reduces ring size
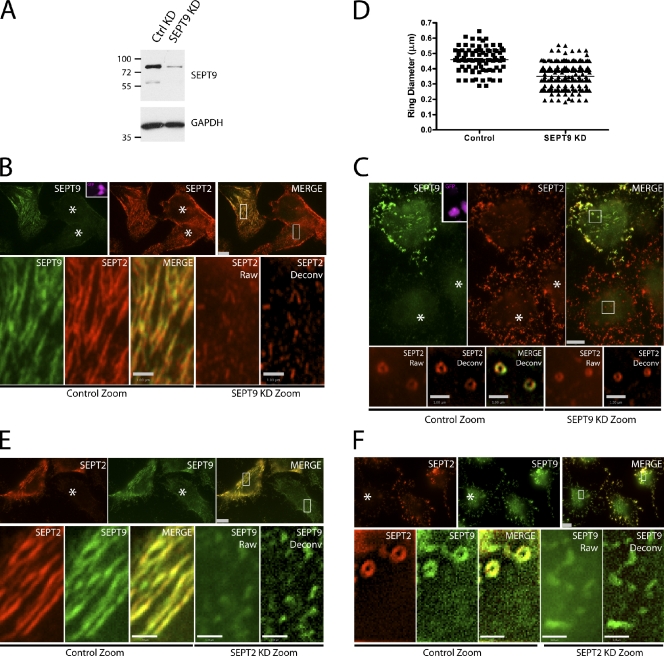
Next, we asked whether depleting individual septins could interfere with septin higher-ordered structures, such as fibers and rings. In HeLa cells, SEPT9 was depleted by targeted gene knockdown using a small hairpin RNA (shRNA) plasmid with a GFP reporter (Fig. 2 A). The remaining septin structures were assessed by immunostaining with SEPT2 antibodies in the presence or absence of cytochalasin D (Fig. 2, B and C). Cells lacking SEPT9 showed disrupted septin fibers when compared with nearby untransfected control cells (Fig. 2 B). A closer examination of SEPT2 fiber structures in these SEPT9-depleted cells revealed short rudimentary filaments that were more clearly resolved when deconvolved (Fig. 2 B, bottom). Induction of septin rings using cytochalasin D resulted in smaller septin rings in SEPT9-depleted cells when compared with surrounding untransfected control cells (Fig. 2 C). Quantitative analysis of these septin rings revealed a 23% reduction in ring diameter in SEPT9-depleted cells when compared with control cells (Fig. 2 D and Fig. S1). Thus, the remaining septins were able to adopt an alternate higher-ordered configuration that supported suboptimal filaments and smaller rings.
Figure 2.
SEPT9 knockdown disrupts septin fibers and rings. (A) Transfection of control (CTRL) or SEPT9 shRNA constructs in HeLa cells immunoblotted with SEPT9 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies. Molecular masses are given in kilodaltons. (B) Disruption of septin fibers with SEPT9 knockdown (KD). HeLa cells transfected with SEPT9 shRNA carrying a GFP reporter. GFP-positive cells are pseudocolored pink in the inset. (top) Immunofluorescent staining of SEPT2 (red) and SEPT9 (green) using rabbit polyclonal and mouse monoclonal antibodies, respectively. The merged image shows reduction of SEPT9 staining in knockdown cells. (bottom) Boxed areas are magnifications of nontransfected and transfected SEPT9 knockdown cells. SEPT9 knockdown cells have disrupted SEPT2 fibers (raw and deconvolved [Deconv] images). (C) Disruption in higher-order structure of septin rings with SEPT9 knockdown upon cytochalasin D treatment. (top) Immunofluorescent staining of SEPT2 (red) and SEPT9 (green) using rabbit polyclonal and mouse monoclonal antibodies, respectively. SEPT9 knockdown cells are GFP positive (pink, inset) and show reduced SEPT9 staining relative to SEPT2. (bottom) Boxed areas are magnifications of nontransfected and transfected SEPT9 knockdown cells. SEPT9 knockdown cells have smaller SEPT2 rings compared with untransfected control (raw and deconvolved images). (D) Quantitation of SEPT2 rings in control (n = 87) and SEPT9 knockdown (n = 230) rings. Mean diameters of 0.35 and 0.45 µm were observed for SEPT9 knockdown and control cells, respectively (horizontal lines). (E and F) The effect of SEPT2 knockdown on SEPT9 fibers (E) and rings (F). Knockdown cells are indicated with asterisks. Bars: (main images) 5 µm; (zoom) 1 µm.
In cells lacking SEPT2, there also appeared to be rudimentary filaments (Fig. 2 E), but these did not assemble into septin rings (Fig. 2 F). The plasticity observed in septin higher-order structures appears to be dependent on the particular septin being removed from the complex. The effects of depletion of SEPT2 and SEPT9 on septin fiber and ring formation may correlate with the positional arrangement of the septins in the unit complex. To further investigate the organizational arrangement of mammalian septins in the unit complex, we sought to determine where SEPT9 might be in the septin complex.
SEPT9 is a terminal septin subunit in the SEPT2–6–7 complex
The mammalian septin hexamer (SEPT7–SEPT6–SEPT2–SEPT2–SEPT6–SEPT7) has been shown to polymerize in vitro and is sufficient to form septin bundles and rings (Kinoshita et al., 2002; Sirajuddin et al., 2007). However, it is possible that these hexamers might represent a suboptimal configuration for mammalian septin assembly and that a septin from the SEPT3 group might be needed for efficient septin polymerization and function. Native mammalian septin complexes comprise a septin from each of the four septin groups and assemble as octamers (Sellin et al., 2011). In addition, we observe distinct effects on septin polymerization when SEPT9 is added in conjunction with SEPT2–6–7 overexpression. Under conditions of septin overexpression, when HeLa cells are cotransfected with SEPT2, SEPT6, and SEPT7, septin filaments are short and stubby. However, when SEPT9 isoforms are cotransfected with the other three septins, septin filaments are much longer and thicker (Fig. S2).
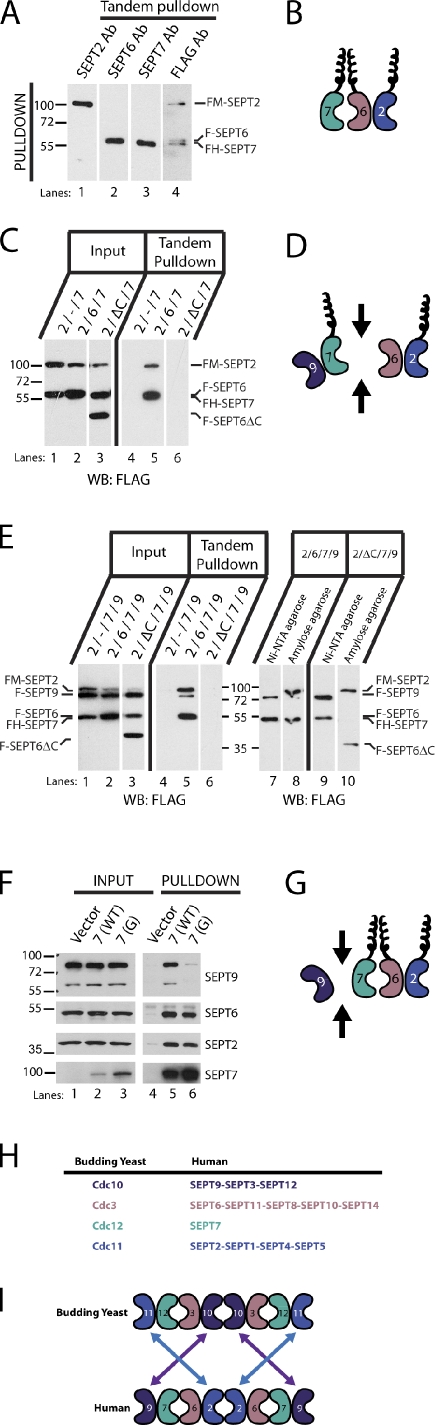
In yeast, removal of the innermost (Cdc10) or outermost (Cdc11) septin in the octamer gives rise to suboptimal filaments (McMurray et al., 2011). These might be similar to the suboptimal filaments that result from depletion of SEPT9 in mammalian cells. Thus, SEPT9 might be found at the innermost or outermost position in the octameric complex. To confirm that the order of the septins matched that predicted by the crystal structure and to determine where SEPT9 fits into this complex, we first developed a small-scale method for purifying equal amounts of SEPT2, SEPT6, and SEPT7. We used a similar tandem affinity purification scheme and retained similar affinity tags on the septins as described previously (Sirajuddin et al., 2007). HeLa cells were triply transfected with three septin plasmids and purified by tandem affinity purification using polyhistidine (His6) and maltose-binding protein (MBP) as affinity tags. One affinity tag, MBP, was fused to SEPT2, and the other affinity tag, His6, was fused to SEPT7, whereas SEPT6 did not contain an affinity tag. All plasmids carried a FLAG epitope to monitor septin complex purification. After tandem affinity purification, the resultant septin complexes contained equimolar amounts of SEPT2, SEPT6, and SEPT7 when immunoblotted for FLAG (Fig. 3 A, lane 4). The presence of each septin was further confirmed by immunoblotting with each individual septin antibody (Fig. 3 A, lanes 1–3). In addition, no endogenous septins copurified with the complex (Fig. S3), and SEPT6 was required for septin complex assembly, consistent with the possibility that it connects MBP-SEPT2 and His6-SEPT7 (Fig. 3 C, lane 4).
Figure 3.
SEPT9 interacts at the ends of the SEPT2–6–7 complex. (A) Small-scale tandem purification of the SEPT2–6–7 complex. Two affinity purification tags, His6 and MBP, are fused to two septins, SEPT7 and SEPT2, respectively. Triple transfection of septin constructs in HeLa cells followed by His6 and then MBP purification. All constructs contain FLAG epitope to monitor stoichiometry of the septin complex. (lane 4) Tandem pull-down contains SEPT2, SEPT6, and SEPT7 in equal amounts. (lanes 1–3) Final eluate immunoblotted with individual septin antibodies and the FLAG antibody (Ab). (B) Schematic representation of a heteromeric SEPT2–SEPT6–SEPT7 complex interacting at two interfaces. The two ends of the jellybean represent an NC interface with a predicted coiled coil projecting orthogonally to the axis of the unit complex. The G interface is represented by the smooth curved end of the jellybean. Only half of the septin unit complex is shown for simplicity. (C, lane 6) SEPT6ΔC disrupts SEPT2–SEPT6–SEPT7 complex assembly. (D) Schematic representation of disruption in the SEPT2–SEPT6–SEPT7–SEPT9 complex upon removal of the SEPT6 coiled coil. (E) Small-scale tandem purification of the SEPT2–SEPT6–SEPT7–SEPT9 complex. Two affinity purification tags, His6 and MBP, are fused to two septins, SEPT7 and SEPT2, respectively. Quadruple transfection of septin constructs in HeLa cells followed by His6 and then MBP purification. All constructs contain the FLAG epitope. (lanes 6, 9, and 10) SEPT6ΔC disrupts SEPT2–SEPT6–SEPT7–SEPT9 complex assembly (lane 6), and intermediate purification products partition the fragmented complex into SEPT2-SEPT6 and SEPT7-SEPT9 (lanes 9 and 10). (F) SEPT7 W249A G mutant disrupts SEPT2–SEPT6–SEPT7–SEPT9 complex assembly in vivo by preventing SEPT9 binding. Transfection of low-expressing GFP-FLAG-His6–wild-type (WT) and G mutant SEPT7 in HeLa cells followed by His6 purification and immunoblotted with FLAG, SEPT2, SEPT6, and SEPT9 antibodies. (G) Schematic representation of disruption in the SEPT2–SEPT6–SEPT7–SEPT9 complex with G mutant in SEPT7. (D and G) Arrows demarcate the septin–septin interface that is disrupted. (H) 14 mammalian septins are grouped into four septin families. (I) Comparison of yeast septin unit complex and the mammalian SEPT2–SEPT6–SEPT7–SEPT9 complex. Inversion of SEPT2 and SEPT9 families from yeast to mammals. Molecular masses are given in kilodaltons. WB, Western blot; F, FLAG epitope; FM, FLAG-MBP affinity tag; FH, FLAG-His6 affinity tag.
To determine the order of the septins within the septin complex, we first developed mutations that blocked interaction at the SEPT6–SEPT7 interface. The interaction of SEPT6 and SEPT7 occurs at an NC interface where their predicted coiled coils are closely apposed and project orthogonally to the hexamer axis. The coiled coils of SEPT6 and SEPT7 interact with each other directly (Low and Macara, 2006; Shinoda et al., 2010), and in yeast, septin coiled coils are necessary and sufficient for septin interaction (Versele et al., 2004), so we deleted the coiled-coil domain of SEPT6. As shown in Fig. 3 C, MBP-SEPT2 and His6-SEPT7 were coexpressed alone or with either full-length SEPT6 or SEPT6 lacking its coiled-coil domain (SEPT6ΔC). Expression of the individual septins was confirmed in the input lanes by blotting for FLAG (Fig. 3 C, lanes 1–3), but tandem purification of SEPT2 and SEPT7 only occurred when full-length SEPT6 was present (Fig. 3 C, lane 5). Although not detected by tandem affinity purification (Fig. 3 C, lane 6), single affinity purification revealed that SEPT6ΔC purified with MBP-SEPT2 on the amylose column, whereas His6-SEPT7 was found alone on the Ni–nitrilotriacetic acid (NTA) agarose column (not depicted). This indicates that the coiled coil of SEPT6 is necessary for its interaction with SEPT7 and that SEPT6 is between SEPT2 and SEPT7, consistent with the organization predicted by the crystal structure.
When SEPT9 was cotransfected with the other septins, a four-septin complex was isolated under the same purification conditions. Similar to the SEPT2–6–7 complex, this complex required the presence of SEPT6 (Fig. 3 E, lane 5). Disrupting the SEPT6–7 interaction using the SEPT6ΔC mutant confirmed the stable association of SEPT2 with SEPT6 and SEPT7 with SEPT9 (Fig. 3 E, lanes 9 and 10). This suggests that SEPT9 occupies the terminal position in the septin unit complex, adjacent to SEPT7 (Fig. 3 D). Because the NC and G interfaces alternate in the septin complex, SEPT9 and SEPT7 would interact with SEPT7 at their G interfaces. To test this possibility, we created a mutation in the G interface of SEPT7 (W249A; Sirajuddin et al., 2007; McMurray et al., 2011). We replaced the strong cytomegalovirus (CMV) promoter with a cloned fragment of the SEPT2 promoter to drive much lower expression of either wild-type GFP-FLAG-His6-SEPT7 or the SEPT7 G mutant. These constructs were then expressed in HeLa cells and pulled down using Ni-NTA agarose beads. Both wild-type and G mutant SEPT7 complexes pulled down similar amounts of endogenous SEPT2 and SEPT6 (Fig. 3 F, lanes 5 and 6). However, endogenous SEPT9 isoforms were pulled down much less efficiently in SEPT7 G mutant complexes compared with wild type (Fig. 3, F [lane 6] and G). Residual amounts of SEPT9 isoforms in the SEPT7 G mutant complex could be caused by binding at the other tetrameric arm of the complex, where endogenous SEPT7 may have incorporated.
This indicates that SEPT9 is located at the ultimate position of the septin complex, which appears to be an octamer. Therefore, our data suggest that mammalian septin complexes have an organizational arrangement similar to yeast, with SEPT9 being the outermost septin, so that the organization is SEPT9–SEPT7–SEPT6–SEPT2–SEPT2–SEPT6–SEPT7–SEPT9. Because mammalian SEPT9 and yeast Cdc10 are both in the same subgroup, which lack coiled coils (Fig. 3 H), it is interesting to note that Cdc10 is located at the middle of the complex, whereas SEPT9 is found at the ends (Fig. 3 I). Any septin that occupies a terminal position in the octameric unit complex is predicted to mediate end-on-end polymerization. To inhibit polymerization, a mutation in the terminal septin that preserves the octameric unit but prevents addition at these ends was created. This mutation was recently characterized in budding yeast (McMurray et al., 2011).
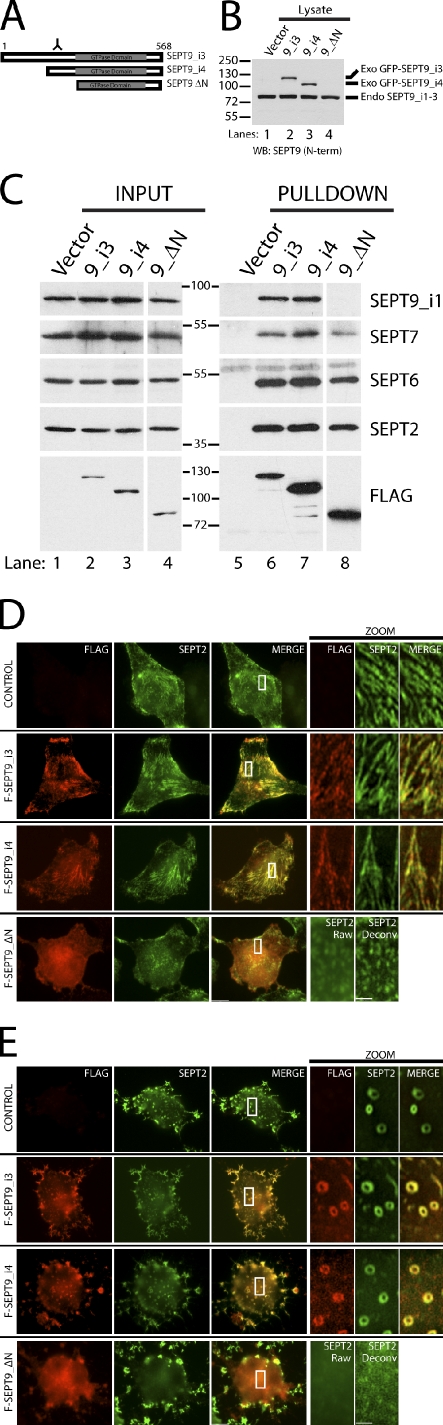
SEPT9ΔN prevents self-association and interferes with septin fiber and ring structures
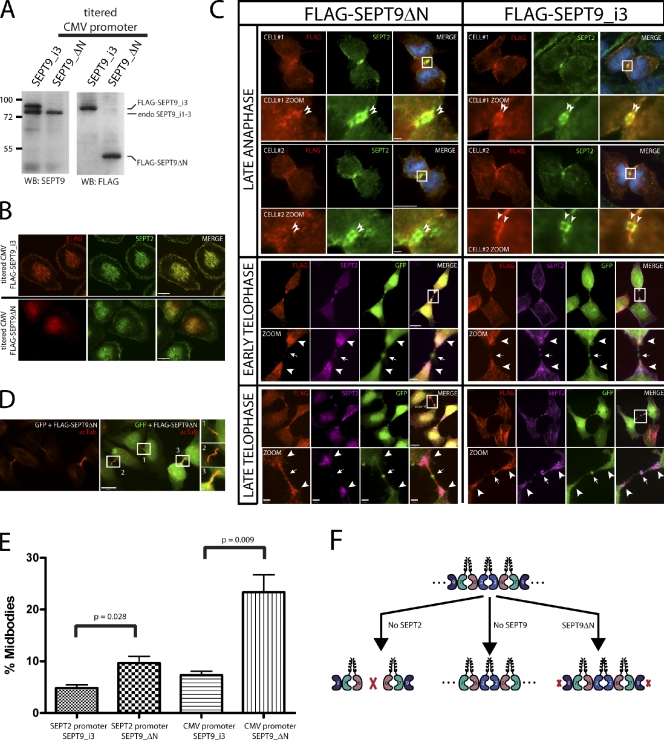
In budding yeast, septin filaments have recently been shown to be required for cytokinesis (McMurray et al., 2011). In that study, a Cdc11 deletion mutant in the α0 helix prevented homotypic Cdc11–Cdc11 interactions at the NC interface. This resulted in septin octamers that were not capable of polymerizing into filaments. To address whether we could disrupt septin higher-order structures by preventing interaction at the SEPT9–SEPT9 interface, we generated a SEPT9 mutant lacking the N-terminal region including the α0 helix (Fig. 4 A). Using the cloned fragment of the SEPT2 promoter, we slightly overexpressed isoforms of SEPT9 (i3 and i4) and the SEPT9ΔN mutant in HeLa cells (Fig. 4, B and C, input lanes; note that SEPT9ΔN is not detected by the SEPT9 N-terminal antibody in Fig. 4 B [lane 4] but is shown at comparable levels when all are detected with the FLAG antibody in Fig. 4 C) or by titrating the levels of CMV promoter-driven plasmids (Fig. 5 A). At lower protein expression levels, both isoforms of SEPT9 localized to septin fibers and rings (Fig. 4, D and E) and pulled down endogenous SEPT2, SEPT6, SEPT7, and the longest isoform of SEPT9, SEPT9_i1 (Fig. 4 C, lanes 6 and 7). However, the SEPT9 mutant pulled down endogenous SEPT2, SEPT6, and SEPT7, but no appreciable endogenous SEPT9 was isolated with the SEPT9 mutant (Fig. 4 C, lane 8). In addition, the SEPT9 mutant localized diffusely to the cytoplasm and disrupted SEPT2 fibers and rings (Fig. 4, D and E). Under conditions of septin overexpression, DsRed-SEPT7 formed short fibers when coexpressed with SEPT2 and SEPT6. Adding the two isoforms of SEPT9 drastically increased the filamentous nature of DsRed-SEPT7 fibers. The SEPT9 mutant was able to potently disrupt DsRed-SEPT7 fibers when coexpressed with the other three septins (Fig. S2). Thus, the N terminus of SEPT9 is required for an interaction with itself, and the SEPT9ΔN mutant disrupts endogenous and exogenous septin higher-ordered structures. Of note, the protein expression of the SEPT9 mutant was carefully controlled so that its expression level was similar to the SEPT9 isoforms. Levels that are too high (for example, CMV-driven septin constructs) cause wild-type SEPT9 isoforms to act in a dominant-negative manner (Fig. S4), and lower levels of the SEPT9 mutant are not sufficient to completely disrupt septin higher-order structures (not depicted).
Figure 4.
SEPT9ΔN caps the SEPT2–SEPT6–SEPT7–SEPT9 complex and interferes with septin fiber and ring structures. (A) Schematic representation of SEPT9 isoforms and SEPT9ΔN mutant. Antigenicity of SEPT9 antibody binding to SEPT9_i3 and i4, but not the SEPT9ΔN mutant, is clearly marked. (B) Low-expressing SEPT9 isoforms and the SEPT9ΔN mutant in HeLa cells. Expression of GFP-FLAG-His6-Stag-SEPT9 constructs is driven by the SEPT2 promoter and immunoblotted with an N-terminal SEPT9 antibody. Quadruple tag (GFP-FLAG-His6-Stag) and intervening protease cleavage and MCS adds ∼45 kD to each fusion protein. Dominant-negative effects of slight overexpression of SEPT9ΔN mutant on septin fibers. (C) SEPT9ΔN mutant prevents SEPT9–SEPT9 interaction in vivo. Transfection of low-expressing GFP-FLAG-His6-Stag-SEPT9_i3, -SEPT9_i4, and -SEPT9ΔN in HeLa cells followed by His6 purification and immunoblotting with FLAG, SEPT2, SEPT6, SEPT7, and SEPT9_i1 antibodies. (D) Transfection of low-expressing SEPT9 isoforms and the SEPT9ΔN mutant on septin fibers in HeLa cells. Transfected cells are immunostained with FLAG (red) and SEPT2 (green) antibodies. (E) Transfection of low-expressing SEPT9 isoforms and SEPT9ΔN mutant on septin rings in HeLa cells. Transfected cells were treated with cytochalasin D and immunostained with FLAG (red) and SEPT2 (green) antibodies. Boxed areas are magnified on the right. Molecular masses are given in kilodaltons. WB, Western blot; Exo, exogenous; Endo, endogenous; Deconv, deconvolved; N-term, N-terminal. Bars: (main images) 5 µm; (zoom) 1 µm.
Figure 5.
SEPT9ΔN and abscission defects. (A) Expression of titrated CMV promoter-driven FLAG-His6-Stag-SEPT9_i3 or -SEPT9ΔN in wild-type HeLa cells. Molecular masses are given in kilodaltons. (B) Localization of SEPT2 fibers in interphase wild-type HeLa cells transfected with FLAG-His6-Stag-SEPT9_i3 or -SEPT9ΔN. (C) Localization of SEPT2 in mitotic wild-type HeLa cells transfected with SEPT9ΔN or SEPT9_i3. Late anaphase (top), early telophase (middle), and late telophase cells (bottom) are transfected with FLAG-SEPT9ΔN (left) or FLAG-SEPT9_i3 (right) and immunostained with FLAG (red) or SEPT2 (green in top and pink in middle and bottom). Two cells in late anaphase are depicted in the first row (cell#1) and third row (cell#2). The localization of SEPT2 as a double band is not altered in the SEPT9 mutant (thin arrowheads in magnification, second and fourth rows). Early and late telophase cells were cotransfected with a GFP reporter plasmid to mark the Fleming body (arrows). SEPT2 localizes predominately at the ends of the spindle midzone (thick arrowheads). Boxes show areas of magnification below. (D) Cotransfection of SEPT9ΔN mutant with a GFP reporter and staining with acetylated tubulin (acTub; red), which reveals enrichment of midbodies. Acetylated tubulin staining is shown in the left, and the middle is the merged image with GFP to identify cotransfected cells and mark the Fleming bodies. Boxes in the merged image are magnified in the right and show midbody structures. (E) Quantitation of midbody enrichment of SEPT9ΔN mutant and control (SEPT9_i3) using two different promoters. Very low expression (SEPT2 promoter) and low expression (titrated CMV promoter) of the SEPT9ΔN mutant and control (SEPT9_i3). Under very low expression (SEPT2 promoter), there was a twofold increase in midbodies in the SEPT9 mutant compared with control (9.7 ± 1.3 vs. 4.8 ± 0.6). Under low expression (titrated CMV promoter), there was a 2.8-fold increase in midbodies in SEPT9 mutant compared with control (20.7 ± 0.9 vs. 7.3 ± 0.7). Error bars show SEM. (F) Model of mammalian septin complex assembly. WB, Western blot. Bars: (B, C [main images], and D) 5 µm; (C, zoom) 1 µm.
Septin higher-order structures and abscission
Previously, our laboratory has demonstrated two major defects in cell division in HeLa cells associated with depleting mammalian septins: binucleation and an increase in the presence of cells connected by midbodies (Estey et al., 2010). Cells lacking SEPT2, SEPT7, or SEPT11 were binucleate after imprecise furrow ingression or contractile ring instability. Cells lacking SEPT9 resulted in arrest later at the abscission step, leaving persistent midbodies. Using the SEPT9ΔN self-association mutant to block septin fibers and rings, we asked whether septin higher-order structures are required for cytokinesis. These cells were transfected with either SEPT9_i3 or SEPT9ΔN for 36 h, and midbodies were visualized by immunostaining with acetylated tubulin and counted (Fig. 5 D). Even when expressed at very low levels, SEPT9_i3 incorporated into septin fibers, whereas the SEPT9ΔN partially disrupted septin fibers (unpublished data). When both SEPT9_i3 and SEPT9ΔN were expressed at levels that were comparable with endogenous SEPT9 (Fig. 5 A), SEPT9ΔN fully disrupted septin fibers in interphase cells (Fig. 5 B) but did not affect the proper localization of SEPT2 to the cleavage furrow in dividing cells (Fig. 5 C). Of the 37 cells examined in late telophase, all showed localization of SEPT2 at the cleavage furrow (unpublished data). In control cells transfected with SEPT9_i3, this wild-type isoform incorporated into normal septin structures in interphase cells (Fig. 5 B) and colocalized strongly with SEPT2 at the cleavage furrow in dividing cells (Fig. 5 C). This suggests that SEPT2 does not require septin higher-order structures to localize to the cleavage furrow. Correct targeting of SEPT2 to the cleavage furrow could occur through the interaction with myosin II (Joo et al., 2007) or through interaction with complexes containing endogenous SEPT9. The late telophase defects associated with SEPT9ΔN were quantified, and a threefold increase in the number of cells with midbodies was observed when compared with wild type (Fig. 5 E). Although we cannot preclude the possibility that SEPT9ΔN fails to scaffold an essential protein to the midbody, our data suggest that septin higher-order structures may be required for later stages of cell division during abscission.
Discussion
Plasticity of septin complexes is a conserved property of septins
Yeast septin octamers are arranged 11–12–3–10–10–3–12–11. Although the order of the yeast septins within the octamer appear fixed, a recent study in budding yeast unequivocally demonstrated plasticity of septin complexes (McMurray et al., 2011). Removing the innermost septin (Cdc10) from the octameric complex reconfigures the remaining septins into hexamers with the following organization: 11–12–3–3–12–11. Similarly, removing the outermost septin (Cdc11) preserves the preexisting hexamer template: 12–3–10–10–3–12. Although not optimal, in both cases, the remaining septins can polymerize in vitro and support yeast viability. However, removal of internal septins that connect the innermost and outermost septins, Cdc3 or Cdc12, does not support septin filaments in vitro and does not support viability. This indicates that plasticity of the yeast septin complex and filament assembly is most tolerant to septins that interact homotypically. Thus, yeast septin complexes can adopt filamentous structures after the removal of the innermost and outermost septin. In mammals, septins can adopt alternate higher-order ring structures when only the outermost septin, SEPT9, is removed. Removal of the innermost septin, SEPT2, may prevent higher-order structures because of the inability of SEPT6 to interact with itself (unpublished data). This could result in septin trimers (SEPT6–SEPT7–SEPT9) that could form hexamers (SEPT6–SEPT7–SEPT9–SEPT9–SEPT7–SEPT6) but not filaments (Fig. 5 F), although the presence of small rudimentary filaments by immunocytochemistry suggests that they may still be capable of limited polymerization. This degree of septin plasticity might explain our earlier data in which SEPT2 and SEPT9 depletion had different effects on cell division, with SEPT2 depletion resulting in early mitotic defects, whereas SEPT9 depletion causes late abscission defects (Estey et al., 2010).
Another mode of septin plasticity in mammals may arise from the substitution of septins within the same subgroup of the unit complex. It has been reported by Kinoshita (2003) that septins within the same subgroup are able to substitute for one another in vitro. Furthermore, Sellin et al. (2011) have shown that complex septin mixtures (containing multiple septins in each subgroup) dissociate into protomeric octamers in vivo. Collectively, septins within the same subgroup are likely substituting for each other at the same position in the unit complex. Both modes of septin plasticity (septin subgroup substitution and generation of suboptimal septin–septin interactions) likely explain the lack of severe phenotypes seen in septin knockout mice, despite the fundamental role septins play in cell division.
Organizational arrangement of septins in the unit complex is a conserved property of septins
In budding yeast, four septins assemble into apolar octamers such that the arrangement is 11–12–3–10–10–3–12–11. In this order, two septins bind homotypically at the NC interface. The innermost septin (Cdc10) joins the tetrameric halves to form an octamer, and the outermost septin (Cdc11) joins with other Cdc11-containing octamers to form filaments. Using the positional arrangement of the septins within the octamer, McMurray et al. (2011) generated two different octamer mutants. The first mutant was a Cdc11 self-association mutant that retained septin octamers but prevented polymerization by inhibiting octamer joining at the ends. The second mutant was a Cdc10 self-association mutant that generated a flipped inside-out octamer such that the organization was 10–3–12–11–11–12–3–10. Both were polymerization defective and disrupted cytokinesis in budding yeast. Thus, septin filaments were required for cytokinesis.
Mammalian septins fall into four subgroups, of which three have been shown to associate as an apolar hexamer in vitro (Sirajuddin et al., 2007). Native mammalian septin complexes disassemble into octamers regardless of the compositional number of septins, suggesting that the four subgroups constitute fundamental building blocks of septin unit complexes (Sellin et al., 2011). In addition, Sandrock et al. (2011) have shown that SEPT9 might also occupy a terminal position based on yeast two-hybrid and yeast three-hybrid analyses. Here, we show that mammalian septin complexes can form octamers that are arranged as 9–7–6–2–2–6–7–9. Thus, the in vitro 7–6–2–2–6–7 hexamer is an incomplete mammalian septin complex, and a septin member of the fourth subgroup satisfies septin octamer assembly, such that the organizational principles of mammalian septin complex assembly are similar to budding yeast. In this arrangement, two septins homotypically interact at NC interfaces: two SEPT2 proteins join the two tetrameric arms of the complex, and two SEPT9 proteins interact to join octamers into larger structures. The N and C termini of SEPT9 undergo extensive alternative splicing, so it will be of great interest to determine whether the different splice forms alter interaction properties. Interestingly, it was reported by Chacko et al. (2005) that when stably overexpressed in cell lines, one of the small isoforms of SEPT9 (SEPT9_i4) was able to disrupt endogenous septin fibers. It was also found that SEPT9_i3, but not SEPT9_i4, could rescue cytokinesis defects caused by depletion of all SEPT9 isoforms (Estey et al., 2010). This raised the possibility that smaller isoforms of SEPT9 (SEPT9_i4–5) might act as chain terminators. Consistent with this idea, the SEPT9ΔN mutant used in this study is shorter than the smallest isoform of SEPT9, SEPT9_i5, and disrupts endogenous septin fibers and rings. However, in contrast to the previous studies, we found that SETP9_i4 was able to incorporate into septin filaments and rings and associated with endogenous septins, inconsistent with a potent capacity for chain termination. It is possible that SEPT9_i4 is weaker in its capacity for chain termination than the shorter forms of SEPT9 and that only in the cases of high expression (Chacko et al., 2005) or when present as the only form of SEPT9 (Estey et al., 2010) can inhibition of septin polymerization be seen, whereas only low levels of SEPT9ΔN (or possibly SEPT9_i5) are sufficient to achieve chain disruption.
Our analyses have taken advantage of the same types of self-association mutants in yeast that generated octamers, and we show that an N-terminal deletion truncation in SEPT9 is a self-association mutant that disrupts septin higher-order structures and is required for the late stages of cytokinesis. It seems unexpected that the SEPT9ΔN mutant does not interact with endogenous SEPT9, even though it appears as though it is expressed at levels similar to endogenous SEPT9 (Fig. 4 B and Fig. 5 A). At levels expected to be at or near endogenous levels, endogenous SEPT9 would be expected to occupy the other tetrameric arm of the unit complex, and a reduction (not a complete absence) of binding to endogenous SEPT9 would be expected. However, because transfection of HeLa cells in our hands is between 50 and 60%, the levels of exogenously expressed septins in the transfected cells is underestimated. The SEPT9ΔN mutant is likely in excess of the level of endogenous SEPT9, thereby capping the octamer at both ends. This mutant also contains a large N-terminal deletion that in addition to disrupting septin higher-order structures, could interfere with other processes. For instance, septin-associated Rho guanine nucleotide exchange factor is an exchange factor for RhoA and has been found to bind to the N terminus of SEPT9 (Nagata and Inagaki, 2005), and deletion up to the α0 helix also removes a putative phospholipid binding site. Thus, we cannot rule out the possibility that lipids or proteins that bind to the N terminus of SEPT9 might be involved in late cytokinetic events. A better approach might be to specifically mutate the α0 helix in an otherwise intact SEPT N terminus (thereby retaining the lipid-binding and protein interaction domains). Alternatively, generating flipped inside-out octamers using SEPT2 self-association mutants would further add to the evidence that mammalian septin filaments are required for cytokinesis.
Requirement of septin filaments in cytokinesis is a conserved property of septins
Septins have two well-established roles as scaffolds and diffusion barriers at the mother–bud neck in yeast. Intuitively, scaffold functions may exist for small assemblies of septin complexes, whereas diffusion barriers may require long, continuous filaments to block membrane protein movement. Yeast septins undergo dramatic changes at the mother–bud neck. FRAP experiments on GFP-septins have demonstrated oscillating dynamic and static properties of the septins through cell division that correlate with posttranslational modification of septins (Dobbelaere et al., 2003). Moreover, a 90° rotation in septin structures at the mother–bud neck was detected with fluorescence anisotropy using fixed GFP moieties to yeast septins (Vrabioiu and Mitchison, 2006). Collectively, these results suggest major structural changes in septins that could increase or decrease the relative contribution of septins as either scaffolds or diffusion barriers. Our data suggest that, at least in some circumstances, SEPT9 would be present at the terminal position of septin complexes and might therefore be responsible for subunit polymerization. We have previously shown that depletion of SEPT9 resulted in defects only at the late stages of cell division but did not affect the early septin role in contractile ring stability. Similarly, in this study, we observed a similar result using mutants of SEPT9 that prevent polymerization. This indicates that subunits that are inefficient at polymerization are still capable of functioning during early stages of cytokinesis (in which they may play a scaffolding role) but that filament-forming septins are required for late stages of cytokinesis (consistent with a diffusion barrier role). In support of a scaffolding role during early stages in cell division, septins have been observed at the metaphase plate and act as a scaffold for mitotic motors and checkpoint proteins to coordinate chromosome congression and separation (Spiliotis et al., 2005). During early anaphase, septins have been shown to act as a scaffold to recruit citron and Rho kinases, and this has been shown to promote efficient phosphorylation of myosin light chain necessary to stabilize the contractile ring (Joo et al., 2007). The notion that SEPT9 may be important for a diffusion barrier–type role at later steps in mitosis is consistent with our previous observation that depletion of SEPT9 resulted in loss of the exocyst complex from the midbody (Estey et al., 2010), which is very similar to the diffusion-limited domain between the split septin rings that localizes the exocyst at the mother–bud neck in yeast (Dobbelaere and Barral, 2004). In summary, the organizational principles that govern septin complex assembly are conserved from yeast to mammals, and as in yeast, septin filaments are required for the final steps of cytokinesis.
Materials and methods
Cell culture, transfection, and drug treatments
HeLa cells stably expressing the tetracycline-controlled transactivator protein (Tet-ONs) and wild-type HeLa cells (American Type Culture Collection) were cultured in DME with 10% FBS and grown at 37°C with 5% CO2. Cells were transfected with either Lipofectamine 2000 (Invitrogen) or jetPRIME (VWR) using the manufacturer’s recommendations. HeLa Tet-ONs were treated for 50 min with 3 µM cytochalasin D (Sigma-Aldrich) to disrupt actin and induce septin ring assembly. Septin fibers were disrupted using the SEPT9 mutant in both HeLa Tet-ONs and wild-type HeLa cells. Any wild-type HeLa cells connected via a microtubule bridge, as judged by acetylated tubulin staining (T7451; Sigma-Aldrich), were counted as being connected by a midbody (Estey et al., 2010).
Plasmid construction
FLAG constructs.
A FLAG vector was generated by oligonucleotide cloning the FLAG epitope into pcDNA3.0(+). This FLAG vector was subsequently modified so that the multiple cloning site (MCS) was altered to include EcoRI and XhoI sites that are downstream and in frame with the FLAG epitope. Additional purification and epitope tags were added between the FLAG epitope and the MCS, resulting in a FLAG-MBP vector and a FLAG-His6-Stag (polyhistidine and S-tag) vector. These vectors were generated by PCR cloning and were used to purify septin complexes on Ni-NTA agarose and amylose-agarose columns. Human septin genes containing any internal EcoRI or XhoI restriction sites were mutated. Septins were PCR cloned or subcloned into respective FLAG, FLAG-His6, or FLAG-MBP vectors with EcoRI and XhoI.
Fluorescent constructs.
The MCS of pEGFP-C1 (Takara Bio Inc.) was altered to include EcoRI and XhoI sites that are downstream and in frame with the enhanced GFP protein. For lower expression of the septin genes, we removed the strong CMV promoter and replaced it with the SEPT2 promoter. To this end, the CMV promoter was cut out with AseI and AgeI, and the SEPT2 promoter was PCR cloned in its place. A >25-kb bacmid containing the full SEPT2 gene was obtained, and a 400-bp region upstream of the transcriptional start site of SEPT2 was generated using PCR. Additional purification and epitope tags were added between the enhanced GFP protein and the MCS, ultimately resulting in a GFP-FLAG-His6-Stag vector driven under the SEPT2 promoter. SEPT7-DsRed was generated by PCR cloning SEPT7 into pDSRed1-C1 (Takara Bio Inc.) using XhoI and EcoRI.
Septin mutants.
All septin mutant genes were PCR cloned or subcloned using EcoRI and XhoI. Human SEPT6 C-terminal truncation (amino acids 1–333) was generated using PCR. Human SEPT9 N-terminal truncation including the α0 helix (amino acids 268–568) was generated using PCR. SEPT7 G mutant (W249A) was generated by PCR mutagenesis in pBluescript SK(−) and subcloned into respective vectors.
Septin complex isolation (tandem affinity purification)
Septin constructs were transfected into HeLa Tet-ONs grown on 6-well tissue-culture plates and harvested 18–24 h later. Cells were washed with cold PBS and lysed using buffer A (PBS, 20 mM imidazole, 1% Triton X-100, and a protease tablet). Lysates were clarified for 40 min at 200,000 g. The clarified supernatant was incubated with Ni-NTA agarose (QIAGEN) for 1 h, washed three times in buffer B (PBS, 20 mM imidazole, and 0.1% Triton X-100), and eluted in buffer C (PBS, 150 mM imidazole, and 0.1% Triton X-100). The eluate was incubated with amylose-agarose (New England Biolabs, Inc.) for 1 h and washed three times in buffer B. SDS-PAGE loading buffer was added to beads and run on 10% SDS-PAGE gels.
Septin complex isolation (single purification)
Septin constructs driven under the SEPT2 promoter were transfected into HeLa Tet-ONs grown on 6-well plates and harvested 18–24 h later. Cells were lysed and washed as in the previous paragraph. SDS-PAGE loading buffer was added to beads, run on 10% SDS-PAGE gels, and probed with septin antibodies.
Immunoblotting
Standard immunoblotting procedure was performed using 1-h incubations of primary and secondary antibodies. Dilutions of antibodies used for immunoblotting were as follows: mouse anti-FLAG at 1:7,500 (M2; Sigma-Aldrich), mouse anti–glyceraldehyde 3-phosphate dehydrogenase at 1:50,000, rabbit anti-SEPT9 at 1:1,000 (Surka et al., 2002), rabbit anti-SEPT2 at 1:500 (Xie et al., 1999), rabbit anti-SEPT6 at 1:100 (N1), rabbit anti-SEPT7 at 1:1,000 (Santa Cruz Biotechnology, Inc.), and rabbit anti-SEPT9_i1 at 1:5,000 (gift from E. Petty, University of Michigan, Ann Arbor, MI). HRP secondary antibodies were used at 1:5,000 (Jackson ImmunoResearch Laboratories, Inc.). Rhodamine-phalloidin was used at 1:500.
Immunofluorescence
Cells were washed with room temperature PBS and then fixed in 1% PFA in PBS for 20 min at room temperature. PFA was inactivated, and cells were permeabilized using buffer D (PBS with 25 mM glycine, 25 mM ammonium chloride, and 0.1% Triton X-100) for 20 min. Cells were blocked in PBS with 5% horse serum for 1 h. Standard immunofluorescence procedures were used with 1-h incubations of primary and secondary antibodies. Dilutions of antibodies used for immunofluorescence were mouse anti-FLAG at 1:1,000 (M2), mouse anti-SEPT9 undiluted (10C10; Estey et al., 2010), rabbit anti-SEPT2 at 1:100 (Xie et al., 1999), and rabbit anti-SEPT7 at 1:10. Fluorescent secondary antibodies conjugated to Cy3 and Cy5 were used at 1:500 (Jackson Immunologicals). Coverslips were mounted using fluorescence mounting medium (Dako). Fixed cells were imaged at room temperature using an inverted fluorescence microscope (DMIRE2; Leica) equipped with an IEEE 1394–based digital camera (ORCA-ERG model C4742-95-12ERG; Hamamatsu Photonics). Images were acquired using a 100× oil immersion objective (HCX Plan Apochromat with numerical aperture of 1.40–0.7; Leica). The equipment was driven by Volocity acquisition software (PerkinElmer) and powered by a Power Mac G5 (Apple). Deconvolved images were processed using the Fast Restorative function in Volocity. This is a quick method that provides images with high contrast by using a single-pass, nearest neighbor subtractive method. Deconvolution using this method relies on a point spread function to determine what light is out of focus and removes that light from the image. A point spread function for a wide-field microscope was created for each channel using the following settings: numerical aperture = 1.4, medium refractive index = 1.518, and emission wavelength of 600 nm (Cy3) and 515 nm (GFP).
Online supplemental material
Fig. S1 shows the raw data for the quantitation performed in Fig. 2 D. Fig. S2 depicts the filamentous nature of septins when SEPT9 isoforms are cotransfected with SEPT2, SEPT6, and SEPT7. Fig. S3 shows the absence of endogenous septins that copurify with tandem affinity–purified, overexpressed septin complexes. Fig. S4 contains low and high expression of different isoforms of SEPT9 and its effect on endogenous SEPT2 higher-order structures. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106131/DC1.
Acknowledgments
We are grateful to all members of the Trimble laboratory. We thank Elizabeth Petty for the purified antibody to SEPT9_i1.
This work was supported by the Canadian Cancer Society. W.S. Trimble holds a Canada Research Chair in Molecular Cell Biology.
Footnotes
Abbreviations used in this paper:
- CMV
- cytomegalovirus
- MBP
- maltose-binding protein
- MCS
- multiple cloning site
- NTA
- nitrilotriacetic acid
- shRNA
- small hairpin RNA
References
- Bertin A., McMurray M.A., Grob P., Park S.S., Garcia G., III, Patanwala I., Ng H.L., Alber T., Thorner J., Nogales E. 2008. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. USA. 105:8274–8279 10.1073/pnas.0803330105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows J.F., Chanduloy S., McIlhatton M.A., Nagar H., Yeates K., Donaghy P., Price J., Godwin A.K., Johnston P.G., Russell S.E. 2003. Altered expression of the septin gene, SEPT9, in ovarian neoplasia. J. Pathol. 201:581–588 10.1002/path.1484 [DOI] [PubMed] [Google Scholar]
- Chacko A.D., Hyland P.L., McDade S.S., Hamilton P.W., Russell S.H., Hall P.A. 2005. SEPT9_v4 expression induces morphological change, increased motility and disturbed polarity. J. Pathol. 206:458–465 10.1002/path.1794 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Barral Y. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 305:393–396 10.1126/science.1099892 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Gentry M.S., Hallberg R.L., Barral Y. 2003. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell. 4:345–357 10.1016/S1534-5807(03)00061-3 [DOI] [PubMed] [Google Scholar]
- Estey M.P., Di Ciano-Oliveira C., Froese C.D., Bejide M.T., Trimble W.S. 2010. Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J. Cell Biol. 191:741–749 10.1083/jcb.201006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier J.A., Wong M.L., Longtine M.S., Pringle J.R., Mann M., Mitchison T.J., Field C. 1998. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143:737–749 10.1083/jcb.143.3.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Bandi M., Nitta M., Ivanova E.V., Bronson R.T., Pellman D. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 437:1043–1047 10.1038/nature04217 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Storchova Z., Pellman D. 2007. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 17:157–162 10.1016/j.gde.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Gonzalez M.E., Peterson E.A., Privette L.M., Loffreda-Wren J.L., Kalikin L.M., Petty E.M. 2007. High SEPT9_v1 expression in human breast cancer cells is associated with oncogenic phenotypes. Cancer Res. 67:8554–8564 10.1158/0008-5472.CAN-07-1474 [DOI] [PubMed] [Google Scholar]
- Hall P.A., Russell S.E.H., Pringle J.R., 2008. The Septins. John Wiley-Blackwell, Oxford, England/Hoboken, NJ: 380 pp [Google Scholar]
- Joo E., Surka M.C., Trimble W.S. 2007. Mammalian SEPT2 is required for scaffolding nonmuscle myosin II and its kinases. Dev. Cell. 13:677–690 10.1016/j.devcel.2007.09.001 [DOI] [PubMed] [Google Scholar]
- Kinoshita M. 2003. Assembly of mammalian septins. J. Biochem. 134:491–496 10.1093/jb/mvg182 [DOI] [PubMed] [Google Scholar]
- Kinoshita M., Kumar S., Mizoguchi A., Ide C., Kinoshita A., Haraguchi T., Hiraoka Y., Noda M. 1997. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 11:1535–1547 10.1101/gad.11.12.1535 [DOI] [PubMed] [Google Scholar]
- Kinoshita M., Field C.M., Coughlin M.L., Straight A.F., Mitchison T.J. 2002. Self- and actin-templated assembly of Mammalian septins. Dev. Cell. 3:791–802 10.1016/S1534-5807(02)00366-0 [DOI] [PubMed] [Google Scholar]
- Kremer B.E., Adang L.A., Macara I.G. 2007. Septins regulate actin organization and cell-cycle arrest through nuclear accumulation of NCK mediated by SOCS7. Cell. 130:837–850 10.1016/j.cell.2007.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low C., Macara I.G. 2006. Structural analysis of septin 2, 6, and 7 complexes. J. Biol. Chem. 281:30697–30706 10.1074/jbc.M605179200 [DOI] [PubMed] [Google Scholar]
- McIlhatton M.A., Burrows J.F., Donaghy P.G., Chanduloy S., Johnston P.G., Russell S.E. 2001. Genomic organization, complex splicing pattern and expression of a human septin gene on chromosome 17q25.3. Oncogene. 20:5930–5939 10.1038/sj.onc.1204752 [DOI] [PubMed] [Google Scholar]
- McMurray M.A., Bertin A., Garcia G., III, Lam L., Nogales E., Thorner J. 2011. Septin filament formation is essential in budding yeast. Dev. Cell. 20:540–549 10.1016/j.devcel.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna C., Lyu M.S., Hunter K., Lukes L., Lowther W., Reppert T., Hissong B., Weaver Z., Ried T. 2003. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 63:2179–2187 [PubMed] [Google Scholar]
- Nagata K., Inagaki M. 2005. Cytoskeletal modification of Rho guanine nucleotide exchange factor activity: identification of a Rho guanine nucleotide exchange factor as a binding partner for Sept9b, a mammalian septin. Oncogene. 24:65–76 10.1038/sj.onc.1208101 [DOI] [PubMed] [Google Scholar]
- Nagata K., Asano T., Nozawa Y., Inagaki M. 2004. Biochemical and cell biological analyses of a mammalian septin complex, Sept7/9b/11. J. Biol. Chem. 279:55895–55904 10.1074/jbc.M406153200 [DOI] [PubMed] [Google Scholar]
- Pan F., Malmberg R.L., Momany M. 2007. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 7:103 10.1186/1471-2148-7-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E.A., Kalikin L.M., Steels J.D., Estey M.P., Trimble W.S., Petty E.M. 2007. Characterization of a SEPT9 interacting protein, SEPT14, a novel testis-specific septin. Mamm. Genome. 18:796–807 10.1007/s00335-007-9065-x [DOI] [PubMed] [Google Scholar]
- Russell S.E.H. 2008. The genomics and regulation of the human septin genes. The Septins. Hall P.A., Russell S.E.H., Pringle J.R., John Wiley-Blackwell, Oxford, England/Hoboken, NJ: 171–185 [Google Scholar]
- Sagona A.P., Stenmark H. 2010. Cytokinesis and cancer. FEBS Lett. 584:2652–2661 10.1016/j.febslet.2010.03.044 [DOI] [PubMed] [Google Scholar]
- Sandrock K., Bartsch I., Bläser S., Busse A., Busse E., Zieger B. 2011. Characterization of human septin interactions. Biol. Chem. 392:751–761 10.1515/BC.2011.081 [DOI] [PubMed] [Google Scholar]
- Scott M., McCluggage W.G., Hillan K.J., Hall P.A., Russell S.E. 2006. Altered patterns of transcription of the septin gene, SEPT9, in ovarian tumorigenesis. Int. J. Cancer. 118:1325–1329 10.1002/ijc.21486 [DOI] [PubMed] [Google Scholar]
- Sellin M.E., Sandblad L., Stenmark S., Gullberg M. 2011. Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell. 22:3152–3164 10.1091/mbc.E11-03-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda T., Ito H., Sudo K., Iwamoto I., Morishita R., Nagata K. 2010. Septin 14 is involved in cortical neuronal migration via interaction with Septin 4. Mol. Biol. Cell. 21:1324–1334 10.1091/mbc.E09-10-0869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M., Farkasovsky M., Hauer F., Kühlmann D., Macara I.G., Weyand M., Stark H., Wittinghofer A. 2007. Structural insight into filament formation by mammalian septins. Nature. 449:311–315 10.1038/nature06052 [DOI] [PubMed] [Google Scholar]
- Spiliotis E.T., Kinoshita M., Nelson W.J. 2005. A mitotic septin scaffold required for mammalian chromosome congression and segregation. Science. 307:1781–1785 10.1126/science.1106823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surka M.C., Tsang C.W., Trimble W.S. 2002. The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis. Mol. Biol. Cell. 13:3532–3545 10.1091/mbc.E02-01-0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M., Thorner J. 2005. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 15:414–424 10.1016/j.tcb.2005.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M., Gullbrand B., Shulewitz M.J., Cid V.J., Bahmanyar S., Chen R.E., Barth P., Alber T., Thorner J. 2004. Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:4568–4583 10.1091/mbc.E04-04-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabioiu A.M., Mitchison T.J. 2006. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 443:466–469 10.1038/nature05109 [DOI] [PubMed] [Google Scholar]
- Xie H., Surka M., Howard J., Trimble W.S. 1999. Characterization of the mammalian septin H5: distinct patterns of cytoskeletal and membrane association from other septin proteins. Cell Motil. Cytoskeleton. 43:52–62 [DOI] [PubMed] [Google Scholar]