A Rac–PAK1–Ajuba feedback loop stabilizes cadherin complexes via coordination of spatiotemporal signaling with actin remodeling at cell–cell contacts.
Abstract
Maintenance of stable E-cadherin–dependent adhesion is essential for epithelial function. The small GTPase Rac is activated by initial cadherin clustering, but the precise mechanisms underlying Rac-dependent junction stabilization are not well understood. Ajuba, a LIM domain protein, colocalizes with cadherins, yet Ajuba function at junctions is unknown. We show that, in Ajuba-depleted cells, Rac activation and actin accumulation at cadherin receptors was impaired, and junctions did not sustain mechanical stress. The Rac effector PAK1 was also transiently activated upon cell–cell adhesion and directly phosphorylated Ajuba (Thr172). Interestingly, similar to Ajuba depletion, blocking PAK1 activation perturbed junction maintenance and actin recruitment. Expression of phosphomimetic Ajuba rescued the effects of PAK1 inhibition. Ajuba bound directly to Rac·GDP or Rac·GTP, but phosphorylated Ajuba interacted preferentially with active Rac. Rather than facilitating Rac recruitment to junctions, Ajuba modulated Rac dynamics at contacts depending on its phosphorylation status. Thus, a Rac–PAK1–Ajuba feedback loop integrates spatiotemporal signaling with actin remodeling at cell–cell contacts and stabilizes preassembled cadherin complexes.
Introduction
In epithelia, biogenesis and maintenance of cell–cell adhesions is a highly organized process that influences cell morphology, initiates polarity, and supports tissue functions. Maintenance of cadherin-dependent junctions between neighboring cells is fundamental to ensure epithelial cell differentiation during morphogenesis and tissue homeostasis (Wirtz-Peitz and Zallen, 2009). Conversely, regulatory circuits that modulate junction dynamics can go awry during pathogen invasion, inflammation, epithelial–mesenchymal conversion, and tumor progression (Tanos and Rodriguez-Boulan, 2008). Understanding the mechanisms via which junctions are stabilized may provide insights into therapeutic strategies to maintain an epithelial phenotype.
Adhesive E-cadherin receptors provide a platform for assembly of macromolecular complexes containing cytoskeletal proteins, actin filaments, and signaling molecules (Braga and Yap, 2005). E-cadherin adhesion triggers specific actin remodeling that enables cell shape changes and stabilization of receptors at junctions (Braga, 2002; Braga and Yap, 2005; Zhang et al., 2005; Mège et al., 2006). Yet, the precise mechanisms leading to local actin reorganization at cell–cell contacts and the repertoire of regulatory proteins involved remain unclear.
A signaling pathway important for junction-dependent actin remodeling is triggered by the small GTPase Rac1 (referred as Rac hereafter), which coordinates cadherin–F-actin association at the plasma membrane. Rac mediates recruitment of actin to clustered cadherin complexes (Braga et al., 1997; Takaishi et al., 1997; Nakagawa et al., 2001; Lambert et al., 2002) and the maintenance of cadherins at mature cell–cell contact sites (Braga et al., 1999). Rac is activated by newly formed cell–cell adhesion sites (Nakagawa et al., 2001; Betson et al., 2002) and its local activation at contacting membranes triggers initiation, expansion, and consolidation of cell–cell adhesion (Yamada and Nelson, 2007). Force measurements reveal that the strength of cadherin-mediated contacts increases with time in an actin cytoskeleton–dependent manner and under the control of Rac (Lambert et al., 2002; Chu et al., 2004). However, how Rac activity is regulated at cadherin-dependent contacts or how it modulates epithelia-specific actin remodeling is not completely understood.
Ajuba is an actin-binding protein of the family of LIM domain proteins containing Ajuba, LIMD1, WTIP, Zyxin, LPP, and Trip6 (Kadrmas and Beckerle, 2004). Members of this family are characterized by two or three C-terminal LIM domains and an N-terminal PreLIM region. Ajuba localizes at focal adhesions, nucleus, and preferentially at cell–cell contacts in epithelial cells. Consistent with its localization at multiple sites, Ajuba is involved in several cellular processes such as cell fate determination (Kanungo et al., 2000; Nagai et al., 2010), repression of gene transcription (Ayyanathan et al., 2007; Hou et al., 2008, 2010a; Langer et al., 2008; Montoya-Durango et al., 2008), mitotic commitment (Hirota et al., 2003), cell–cell adhesion (Marie et al., 2003), and migration (Kisseleva et al., 2005; Pratt et al., 2005). Underlying these distinct functions is the ability of Ajuba to interact with signaling and scaffolding molecules such as PIPKIα (Kisseleva et al., 2005), Grb2 (Goyal et al., 1999), and 14-3-3 proteins (Hou et al., 2010b), and to modulate Wnt (Haraguchi et al., 2008) and Rac signaling (Pratt et al., 2005).
The regulation of Rac function by Ajuba is particularly interesting. In fibroblasts from Ajuba-null mice, wound healing is delayed due to reduced Rac activation at the leading edge, thereby interfering with forward movement (Pratt et al., 2005). Interestingly, Ajuba is not required for PDGF-dependent Rac activation, indicating the specificity of this process (Pratt et al., 2005). Taken together, there is the intriguing possibility that Ajuba can modulate Rac function in focal adhesions by providing spatiotemporal clues. An important point to address is whether the regulation of Rac activation by Ajuba is relevant for cell–cell junctions and associated actin reorganization.
Ajuba-null mouse keratinocytes display abnormal cell–cell junction formation and/or stability (Marie et al., 2003). Yet, the molecular mechanisms that underpin stabilization of junctions by Ajuba are far from elucidated. We envisage two potential mechanisms: a potential cross talk with Rac function, similar to what is described for fibroblast migration (Pratt et al., 2005), or a direct participation of Ajuba on actin remodeling at cell–cell contacts. These two mechanisms are not mutually exclusive and may cooperate with each other.
Although Ajuba is an actin-binding protein (Marie et al., 2003), the specific actin activities regulated by Ajuba to remodel the actin cytoskeleton are not currently known. Ajuba localizes at junctions via a direct interaction between its LIM domains and α-catenin, while the PreLIM domain binds directly to F-actin (Marie et al., 2003). Thus, Ajuba could stabilize junctions by remodeling F-actin at cell–cell contacts and/or by connecting cadherin complexes with the underlying cytoskeleton. An interesting point to test is whether Rac signaling modulates any of these two possibilities.
The fact that the functions of Ajuba and Rac may be entwined in cell–cell junctions defines a potential wide-spread mechanism for the dynamic regulation of adhesive sites by Rac (cell–substratum and cell–cell adhesion). In this paper, we set out to test the hypothesis that Ajuba participates in Rac activation at junctions and contributes to cytoskeletal reorganization necessary for cadherin adhesion. We find that Ajuba is not required for assembly, but rather for the maintenance of stable contacts. In the absence of Ajuba, Rac activation by cell–cell adhesion is perturbed and cadherin complexes are severely compromised in F-actin recruitment. We unravel the mechanisms underlying Ajuba function in Rac activation and identify a key role for the kinase PAK1, a known Rac effector, in this process.
Results
Ajuba is necessary for the maintenance of stable cell–cell adhesion
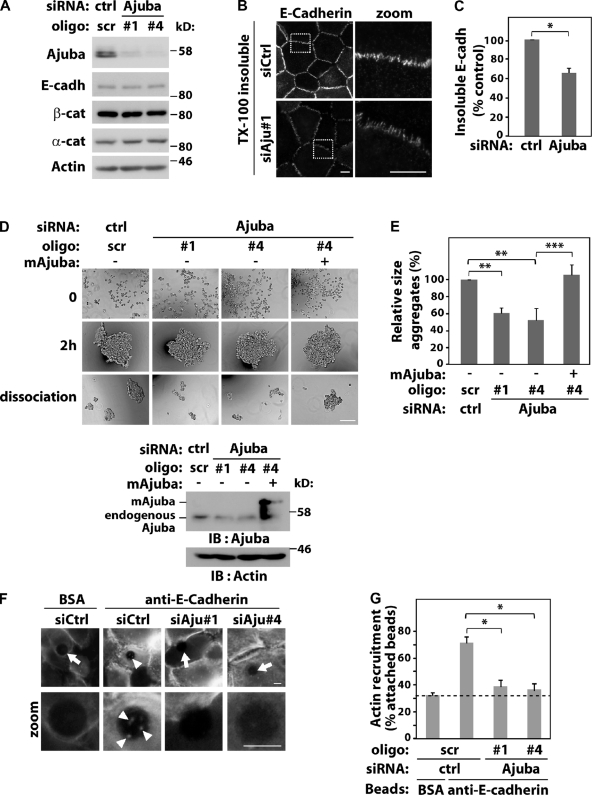
Human keratinocytes were transfected with different siRNA oligos targeting Ajuba (Fig. 1). Depletion of Ajuba did not affect the expression levels of junctional proteins such as E-cadherin, or α- and β-catenins (Fig. 1 A). No difference in E-cadherin levels at newly formed cell–cell contacts was observed in the absence of Ajuba by standard immunostaining (unpublished data). When keratinocytes were preextracted with detergent before fixation, an insoluble pool of E-cadherin was found in controls, as shown in our previous work (Braga et al., 1995a). Interestingly, a significant reduction on insoluble E-cadherin levels is observed after Ajuba RNAi (Fig. 1, B and C).
Figure 1.
Ajuba regulates junction maintenance. Keratinocytes were transfected with control scramble oligos (scr, ctrl) or Ajuba RNAi oligos with or without expression of siRNA-resistant Ajuba (mAjuba). (A) Lysates were analyzed by Western blotting with the antibodies indicated on the left. (B and C) Keratinocytes were preextracted with 0.5% Triton X-100–containing buffer, fixed, and stained for E-cadherin. The amount of E-cadherin insoluble pool at junctions was quantified and expressed relative to control (arbitrarily set as 100). (D and E) Aggregation of RNAi-treated cells was tested in hanging-drop suspension. Representative images are shown before (time 0), after addition of calcium ions for 2 h, and after moderate trituration (dissociation). Knock-down and expression of exogenous RNAi-resistant Ajuba were confirmed for each experiment. (E) Average size of all disaggregates after trituration was corrected for the size of each aggregate before trituration (2 h) and expressed relative to control samples (Scr., set as 100%). (F) siRNA-transfected cells were incubated with beads coated with BSA or anti–E-cadherin antibody. After washes, cells were fixed, stained for F-actin, and imaged. Arrows point at beads without F-actin accumulation; arrowheads show actin recruitment (zoom). (G) Quantification of data shown in F. Beads containing F-actin were visually scored and expressed as percentage of total attached beads in each sample. Dashed line represents nonspecific binding observed with control BSA-coated beads. Between 40 and 200 beads were counted for each group per experiment. Data represent mean and SD of three independent experiments (hereafter n = 3). Molecular weight markers are shown as kD (A and D). *, P < 0.03; **, P < 0.009; ***, P < 0.004. Bars: 10 µm or 200 µm.
To assess changes in cadherin adhesion quantitatively, we used aggregation assays (Thoreson et al., 2000; Ehrlich et al., 2002) in which the size of aggregates correlates with the number and strength of cell–cell contacts (Takeichi, 1977). Formation of junctions as well as their maintenance can be evaluated in this assay (resistance to shear stress upon trituration; Ehrlich et al., 2002). The size of aggregates before trituration (2 h) was not affected by Ajuba RNAi, suggesting that Ajuba did not regulate assembly of junctions. Compared with control cells, RNAi depletion of Ajuba approximately halved the size of aggregates after dissociation (Fig. 1, D and E). This effect was rescued significantly by overexpression of Ajuba mouse homologue that is siRNA resistant (mAjuba; Fig. 1, D and E). Our results indicate that Ajuba contributes to stabilization of preformed junctions.
A potential mechanism for junction stabilization is via F-actin recruitment to cadherin complexes. To determine whether Ajuba participates in this process, we used latex beads coated with an anti–E-cadherin antibody to cluster cadherins and trigger F-actin accumulation (Braga et al., 1997). β-Catenin is recruited to cadherin beads but not talin (Braga et al., 1997), indicating the specificity of this assay. Beads coated with BSA gave the nonspecific binding/recruitment levels (Fig. 1, F and G, dashed line). Ajuba depletion reduced significantly the proportion of anti-cadherin beads containing actin when compared with control oligos (Fig. 1 G). As cadherins are clustered by antibody-coated beads, we concluded that Ajuba participates in downstream events from cadherin receptors rather than clustering, by itself. Taken together, these data demonstrate that Ajuba regulation of F-actin recruitment to cadherin receptors (Fig. 1 G) may explain in part its stabilization of preformed cell–cell contacts (Fig. 1, B–E).
Ajuba interacts in vitro directly and specifically with Rac
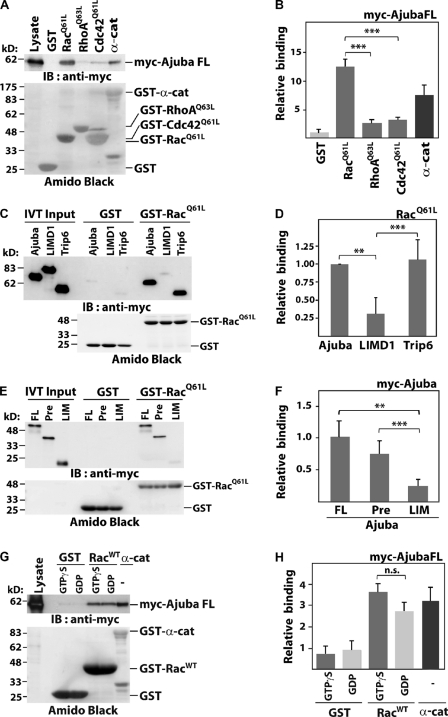
The participation of Ajuba in F-actin recruitment and maintenance of junctions is intriguing, considering that in keratinocytes Rac is necessary for these events (Braga et al., 1997). We investigated whether Rac and Ajuba can interact (Fig. 2). Interestingly, Ajuba bound to active Rac (RacQ61L), but not to active Rho (RhoQ63L) or Cdc42 (Cdc42Q61L; Fig. 2, A and B). Using in vitro–translated LIM domain–containing proteins, active Rac bound selectively to Ajuba and Trip6, but not to LIMD1 (Fig. 2, C and D; IVT). These results indicate selectivity of Rac interaction with some LIM proteins.
Figure 2.
Ajuba PreLIM interacts directly with Rac in vitro. GST-tagged small GTPases were incubated with (A and G) COS-7 lysates expressing full-length Ajuba, (C and E) in vitro translated Ajuba, or other LIM proteins. Immunoblot of myc-tagged constructs (IB: anti-myc) and GST-proteins as Amido black staining are shown (A, C, E, and G). α-Catenin was used as positive control (A and G). Quantification of experiments is shown as binding relative to GST (B and H) or to Ajuba full-length (D and F). (A and B) Activated forms of Rac (GST-RacQ61L), RhoA (GST-RhoAQ63L), and Cdc42 (GST-Cdc42 [GST-Cdc42Q61L]) were incubated with lysates expressing full-length Ajuba (myc-Ajuba FL). (C and D) Specificity of Ajuba interaction with active Rac. Ajuba, LIMD1, or Trip6 were allowed to interact with GST or GST-RacQ61L. (E and F) Full-length (FL) and Ajuba fragments PreLIM (Pre) or LIM were tested for binding to activated Rac. (G and H) Wild-type Rac (GST-RacWT) was loaded with GTPγS or GDP and incubated with lysates expressing myc-tagged full-length Ajuba. Histograms represent the mean and SD. n = 3. **, P < 0.009; ***, P < 0.005; n.s., nonsignificant.
Consistent with the above data, we mapped the Ajuba PreLIM region as the site of interaction with active Rac (Fig. 2, E and F; Fig. S1), which is the most divergent region of LIM proteins. To investigate whether Ajuba interaction depends on Rac activation, we quantified its binding to wild-type Rac (WT) loaded with GDP or GTPγS, a nonhydrolysable form (Fig. 2, G and H). No significant difference was observed in the interaction with Rac·GDP or Rac·GTPγS. Altogether, these results demonstrate that (a) Ajuba and Trip6 bind to Rac, (b) Ajuba binds to Rac independently of its activation status, and (c) Ajuba PreLIM region mediates a direct interaction with Rac.
Ajuba is required for Rac activation induced by cell–cell contacts
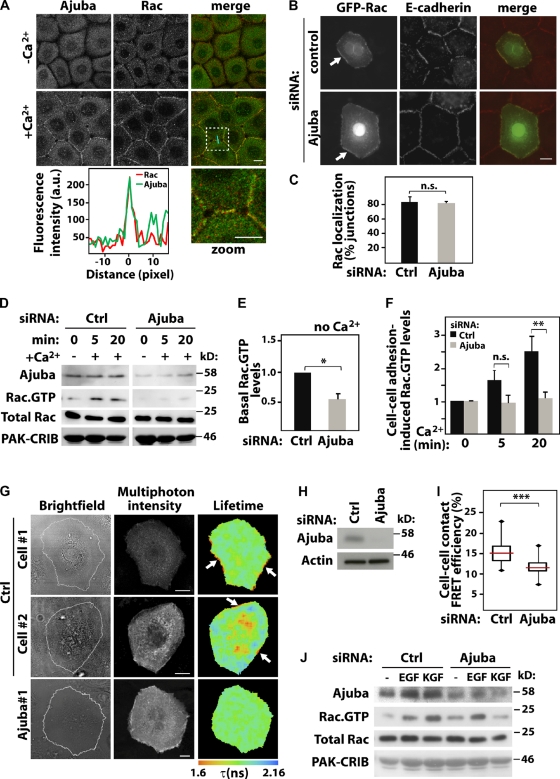
The fact that Rac interacts directly with Ajuba PreLIM region (Fig. 2, E and F) raises the question of whether Ajuba colocalizes with Rac at junctions. In the absence of cell–cell contacts, cells displayed a mainly diffuse localization of endogenous Rac and endogenous Ajuba (Fig. 3 A, −Ca2+). Ajuba and Rac clearly colocalized at newly formed junctions (+Ca2+). Time-lapse experiments showed that both proteins were concomitantly enriched as soon as 4 min after junction formation (Fig. S2). The above results suggest that Ajuba may facilitate Rac recruitment to junctions. However, Ajuba siRNA did not significantly perturb GFP-RacWT relocalization to cell–cell contacts (Fig. 3, B and C). These data indicate that Ajuba colocalizes with Rac at junctions, but Ajuba is not required for Rac recruitment to newly formed cell–cell contacts.
Figure 3.
Ajuba is required for Rac activation at cell–cell contacts. Keratinocytes grown in the absence of contacts (−Ca2+) were induced to form junctions by addition of calcium for 20 min (+Ca2+). Cells were stained, imaged, and localization of different constructs at junctions quantified. (A) Endogenous Ajuba and endogenous Rac relocalize upon junction assembly. Merged images and zoom of the boxed area are shown on the right. A line scan plot shows the fluorescence intensity of Rac and Ajuba at junctions. (B and C) Cells were treated with control (ctrl) or Ajuba RNAi oligo, transfected with GFP-RacWT, and stained for E-cadherin. Junctions containing GFP-RacWT were expressed as the percentage of total number of junctions in each sample. (D–I) Rac activity induced by cell–cell contact assembly was evaluated biochemically (D–F) or in situ (G–I). (D) Cells depleted of Ajuba were induced to assemble junctions for up to 20 min, and lysates used in pull-downs to determine Rac activation. Samples were probed with anti-Ajuba and anti-Rac antibodies to detect active Rac (Rac·GTP) and total levels of Rac in lysates (Total Rac). GST-PAK-CRIB is shown by Amido Black staining. Quantification of basal (without calcium, E) and junction-induced Rac·GTP levels (with calcium, F) are shown. Values are expressed relative to time-zero control in each group (scramble or Ajuba siRNA). (G) Cells depleted of Ajuba were transfected with the unimolecular biosensor mRFP1-Raichu-Rac-GFP. Multi-photon FLIM was used to image intramolecular FRET between GFP and mRFP1 upon Rac activation. Pseudocolored images show shortening of GFP fluorescence lifetime (τ, expressed in ns) as Rac activation. Two representative distributions of active Rac are shown for controls (cell #1 and #2). Arrows show Rac activation at junctions. (H) Representative blots showing Ajuba depletion. (I) FRET efficiency was measured at junctions (see Materials and methods) and shown as box plots (25th and 75th percentiles, medians as horizontal lines, and minimal/maximal values as vertical bars). About 100 contacts per condition across independent experiments were quantified. (J) Keratinocytes were stimulated with EGF or KGF and Rac activation assessed as described for D above. Graphs show the mean and standard error. n > 3. *, P < 0.04; **; P < 0.02; ***, P < 0.001. n.s., nonsignificant. Bars, 10 µm.
Although Ajuba and Rac may be recruited to junctions independently, Ajuba RNAi could impair cadherin-induced Rac activation in keratinocytes or behave as a scaffolding protein for Rac at cell–cell contacts. We tested the first possibility using GST-PAK-CRIB pull-down and FRET assays (Fig. 3, D–I). Basal levels of Rac·GTP in Ajuba-depleted cells were clearly decreased compared with control (Fig. 3 E). However, this effect is not related to junctions and could be an artifact due to disruption of the cytoskeleton (Nakamura et al., 2011). After junction assembly, Rac was activated in controls treated with scrambled oligos (Fig. 3 F). When expressed relative to basal levels (time-zero Ajuba RNAi) a significant difference in Rac activity in Ajuba-depleted cells was observed at 20 min, but not earlier (Fig. 3 F). We concluded that Ajuba is not essential for initial Rac activation by junctions, but contributes to its activation at later time points.
We reasoned that Ajuba may facilitate local Rac activation at junctions. To test this, cells were transfected with control or Ajuba siRNA oligos and the unimolecular FRET biosensor mRFP1-Raichu-Rac-GFP (Makrogianneli et al., 2009). Intramolecular FRET between GFP donor and mRFP1 acceptor occurs when Rac binds to GTP. This is detectable by a shortening of the donor GFP fluorescence lifetime (τ) as measured by multi-photon FLIM (Fig. 3, G–I). After 20 min of junction assembly, Rac activation was particularly visible at cell–cell contacts by FRET in control keratinocytes (cell #1 and cell #2). We also observed a concomitant cytoplasmic Rac activation in about half of the cells analyzed (i.e., cell #2). We selectively measured FRET efficiency at junctions (∼100 junctions per sample; see Materials and methods) and observed a significant decrease of FRET efficiency in Ajuba-depleted cells (Fig. 3 I). Thus, biochemical and in situ analyses demonstrate that Ajuba is necessary for appropriate levels of Rac activation by junctions. Importantly, this regulation is specific, as Ajuba is not required for Rac activation after EGF (Fig. 3 J) or PDGF stimulation (Pratt et al., 2005). However, KGF-dependent activation of Rac was reduced in the absence of Ajuba, indicating selectivity for growth factor signaling.
PAK1 inhibition phenocopies depletion of Ajuba
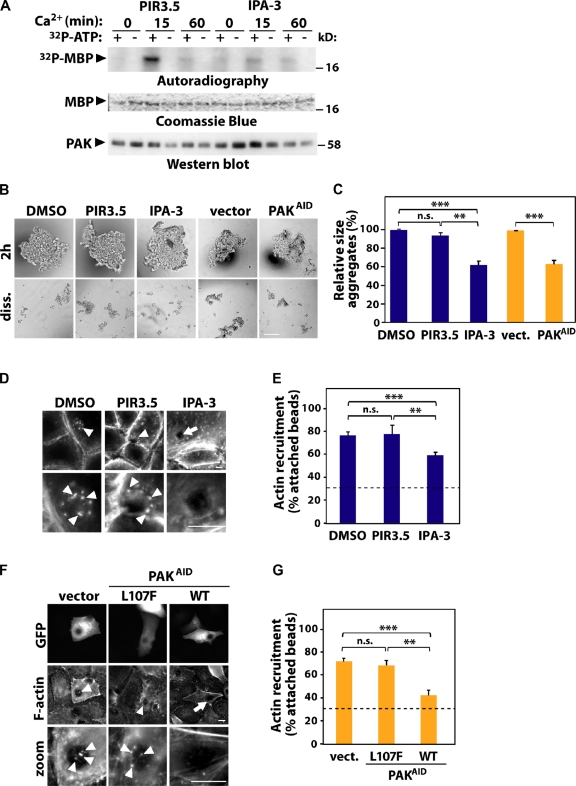
PAK1 is a serine-threonine kinase Rac effector that has different cellular functions, including actin cytoskeleton rearrangements (Bokoch, 2003). PAK1 localizes at junctions and has been implicated in HGF-dependent junction destabilization (Zegers et al., 2003). PAK1 is the only member of the Group I PAKs (PAKs 1–3) expressed in cultured keratinocytes (Lozano et al., 2008). We show here that PAK1 is transiently activated by junction assembly in controls treated with the inactive compound PIR-3.5 (Fig. 4 A; Flaiz et al., 2009). Interestingly, this effect requires PAK1 auto-activation as it is prevented after treatment with IPA3, a specific inhibitor of Group I PAKs (Fig. 4 A; Deacon et al., 2008).
Figure 4.
PAK contributes to junction stabilization and F-actin recruitment. (A–E) PAK activity was inhibited in keratinocytes by treatment with IPA-3; treatment with DMSO or the inactive compound PIR-3.5 was used as control. (B, C, F, and G) Alternatively, endogenous PAK1 was blocked by expression of the auto-inhibitory domain (PAKAID). As controls, empty vector or PAKAID mutated to abolish interaction with PAK1 was used (L107F). (A) Cells were induced to form new junctions, endogenous PAK was immunoprecipitated and subjected to in vitro kinase assay using MBP as substrate and [32P]ATP (+/−). Levels of PAK1 are shown by Western blot and MBP by Coomassie blue. (B) Cells were allowed to aggregate for 2 h and mildly dissociated by pipetting (diss.). (C) Relative size of remaining aggregates was calculated and expressed as percentage of their control as described in Fig. 1. (D–G) F-actin recruitment to beads coated with E-cadherin antibody was tested. Arrow points at attached beads without F-actin; arrowheads show F-actin recruitment. (E and G) The percentage of attached beads containing F-actin was quantified as described in Fig. 1. Dashed line represents baseline of control BSA-coated beads. n = 3. *, P < 0.003; **, P < 0.009; ***, P < 0.0002. n.s., nonsignificant. Bars: (B) 200 µm, (D and F) 10 µm.
To address whether PAK1 is required for junction stabilization, aggregation was performed using cells incubated with IPA3, PIR3.5, or transfected with PAK1 auto-inhibitory domain (PAKAID; Fig. 4, B and C; Zhao et al., 1998). Interfering with PAK1 activity did not perturb formation of aggregates (Fig. 4 B; 2 h), consistent with our previous work on PAK depletion and junction assembly (Lozano et al., 2008). Instead, prevention of PAK activation increased aggregate size slightly (unpublished data). Interestingly, after trituration, we found that the sizes of the dissociated aggregates (diss.; normalized to 2 h initial aggregate size) were notably reduced upon PAK1 inhibition. The latter was significantly different when compared with controls treated with DMSO, PIR-3.5, or empty vector (Fig. 4 C). This result suggests an involvement of PAK1 in junction maintenance rather than assembly.
Furthermore, inhibition of PAK1 using IPA-3 significantly decreased the proportion of anti-cadherin beads able to recruit F-actin compared with control (Fig. 4, D and E). Similar results were seen after expression of PAK-AID, but not a mutant PAK-AIDL107F that does not interact with PAK kinase domain (Fig. 4, F and G; Arias-Romero et al., 2010). Both effects (reduced aggregate size upon dissociation and perturbed actin recruitment) were not as strong as observed after Ajuba RNAi (Fig. 1). We concluded that PAK1 inhibition partially phenocopies Ajuba depletion (Fig. 1) and Rac1 inhibition in keratinocytes (Braga et al., 1997).
PAK1 phosphorylates Ajuba
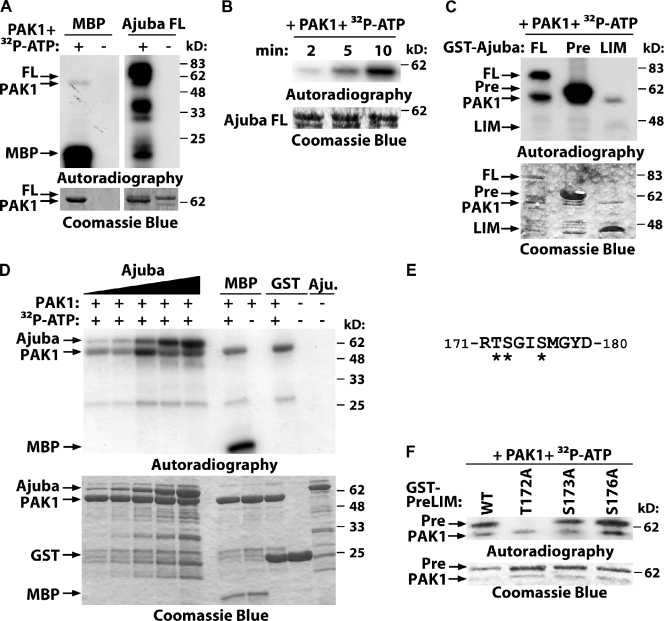
Previous studies indicate that Ajuba may be phosphorylated (Daub et al., 2008; Haraguchi et al., 2008; Oppermann et al., 2009). However, no kinase or the functional consequences of Ajuba phosphorylation was identified. Due to the similarities of Ajuba depletion and PAK1 inhibition on cell–cell adhesion, we addressed whether Ajuba is a substrate of PAK1. In vitro phosphorylation assays using purified proteins showed that full-length Ajuba can be readily phosphorylated by PAK1 (Fig. 5 A). Phosphorylation occurs at the PreLIM region in a time- and concentration-dependent manner (Fig. 5, B–D). Quantification of the relative levels of 32P incorporation demonstrated that Ajuba phosphorylation is readily saturated at 30 pmol after 5 min incubation (Fig. 5 D; Fig. S4, D–F). Relative levels of [32P]Ajuba were comparable to PAK1 auto-phosphorylation or [32P]MBP (positive control; Fig. S4, D–F), suggesting that Ajuba is efficiently phosphorylated by PAK1.
Figure 5.
PAK phosphorylates Ajuba. (A–C) Purified full-length Ajuba (FL) and the truncation mutants PreLIM (Pre) or LIM (LIM) were incubated in vitro with PAK1 kinase domain with or without radioactive ATP (32P-ATP). Phosphorylated bands are shown by autoradiography and fusion proteins by Coomassie blue staining. (A) Full-length Ajuba is phosphorylated by PAK1; MBP was used as positive control; PAK1 auto-phosphorylates itself. (B) Time course of Ajuba phosphorylation. (C) Mapping of Ajuba-phosphorylated region. (D) Different concentrations of Ajuba PreLIM (7.5, 15, 30, 60, and 120 pmol), GST, or MBP were incubated with PAK kinase domain (80 pmol) in an in vitro kinase assay for 5 min. (E) Phosphopeptide isolated after Ajuba phosphorylation by PAK1 using mass spectrometry. Three putative phosphorylation sites are shown (asterisks). (F) PAK1 phosphorylates Ajuba at residue 172. Different alanine mutations were prepared in Ajuba PreLIM region (T172A, S172A, or S176A) and GST fusion proteins phosphorylated in vitro by PAK1.
PAK1 is a promiscuous kinase that phosphorylates serine and threonine residues, but consensus sequences and preferred flanking residues are known (Fig. S4 A; Miller et al., 2008). In vitro phosphorylation of the PreLIM region followed by mass spectrometry identified a single phosphopeptide with three putative PAK1 phosphorylation site(s) (Fig. 5 E). Mutations of these different amino acids to alanine showed that PAK1 phosphorylates Ajuba at a single site, threonine 172 of mouse Ajuba (human residue 161; Fig. 5 F). This motif is not picked up by online searches for kinase substrates, as it does not fit precisely the consensus motif RRxS/T (conserved arginine at position −1 in Ajuba protein rather than −2; Miller et al., 2008).
Ajuba bundles actin filaments independently of its phosphorylation status
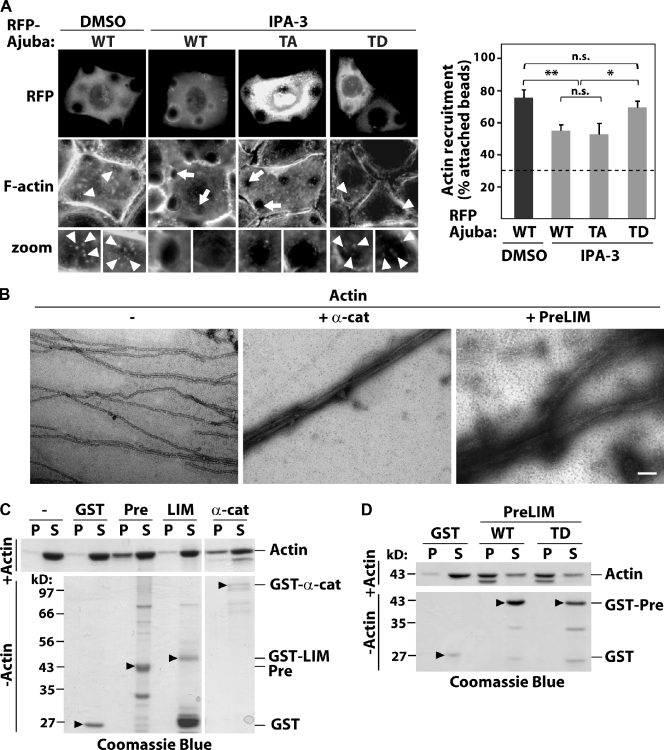
As the PreLIM region interacts with F-actin and Ajuba depletion reduces F-actin recruitment to cadherins (Fig. 1 G), we envisaged that phosphorylation at T172 may modulate actin reorganization downstream of PAK1. We tested whether PAK1 inhibition of F-actin recruitment to cadherins is rescued by a phosphomimetic mutant of Ajuba (AjubaT172D). Blocking PAK1 activation by IPA-3 reduced the percentage of cadherin beads containing F-actin on cells expressing wild-type Ajuba (Fig. 6, A and B), similar to what was observed for endogenous Ajuba (Fig. 4 D). Expression of a nonphosphorylatable Ajuba (AjubaT172A, TA) exhibited a comparable effect. Interestingly, expression of AjubaT172D (TD) significantly rescued PAK1 inhibition and reversed the decrease in the number of beads labeled with F-actin (Fig. 6 B). Although PAK1 inhibition did not prevent actin recruitment completely, these data indicate that phosphorylation of threonine 172 on Ajuba contributes to the regulation of actin remodeling triggered by cadherin clustering.
Figure 6.
Ajuba bundles F-actin independently of phosphorylation. (A) Cells expressing RFP-tagged Ajuba (WT), Ajuba T172A (TA), or T172D (TD) were treated with DMSO or IPA-3 to inhibit PAK1 for 10 min. F-actin recruitment to anti-cadherin–coated beads was detected by phalloidin staining. Arrowheads show actin-recruited beads; arrows point beads with no F-actin accumulation. (B) Quantification of the percentage of attached beads containing F-actin and attached to expressing cells. Dashed line shows baseline (control BSA-coated beads). (C) Negative staining electron micrographs of actin filaments alone (−, 5 µM) or mixed with α-catenin as positive control (+ α-cat, 1.8 µM) or Ajuba PreLIM (+ PreLIM, 2 µM). (D and E) Low speed cosedimentation assays. Indicated GST fusion proteins were incubated with polymerized actin in vitro, centrifuged to separate bundles (pellet, P) from short filaments (supernatant, S), resolved on SDS-PAGE, and stained with Coomassie blue. Actin (−) and GST alone were used as negative controls, α-catenin as positive control. As additional control, fusion proteins were processed as above but without actin (−Actin; arrowheads on bottom panels). n = 3. *, P < 0.025; **, P < 0.01. n.s., nonsignificant. Bars: (A) 20 µm; (C) 100 nm.
It is unclear which actin reorganization process Ajuba regulates to stabilize junctions. Using electron microscopy (Fig. 6 C), we observed that Ajuba PreLIM region induced the formation of thick actin bundles. The latter were less compact and straight than bundles induced by α-catenin (Rimm et al., 1995). Low speed actin sedimentation assays confirmed these results (Fig. 6 D), as addition of Ajuba PreLIM region to preassembled actin filaments enhanced the sedimentation of actin bundles and long filaments (pellet fraction, P; Fig. 6 D) compared with GST (Fig. 6 D). Ajuba LIM domains do not bind F-actin (Marie et al., 2003) and behaved as expected (i.e., no bundling activity; Fig. 6 D).
To address whether F-actin bundling is regulated by Ajuba phosphorylation, Ajuba mutants were tested. No major difference was detected between the ability of wild-type PreLIM (WT) or its phosphomimetic counterpart (TD) to bundle filaments (Fig. 6 E). As controls, none of the proteins used precipitated by themselves in the absence of F-actin (Fig. 6, D and E; bottom panels). Collectively, these complementary data reveal that the PreLIM region of Ajuba bundles actin filaments and that this function is not regulated by phosphorylation. The above results suggest that alternative mechanisms should be in place to explain the role of PAK>Ajuba in junction stabilization.
Phosphorylation of Ajuba enhances its binding to active Rac
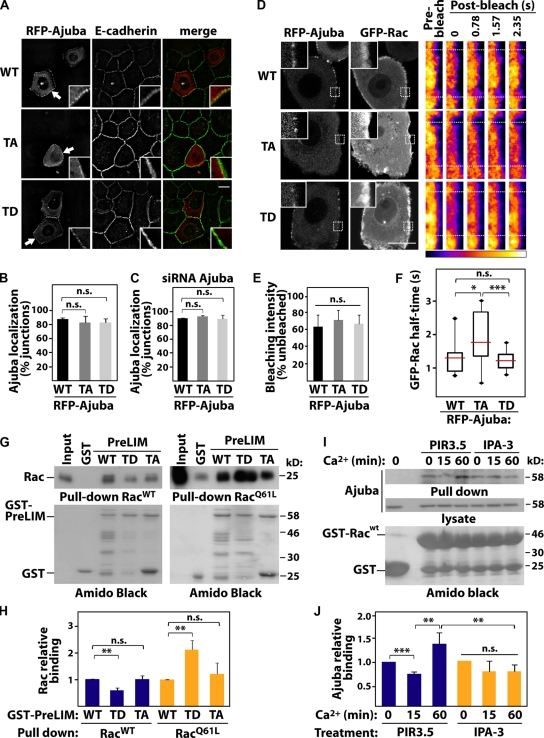
In the context of epithelial junctions, we envisioned two other processes regulated by phosphorylation: Ajuba localization at cell–cell contacts or its interaction with Rac. To address the former, different RFP-Ajuba mutants were expressed and junctions induced for 20 min (Fig. 7, A–C). No difference in junctional distribution of full-length proteins was observed among AjubaWT (WT), AjubaT172A (TA), or AjubaT172D (TD, Fig. 7, A and B). Ajuba is able to dimerize (Fig. S3) and this could interfere with the distribution of exogenous proteins. However, similar localization of mutants was obtained when transfected in Ajuba-depleted cells (Fig. 7 C). Thus, Ajuba phosphorylation at the PreLIM region does not regulate its localization at cell–cell contacts. These results are in accordance with the requirement of the LIM domains for localization at junctions (Marie et al., 2003).
Figure 7.
PAK phosphorylation increases binding of Ajuba to active Rac. (A) Keratinocytes transfected with full-length RFP-Ajuba (wild-type, WT), nonphosphorylatable (T172A, TA), or phosphomimetic (T172D, TD) mutants were induced to form junctions, fixed, and stained for E-cadherin. Arrows point at Ajuba at junctions (boxed magnifications). (B and C) Quantification of exogenous Ajuba localization in normal (B) and Ajuba-depleted keratinocytes (C). Values are expressed as percentage of junctions containing Ajuba WT. (D–F) Keratinocytes expressing GFP-RacWT and RFP-Ajuba (WT, TA, or TD) were induced to form junctions for 15–20 min (left) and subjected to fluorescence recovery after photobleaching (FRAP). (D) Pseudocolored fluorescence intensity of GFP-Rac at cell–cell contact before and after bleaching is shown on right panels. Dotted boxes indicate bleached region. (E) Quantification of GFP-RacWT bleaching efficiency at time zero (average of at least 11 cells per sample). (F) Quantification of GFP-Rac after bleaching expressed according to half-time of fluorescence recovery. The numbers of junctions quantified were: RFP-Ajuba WT (15), TA (12), TD (18). (G and H) GST pull-down was performed using immobilized GST, GST-PreLIM WT, TD, or TA mutants and RacWT or active Rac (RacQ61L). Bound Rac and GST fusion proteins were detected by immunoblotting (IB) and Amido Black staining, respectively. (H) Relative Rac binding with indicated proteins was quantified and normalized to levels of GST-PreLIM WT. (I and J) Cells were pretreated with IPA-3 to inhibit PAK1 or PIR-3.5 as control. After junction assembly, the ability of endogenous Ajuba to bind to GST-RacWT or GST was evaluated with pull-downs. Precipitated proteins and lysates were probed with anti-Ajuba antibodies. (J) Quantification of the above experiments. Error bars represent SEM (n = 3). *, P < 0.03; **, P < 0.02; ***, P < 0.003. n.s., nonsignificant. Bars, 20 µm.
It is feasible that Ajuba interaction with Rac at junctions may be modulated by phosphorylation of the PreLIM region (Fig. 5), which also binds to Rac (Fig. 2, G and H). Using FRAP, we investigated Rac dynamics at newly formed junctions (Fig. 7, D–F). GFP-RacWT was used, as expression of active Rac can destabilize cell–cell adhesion in keratinocytes (Braga et al., 2000). The majority of RacWT is found GDP bound, as only a small proportion is activated upon a stimulus (∼1%; Ren et al., 1999). Therefore, the readout of FRAP experiments reflects the dynamics of Rac·GDP, as Rac·GTP levels induced by junction assembly are not detected under these conditions. The bleaching efficiency was equivalent among samples and across different experiments (60–70%, Fig. 7 E). When coexpressed with AjubaWT or phosphomimetic AjubaT172D (TD), RacWT had a fast recovery time (Fig. 7 F). However, expression of AjubaT172A (TA) led to a significant increase in the half-life of RacWT at junctions. These data indicate that replacement of the pool of Rac·GDP at junctions is slower in the presence of AjubaT172A (Fig. 7 F).
These results could be explained if phosphorylation at T172 (or lack of) affects the ability of Ajuba to interact with Rac and thus interfere with the release or retention of Rac at cell–cell contacts. Using purified proteins and in vitro phosphorylation, phosphorylated Ajuba PreLIM was able to pull down 50% more activated Rac than controls (Fig. S4, B and C). These results were confirmed by a preferential interaction of AjubaT172A to RacWT, as a read-out for Rac·GDP (Fig. 7, G and H). Conversely, AjubaT172D (TD) was able to pull down twofold more active Rac than AjubaT172A or AjubaWT (WT, RacQ61L; Fig. 7, G and H).
In vivo, we predict that new junction assembly activates Rac (Fig. 3), triggers PAK auto-activation (Fig. 4), and increases Rac binding to Ajuba in a PAK-dependent manner. To test this prediction, junctions were induced in the presence of IPA3 or PIR3.5 and pull-downs were performed using GST-RacWT that mimics inactive Rac (Fig. 7, I and J). RacWT was used to be consistent with FRAP experiments and avoid competition with effectors for binding to active Rac. Endogenous Ajuba interacted with RacWT at steady state (Fig. 7, I and J; time zero) because of its ability to bind to Rac·GDP. When junctions are induced for 15 min, Ajuba binding levels are reduced, consistent with the transient peak of PAK activation (Fig. 4 A) and decreased affinity of phosphorylated Ajuba to Rac·GDP (Fig. 7, G and H). Conversely, Ajuba-RacWT interaction is increased after 60 min, which correlates with PAK1 inactivation (Fig. 4 A). These effects are abrogated when PAK1 is inhibited by IPA3. Taken together, these results reveal that, rather than perturbing Ajuba localization at junctions, phosphorylation by PAK1 increases the affinity of endogenous Ajuba for active Rac.
Discussion
Rac activation at cadherin adhesive sites induces membrane expansion during contact assembly, actin recruitment, or trafficking of cadherins to and from junctions (Nelson, 2008; Delva and Kowalczyk, 2009). We show that Ajuba fine-tunes Rac activation at junctions, thereby contributing to cell–cell adhesion maintenance. In addition, PAK1, a serine-threonine kinase Rac effector (Bokoch, 2003), phosphorylates Ajuba. We unravel a novel PAK1>Ajuba cross talk that modulates actin reorganization and Rac dynamics, leading to junction stabilization.
Ajuba depletion does not prevent junction assembly, but rather the maintenance of preassembled cell–cell contacts. Our data point to differences in the molecular regulation of formation versus stabilization of junctions, which is poorly understood (Braga et al., 1999). Clearly, Ajuba has an auxiliary role as revealed when junctions are stressed, i.e., upon trituration. Yet, Ajuba may be important to other physiological events in which junctions are challenged by mechanical stress such as cytokinesis. Other LIM proteins (LPP and Zyxin) also localize at cell–cell and focal adhesions (Reinhard et al., 1995; Drees et al., 1999). However, they bind indirectly to F-actin and appear to reduce contacts by perturbing VASP function (Hansen and Beckerle, 2006).
The mechanisms underlying F-actin reorganization at cadherin complexes are largely unexplored. Recent evidence suggests that actin polymerization cannot be the sole contributor to the F-actin pool at junctions. Theoretically, additional mechanisms could involve binding/bundling of preassembled filaments and/or cross-linking with underlying cortical cytoskeleton. That additional actin remodeling events occur is supported by Mège et al. (2006): (a) regulators of capping and linear filaments stabilize cadherin adhesion, (b) actin cross-linking and bundling proteins are found at junctions, and (c) α-catenin inhibits actin polymerization (Drees et al., 2005). We show that Ajuba is able to bundle F-actin, which could accumulate linear actin filaments at cadherins.
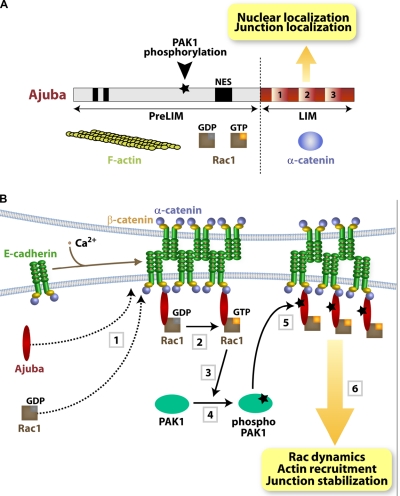
Ajuba may contribute to the interaction of cadherin complexes with the cortical cytoskeleton: it binds directly to α-catenin via the LIM domains and F-actin via the PreLIM region (Fig. 8 A; Marie et al., 2003). After Ajuba RNAi, the reduced pools of insoluble E-cadherin and the strong inhibition of actin recruitment suggest that cell–cell contact stabilization by Ajuba may be explained by actin remodeling. Yet, our data indicate that this is not the full explanation. Three lines of evidence suggest that Ajuba and Rac signaling are entwined. First, inhibiting PAK1 activation leads to smaller aggregates and reduced actin recruitment to clustered cadherins, similar to Ajuba depletion (this paper) or Rac inhibition (Braga et al., 1997; Ehrlich et al., 2002; Lambert et al., 2002).
Figure 8.
Summary diagrams. (A) Ajuba domain organization: PreLIM region, three LIM domains, nuclear export signal (NES), and PAK1 phosphorylation site. Protein partners are shown underneath each region. (B) Summary of the data presented. Upon cell–cell contact assembly, Ajuba and Rac are recruited independently to junctions (1). Ajuba interacts with both active (Rac·GTP) and inactive Rac (Rac·GDP) (2). Upon Rac activation by junctions, PAK1 is activated by auto-phosphorylation (3 and 4). Active PAK phosphorylates Ajuba, which increases its affinity to active Rac. Thus, active Rac is stabilized at cell–cell adhesion sites depending on Ajuba phosphorylation levels, thereby fine-tuning locally Rac activation and dynamics (5). The outcome is sustained remodeling of actin filaments and junction stabilization (6). In the absence of Ajuba, Rac can still be recruited and activated by junction assembly, but this is not sufficient to ensure resistance to mechanical stress.
PAK1 is a serine-threonine kinase that regulates a variety of different processes (Bokoch, 2003). PAK1 localizes at junctions in epithelial cells (Zegers et al., 2003) and has an emerging role in tumor progression, epithelial morphogenesis, and cell–cell contact inhibition of motility (Bokoch, 2003; Zegers et al., 2003). Interestingly, PAK1 is transiently activated by junction assembly and specifically phosphorylates Ajuba (Fig. 8 A). PAK1 is known to phosphorylate other proteins (Bokoch, 2003) that have been shown to stabilize cell–cell contacts (Ivanov et al., 2007; Smutny et al., 2010). Yet, expression of Ajuba phosphomimetic mutant (AjubaT172D) rescues actin recruitment to cadherins after PAK1 inhibition. Thus, Ajuba appears to be the main PAK1 substrate that regulates actin remodeling at cadherins.
Second, Rac requires Ajuba to maintain its activation at junctions. The fact that the initial Rac activation (5 min) is not disrupted is consistent with (a) the role of Ajuba in junction maintenance rather than assembly and (b) a prediction that Rac may be activated by multiple mechanisms upon junction assembly (Braga and Yap, 2005). Interestingly, the interplay between Ajuba and Rac is specific for cell–cell contact formation: Ajuba depletion does not impair Rac activation after stimulation with EGF (this work) or PDGF (Pratt et al., 2005).
Third, Ajuba binds to Rac, but not RhoA or Cdc42. Similar to the interaction of Rac with PRK2 and PIP-5K (Vincent and Settleman, 1997; Tolias et al., 2000), Ajuba binds to both Rac·GDP and Rac·GTP (Fig. 8 A), indicating that Ajuba is not a Rac effector. The fact that Ajuba modulates Rac activity in vivo suggests a unique role of Ajuba in integrating spatiotemporal Rac signaling with cytoskeletal remodeling at junctions.
We envisage three mechanisms for how Ajuba regulates junction-induced Rac activation: Ajuba may facilitate Rac recruitment to cell–cell contacts, recruit an exchange factor to activate Rac locally, or prevent Rac inactivation at junctions (Vigil et al., 2010). However, Rac localizes at junctions independently of Ajuba and thus Ajuba regulates the activity levels of a Rac pool already found at sites of adhesion (Fig. 8 B). At present, we cannot formally exclude that Ajuba may recruit a Rac exchange factor to cell–cell contacts. Instead, our data suggest that Ajuba maintains active Rac at junctions, either as a scaffold protein or by preventing Rac inactivation by a GAP protein (i.e., via steric hindrance).
We hypothesize that cadherin-dependent Rac signaling triggers PAK1 activation and Ajuba phosphorylation, leading to a dynamic regulation of active Rac at junctions (Fig. 8 B). Our data strongly support our hypothesis: Ajuba localization at cell–cell contacts occurs independently of its phosphorylation status and phosphorylated Ajuba interacts preferentially with Rac·GTP. The outcome of the change in affinity is the local regulation of Rac activity at junctions. In vivo, expression of AjubaT172A stabilizes inactive Rac at cell–cell contacts, which results in slower recovery fluorescence time of Rac·GDP in FRAP experiments. In support of our findings, junction assembly transiently reduces binding of endogenous Ajuba to RacWT, a process that requires PAK1 activity and correlates with the profile of PAK1 activation by junctions.
Serine-threonine phosphorylation of small GTPases (Ozdamar et al., 2005; Riento et al., 2005; Rolli-Derkinderen et al., 2010) or their upstream regulators (DerMardirossian et al., 2004; Vigil et al., 2010) is a well-established mechanism to modulate GTPase function. Although other cytoskeletal proteins such as ERM have been shown to regulate GTPase activity, the latter occurs indirectly via modulation of Rho regulators (Fehon et al., 2010). In contrast, Ajuba interacts directly with Rac and phosphorylated Ajuba helps to maintain transient levels of active Rac at junctions. This mechanism resembles the change in specificity of RhoGDI by phosphorylation, resulting in lower affinity for Rac (DerMardirossian et al., 2004). Junction assembly and relocalization of Ajuba could provide a feedback loop and quick switch for retention of Rac·GTP or Rac·GDP at adhesive sites, depending on Ajuba phosphorylation (Fig. 8 B). Therefore, fine-tuning of Rac activation after junction assembly is achieved.
Our data are consistent with a wide-spread interplay between signaling and cytoskeletal proteins to ensure spatiotemporal coordination of Rac function in the stabilization of junctions. As similar regulation occurs during wound healing (Pratt et al., 2005), our results highlight the broad implications of Ajuba function for Rac signaling at adhesive events. It will be interesting to address whether Ajuba and PAK1 can also cooperate in cellular processes at other sites where both proteins localize (Kanungo et al., 2000; Hirota et al., 2003; Hou et al., 2010a).
Aberrant expression of PAK1 and Rac are critical events during tumor invasion (Lozano et al., 2003; Kumar et al., 2006). Although a putative role of Ajuba during tumorigenesis has not been investigated, Ajuba regulates proliferation and cell fate specification (Kanungo et al., 2000; Ayyanathan et al., 2007), similar to its binding partners Rac and α-catenin (Vasioukhin et al., 2001). We argue that Ajuba helps to maintain an epithelial phenotype due to its role on F-actin bundling and channeling Rac function to junctions. Furthermore, Ajuba may be an important component in the cross talk between cell–cell adhesion (this paper) and migration (Pratt et al., 2005). When junctions are disrupted during tumor progression, we speculate that de-regulated Ajuba localization may influence the level and specificity of Rac signaling pathways and contribute to epithelial-to-mesenchymal transition (Ayyanathan et al., 2007; Hou et al., 2008; Langer et al., 2008). Thus, our results have considerable implications to the regulation of epithelial homeostasis and tumorigenesis at the molecular, cellular, and tissue levels.
Materials and methods
Cell treatment, RNAi, and transfections
Normal human keratinocytes from neonatal foreskin (strain SF, passages 3–6) were routinely grown on a mitomycin C–treated monolayer of 3T3 fibroblasts at 37°C and 5% CO2 in standard medium (containing 1.8 mM CaCl2; Rheinwald and Green, 1975). Cells were switched to low calcium medium (0.1 mM CaCl2) when small colonies were visible and grown until confluent (Hodivala and Watt, 1994). For induction of cell–cell contacts, 1.8 mM CaCl2 was added for different periods of times. For Triton X-100 insolubility of cadherin receptors, cells were preextracted with a CSK buffer (10 mM Pipes, pH 6.8, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 0.5% Triton X-100) for 10 min at room temperature before fixation as described previously (Braga et al., 1995b).
Keratinocytes were preincubated with 20 µM IPA-3 (I2285; Sigma-Aldrich) to inhibit PAK activity, as controls the inactive PIR-3.5 compound (TCS9528; TCris) or an equivalent volume of DMSO for 10 min. Human keratinocytes were transfected with cDNA or siRNA oligos with TransIT-keratinocyte (MirusBio; Cambridge Biosciences) or RNAiFect (QIAGEN), respectively. Following manufacturer’s instructions, redesigned siRNA oligos were bought from Thermo Fisher Scientific (Ajuba D-021473-01 and D-021473-04 or control scrambled D-001206-13). Depletion of proteins was monitored for each experiment and in case depletion was less than 70%, experiments were excluded.
Aggregation assays were performed as described previously (Thoreson et al., 2000) with minor modifications. In brief, confluent keratinocytes in low calcium medium were detached from culture dishes using 0.1% trypsin in Versene with 1 mM CaCl2 to prevent E-cadherin degradation. Cells in suspension (5 × 104/ml) were incubated for 2 h in standard calcium medium as 20-µl hanging drops. Cells were dissociated by pipetting 10 times through a 200-µl tip.
COS-7 cells were grown in DME medium (Sigma-Aldrich) supplemented with 10% fetal calf serum (Serum Laboratories Ltd). COS-7 cells were transfected by Lipofectamine (Invitrogen) according to the manufacturer’s recommendations. Transfected COS-7 cells were lysed in lysis buffer (10 mM Tris-HCl, pH 7.5, 1% NP-40, 150 mM NaCl, 1 mM MgCl2, 50 mM NaF, 1 mM DTT, 5 µM leupeptin, and 1 mM PMSF) and centrifuged at 8,800 g for 10 min at 4°C. Pellet was discarded and supernatant used in different assays.
Constructs, cloning, and mutagenesis
Auto-inhibitory domain of PAK (PAKAID) in pRK5myc, PAK kinase domain in pGEX-2T (GST-PAKKD), and the activated mutants RacQ61L, Cdc42Q61L, and RhoQ63L (pRK5myc and/or pGEX-2T vectors) were gifts from A. Hall (Sloan Kettering Institute, New York, NY). pEGFC1 containing the PAK1 auto-inhibitory domain (PAKAID, amino acids 83–149) or mutated to prevent binding to endogenous PAK1 (PAKAID L107F; Arias-Romero et al., 2010) were provided by J. Chernoff (Fox Chase Center, Philadelphia, PA). pGEX2T-PAK-CRIB was a gift from J. Collard (Netherlands Cancer Institute, Amsterdam, The Netherlands). Myc-tagged Ajuba full-length (Kanungo et al., 2000), pGEX-2T-full-length Ajuba or truncation mutants containing PreLIM or LIM regions (Goyal et al., 1999), and mRFP1-full-length Ajuba (RFP-Ajuba; Pratt et al., 2005) were used. GST–α-catenin was provided by D. Rimm (Yale University, New Haven, CT). pEGFP-Rac1 wild-type was provided by M. Cebecauer (Imperial College London, London, UK). mRFP1-Raichu-Rac-GFP FRET plasmid containing Rac1 CAAX box (Makrogianneli et al., 2009) was provided by T. Ng (King’s College London, London, UK).
Site-directed mutagenesis was performed using the QuikChange II kit (Agilent Technologies) according to the manufacturer’s recommendations to obtain the mutations T172D or T172A in RFP-Ajuba (full length) or GST-PreLIM mutants T172A, T172D, S173A, and S176A. Mutations were confirmed by sequencing.
Antibodies and immunostaining
The following primary mouse monoclonal antibodies were used against: E-cadherin (HECD-1; Cancer Research UK, London), Myc (9E10; Cancer Research UK), actin (C4; MP Biomedicals), and Rac (23A8; Millipore). Rabbit polyclonal antiserum used were anti–α-catenin (VB1), anti–β-catenin (VB2; Braga et al., 1995a), anti-PAK1 (sc-882; Santa Cruz Biotechnology, Inc.), and affinity-purified anti-Ajuba (4897; Cell Signaling Technology and 9104). Alexa Fluor 488–Phalloidin (Invitrogen) and Fluorophore-conjugated antibodies (Jackson ImmunoResearch Laboratories, Inc.) and horseradish peroxidase–coupled antibodies (Dako) were purchased.
Immunofluorescence was performed essentially as described previously (Braga et al., 1997). In brief, cells were fixed in 3% paraformaldehyde for 10 min at RT and permeabilized with 0.1% Triton X-100 in 10% FCS/PBS for 10 min before sequential incubations with the primary and secondary antibodies. In some experiments, cells were simultaneously fixed and permeabilized in 3% paraformaldehyde with 0.5% Triton X-100 to reduce cytoplasmic signal. Coverslips were mounted in Mowiol. For endogenous Rac staining, cells were fixed in 10% trichloroacetic acid/PBS, washed in 30 mM glycine in PBS and blocked with 3% BSA/0.1% Triton X-100 in PBS for 1 h.
Microscopy
Cells were imaged using an inverted microscope (Axiovert; Carl Zeiss) with a EC Plan-Neofluor 10x/0.3 NA phase-contrast objective (aggregation assays; Carl Zeiss) or an upright fluorescence microscope (Provis AX70; Olympus) coupled to a monochrome camera (SPOT RT; SPOT Imaging Solutions), using an oil immersion PlanApo 60x/1.40 NA objective (Olympus), and controlled by SimplePCI software (Hamamatsu Photonics). Live cell imaging and fluorescence recovery after photobleaching (FRAP) experiments were performed on a confocal laser scanning microscope (LSM 510; Carl Zeiss) equipped with a 37°C incubation chamber and using an oil-immersion PlanApochromat 63x/1.40 NA differential interference contrast or an oil-immersion PlanApochromat 100x/1.40 NA Ph3 objective. For FRAP, bleaching was performed using 10 iterations of 75% laser power and recovery was monitored every 783 ms for 38 s.
Fluorescent lifetime measurements were performed via time-correlated single-photon counting using a multiphoton microscope system comprising an upright microscope (TCS SP5; Leica) with a krypton-argon laser (Leica), a Mai Tai tunable infrared laser (set at 890 nm, 80 MHz; Newport Spectrophysics), and a single photon counter from Becker & Hickl GmbH. An oil-immersion PlanApochromat HCX 63x/1.40-0.6 NA objective (Leica) was used throughout. Acquisition was performed using LAS AF (Leica) and SPCM (Becker & Hickl GmbH) software. Pictures were processed using Adobe Photoshop or ImageJ (http://rsb.info.nih.gov/ij).
Protein purification and protein–protein interaction assays
In vitro translation of different proteins was performed using the SuperScript II kit from Invitrogen. Fusion proteins were induced and purified using standard techniques. For insoluble GST-Ajuba full-length, the pellet was sequentially washed in 10 mM Tris-HCl, pH 8, 1 mM EDTA, 2% Triton X-100, 10 mM DTT, and distilled water containing 10 mM DTT. Extraction was performed by incubating the pellet in 8 M urea with 10 mM DTT with agitation at 30°C for 1 h. After centrifugation (25,000 g, 20 min, 4°C), supernatant was dialyzed using buffers containing 20 mM Tris-HCl, pH 7.5, 200 mM KCl, and gradual dilutions of urea (from 4 M to 0.2 mM) and DTT (from 10 mM to 0.5 mM) for refolding. Depending on the experiment, fusion proteins were cleaved using Thrombin (Sigma-Aldrich) or Precission enzyme as required and then dialyzed (20 mM Tris-HCl, pH 7.5, and 0.5 mM DTT).
For pull-down assays, fusion proteins immobilized in GSH-Sepharose were incubated with in vitro–translated products or cleaved proteins. Supernatants were incubated in a buffer containing 10 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 1 mM DTT, with GST or the different GST fusion proteins bound to the GSH-Sepharose beads, for 30 min at 4°C with rotation. Beads were then washed four times with the same buffer and bound proteins detected by enhanced chemiluminescence (ECL; GE Healthcare). For interaction of endogenous Ajuba with wild-type Rac, keratinocytes were lysed (10 mM Tris-HCl, pH 7.5, 1% NP-40, 150 mM NaCl, 1 mM MgCl2, 50 mM NaF, 1 mM DTT, 5 µM Leupeptin, and 1 mM PMSF) and centrifuged at 8,800 g for 10 min at 4°C. Supernatant was incubated with 20 µg GST-RacWT for 1h. Precipitates were washed with lysis buffer three times.
Rac activation assays
GDP/GTP loading of Rac on glutathione–Sepharose beads was performed as reported previously (Self and Hall, 1995). Rac·GTP levels were assessed in vivo by pull-down assays with GST-PAK-CRIB fusion protein as described previously (Sander et al., 1998; Betson et al., 2002). Junctions were induced for different periods of time by addition of calcium ions to avoid stimulation of GTPases with fresh serum, and lysates incubated with GST-PAK-CRIB to fish out Rac activated by cell–cell contacts. To study the localization of active Rac in situ, keratinocytes expressing the biosensor mRFP1-Raichu-Rac-GFP (Makrogianneli et al., 2009) were induced to form junctions for 20 min (CaCl2 addition), fixed in 3% paraformaldehyde, and imaged. Fluorescent resonance energy transfer (FRET) was determined by fluorescence lifetime imaging microscopy (FLIM). FLIM analysis was performed with SPCImage software (Becker & Hickl GmbH) as reported previously (Lleres et al., 2007). To restrict the lifetime measurements to the junctions, each straight cell–cell contact, regardless of signal intensity of the probe at the membrane, was manually selected. A bi-exponential fluorescence decay fitting was applied and the mean FRET efficiency per contact was calculated by the equation FRET efficiency = 1 – τda/τd, where τda is the lifetime of donor (GFP) interacting with acceptor (RFP) molecules and τd is GFP lifetime in the absence of acceptor (GFP-Rac control).
F-actin analyses
For F-actin clustering experiments, latex beads (15 µm; Polysciences) were coated with a monoclonal anti–E-cadherin antibody or bovine serum albumin (Braga et al., 1997; Betson et al., 2002). Attached beads were scored for actin recruitment when more than three distinct dots of F-actin were visible around the beads and expressed as a percentage of total attached beads. Around 30% of the few BSA-coated beads able to attach to keratinocytes showed some weak phalloidin staining (negative or nonspecific binding).
In low speed actin cosedimentation assays, monomeric G-actin (7 µM final, rabbit skeletal muscle) was polymerized into F-actin by incubation for 25 min at RT with G-buffer (2 mM Tris HCl, pH 7.5, 2 mM CaCl2, 200 mM KCl, 0.2 mM ATP, 0.5 mM dithiothreitol). Purified proteins (1 µM final) and MgCl2 (2 mM final) were then added to the F-actin solution and incubated for 20 min at 4°C with rotation. Samples were then centrifuged (8,800 g, 10 min, RT). Short filaments and monomeric actin were present in supernatant whereas resulting pellet containing long filaments and bundles was washed twice in G-buffer with 2 mM MgCl2 (100,000 g, 20 min, RT). Equivalent amounts of the resulting fractions were analyzed by Coomassie blue staining.
F-actin negative staining (Bailly et al., 2001) was performed with the following modifications: G-actin (5 µM, in G-buffer) was polymerized for 50 min at RT by the addition of 50 mM KCl, 2 mM MgCl2, and 10 mM Pipes, pH 7.0 in the absence or presence of Ajuba PreLIM domain (2 µM), or α-catenin (1.8 µM). Polymerization mixtures were diluted (1:10) in the same buffer, blotted on carbon-coated grids, and negatively stained with 1% uranyl acetate (Bailly et al., 2001). Samples were imaged using a transmission electron microscope (model 1010; JEOL). Similar co-sedimentation of F-actin was obtained when the proteins were incubated for 20 min with preformed actin filaments.
PAK1 kinase assays
Recombinant PAK1 kinase domain was incubated with 10 µCi γ-[32P]ATP (PerkinElmer) and different GST fusion proteins (0.4 nmol) trapped on glutathione–Sepharose beads in phosphorylation buffer (50 mM Hepes, pH 7.3, 10 mM MgCl2, 10 mM NaF, and 2 mM MnCl2) containing 40 µM cold ATP. After 5 min incubation at 30°C and washing in phosphorylation buffer, Laemmli buffer was added to the samples. Proteins were separated in SDS-PAGE gel and phosphorylation was visualized by autoradiography. Ajuba phospho-peptides were identified by mass spectroscopy commercially (FingerPrints Proteomics Facility, University of Dundee, Dundee, UK).
For kinase assays on immunoprecipitated PAK1, cells treated with IPA3 or PIR-3.5 were lysed in ice-cold buffer (50 mM Hepes, pH 7.3, 150 mM NaCl, 15 mM NaF, 10% glycerol, 1% Triton X-100, 20 mM β-glycerophosphate, 1 mM orthovanadate, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 µg/ml leupeptin). Lysates were precleared by centrifugation at 8,800 g for 5 min at 4°C and immunoprecipitated with 2 µg of PAK1 antibody for 2 h at 4°C. Precipitates were washed three times in 50 mM Hepes, pH 7.3, 150 mM NaCl, 15 mM NaF, 2 mM EDTA, 10% glycerol, and 0.2% Triton X-100, and once with phosphorylation buffer. PAK1 immunoprecipitates were subjected to in vitro kinase assays with the following modifications: samples were incubated with 20 µCi γ-[32P]ATP (PerkinElmer) in phosphorylation buffer containing 20 µM cold ATP. After 10 min incubation, Laemmli buffer was added to samples and proteins were separated and phosphorylation was visualized as described above.
Quantifications
For aggregation assays, the area of aggregates across six drops per sample and per experiment was determined using ImageJ software. After mechanical disruption, all disaggregates in a drop were imaged. Aggregates of less than three cells were excluded from the quantification. Results are depicted as mean area of each aggregate after dissociation, normalized with area of respective aggregates before trituration. For quantification of cadherin levels upon detergent extraction, background of images was subtracted using dedicated function in ImageJ. E-cadherin intensity was thresholded in order to mainly select signal at junctions. Area of the resulting binary mask was measured and divided by total area of the picture (in pixel2) for normalization. Ratio values were expressed relative to control, which was set as 100%. More than three random fields of view were computed per sample and per experiment (n = 3).
For FRAP, fluorescence intensity was measured with ImageJ. To calculate bleaching efficiency, GFP-Rac intensity was measured before and immediately after bleaching and normalized to initial levels (nonbleached, arbitrarily set as 100%). Values were averaged for each condition across different experiments and expressed as bleaching intensity. Fluorescence recovery time was assessed by measuring intensity values of bleached areas for each time point and correcting for background and acquisition bleaching (Goodwin and Kenworthy, 2005). Normalized percentage of recovery was plotted against time and half-time of fluorescence recovery was deducted from a nonlinear regression analysis (one-phase association exponential fitting curve) with GraphPad Prism software.
For live imaging, pixel intensity for each time point was measured across cell–cell contact of interest and divided by corresponding whole image intensity for photobleaching correction. The integral of the resulting curves was computed for each time point, normalized by time-zero values and plotted as “intensity at membrane” across time. For quantification of protein–protein interactions, X-ray films exposed in the linear range were scanned and specific bands quantified using ImageJ. Values were expressed relative to controls as stated in figure legends. Statistical significance was assessed using Student’s t test.
Online supplemental material
Fig. S1 shows the mapping of active Rac binding site on Ajuba. Fig. S2 shows the kinetics of Rac and Ajuba recruitment to newly formed contacts. Fig. S3 demonstrates the ability of Ajuba to dimerize via interaction of the LIM with PreLIM domain. Fig. S4 shows supporting evidence for Ajuba as a substrate for PAK1 (alignment of sequences, changes in affinity for active Rac binding, and quantification of relative 32P incorporation on Ajuba). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201107162/DC1.
Acknowledgments
We are thankful to Drs. Valentina Caorsi and Martin Spitaler for advice on FRET-FILM and Y. Fujita for advice with phosphorylation assays.
Work in the Braga Laboratory was generously funded by The Wellcome Trust (S. Nola and R. Daigaku; GR081357MA) and the Medical Research Council (K. Smolarczyk; G0600791). The Cancer Research UK (B. Martin-Martin; C10566/A8425) supported work in M. Bailly’s laboratory.
The authors declare no conflict of interest.
References
- Arias-Romero L.E., Villamar-Cruz O., Pacheco A., Kosoff R., Huang M., Muthuswamy S.K., Chernoff J. 2010. A Rac-Pak signaling pathway is essential for ErbB2-mediated transformation of human breast epithelial cancer cells. Oncogene. 29:5839–5849 10.1038/onc.2010.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanathan K., Peng H., Hou Z., Fredericks W.J., Goyal R.K., Langer E.M., Longmore G.D., Rauscher F.J., III 2007. The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic shuttling. Cancer Res. 67:9097–9106 10.1158/0008-5472.CAN-07-2987 [DOI] [PubMed] [Google Scholar]
- Bailly M., Ichetovkin I., Grant W., Zebda N., Machesky L.M., Segall J.E., Condeelis J. 2001. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr. Biol. 11:620–625 10.1016/S0960-9822(01)00152-X [DOI] [PubMed] [Google Scholar]
- Betson M., Lozano E., Zhang J., Braga V.M. 2002. Rac activation upon cell-cell contact formation is dependent on signaling from the epidermal growth factor receptor. J. Biol. Chem. 277:36962–36969 10.1074/jbc.M207358200 [DOI] [PubMed] [Google Scholar]
- Bokoch G.M. 2003. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72:743–781 10.1146/annurev.biochem.72.121801.161742 [DOI] [PubMed] [Google Scholar]
- Braga V.M. 2002. Cell-cell adhesion and signalling. Curr. Opin. Cell Biol. 14:546–556 10.1016/S0955-0674(02)00373-3 [DOI] [PubMed] [Google Scholar]
- Braga V.M., Yap A.S. 2005. The challenges of abundance: epithelial junctions and small GTPase signalling. Curr. Opin. Cell Biol. 17:466–474 10.1016/j.ceb.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Braga V.M., Hodivala K.J., Watt F.M. 1995a. Calcium-induced changes in distribution and solubility of cadherins, integrins and their associated cytoplasmic proteins in human keratinocytes. Cell Adhes. Commun. 3:201–215 10.3109/15419069509081287 [DOI] [PubMed] [Google Scholar]
- Braga V.M.M., Hodivala K.J., Watt F.M. 1995b. Calcium-induced changes in distribution and solubility of cadherins, integrins and their associated cytoplasmic proteins in human keratinocytes. Cell Adhes. Commun. 3:201–215 10.3109/15419069509081287 [DOI] [PubMed] [Google Scholar]
- Braga V.M., Machesky L.M., Hall A., Hotchin N.A. 1997. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J. Cell Biol. 137:1421–1431 10.1083/jcb.137.6.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga V.M., Del Maschio A., Machesky L., Dejana E. 1999. Regulation of cadherin function by Rho and Rac: modulation by junction maturation and cellular context. Mol. Biol. Cell. 10:9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga V.M., Betson M., Li X., Lamarche-Vane N. 2000. Activation of the small GTPase Rac is sufficient to disrupt cadherin-dependent cell-cell adhesion in normal human keratinocytes. Mol. Biol. Cell. 11:3703–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y.S., Thomas W.A., Eder O., Pincet F., Perez E., Thiery J.P., Dufour S. 2004. Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J. Cell Biol. 167:1183–1194 10.1083/jcb.200403043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H., Olsen J.V., Bairlein M., Gnad F., Oppermann F.S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. 2008. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell. 31:438–448 10.1016/j.molcel.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Deacon S.W., Beeser A., Fukui J.A., Rennefahrt U.E., Myers C., Chernoff J., Peterson J.R. 2008. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem. Biol. 15:322–331 10.1016/j.chembiol.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delva E., Kowalczyk A.P. 2009. Regulation of cadherin trafficking. Traffic. 10:259–267 10.1111/j.1600-0854.2008.00862.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerMardirossian C., Schnelzer A., Bokoch G.M. 2004. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol. Cell. 15:117–127 10.1016/j.molcel.2004.05.019 [DOI] [PubMed] [Google Scholar]
- Drees B.E., Andrews K.M., Beckerle M.C. 1999. Molecular dissection of zyxin function reveals its involvement in cell motility. J. Cell Biol. 147:1549–1560 10.1083/jcb.147.7.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees F., Pokutta S., Yamada S., Nelson W.J., Weis W.I. 2005. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 123:903–915 10.1016/j.cell.2005.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich J.S., Hansen M.D., Nelson W.J. 2002. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev. Cell. 3:259–270 10.1016/S1534-5807(02)00216-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon R.G., McClatchey A.I., Bretscher A. 2010. Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11:276–287 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaiz C., Chernoff J., Ammoun S., Peterson J.R., Hanemann C.O. 2009. PAK kinase regulates Rac GTPase and is a potential target in human schwannomas. Exp. Neurol. 218:137–144 10.1016/j.expneurol.2009.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J.S., Kenworthy A.K. 2005. Photobleaching approaches to investigate diffusional mobility and trafficking of Ras in living cells. Methods. 37:154–164 10.1016/j.ymeth.2005.05.013 [DOI] [PubMed] [Google Scholar]
- Goyal R.K., Lin P., Kanungo J., Payne A.S., Muslin A.J., Longmore G.D. 1999. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol. Cell. Biol. 19:4379–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M.D., Beckerle M.C. 2006. Opposing roles of zyxin/LPP ACTA repeats and the LIM domain region in cell-cell adhesion. J. Biol. Chem. 281:16178–16188 10.1074/jbc.M512771200 [DOI] [PubMed] [Google Scholar]
- Haraguchi K., Ohsugi M., Abe Y., Semba K., Akiyama T., Yamamoto T. 2008. Ajuba negatively regulates the Wnt signaling pathway by promoting GSK-3β-mediated phosphorylation of β-catenin. Oncogene. 27:274–284 10.1038/sj.onc.1210644 [DOI] [PubMed] [Google Scholar]
- Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 114:585–598 10.1016/S0092-8674(03)00642-1 [DOI] [PubMed] [Google Scholar]
- Hodivala K.J., Watt F.M. 1994. Evidence that cadherins play a role in the downregulation of integrin expression that occurs during keratinocyte terminal differentiation. J. Cell Biol. 124:589–600 10.1083/jcb.124.4.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Peng H., Ayyanathan K., Yan K.P., Langer E.M., Longmore G.D., Rauscher F.J., III 2008. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol. Cell. Biol. 28:3198–3207 10.1128/MCB.01435-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Peng H., White D.E., Negorev D.G., Maul G.G., Feng Y., Longmore G.D., Waxman S., Zelent A., Rauscher F.J., III 2010a. LIM protein Ajuba functions as a nuclear receptor corepressor and negatively regulates retinoic acid signaling. Proc. Natl. Acad. Sci. USA. 107:2938–2943 10.1073/pnas.0908656107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Peng H., White D.E., Wang P., Lieberman P.M., Halazonetis T., Rauscher F.J., III 2010b. 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial-mesenchymal differentiation. Cancer Res. 70:4385–4393 10.1158/0008-5472.CAN-10-0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A.I., Bachar M., Babbin B.A., Adelstein R.S., Nusrat A., Parkos C.A. 2007. A unique role for nonmuscle myosin heavy chain IIA in regulation of epithelial apical junctions. PLoS ONE. 2:e658 10.1371/journal.pone.0000658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas J.L., Beckerle M.C. 2004. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5:920–931 10.1038/nrm1499 [DOI] [PubMed] [Google Scholar]
- Kanungo J., Pratt S.J., Marie H., Longmore G.D. 2000. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell. 11:3299–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva M., Feng Y., Ward M., Song C., Anderson R.A., Longmore G.D. 2005. The LIM protein Ajuba regulates phosphatidylinositol 4,5-bisphosphate levels in migrating cells through an interaction with and activation of PIPKI alpha. Mol. Cell. Biol. 25:3956–3966 10.1128/MCB.25.10.3956-3966.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Gururaj A.E., Barnes C.J. 2006. p21-activated kinases in cancer. Nat. Rev. Cancer. 6:459–471 10.1038/nrc1892 [DOI] [PubMed] [Google Scholar]
- Lambert M., Choquet D., Mège R.M. 2002. Dynamics of ligand-induced, Rac1-dependent anchoring of cadherins to the actin cytoskeleton. J. Cell Biol. 157:469–479 10.1083/jcb.200107104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer E.M., Feng Y., Zhaoyuan H., Rauscher F.J., III, Kroll K.L., Longmore G.D. 2008. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev. Cell. 14:424–436 10.1016/j.devcel.2008.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lleres D., Swift S., Lamond A.I. 2007. Detecting protein-protein interactions in vivo with FRET using multiphoton fluorescence lifetime imaging microscopy (FLIM). Curr. Protoc. Cytom. Chapter 12:Unit12 10 [DOI] [PubMed] [Google Scholar]
- Lozano E., Betson M., Braga V.M. 2003. Tumor progression: Small GTPases and loss of cell-cell adhesion. Bioessays. 25:452–463 10.1002/bies.10262 [DOI] [PubMed] [Google Scholar]
- Lozano E., Frasa M.A., Smolarczyk K., Knaus U.G., Braga V.M. 2008. PAK is required for the disruption of E-cadherin adhesion by the small GTPase Rac. J. Cell Sci. 121:933–938 10.1242/jcs.016121 [DOI] [PubMed] [Google Scholar]
- Makrogianneli K., Carlin L.M., Keppler M.D., Matthews D.R., Ofo E., Coolen A., Ameer-Beg S.M., Barber P.R., Vojnovic B., Ng T. 2009. Integrating receptor signal inputs that influence small Rho GTPase activation dynamics at the immunological synapse. Mol. Cell. Biol. 29:2997–3006 10.1128/MCB.01008-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie H., Pratt S.J., Betson M., Epple H., Kittler J.T., Meek L., Moss S.J., Troyanovsky S., Attwell D., Longmore G.D., Braga V.M. 2003. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J. Biol. Chem. 278:1220–1228 10.1074/jbc.M205391200 [DOI] [PubMed] [Google Scholar]
- Mège R.M., Gavard J., Lambert M. 2006. Regulation of cell-cell junctions by the cytoskeleton. Curr. Opin. Cell Biol. 18:541–548 10.1016/j.ceb.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Miller M.L., Jensen L.J., Diella F., Jørgensen C., Tinti M., Li L., Hsiung M., Parker S.A., Bordeaux J., Sicheritz-Ponten T., et al. 2008. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 1:ra2 10.1126/scisignal.1159433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya-Durango D.E., Velu C.S., Kazanjian A., Rojas M.E., Jay C.M., Longmore G.D., Grimes H.L. 2008. Ajuba functions as a histone deacetylase-dependent co-repressor for autoregulation of the growth factor-independent-1 transcription factor. J. Biol. Chem. 283:32056–32065 10.1074/jbc.M802320200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y., Asaoka Y., Namae M., Saito K., Momose H., Mitani H., Furutani-Seiki M., Katada T., Nishina H. 2010. The LIM protein Ajuba is required for ciliogenesis and left-right axis determination in medaka. Biochem. Biophys. Res. Commun. 396:887–893 10.1016/j.bbrc.2010.05.017 [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Fukata M., Yamaga M., Itoh N., Kaibuchi K. 2001. Recruitment and activation of Rac1 by the formation of E-cadherin-mediated cell-cell adhesion sites. J. Cell Sci. 114:1829–1838 [DOI] [PubMed] [Google Scholar]
- Nakamura F., Stossel T.P., Hartwig J.H. 2011. The filamins: organizers of cell structure and function. Cell Adh Migr. 5:160–169 10.4161/cam.5.2.14401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W.J. 2008. Regulation of cell-cell adhesion by the cadherin-catenin complex. Biochem. Soc. Trans. 36:149–155 10.1042/BST0360149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann F.S., Gnad F., Olsen J.V., Hornberger R., Greff Z., Kéri G., Mann M., Daub H. 2009. Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics. 8:1751–1764 10.1074/mcp.M800588-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdamar B., Bose R., Barrios-Rodiles M., Wang H.R., Zhang Y., Wrana J.L. 2005. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 307:1603–1609 10.1126/science.1105718 [DOI] [PubMed] [Google Scholar]
- Pratt S.J., Epple H., Ward M., Feng Y., Braga V.M., Longmore G.D. 2005. The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. J. Cell Biol. 168:813–824 10.1083/jcb.200406083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M., Jouvenal K., Tripier D., Walter U. 1995. Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein). Proc. Natl. Acad. Sci. USA. 92:7956–7960 10.1073/pnas.92.17.7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X.-D., Kiosses W.B., Schwartz M.A. 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18:578–585 10.1093/emboj/18.3.578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennefahrt U.E., Deacon S.W., Parker S.A., Devarajan K., Beeser A., Chernoff J., Knapp S., Turk B.E., Peterson J.R. 2007. Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J. Biol. Chem. 282:15667–15678 10.1074/jbc.M700253200 [DOI] [PubMed] [Google Scholar]
- Rheinwald J.G., Green H. 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 6:331–343 10.1016/S0092-8674(75)80001-8 [DOI] [PubMed] [Google Scholar]
- Riento K., Totty N., Villalonga P., Garg R., Guasch R., Ridley A.J. 2005. RhoE function is regulated by ROCK I-mediated phosphorylation. EMBO J. 24:1170–1180 10.1038/sj.emboj.7600612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimm D.L., Koslov E.R., Kebriaei P., Cianci C.D., Morrow J.S. 1995. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. USA. 92:8813–8817 10.1073/pnas.92.19.8813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolli-Derkinderen M., Toumaniantz G., Pacaud P., Loirand G. 2010. RhoA phosphorylation induces Rac1 release from guanine dissociation inhibitor alpha and stimulation of vascular smooth muscle cell migration. Mol. Cell. Biol. 30:4786–4796 10.1128/MCB.00381-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander E.E., van Delft S., ten Klooster J.P., Reid T., van der Kammen R.A., Michiels F., Collard J.G. 1998. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J. Cell Biol. 143:1385–1398 10.1083/jcb.143.5.1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self A.J., Hall A. 1995. Measurement of intrinsic nucleotide exchange and GTP hydrolysis rates. Methods Enzymol. 256:67–76 10.1016/0076-6879(95)56010-6 [DOI] [PubMed] [Google Scholar]
- Smutny M., Cox H.L., Leerberg J.M., Kovacs E.M., Conti M.A., Ferguson C., Hamilton N.A., Parton R.G., Adelstein R.S., Yap A.S. 2010. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat. Cell Biol. 12:696–702 10.1038/ncb2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi K., Sasaki T., Kotani H., Nishioka H., Takai Y. 1997. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J. Cell Biol. 139:1047–1059 10.1083/jcb.139.4.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. 1977. Functional correlation between cell adhesive properties and some cell surface proteins. J. Cell Biol. 75:464–474 10.1083/jcb.75.2.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos B., Rodriguez-Boulan E. 2008. The epithelial polarity program: machineries involved and their hijacking by cancer. Oncogene. 27:6939–6957 10.1038/onc.2008.345 [DOI] [PubMed] [Google Scholar]
- Thoreson M.A., Anastasiadis P.Z., Daniel J.M., Ireton R.C., Wheelock M.J., Johnson K.R., Hummingbird D.K., Reynolds A.B. 2000. Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J. Cell Biol. 148:189–202 10.1083/jcb.148.1.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias K.F., Hartwig J.H., Ishihara H., Shibasaki Y., Cantley L.C., Carpenter C.L. 2000. Type Ialpha phosphatidylinositol-4-phosphate 5-kinase mediates Rac-dependent actin assembly. Curr. Biol. 10:153–156 10.1016/S0960-9822(00)00315-8 [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Degenstein L., Wise B., Fuchs E. 2001. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 104:605–617 10.1016/S0092-8674(01)00246-X [DOI] [PubMed] [Google Scholar]
- Vigil D., Cherfils J., Rossman K.L., Der C.J. 2010. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer. 10:842–857 10.1038/nrc2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S., Settleman J. 1997. The PRK2 kinase is a potential effector target of both Rho and Rac GTPases and regulates actin cytoskeletal organization. Mol. Cell. Biol. 17:2247–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Zallen J.A. 2009. Junctional trafficking and epithelial morphogenesis. Curr. Opin. Genet. Dev. 19:350–356 10.1016/j.gde.2009.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Nelson W.J. 2007. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J. Cell Biol. 178:517–527 10.1083/jcb.200701058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers M.M.P., Forget M.-A., Chernoff J., Mostov K.E., ter Beest M.B.A., Hansen S.H. 2003. Pak1 and PIX regulate contact inhibition during epithelial wound healing. EMBO J. 22:4155–4165 10.1093/emboj/cdg398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Betson M., Erasmus J., Zeikos K., Bailly M., Cramer L.P., Braga V.M. 2005. Actin at cell-cell junctions is composed of two dynamic and functional populations. J. Cell Sci. 118:5549–5562 10.1242/jcs.02639 [DOI] [PubMed] [Google Scholar]
- Zhao Z.S., Manser E., Chen X.Q., Chong C., Leung T., Lim L. 1998. A conserved negative regulatory region in alphaPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol. Cell. Biol. 18:2153–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]