Abstract
In this study, we initiated experiments to address the structure-function relationship of Rin1. A total of ten substitute mutations were created, and their effects on Rin1 function were examined. Of the ten mutants, four of them (P541A, E574A, Y577F, T580A) were defective in Rab5 binding, while two other Rin1 mutants (D537A, Y561F) partially interacted with Rab5. Mutations in several other residues (Y506F, Y523F, T572A, Y578F) resulted in partial loss of Rab5 function. Biochemical studies showed that six of them (D537A, P541A, Y561F, E574A, Y577F, T580A) were unable to activate Rab5 in an in vitro assay.
In addition, Rin1: D537A and Rin1: Y561F mutants showed dominant inhibition of Rab5 function. Consistent with the biochemical studies, we observed that these two Rin1 mutants have lost their ability to stimulate the endocytosis of EGF, form enlarged Rab5-positive endosomes, or support in vitro endosome fusion. Based on these data, our results showed that mutations in the Vps9 domain of Rin1 lead to a loss-of-function phenotype, indicating a specific structure-function relationship between Rab5 and Rin1.
INTRODUCTION
The EGF-receptor plays a central role in cell proliferation, differentiation, survival and migration. As such it has served as a prototype in growth factor receptor trafficking [1, 2]. Following activation, the tyrosine phosphorylated EGF-receptor is rapidly internalized from the cell surface to early endosomes, then transported to lysosomes for degradation. This process is generally known as “receptor down- regulation” and is also considered to be an important cellular strategy for signal attenuation [3–6]. Thus, the phosphotyrosine residues on the tail of the EGF-receptor mediate the recruitment of several effectors [7–15], including Ras interference 1 (Rin1) [16].
Rin1 was originally identified as a Ras effector protein [17] and found to contain several functional domains: SH2 and proline-rich (PR) domains in the N-terminal region, and the Ras association (RA) domains in the C-terminal region [18]. In addition, it has been shown that Rin1 has an additional domain that exhibits Rab5-guanine nucleotide exchange factor (GEF) activity and is known as the Vps9 catalytic domain [19]. Rin1 has been shown to interact with 14-3-3, Bcr-Abl and STAM proteins [20–22], which in turn may regulate the sub-cellular localization and function of Rin1. Thus, the interaction of Rin1 with 14-3-3 seems to affect the ability of Raf to bind Rin1. Rin1 also interacts, through the proline-rich domain, with Abl and STAM proteins. The interaction of Rin1 with Abl tyrosine kinase has been suggested to mediate actin cytoskeleton remodeling associated with cell migration [23]. The interaction of Rin1 with STAM proteins may regulate EGF-receptor degradation [22].
Previous work suggested that Rin1 was a guanine-nucleotide exchange factor for the small GTPase Rab5 [19]. Overexpression of Rab5 was shown to stimulate EGF-mediated endocytosis [24] and its Vps9 activity was shown to be potentiated by a GTP-bound form of Ras [19]. The Rin family now has at least four members, all of which have an active Rab5 Vps9 domain [25]. Subsequent work showed that Rin1 was targeted to the EGF- and insulin-receptors via its SH2 domain [19, 26].
Interestingly, the putative Vps9 catalytic domain of Rin1 is found in the N-terminal of the Ras association domain. The Vps9 domain mediates an association with the Rab5 protein in a nucleotide- dependent manner [19]. This Vps9 domain has been identified in several proteins, including Rabex-5, ALS2, RAP6/RME6/Gapex-5, and VARP, in addition to the Rin family [27–32]. RME6 was originally described in C. elegans [29]. The mammalian ortholog was initially identified by Hunker [30] and called RAP6; a mouse ortholog of RME6 has also been identified, and shows affinity for Rab31 and Rab5 [31]. The VARP proteins also contain a Vps9 domain, but show greater affinity for Rab21 than for Rab5 [32]. Rin1 knockout mice show inhibited neuronal plasticity in aversive memory formation [33]. However, Rabex-5-deficient mice developed severe skin inflammation, and the ALS gene has been identified in individuals with an autosomal recessive juvenile-onset syndrome related to ALS [34].
Recent studies have described the three-dimensional structure of Rabex-5 [35] as an approximately 60-kDa protein containing Vps9 domain and other domains for interaction with its regulators and effectors [36–40]. On one hand, its effector Rabaptin-5, forms a complex with Rabex-5 and increases its weak exchange activity [37]. On the other hand, Rabex-5 contains two distinct ubiquitin-binding sites which are important in interaction with ubiquitinated EGF-receptor [40]. Interestingly, these two cytosolic factors (i.e., Rabex-5 and Rin1) are recruited onto the activated EGF-receptor tail via two different mechanisms: Rabex-5 require ubiquitination [40], while Rin1 require tyrosine phosphorylation of the EGF-receptor [16]. Thus, both Vps9 containing proteins are likely to associate with the early endosomes and the plasma membrane. Accumulating evidence suggests that Rin1 is involved in the early steps of vesicle transport from the plasma membrane to endosomes [16, 19], although the exact mechanism by which Rin1 regulates this process has not been established.
In this study, we have investigated the structural features of Rin1 that are necessary for its function. Several point mutants were created in the Vps9 domain of Rin1, and the effects on its biological activity were examined using several in vitro and in vivo assays. The results suggest a structure-function relationship of Rin1 and also indicate the structural features that are required for the inhibitory effect of these mutants.
MATERIAL AND METHODS
Reagents
[125I] Epidermal growth factor (EGF) was purchased from Amersham Biosciences (New Jersey, NJ). Dako fluorescent mounting medium was purchased from Dako Corporation (Copenhagen, Denmark). FuGENE6 was purchased from Roche Molecular Biochemicals (Indianapolis, IN). Monoclonal anti-HisG and anti-GST polyclonal antibodies were purchased from Invitrogen (Carlsbad, CA). Mouse monoclonal and polyclonal anti-Rab5 and anti-Rin1 antibodies were purchased from BD Biosciences and from Novus Biological. Paraformaldehyde was purchase from Electron Microscopy Sciences. All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). All other reagents were from Sigma unless otherwise noted.
Construction of recombinant retroviruses and stable cell lines expressing Rin1 constructs
Rin1: wild type, and Rin1 constructs were subcloned into EcoRI restriction sites of the pMX-puro vector as described [19]. Amino acid substitutions in Rin1 (i.e. Rin1: Y506F, Rin1: Y523F, Rin1: D537A, Rin1: P541A, Rin1: Y561F, Rin1: T572A, Rin1: E574A, Rin1: Y577F, Rin1: Y578F, Rin1: T580A) were made with the QuickChange mutagenesis kit (Stratagene, La Jolla, CA). To make stable cell lines, each expressing a Rin1 construct, Plat-E packaging cells were cultured in Dulbecco medium containing 10% feta bovine serum and transfected as described [19, 41]. Rab5 (wild type and S34N) and Rin1 constructs (Rin1: SH2, Rin1: Vps9-RA) were also sub-cloned into the pGEX-2T and pB42AD vector as described [19].
Protein Purification
His-Rab5 (wild type and S34N) proteins were expressed in M15 pREP4 E. coli (Novagen, Inc) and His-Rin1: wild type and its mutants were expressed in NR6 cells [20]. Recombinant His-tagged proteins were purified from either E. coli or NR6 cells using Ni2+-NTA-agarose according to the manufacturer’s protocol (Qiagen, Inc.) Protein concentrations were determined by Bio-Rad protein assay, and verified by band intensities on SDS-PAGE gels stained with Coomassie Blue. Recombinant proteins were expressed in large quantity as glutathione S-transferase (GST) fusion proteins in the E. coli strain (DE3) according to the manufacturer’s protocol (Thermo Scientific). The fusion proteins were affinity purified and used in vitro pull-down, exchange activity and endosome fusion assays.
Yeast Two-Hybrid assay
Two-hybrid LexA DNA binding domain (LexA-BD) and B42 transcription activation domain (B42AD) fusion constructs were prepared by ligating cDNAs for different proteins or protein fragments into the pLexA (His) and pB42AD (Trp) plasmids, respectively (Clontech, Palo Alto, CA). Mating and transformation of Yeast-EGY48 cells were done as described in the MATCHMAKER two-hybrid manual (Clontech). The Rin1 constructs were expressed as pB42AD fusion constructs whereas Rab5: S34N mutant was expressed as pLexA fusion constructs. Yeast-EGY48 co-transformants were plated on medium without His (H), Trp (W), Leu (L) or Ura (U) (-HWUL) to detect LacZ reporter gene activation due to interaction of constructs and on medium without His, Trp, or Ura (-HWU) as a control for loading and growth on galactose containing medium. All Rin1 and Rab5 constructs were detected by Western blot analysis. For liquid β-galactosidase (β-gal) assays, yeast was grown on appropriate selective media and assayed as described in the yeast β-gal assay manual (Pierce). Results are expressed in Units: one unit of β-gal was defined as 1000 xOD420/[time × vol xOD660].
Ligand internalization
For EGF uptake, cells expressing Rin1, Rin1 mutants or GFP (control) were serum-starved for 16 h. Cells were then incubated at 4°C for 2h with 100 pM [125I]-EGF (750 Ci/mmol), washed with ice-cold Ringer buffer and warmed for 6 min. At incubation, unbound ligand was removed by washing the monolayer 3 times with ice-cold Ringer buffer. Surface-bound and internalized ligands were measured as described [24]. For HRP uptake, cells expressing different constructs were washed three times with serum-free Dulbecco’s Modified Eagle medium (DMEM). HRP uptake was then initiated by addition of 1 ml of DMEM containing 2 mg/ml HRP (Sigma) and 0.2% bovine serum albumin (BSA) for 30 min at 37°C. For EGF-stimulated HRP uptake, cells were processed as described above, except that HRP uptake was carried out in the absence or presence of 100 ng/ml EGF. The uptake was conducted at 37°C for the indicated time in Figure 4. After uptake, the cells were washed, scraped and resuspended in 1 ml of PBS. The dishes were rinsed once with PBS, and the cell suspensions were pooled. Cells were centrifuged, and washed twice by resuspension in 0.5 ml of PBS. Each cell pellet was lysed in 500 ml of PBS containing 0.75% Triton X-100 and assayed for HRP activity as described [24].
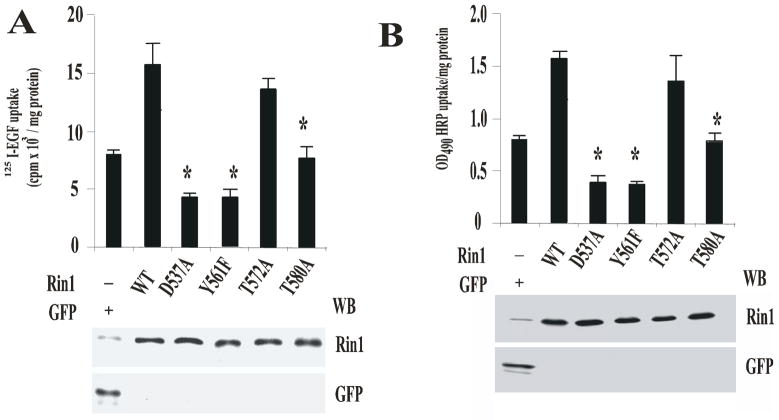
Figure 4. Effect of Rin1 mutants on the endocytosis of HRP and EGF.
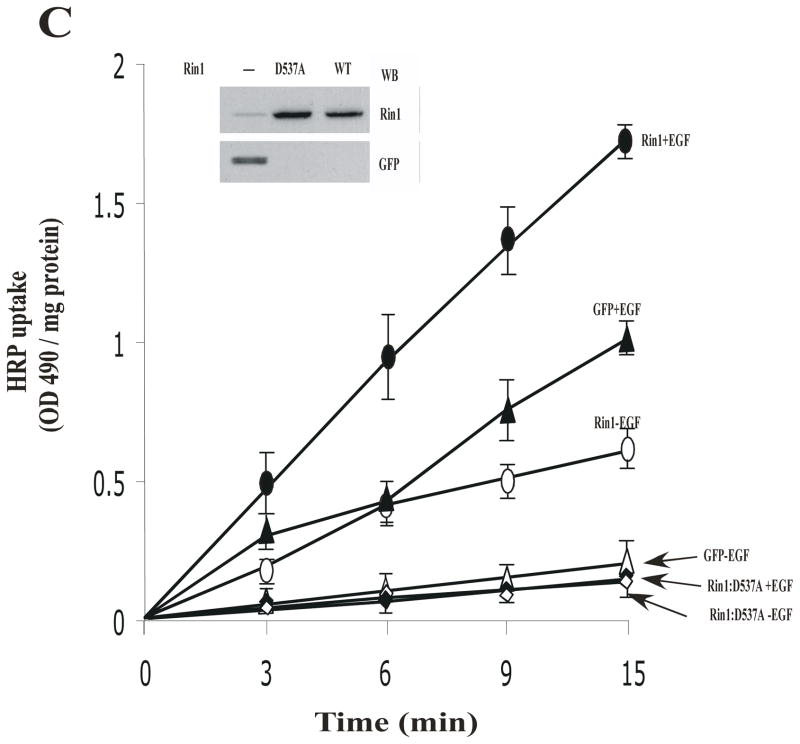
Stable NR6 cell lines over- expressing either Rin1 constructs or GFP were incubated with 125I-EGF (A) or HRP (B) at 37°C. After incubation, the internalized ligand was determined as described in Material and Methods. The data are presented as means ± SD of three independent experiments, n=3 *P < 0.001. Inset: Stable NR6 cell lines over-expressing either GFP or Rin1 constructs were separated by SDS-PAGE, blotted to nitrocellulose, and were visualized with anti-GFP and anti-Rin1 antibodies, respectively. This experiment was repeated three times with similar results. (C) Stable NR6 cell lines over-expressing GFP alone (→,←), Rin1: wild type (
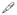 ,
,
 ), or Rin1: D537A (
), or Rin1: D537A (
 ,
,
 ) were incubated with HRP at 37°C for the time indicated in the absence (
) were incubated with HRP at 37°C for the time indicated in the absence (
 ,
,
 ,→) or presence (
,→) or presence (
 ,
,
 ,←) of 100 ng/ml EGF. After incubation, the internalized HRP was determined as described in Material and Methods. Relative Units (R.U.) indicates the HRP uptake (OD440/mg protein). The data are presented as means ± SD of three independent experiments, n=3. Inset: Stable NR6 cell lines over-expressing either GFP, Rin1: wild type, or Rin1: D537A mutants were separated by SDS-PAGE, blotted to nitrocellulose, and these proteins were visualize with anti-GFP and anti-Rin1 antibodies. This experiment was repeated three times with similar results.
,←) of 100 ng/ml EGF. After incubation, the internalized HRP was determined as described in Material and Methods. Relative Units (R.U.) indicates the HRP uptake (OD440/mg protein). The data are presented as means ± SD of three independent experiments, n=3. Inset: Stable NR6 cell lines over-expressing either GFP, Rin1: wild type, or Rin1: D537A mutants were separated by SDS-PAGE, blotted to nitrocellulose, and these proteins were visualize with anti-GFP and anti-Rin1 antibodies. This experiment was repeated three times with similar results.
GST-Pull-down Assay
For in vitro studies, fusion proteins were expressed and purified as indicated above. Ten μg of immobilized GST or GST-Rab5: S34N to glutathione-Sepharose beads were incubated with different amounts of His-Rin1 proteins for 1h at 4°C in binding buffer containing 20 mM Tris-Cl, pH 7.4, 50 mM NaCl, 2 mM EDTA, 5 mM MgCl2, 5% glycerol and 0.01% Triton X-100. After incubation, the beads were washed three times by centrifugation and bound proteins were eluted by the addition of SDS sample buffer. Solubilized proteins were separated by SDS-PAGE, transferred to nitrocellulose filters, and probed with specific antibodies.
Guanine Nucleotide Exchange Assays
Exchange kinetics were measured by monitoring the quenching of fluorescence after release of the nucleotide analog 2′-(3′)-bis-O-(N-methylanthraniloyl)-guanosine 5′-diphosphate (mant-GDP) [42]. The exchange reaction was started by using 0.01–1.0 μM of purified Rin1 proteins in 50 mM Tris (pH 8.0), 37 mM NaCl and 2 mM MgCl2 in the presence of 100 pmol Rab5-mant-GDP. Samples were excited at 360 nm and the emission monitored at 440 nm. Data were collected using a multimode microplate spectrophotometer (Tecan).
Lysate preparation, SDS-PAGE and Western blotting
To prepare whole cell lysates, cell monolayers were washed with phosphate buffer saline and lysed in ice-cold lysis buffer containing protease inhibitor cocktail (EDTA-free, Roche). The lysates were clarified by centrifugation, and protein concentrations were determined by Bio-Rad protein assay. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes, blocked and probed with the indicated antibodies. To determine relative protein amounts, four representative exposures for each sample were quantitated by densitometry analysis.
Immunoprecipitation
Cells were lysed in ice-cold lysis buffer containing protease inhibitor cocktail (EDTA-free, Roche) and then used for immunoprecipitation reactions. After clarification, protein concentrations were measured and 1.0 mg/ml extracts were immunoprecipitated by incubation with the appropriate antibody, followed by immobilization on Protein G-Sepharose beads (Sigma). Beads were resuspended in sample buffer and analyzed by SDS-PAGE, and Western blotted with the indicated antibodies.
Immunofluorescence
Cells stably transfected with Rin1 constructs were seeded onto glass coverslips. The cells were then washed with ice-cold Ringer buffer and then fixed for 20 min in Ringer buffer containing 4% (v/v) paraformaldehyde. The cells were washed and mounted onto slides with Fluorescent Mounting Medium (DakoCytomation, CA). The cells were examined with a Leica TCS SP2 laser scanning confocal unit attached to an upright fluorescence microscope (Leica DM RXE) with a 63x, 1.4 plan Apochromat objective (Leica) by using the appropriate filter for GFP and RFP fluorochromes. Images were merged and aligned using Adobe Photoshop 7.0 (Adobe Systems, Mountain View, CA, USA).
In vitro endosome fusion assay
Early endosome loaded with dinitrophenol-derivatized beta- glucuronidase or mannosylated anti-dinitrophenol IgG by 5 min uptake at 37°C in the macrophage cell line J774E [43]. Cells were washed and resuspended in 2 ml of homogenization buffer (250 mM sucrose, 0.5 mM EGTA, 20 mm Hepes-KOH pH7.0). The cells were homogenized and the homogenates were centrifuged at 800 × g for 5 min as described [43]. The supernatants were centrifuged at 37,000 × g for 1 min, following by another centrifugation at 50,000 × g for 5 min. The pellets, enriched in early endosomes, were resuspended in homogenization buffer supplemented with 1 mM dithiothreitol, 1.5 mM MgCl2, 50 μg/ml dinitrophenol-BSA, 50 mM KCl containing Sephadex G25-filtered cytosol, ATP regenerated system and purified recombinant proteins (His-Rab5 and His-Rin1 constructs). Fusion reactions were conducted at 37°C for 45 min and quantified by the β-glucuronidase activity in the immunocomplex [43]. His-tagged Rin1: wild type and mutants were expressed using pMX retrovirus system in NR6 cells as described early [19]. His-tagged Rin1 proteins were then purified from NR6 cell lysates on 1 ml of Hi-trap Ni2+-Agarose columns (Pharmacia) according to the instruction of the manufacturer. Purified recombinant proteins (Rin1, Rab5, REP-1) from mammalian NR6 and SF9 baculorirus-infected cells were then used fresh for fusion or stored frozen in liquid nitrogen for later use. The complexes of Rab5 proteins with REP-1 were prepared as previously described [45, 52] and then added to the fusion assay.
Image analysis
Fluorescent images of Rab5-positive endosomes were digitized by using Leica TCS software and projection images from a series of 15 focal planes were created in cells co-expressing both Rin1 and Rab5 constructs as described in Figure 5. Size of the endosomes was analyzed with the NIH Image software which can be accessed on the website (http://rsb.info.nih.gov/nih-image/).
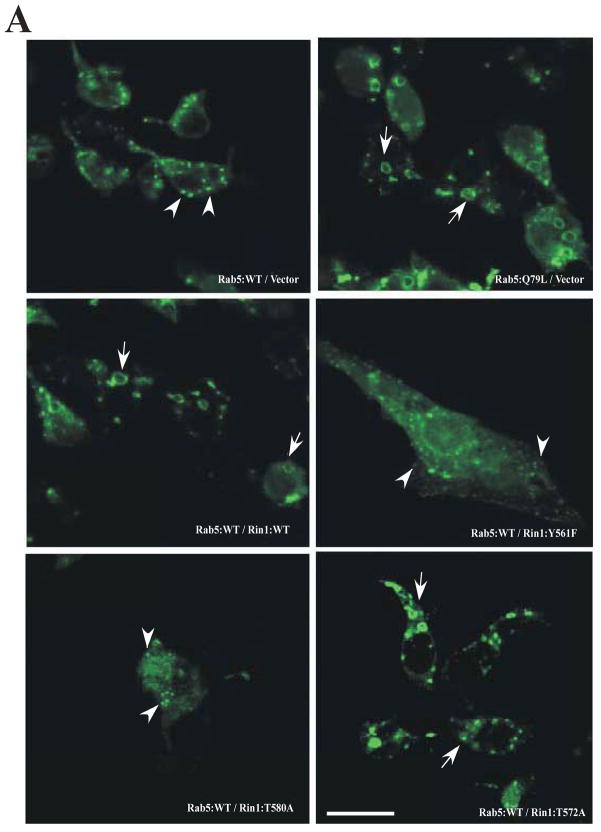
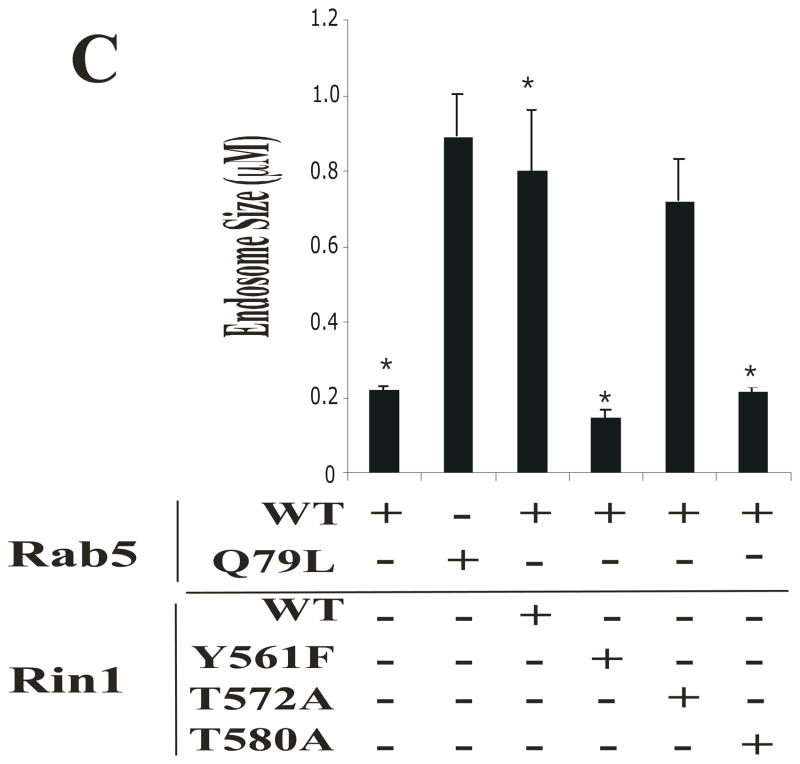
Figure 5. Rin1 mutants affect the formation of enlarged Rab5-positive endosomes.
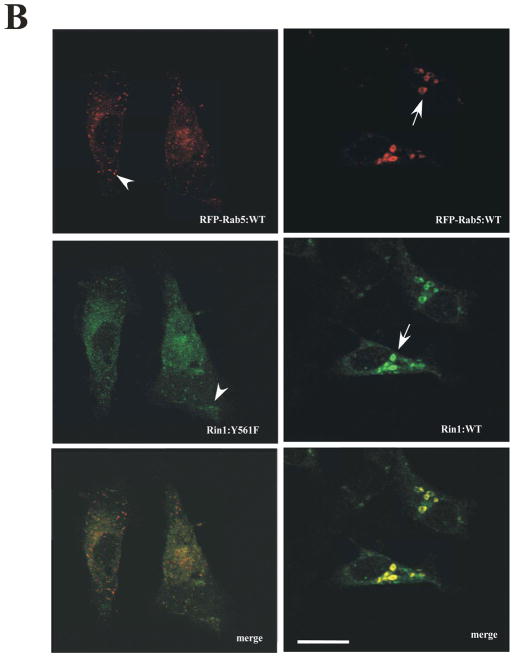
(A) Stable NR6 cell lines over-expressing either vector alone (control), Rin1: wild type, or Rin1 mutants were transiently transfected with GFP-Rab5: wild type and GFP-Rab5: Q79L mutant. After transfection, cells were processed for immunofluorescence as described in Material and Methods. Confocal fluorescence microscopy images show the morphological changes of Rab5-labeled endosom es in NR6 cells coexpressing both Rin1 and Rab5 constructs as indicated in the Figure. This experiment was repeated three times with similar results. Bar: 10 μm. (B) Stable NR6 cell lines over-expressing either Rin1: wild type or Rin1: Y561F mutant were transiently transfected with RFP-Rab5: wild type. After transfection, the cells were fixed and processed as described in Material and Methods. This experiment was repeated three times with similar results. Bar: 10 μm. The arrowheads and arrows denote the small and enlarged Rab5-positve endosomes, respectively. (C) Different sizes of Rab5-positive endosomes were quantified in control cells (i.e., cells co-expressing Rab5 constructs and vector alone) and cells co-expressing both Rin1 and Rab5 constructs. A total of 2000 Rab5-positive endosomes from 7 cells were used to determine endosome sizes in each experiment. The data are presented as means ± SD of three independent experiments, n=3 *P < 0.001.
Statistical analysis
All experiments presented were repeated a minimum of three times. The data represent the mean ± SD. Student’s t test was performed to calculate statistical significance.
RESULTS
Analysis of the interaction between Rab5 and Rin1 constructs
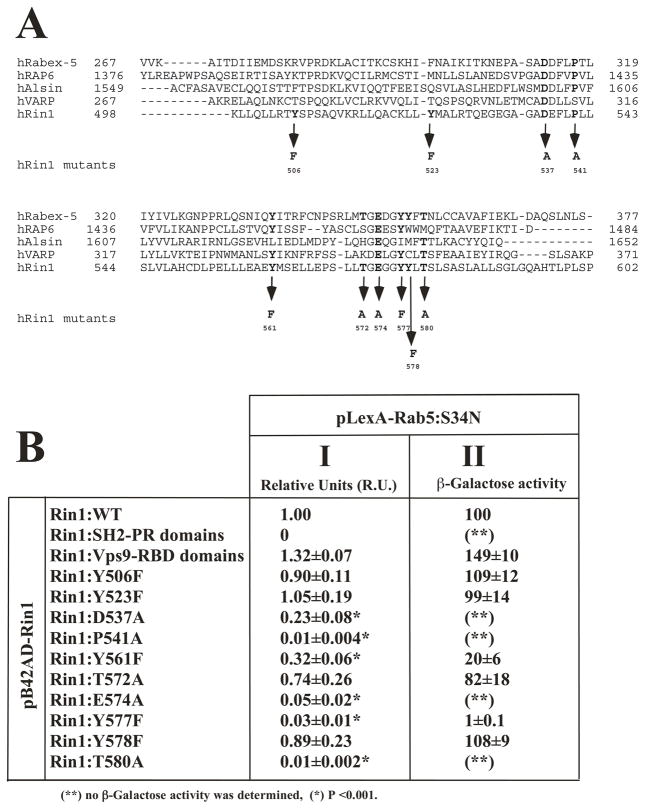
In order to investigate the structure- function relationship of the Vps9 domain of Rin1, it was necessary to use several functional in vitro and in vivo assays. The most noticeable characteristic of all Vps9 sequences is the presence of several conserved residues [25]. Mutations in these residues dramatically influence the GEF activity of Rabex-5 [35]. Corresponding mutations in other Vps9 domain are expected to have similar effects on their biochemical functions. We have used oligonucleotide-directed mutagenesis on several residues to introduce ten mutations into the Vps9 catalytic domain of Rin1. The residue substitutions used in this study were: Y506F, Y523F, D537A, P541A, Y561F, T572A, E574A, Y577F, Y578F, T580A (Figure 1A). It is important to notice that Asp 537, Pro 541 and Glu 574 are conserved among several Vps9 containing proteins (Figure 1A). However, the amino acid residue corresponding to Pro 541 is Ser 314 in VARP proteins. Also, the amino acid residues corresponding to Tyr 561 and Tyr 577 in the sequence of Alsin proteins are Leu 1224 and Ile 1640, respectively (Figure 1A). The amino acid residues corresponding to Thr 572 and Tyr 578 are not present in all the Vps9 containing proteins, except in Rabex-5 proteins where the amino acid residues corresponding to Thr 572 and Tyr 578 are Thr 349 and Tyr 355, respectively. Finally, the amino acid Thr 580 is conserved in the Vps9 containing proteins shown in Figure 1A, except in RAP6, in which the amino acid residue corresponding to Thr 580 is Met 580.
Figure 1. Substitution mutations in the Vps9 domain of Rin1, and their effects on the interaction with Rab5: S34N.
(A) Alignment of the humanVps9 domain containing proteins (i.e., Rabex-5 [hRabex-5], Rin1 [hRin1], RAP6 [hRAP6], Alsin [hAlsin], and VARP- [hVARP]), and localization of the mutations in the Vps9 domain of Rin1 are shown. (B) Yeast cells were co-transformed with plasmids pB42AD fused to the Rin1 sequences indicated with pLexA fused to the Rab5: S34N mutant. Co- transformed cells were spotted onto plates containing selected media, and incubated at 30°C as described in Material and Methods. (I) Growth on deficient interaction media indicates interaction between Rin1 and Rab5 proteins. (II) Liquid cultures in selective media were also assayed for β-Galactosidase (β-Gal) to verify and quantify two-hybrid interactions. Relative Units (RU): number of colonies in interaction media/number of colonies in expression media. The data are presented as means ± SD of five independent experiments, n=5 *P < 0.001. Each independent experiment was performed in quintuplicate.
The yeast two-hybrid system was initially used to determine whether the Rin1-Rab5 interaction was mediated by specific sequences in the Vps9 domain of Rin1. As expected, Rin1: wild type prey fusion protein interacted with Rab5: S34N bait fusion protein, a Rab5 GTP-defective mutant protein. A deletion mutant of Rin1 two-hybrid prey (i.e., Rin1: Vps9-RA deletion mutant) strongly interacted with Rab5. However, other deletion mutant of Rin1 two-hybrid prey (i.e., Rin1: SH2-PR) did not interact with Rab5 (Figure 1B). Furthermore, mutation of four residues (Pro 541, Glu 574, Tyr 577 and Thr 580) severely impaired Rin1-Rab5 interaction. In addition, mutation of two residues (Asp 537 and Tyr 561) also resulted in significant defect, whereas mutations of other residues including Tyr 506, Tyr 523, Tyr 578, and Thr 572 had moderate, but not statistically significant effect (Figure 1B).
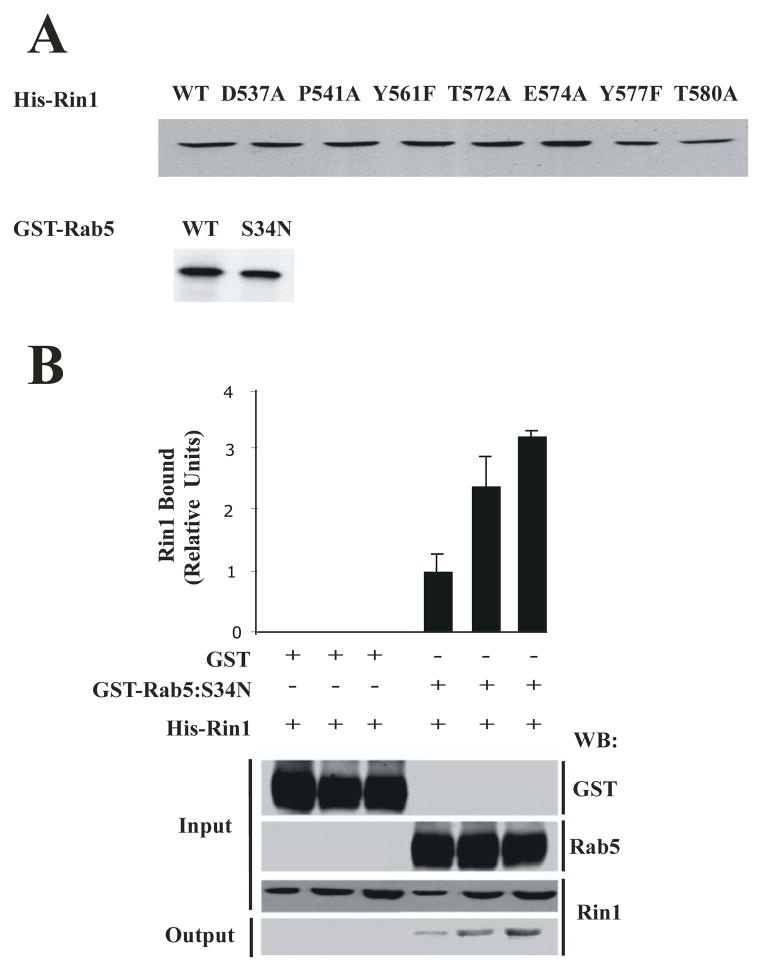
We then characterized the interaction of Rab5 with Rin1 mutants by using a pull-down in vitro assay. First, we investigated the structural instability of the protein mutants. One indication of structural instability introduced by the loss or substitution of residues important for folding is a large decrease in soluble polypeptide upon expression in cells. The level of soluble expression of each Rin1 mutant was measured by growing small volumes of transformed E. coli under identical conditions, followed by purification of the tagged fusion protein on appropriate beads. The results of one experiment are shown in Figure 2A. Compared with Rin1: wild type, five Rin1 mutants (D537A, P541A, Y561F, T572A, and E574A) showed the same level of expression and were considered structurally stable. Two mutants (Y577A, and T580A) were expressed about ~0.87-fold less than Rin1: wild type, suggesting a minor effect on protein stability. Similar results were observed when the expression of GST-Rab5 protein was analyzed in E. coli by SDS-PAGE analysis (Figure 2A). We also found that the expression levels of GST- Rin1 mutants were similar to the expression level of Rin1: wild type in E. coli (data not shown). Taken together, these results indicate that the fusion proteins were expressed in a soluble form at wild-type levels and were well behaved.
Figure 2. Expression and interaction of Rab5: S34N with Rin1: wild type and its mutants.
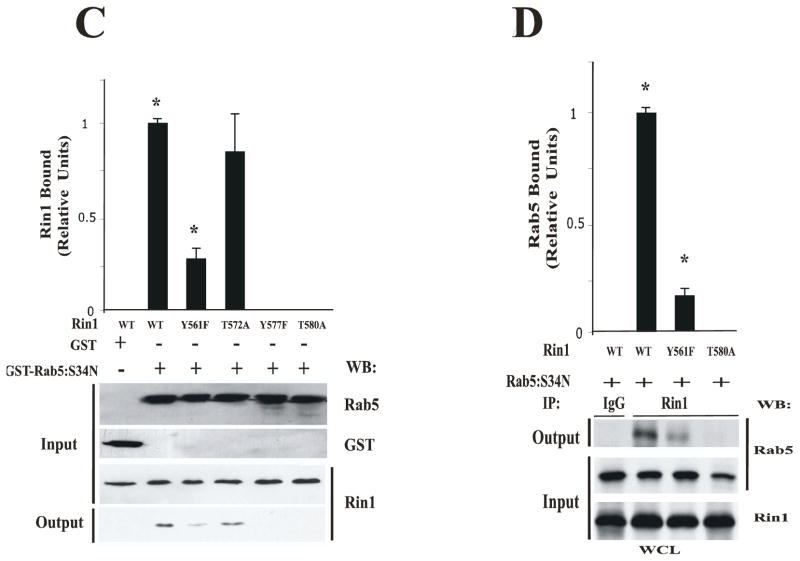
(A) SDS- PAGE showing the expression levels of Rin1 and Rab5 proteins. Recombinant fusion proteins were precipitated from equivalent bacterial cell extracts with glutathione-Sepharose (GST-Rab5) or with Ni2+Agarose (His-Rin1) beads. The beads were washed and fusion proteins visualized on SDS-PAGE gels stained with Coomassie Blue. This experiment was repeated three times with similar results. (B) Recombinant GST alone (10 μg) and GST-Rab5: S34N mutant (10 μg) were incubated with increasing amounts of recombinant His-Rin1: wild type proteins (0.01 μg, 0.1 μg, 1 μg), and the amount of Rin1 bound to Rab5 proteins were detected by Western blotting with anti-Rin1 antibodies. Input: total amount of GST, GST-Rab5: S34N, or His-Rin1 added to the in vitro pull-down assay. Output: the amount of Rin1 bound to GST-Rab5 was visualized by Western blot analysis. Proteins were quantified by densitometry of Westerns blots. The data are presented as means ± SD of four independent experiments, n=4. (C) 10 μg Recombinant GST-Rab5: S34N was incubated with 0.01 μg recombinant His-Rin1: wild type and its mutants (input). Relative levels of Rin1 proteins bound to Rab5: S34N were determined by densitometry of Westerns blots with anti-Rin1 antibodies (output). The data are presented as means ± SD of three independent experiments, n=3 *P < 0.001. (D) Stable NR6 cell lines over-expressing either Rin1: wild type or Rin1 constructs were transiently transfected with Rab5: S34N mutant. After transfection, cells were lysed and immunoprecipitated as described in Material and Methods. Whole cell lysates (WCL) containing Rab5: S34N and Rin1 constructs (input) were immunoprecipitated with anti-Rin1 antibodies, and the presence of Rab5 bound to Rin1 constructs (output) was visualized with anti-Rab5 antibodies. Proteins were quantified by densitometry of Westerns Blots. The data are presented as means ± SD of three independent experiments, n=3 *P < 0.001.
Second, we investigated the ability of Rin1 mutants to interact with Rab5 proteins in an in vitro pull- down assay. GST-Rab5: S34N protein was bound to glutathione-Sepharose beads. Then, the beads containing Rab5: S34N proteins were incubated with different concentrations of Rin1: wild type for 1 hat 4°C. After the incubation, the beads were washed, and the presence of Rin1 bound to Rab5 was visualized with SDS-PAGE followed by Western blotting analysis with anti-Rin1 antibodies. We observed that Rin1: wild type interacted with Rab5: S34N mutant in a concentration dependent manner (Figure 2B). As expected, Rin1: wild type did not interact with GST alone (Figure 2B). These results indicated that Rin1: wild type specifically interacts with Rab5: S34N mutant, which is consistent with previously published data [19]. Quantification analysis showed that two Rin1 mutants (Y577F and T580A) severely inhibited the interaction with Rab5. However, Rin1: Y561F mutant was shown to only partially affect the interaction with Rab5 whereas Rin1: T572A mutant showed a moderate but not statistically significant inhibitory effect according to quantitative analysis (Figure 2C). In addition, we observed that Rin1: Y577F, Rin1: T580A (Figure 2C), Rin1: P541A and Rin1: E574A mutants failed to interact with Rab5 (data not shown). These observations were further confirmed when Rin1 was immunoprecipitated from cells co-expressing both Rab5 and Rin1 constructs (Figure 2D). Specifically, we observed that Rin1: wild type, but not Rin1: T580A mutant, co-immunoprecipitated with Rab5: S34N protein. Rin1: Y561F mutant appeared to partially interact with Rab5: S34N mutant (Figure 2D). Collectively, these results suggest that several key amino acids of Rin1 are critical for its interaction with Rab5.
Analysis of the GEF activity of Rin1
To identify key residues in the Vps9 domain of Rin1 that are required for the release of GDP from Rab5, we used an established quantitative approach for measuring the kinetics of the release of fluorescent mant-GDP from Rab5 [42, 44]. In our experiments, Rab5: wild type loaded with mant-GDP was incubated with different amount of Rin1 as described in Material and Methods. Then, the GEF activity profile of several Rin1 mutants was compared with that of Rin1: wild type proteins. The fusion-proteins were purified to >85% homogeneity as determined by SDS-PAGE and, more importantly, all of the Rin1 mutant proteins were expressed in a soluble form at Rin1: wild type levels.
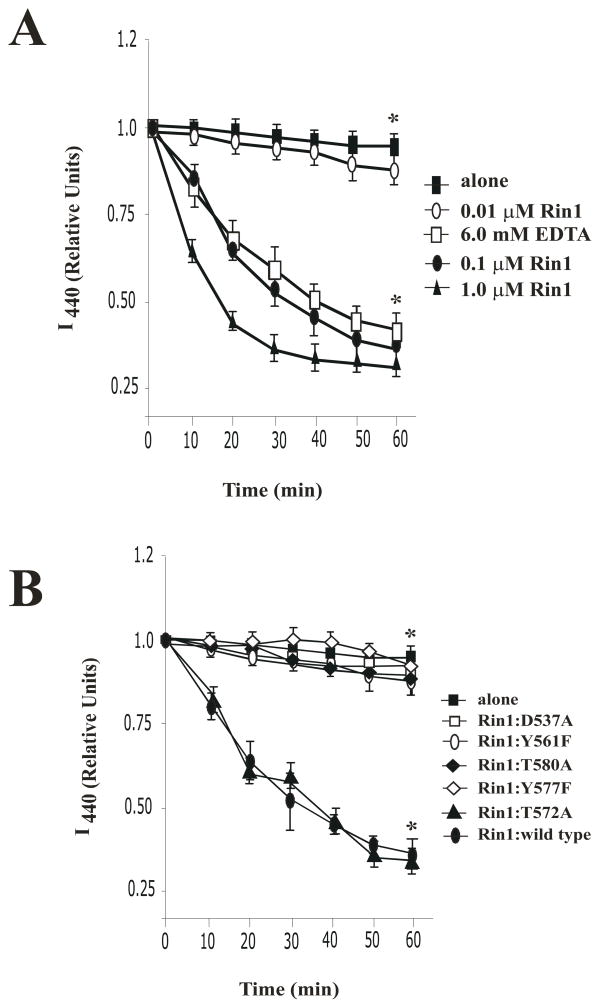
In Figure 3, we show the ability of Rin1: wild type and its mutants to catalyze the release of mant-GDP from Rab5. First, under our experimental conditions, we observed a concentration dependent release of mant-GDP from Rab5 (Figure 3A). Furthermore, the addition of 6 mM EDTA, which reduces the free Mg2+ concentration, greatly stimulates the rate of nucleotide release, indicating that the absence of detectable activity was not due to an inability to release mant-GDP, but instead reflected the high specificity of Rin1 proteins (Figure 3A).
Figure 3. Rin1 mutants affect the release of mant-GDP bound to Rab5.
(A) Kinetics of mant-GDP release from 100 pM His-Rab5 in the absence (
 ) and presence of 0.01 μM (
) and presence of 0.01 μM (
 ), 0.1 μM (
), 0.1 μM (
 ), 1 μM (←) Rin1: wild type proteins or following the addition of 6 mM EDTA (
), 1 μM (←) Rin1: wild type proteins or following the addition of 6 mM EDTA (
 ) for the indicated time at 30°C. The data are presented as means ± SD of three independent experiments, n=3 *P < 0.001. (B) Release of mant-GDP from 100 pM His-Rab5 alone (
) for the indicated time at 30°C. The data are presented as means ± SD of three independent experiments, n=3 *P < 0.001. (B) Release of mant-GDP from 100 pM His-Rab5 alone (
 ) or in the presence of 0.1 μM Rin1 constructs for the indicated time at 30°C as described in Material and Methods. The data are presented as means ± SD of three independent experiments, n=3 *P < 0.001. Each independent experiment was carried out in duplicate. Relative Units (R.U.) indicate the amount of fluorescence measured at 440 nm (I440 nm).
) or in the presence of 0.1 μM Rin1 constructs for the indicated time at 30°C as described in Material and Methods. The data are presented as means ± SD of three independent experiments, n=3 *P < 0.001. Each independent experiment was carried out in duplicate. Relative Units (R.U.) indicate the amount of fluorescence measured at 440 nm (I440 nm).
Second, we observed a selective effect of Rin1 mutants on the release of mant-GDP from Rab5. Mutation of four residues (Asp 537, Tyr 561, Tyr 577 and Thr 580) severely impaired catalytic efficiency (Figures 3B). Moreover, mutation of two more residues showed a strong deleterious effect on the catalytic activity of Rin1 (i.e., Rin1: P541A and Rin1: E574A mutants) (data not shown). However, mutation on residue Thr 572 resulted in no significant defects (Figures 3B), whereas mutation of other residues had minimal effect (Tyr 506, Tyr 523, and Tyr 578) (data not shown). Collectively, these inhibitory effects suggest that several key amino acids are critical in Rin1’s GEF activity for Rab5.
Rin1 mutants selectively regulate the endocytosis of EGF
To further investigate the structure-function relationship of Rin1, we decided to investigate the role of these Rin1 mutants on the internalization of several ligands. Based on earlier work that showed that overexpression of Rin1 in cultured NR6 fibroblasts and HepG2 cells stimulated the internalization of EGF [19] and insulin [26] as well as the uptake of fluid phase marker (i.e., HRP) [26], we were able to confirm this biological activity of Rin1 by expressing several Rin1 mutants in mammalian cells by using a pMX-retrovirus expression system [19]. This experiment demonstrated the specificity and feasibility of a functional assay for Rin1 and its mutants (Figure 4).
First, to determine whether Rin1 mutants affected EGF receptor-mediated endocytosis, we examined the uptake of EGF in NR6 cells expressing various Rin1 mutants. Our results indicate that Rin1 constructs were expressed on approximately the same level and Rin1 was 3.4-fold over-expressed in retrovirus-infected NR6 cells (Figure 4). Cells were treated with 125I-EGF, washed, incubated at 37°C for 6 min, and the amount of internalized 125I-EGF was determined as described in Material and Methods. Overexpression of Rin1: wild type stimulated the uptake of EGF in a time dependent manner (data not shown), whereas the expression of Rin1 mutants failed to stimulate the internalization of EGF (Figure 4A). Specifically, we found that the expression of Rin1: wild type stimulated the internalization of EGF (Rin1: wild type=16.1± 2.6 EGF-uptake) as compared with the uptake of EGF in GFP-control cells (GFP=7.95± 1.9 EGF-uptake) Furthermore, the expression of the Rin1: T572A mutant stimulated the uptake of EGF similar to Rin1: wild type (Rin1: T572A=13.4± 1.6 EGF-uptake, Figure 4A). Interestingly, we observed that the expression of Rin1: Y561F, Rin1: T580A and Rin1: D537A mutants showed a significant decrease in the internalization of EGF. However, we found that both Rin1: D537A and Rin1: Y561F mutants were more potent inhibitors of the uptake of EGF than Rin1: T580A mutant (Figure 4A, compare the uptake of Rin1: T580A [8.1± 2 EGF-uptake] with the uptake of Rin1: D357A [4± 0.1 EGF- uptake] and Rin1: Y561F mutants [4.1± 1.2 EGF-uptake]). Although these three Rin1 mutants failed to activate Rab5, Rin1: D537A and Rin1: Y561F mutants partially interacted with Rab5. In contrast, Rin1: T580A failed to interact with Rab5. Thus, it is possible to speculate that the strong inhibitory effect observed with Rin1: D537A and Rin1: Y561F mutants on the uptake of EGF can be attributed, at least in part, to a partial interaction of these two mutants with Rab5.
Second, we also examined the effect of Rin1 mutants on HRP accumulation in NR6 cells (Figure 4B). Upon expression of Rin1: wild type, the uptake of HRP was increased by approximately 2.1-fold. The expression of Rin1: T572A mutant also stimulated HRP uptake approximately 1.73-fold higher than control cells. In contrast, the expression of Rin1: D537A, Rin1: T580A and Rin1: Y561F mutants showed a significant inhibition of HRP uptake as compared with control cells (Figure 4B). Interestingly, we also observed that Rin1: D537A and Rin1: Y561F mutants significantly inhibited the HRP uptake as compared with the uptake of Rin1: wild type and Rin1: T580A mutant.
Finally, we investigated the effect of Rin1 mutants on fluid phase endocytosis stimulated by EGF. NR6 cells were incubated with 2 mg/ml of HRP in either the absence or presence of 100 ng/ml EGF for different times (Figure 4C). After each incubation, the HRP activity was measured in control and EGF- stimulated cells. As expected, the expression of Rin1: wild type significantly increased the uptake of HRP either in the absence or presence of EGF with respect to control cells. However, when the effect of Rin1: D537A was examined in this assay, we observed a significant inhibition of HRP uptake after the addition of EGF (Figure 4C). Collectively, these observations suggest that Rin1: D537A and Rin1: Y561F mutants selectively inhibited both receptor and fluid phase endocytosis.
Expression of Rin1 mutants inhibited the enlargement of Rab5-positve endosomes
Previous studies have shown that expression of Rab5: Q79L, a GTPase defective mutant, activated endocytosis and induced the formation of enlarged endosomes in intact cells [45, 46]. It has also been shown that either the addition of EGF to cells expressing Rab5: wild type or the co-expression of Rin1 and Rab5 wild type proteins induced the formation of enlarged Rab5-positive endosomes [19, 24]. Interestingly, the sizes of the enlarged Rab5-positive endosomes were similar to those endosomes observed when activated Rab5: Q79L was expressed [19]. Based on these observations, we then examined the effect of the expression of Rin1 mutants on the formation of enlarged Rab5-positive endosomes in intact cells. Cells expressing Rin1: wild type and its mutants were transfected with Rab5: wild type. After transfection, the cells were fixed and prepared for confocal microscopy. Endosome morphology was examined as described in Material and Methods.
When control cells were transfected with Rab5: wild type alone, we observed a typical punctuate distribution of Rab5 in the cytosol of the cells, which was consistent with previously reported data [46]. In contrast, Rab5-positive endosomes were significantly enlarged in cells co-expressing both Rin1 and Rab5 wild type proteins (Figures 5A). Interestingly, the size of the enlarged Rab5-positve endosomes observed in the later experimental conditions (i.e., cells co-expressing both Rin1 and Rab5 proteins) was comparable to the enlarged Rab5-positive endosomes found in cells expressing the Rab5: Q79L mutant alone. These results suggest that the presence of Rin1: wild type induced the formation of enlarged Rab5- positive endosomes (i.e., fusion between endosomes), which is consistent with previously published data [19]. Therefore, we next examined the effects of several Rin1 mutants on the formation of enlarged Rab5- positive endosomes. Specifically, we found that the expression of Rin1: Y561F and Rin1: T580A mutants was unable to induce enlarged Rab5-positive endosomes (Figure 5A), which is consistent with the fact that these two mutants were unable to activate Rab5 (Figure 3B). Moreover, when Rab5: wild type was co-expressed with Rin1: T572A, a typical punctuate of endosomal distribution with a significant increased size of Rab5-positive endosomes was observed (Figure 5A). These results are not surprising since the Rin1: T572A mutant showed a GEF activity similar to Rin1: wild type (Figure 3B).
In addition, we found that Rab5 co-localized with Rin1: wild type on enlarged endosomes. However, Rin1: Y561F mutant partially co-localized with Rab5: wild type on endosomes. Specifically, we found that Rin1: Y561F partially co-localized on smaller Rab5-positive endosomes rather than enlarged Rab5- positive endosomes induced by the co-expression of Rin1 and Rab5 wild type proteins (Figure 5B). In addition, these two Rin1 mutants (i.e., Rin1: Y561F and Rin1: T580A) showed a more diffuse pattern throughout the cells (Figure 5A, B). Collectively, these results suggest that specific residues of the Vps9 domain of Rin1 regulate the formation of enlarged Rab5-positve endosomes.
To confirm and quantify our morphological observations, we then determined the size of enlarged Rab5-positive endosomes in the presence of Rin1 constructs. To examine how the expression of Rin1 mutants affected the formation of enlarged Rab5-positive endosomes, cells were co-transfected with Rab5: wild type and several Rin1 mutants. After the transfection, the cells were processed for confocal microscopy as described in Material and Methods. Rab5-positive endosomes were identified with confocal immunofuorescence microscopy and the digitized projection images of endosomes were analyzed using the NIH Image program. As shown in Figure 5C, the expression of Rab5: wild type alone created endosomes with a mean diameter of 0.22±0.02 μm, while the expression of GFP-Rab5: Q79L showed endosomes with a mean diameter of 0.89±0.15 μm. These results were consistent with previously reported data [46].
In Figure 5C, we found that cells co-expressing Rab5 and Rin1 wild type proteins showed endosomes with a mean diameter of 0.80±0.16 μm. Similar results were also observed when the Rin1: T572A mutant was co-expressed with Rab5: wild type (0.725±0.11 μm). In contrast, the expression of Rin1: T580A and Rin1: Y561F did not induce formation of enlarged Rab5-positive endosomses [i.e., Rin1: T580A (0.189 ± 0.01 μm) and Rin1: Y561F (0.145±0.01 μm)]. Other Rin1 mutants also showed a significant reduction in the diameter of Rab5-positive endosomes [i.e., Rin1: D537A (0.142±0.01 μm), Rin1: P541A (0.195±0.02 μm), Rin1: T574A (0.192±0.01 μm), Rin1: Y577F (0.195±0.01 μm)], which is consistent with the inability of these residues to activate Rab5. Collectively, these morphological observations suggest that several key residues in the Vps9 domain of Rin1 were required to induce enlarged Rab5-positive endosomes, which have also been shown to be critical in Rin1’s GEF for Rab5.
Effect of Rin1 mutants on endosome fusion in vitro
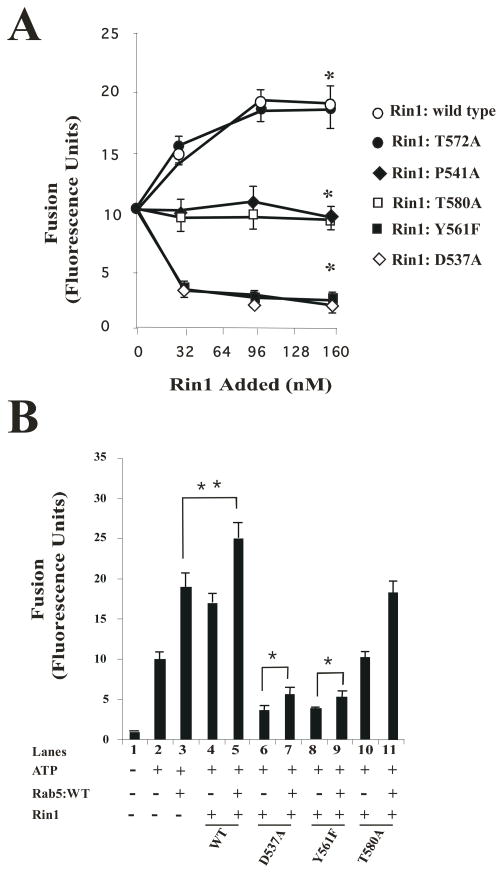
Based on previous data, in which Rab5 played a key role in regulating fusion between endosomes [47], is activated by Rin1 [19], and the results reported here, we investigated the role of Rin1 mutants on the in vitro endosome fusion reaction. The in vitro endosome fusion assay was conducted as described in Material and Methods. Early endosome and cytosol preparations were incubated under fusogenic conditions in the presence of different amounts of Rin1 constructs. We found that the addition of purified Rin1: wild type and Rin1: T572A mutant significantly stimulated endosome fusion in a concentration-dependent manner with respect to control (Figure 6A). However, the addition of purified Rin1: T580A and Rin1: P541A mutants failed to stimulate the fusion reaction. Also, we observed that the addition of purified Rin1: D537A and Rin1: Y561A strongly inhibited endosome fusion as compared to control and Rin1: wild type. In addition, we also found that both Rin1: D537A and Rin1: Y561F mutants significantly inhibited the fusion reaction as compared to control, Rin1: P541A and Rin1: T580A mutants (Figure 6A). Thus, our results confirm the role of Rin1 in the fusion reaction and also suggest that specific residues of Rin1 were required to regulate the endosome fusion in vitro.
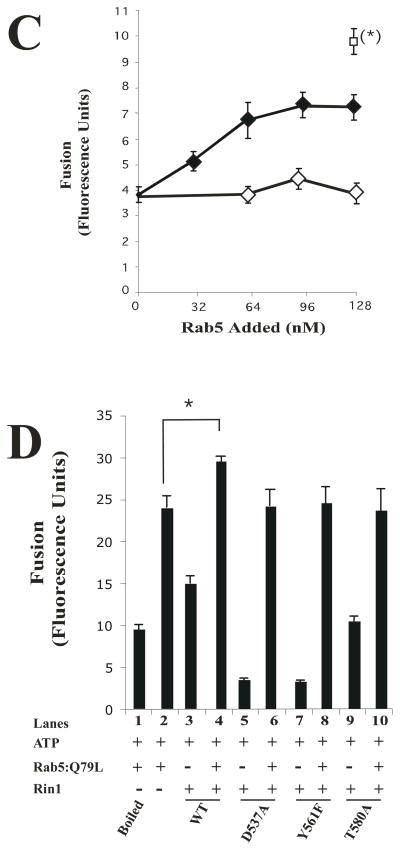
Figure 6. Effect of Rin1 mutants on the endosome fusion reaction.
(A) Fusion assays were performed under standard conditions as described in Material and Methods in the presence of 1.5 mg/ml cytosol alone, or supplemented with different amount (0 to 160 nM) of Rin1 constructs (Rin1: wild type, Rin1: T572A, Rin1: P541A, Rin1: T580A, Rin1: Y561F, or Rin1: D537A). Values are means ± SD, n=3 *P< 0.001. (B) Endosomes were incubated in the presence of 1.5 mg/ml cytosol (Lanes 1 to 11) without (Lane 1, −ATP) or with (Lanes 2 to 11, +ATP) ATP regenerating system and subsequently allowed to fuse as indicated in the graph. Lanes 3 to 11 show the fusion reaction either in the absence (Lanes 1, 2, 4, 6, 8 and 10) or in the presence of purified Rab5: wild type (Lanes 3, 5, 7, 9, and 11). Lane 3 shows the fusion reaction in the presence of cytosol supplemented only with 32 nM Rab5: wild type. Lanes 5, 7, 9 and 11 show the fusion reaction in the presence of 32 nM Rab5: wild type supplemented with 32 nM Rin1: wild type (Lane 5), Rin1: D537A (Lane 7), Rin1: Y561F (Lane 9), or Rin1: T580A (Lane 11). Lanes 4, 6, 8, and 10 show the fusion reaction in the presence of 32 nM Rin1 proteins alone. Values are means ± SD, n=3, *P <0.005, **P <0.001, . (C) Fusion assays were performed in the presence of 1.5 mg/ml cytosol containing 32 nM Rin1: D537A mutants supplemented with different amounts of purified boiled (
 ) and non-boiled Rab5: wild type (
) and non-boiled Rab5: wild type (
 ). The asterisk (*) indicates the fusion activity using complete cytsosol (
). The asterisk (*) indicates the fusion activity using complete cytsosol (
 ). Values are means ± SD, n=4 *P <0.001. (D) Endosomes were incubated with of 1.5 mg/ml cytosol in the presence of ATP-regenerating system (Lanes 1 to 10) and subsequently allowed to fuse as indicated in the graph. Lane 1 and 2 show the fusion reaction in the presence of boiled and non-boiled 32 nM Rab5: Q79L mutant, respectively. Lanes 3 to 10 show the fusion reaction in the presence of 32 nM Rin1 proteins as shown in the graph either in the absence (Lanes 3, 5, 7, and 9) or in the presence of purified 32 nM Rab5: Q79L mutant (Lanes 2, 4, 6, 8 and 10). Lanes 3, 5, 7, and 9 show the fusion reaction in the presence of 32 nM Rin1 proteins alone. Values are means ± SD, n=3 *P <0.001. Purified Rab5 proteins were complexed with REP-1 and then added to the fusion reaction as indicated in Material and Methods.
). Values are means ± SD, n=4 *P <0.001. (D) Endosomes were incubated with of 1.5 mg/ml cytosol in the presence of ATP-regenerating system (Lanes 1 to 10) and subsequently allowed to fuse as indicated in the graph. Lane 1 and 2 show the fusion reaction in the presence of boiled and non-boiled 32 nM Rab5: Q79L mutant, respectively. Lanes 3 to 10 show the fusion reaction in the presence of 32 nM Rin1 proteins as shown in the graph either in the absence (Lanes 3, 5, 7, and 9) or in the presence of purified 32 nM Rab5: Q79L mutant (Lanes 2, 4, 6, 8 and 10). Lanes 3, 5, 7, and 9 show the fusion reaction in the presence of 32 nM Rin1 proteins alone. Values are means ± SD, n=3 *P <0.001. Purified Rab5 proteins were complexed with REP-1 and then added to the fusion reaction as indicated in Material and Methods.
Because the expression of different Rin1 mutants altered the formation of enlarged Rab5-positive endosomes (Figure 5C), affected the endosome fusion (Figure 6A), and Rab5 was shown to be a regulator of early endosome fusion [47], our data suggest that specific residues of Rin1 were involved in the fusion reaction. To confirm this contention, we examined the requirement of Rab5 in the endosome fusion reaction regulated by Rin1. As expected, when Rin1: wild type was supplemented with an equimolar ratio of Rab5: wild type (i.e., 1:1 molar ratio of Rab5 to Rin1), we observed a significant stimulation of the endosome fusion reaction as compared to either control, Rin1: wild type alone, or Rab5: wild type alone (Figure 6B). Interestingly, supplementation of Rab5: wild type with an equimolar ratio of either Rin1 mutants (i.e., Rin1: D537A and Rin1: Y561F) partially rescued the fusion reaction (Figure 6B). These observations suggest that the inhibitory effect on the endosome fusion by Rin1: D537A and Rin1: Y561F may be associated by a direct sequestration of Rab5 by these two mutants. Thus, an excess of Rab5 should rescue the inhibitory effect of these Rin1 mutants. Consistent with idea, we observed a recovery (~60% of control) of the endosome fusion when Rin1 constructs were supplemented with an excess of Rab5: wild type (Figure 6C). As expected, heat inactivated Rab5: wild type failed to support he fusion reaction (Figure 6C). Therefore, the excess of Rab5 added to the fusion reaction that did not interact with Rin1 mutants may be available for other factors (i.e., GDI, Rab5-GEF) existing in the in vitro system. Thus, it is possible that Rab5-GEFs (i.e., Rabex-5 and Alsin) may further regulate the activation of Rab5 in the fusion reaction since these two Rab5-GEFs have been shown to be required in the fusion reaction [48, 49]. In addition, we observed that the addition of Rin1: T580A mutant did not block the fusogenic activity of Rab5: wild type on the fusion reaction (Figure 6B), which is consistent with the observation that Rin1: T580A did not interact with Rab5 (Figures 1B, 2C and 2D) and failed to induce the formation of enlarger of Rab5-positve endosomes (Figure 5C). Taken together, these results strengthen the possibility that the inhibitory effect of Rin1 mutants of the endosome fusion is associated to specific residues of Rin1 involved in the interaction and activation of Rab5.
Additionally, the supplementation of the fusion reaction with Rab5: Q79L GTP hydrolysis-defective mutant reversed the inhibitory effect of Rin1 mutants and also stimulated endosome fusion (Figure 6D). Boiled Rab5: Q79L as control had no effect (Figure 6D). In other control experiments, the fusion reaction was carried out in the absence of ATP (−ATP, Figure 6B). These results made it unlikely that inhibition by Rin1 mutants was due to nonspecific effects. Therefore, this strong inhibitory effect of Rin1: D537A and Rin1: Y561F mutants on endosome fusion may be explained by the fact that, in addition to their ability to partially interact with Rab5, these two Rin1 mutants failed to activate Rab5. Collectively, these in vitro results suggest that several amino acids played an important role in the fusion reaction, and have also been shown to be critical in Rin1’s GEF for Rab5 and for the formation of enlarged Rab5-positive endosomes.
DISCUSSION
We have investigated the structure-function relationship of Rin1 by mutational, biochemical and morphological analysis. On one hand, the analysis of the three dimensional structure of Rabex-5 provides a guide for the characterization of the Vps9 domain of Rin1 [35]. On the other hand, the Vps9 domain described in several proteins may differ both structurally and functionally in specific cells. For example, Rabex-5 knockout mice develop skin inflammation [34], while deletion of part or the entire Vps9 domain of Alsin protein may be involved in a mechanism for motor-neuron degeneration [28]. In addition, Rin1 knockout mice showed an alteration in the long-term potentiation and formation of aversive memories [33]. Therefore, we investigated the relationship between the structure, function, and cellular response of different Rin1 mutants.
First, we observed that of the ten Rin1 mutants created, four (P541A, E574A, Y577F, T580A) were unable to interact with Rab5, whereas two others (D537A, Y561F) partially interacted with Rab5 (Figure 1B). Furthermore, four others (T572A, Y506F, Y523F, Y578F) interacted when compared to interaction between Rin1 and Rab5 wild type proteins. Consistent with these results, we found that the Rin1 mutants that failed to interact with Rab5 were unable to activate it. Interestingly, we also observed that both Rin1: D537A and Rin1: Y561F mutants that partially interacted with Rab5, also failed to activate Rab5. Collectively, these results suggest that specific residues of Rin1 play a more relevant role either in the interaction with Rab5, in the GEF activity for Rab5, or both.
Second, we found that the expression of Rin1: D537A and Rin1: Y561F mutants, but not the Rin1: T580A mutant, significantly inhibited the endocytosis of HRP and EGF (Figure 4). Consistent with these observations, we found that these two mutants (i.e., Rin1: D537A and Rin1: Y561F) that showed a reduced affinity for Rab5 also failed to activate Rab5. Strikingly, the expression of Rin1 T580A mutant did not stimulate the endocytosis of either ligand, which can be attributed, at least in part, to the fact that this mutant failed to interact with Rab5. In addition, we observed that Rin1: wild type induced the formation of enlarged Rab5-positive endosomes and as expected, co-localized with Rab5 on enlarged Rab5-positive endosomes (0.80± 0.16μm). Remarkably, we also found that Rin1: Y561F did not induce the formation of enlarged Rab5-positive endosomes (Figure 5C) and also showed a more diffuse pattern than Rin1: wild type through the cells (Figure 5B). Moreover, Rin1: Y561F partially co-localized with Rab5 on significantly smaller endosomes (0.145± 0.01μm). Furthermore, Rin1: T580A, but not Rin1: T572A, did not support the formation of enlarged Rab5-positive endosomes (Figure 5C). This inhibition by these Rin1: D537A and Rin1: Y561F mutants may result from a direct sequestration of Rab5, which may help to explain their strong dominant inhibitory effect in our in vitro and in vivo assays.
Third, as further confirmation of our observations, we examined the effect of several Rin1 mutants on the in vitro endosome fusion assay. We observed that the addition of purified Rin1: wild type and Rin1: T572A mutant, but not the addition of Rin1: D537A, Rin1: Y561F, Rin1: P541A, and Rin1: T580A mutants, significantly stimulated the fusion reaction (Figure 6). In addition, the stimulation of the endosome fusion by Rin1: wild type was concentration-dependent. Interestingly, we also observed that the addition of purified Rin1: D537A and Rin1: Y561A mutants strongly inhibited endosome fusion as compared to Rin1: T580A. Thus, the evidence suggests that Rin1 mutants selectively regulated the endosome fusion reaction because it affected the in vitro activation of Rab5 and their expression regulated the formation of enlarged Rab5-positive endosomes in intact cells. Our observations suggest a specific role of Rin1 mutants in the endosome fusion in vitro since the addition of exogenous Rab5: Q79L rescued the fusion reaction inhibited by the Rin1 mutants. Furthermore, the supplementation of Rab5: wild type to Rin1: wild type, but not to Rin1: D537A and Rin1: Y561F mutants, stimulated the fusion reaction. However, the fusion reaction inhibited by Rin1: Y561F and Rin1: D537A mutants were partially rescued by the addition of an excess of Rab5 proteins, suggesting the possibility that, in addition to Rin1, other Rab5-GEF proteins may also be involved in the fusion reaction. Consistent with this observation, it has been shown that Rabex-5 and Alsin proteins were also involved in the endosome fusion reaction in vitro [48, 49]. Clearly, additional work should be carried out to further investigate the role of these Rab5-GEFs in the endosome fusion. Therefore, this strong inhibitory effect of Rin1: D537A and Rin1: Y561F mutants on endosome fusion may be explained by the fact that, in addition to their ability to partially interact with Rab5, these two Rin1 mutants failed to activate Rab5, suggesting a mechanism by which Rin1 mutants modulate in vivo and in vitro Rab5 function.
We also observed that the addition of Rin1: T580A did not block the endosome fusion stimulated by Rab5: wild type, which is consistent with the observation that Rin1: T580A failed to interact with Rab5. Furthermore, this observation was also supported by the fact that this mutant did not stimulate the formation of enlarged Rab5-positve endosomes (Figure 5C), or endocytosis of EGF (Figure 4A).
Noticeably, it has been reported that the Rabex-5 equivalent of Rin1: D537A and Rin1: T580A (Rabex-5: D313A and Rabex-5: T357A) inhibited Rab5 function [35]. It has been shown that in Rabex-5 the residue Asp313 contacted both the invariant P-loop lysine and switch II backbone, whereas Thr357 contributed to binding pockets in the interswitch and switch II of Rab5 [35]. Therefore, it is reasonable to speculate that in Rin1 residues, Asp537 and Thr580 may behave similarly to their equivalent residues in Rabex-5. Consistent with this prediction, we have shown that Rin1: D537A and Rin1: T580A were critical determinants for exchange activity. At present, the residues Asp 537 may be critical for exchange activity since the corresponding residue (Asp 313) in Rabex-5 is involved in GEF activity as well [35]. However, it is possible that the Asp 537 residue in the Vps9 domain of Rin1 may play a key role in the interaction and/or activation of Rab5. In contrast, residue Thr 580 in the Vps9 domain of Rin1 may play a more prominent role in the interaction of Rin1 with Rab5 rather than in the activation of Rab5 since a substitution mutation at residue of 580 from threonine (Thr) to alanine (Ala) completely inhibited its interaction with Rab5 (Figure 1B and 2C). Furthermore, Rin1: D537A, Rin1: Y561F and to a lesser extent Rin1: T580A, (i.e., approximately 87%) were expressed to the same level as Rin1: wild type, which suggest a minor effect on protein stability. Therefore, at least two possibilities may account for the observations that these residues critically affect the exchange activity of Rin1 for Rab5, and therefore Rab5 function. First, these residues may be directly involved in the interaction with Rab5. Second, these Rin1 mutants (i.e., Rin1: D537A and Rin1: Y561F mutants) may cause changes in other regions of the molecule that contribute to the disruption of protein-protein interaction between Rab5 and Rin1. While the latter possibility cannot be ruled out, it seems less likely since we have found that the Rin1 mutants investigated in this paper were expressed approximately to the same level as Rin1: wild type, which suggest a minor effect on protein stability. Based on structural models of the Vsp9, SH2 and RA domains of Rabex-5 [35], Src [50] and Ste50 adaptor [51] proteins, 3-D structural figures of Rin1 were constructed and view with I-tasser (http://zhang.bioinformatics.ku.edu//I-tasser/) and Pymol (http://www.pymol.com), respectively. Our observations suggest that the introduction of a single substitution (i.e., Y561F) only causes limited local changes rather altering the overall conformation of the molecule Rin1 (Galvis, A and Barbieri M.A., unpublished data). Alternatively, it is also possible that these Rin1 mutants may also affect the interaction of Rin1 with other factors, including proteins that interact though the SH2 domain of Rin1 [16, 23, 26], and particularly Ras, which has been shown to potentiate the Rab5-GEF activity of Rin1 and alter its function in endosome fusion [19]. Future experiments using appropriate approaches will be required to examine whether Rin1: D537A and Rin1: Y561F mutations affect the interaction of Rin1 with Ras. Collectively, our data support a critical role of residues Asp 537 and Tyr 561 on Rin1 function since these Rin1 residues were involved in the in vitro activation of Rab5 and therefore blocked Rab5 function (i.e., internalization of EGF, formation of enlarged Rab5-positive endosomes and endosome fusion).
In addition, we found that Rin1: T572A (Figures 3B, 4A, 5A and 5C), Rin1: Y506F, Rin1: Y523F, and Rin1: Y578F mutants (data not shown) were able to activate the small GTPase Rab5 in vitro, to stimulate the endocytosis of EGF, and induce the formation of enlarged Rab5-positve endosomes as compared to Rin1: wild type, which suggests that none of these residues abolished the biological function of Rin1 (Supplementary Figure 1). Finally, the identification of these key structural features of the Vps9 domain of Rin1 provide novel information on the mechanism of how Rin1 regulates endocytosis, endosome fusion and also indicate the structural features that are required for the inhibitory effect of these mutants.
Acknowledgments
The authors gratefully acknowledge Dr. D. Lambright, Dr. P. Stahl, Dr. G. Blanco, Dr. Somarelli, and Dr. B. Horazdovsky for providing reagents. This work was supported by National Institutes of Health Grant S06GM8205 (to MAB). We thank Travis Murphy and Jacqueline Zayas for helpful suggestions on the manuscript. We also thank Florida International University Foundation.
The abbreviations used are
- EGF-receptor
Epidermal growth factor-receptor
- Vps9
vacuolar protein sorting 9
- Rin1
Ras interference 1
- Gapex-5
RasGAP exchange factor Rab5
- ALS2
amyotrophic lateral sclerosis 2
- RAP-6
Rab5 Activator Protein-6
- EEA1/NT
Early Endosomal Antigen 1/N-terminus
- VARP
Vps9 ankyrin repeat protein
- STAM
signal transduction adaptor molecule
- HRP
horseradish peroxidase, RME6, Receptor mediated endocyosis 6
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Honegger AM, Schmidt A, Ullrich A, Schlessinger J. Separate endocytic pathways of kinase- defective and -active EGF receptor mutants expressed in same cells. J Cell Biol. 1990;110:1541–1548. doi: 10.1083/jcb.110.5.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hopkins CR, Miller K, Beardmore JM. Receptor-mediated endocytosis of transferrin and epidermal growth factor receptors: a comparison of constitutive and ligand-induced uptake. J Cell Sci Suppl. 1985;3:173–186. doi: 10.1242/jcs.1985.supplement_3.17. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter G. The EGF receptor: a nexus for trafficking and signaling. Bioessays. 2000;22:697–707. doi: 10.1002/1521-1878(200008)22:8<697::AID-BIES3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 4.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 5.Burke P, Schooler K, Wiley HS. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol Biol Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taub N, Teis D, Ebner HL, Hess MW, Huber LA. Late endosomal traffic of the epidermal growth factor receptor ensures spatial and temporal fidelity of mitogen-activated protein kinase signaling. Mol Biol Cell. 2007;18:4698–4710. doi: 10.1091/mbc.E07-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meisenhelder J, Suh PG, Rhee SG, Hunter T. Phospholipase C-gamma is a substrate for the PDGF and EGF receptor protein-tyrosine kinases in vivo and in vitro. Cell. 1989;57:1109–1122. doi: 10.1016/0092-8674(89)90048-2. [DOI] [PubMed] [Google Scholar]
- 8.Wahl MI, Nishibe S, Kim JW, Kim H, Rhee SG, Carpenter G. Identification of two epidermal growth factor-sensitive tyrosine phosphorylation sites of phospholipase C-gamma in intact HSC-1 cells. J Biol Chem. 1990;265:3944–3948. [PubMed] [Google Scholar]
- 9.Kudlow JE, Buss JE, Gill GN. Anti-pp60src antibodies are substrates for EGF-stimulated protein kinase. Nature. 1981;290:519–521. doi: 10.1038/290519a0. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X, Sorkin A. Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol Biol Cell. 2002;13:1522–1535. doi: 10.1091/mbc.01-11-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu P, Margolis B, Skolnik EY, Lammers R, Ullrich A, Schlessinger J. Interaction of phosphatidylinositol 3-kinase-associated p85 with epidermal growth factor and platelet-derived growth factor receptors. Mol Cell Biol. 1992;12:981–990. doi: 10.1128/mcb.12.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fazioli F, Minichiello L, Matoska V, Castagnino P, Miki T, Wong WT, Di Fiore PP. Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. Embo J. 1993;12:3799–3808. doi: 10.1002/j.1460-2075.1993.tb06058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fazioli F, Minichiello L, Matoskova B, Wong WT, Di Fiore PP. eps15, a novel tyrosine kinase substrate, exhibits transforming activity. Mol Cell Biol. 1993;13:5814–5828. doi: 10.1128/mcb.13.9.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galisteo ML, Dikic I, Batzer AG, Langdon WY, Schlessinger J. Tyrosine phosphorylation of the c-cbl proto-oncogene protein product and association with epidermal growth factor (EGF) receptor upon EGF stimulation. J Biol Chem. 1995;270:20242–20245. doi: 10.1074/jbc.270.35.20242. [DOI] [PubMed] [Google Scholar]
- 15.Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 16.Barbieri MA, Kong C, Chen PI, Horazdovsky BF, Stahl PD. The SRC homology 2 domain of Rin1 mediates its binding to the epidermal growth factor receptor and regulates receptor endocytosis. J Biol Chem. 2003;278:32027–32036. doi: 10.1074/jbc.M304324200. [DOI] [PubMed] [Google Scholar]
- 17.Han L, Colicelli J. A human protein selected for interference with Ras function interacts directly with Ras and competes with Raf1. Mol Cell Biol. 1995;15:1318–1323. doi: 10.1128/mcb.15.3.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han L, Wong D, Dhaka A, Afar D, White M, Xie W, Herschman H, Witte O, Colicelli J. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc Natl Acad Sci U S A. 1997;94:4954–4959. doi: 10.1073/pnas.94.10.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cel. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Waldron RT, Dhaka A, Patel A, Riley MM, Rozengurt E, Colicelli J. The RAS effector RIN1 directly competes with RAF and is regulated by 14-3-3 proteins. Mol Cell Biol. 2002;22:916–926. doi: 10.1128/MCB.22.3.916-926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afar DE, Han L, McLaughlin J, Wong S, Dhaka A, Parmar K, Rosenberg N, Witte ON, Colicelli J. Regulation of the oncogenic activity of BCR-ABL by a tightly bound substrate protein RIN1. Immunity. 1997;6:773–782. doi: 10.1016/s1074-7613(00)80452-5. [DOI] [PubMed] [Google Scholar]
- 22.Kong C, Su X, Chen PI, Stahl PD. Rin1 interacts with signal-transducing adaptor molecule (STAM) and mediates epidermal growth factor receptor trafficking and degradation. J Biol Chem. 2007;282:15294–15301. doi: 10.1074/jbc.M611538200. [DOI] [PubMed] [Google Scholar]
- 23.Hu H, Bliss JM, Wang Y, Colicelli J. RIN1 is an ABL tyrosine kinase activator and a regulator of epithelial-cell adhesion and migration. Curr Biol. 2005;15:815–823. doi: 10.1016/j.cub.2005.03.049. [DOI] [PubMed] [Google Scholar]
- 24.Barbieri MA, Roberts RL, Gumusboga A, Highfield H, Alvarez-Dominguez C, Wells A, Stahl PD. Epidermal growth factor and membrane trafficking. EGF receptor activation of endocytosis requires Rab5a. J Cell Biol. 2000;151:539–550. doi: 10.1083/jcb.151.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carney DS, Davies BA, Horazdovsky BF. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Hunker CM, Giambini H, Galvis A, Hall J, Kruk I, Veisaga ML, Barbieri MA. Rin1 regulates insulin receptor signal transduction pathways. Exp Cell Res. 2006;312:1106–1118. doi: 10.1016/j.yexcr.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Horiuchi H, Lippe R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, Zerial M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 28.Otomo A, Hadano S, Okada T, Mizumura H, Kunita R, Nishijima H, Showguchi-Miyata J, Yanagisawa Y, Kohiki E, Suga E, Yasuda M, Osuga H, Nishimoto T, Narumiya S, Ikeda JE. ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum Mol Genet. 2003;12:1671–1687. doi: 10.1093/hmg/ddg184. [DOI] [PubMed] [Google Scholar]
- 29.Sato M, Sato K, Fonarev P, Huang CJ, Liou W, Grant BD. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat Cell Biol. 2005;7:559–569. doi: 10.1038/ncb1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunker CM, Galvis A, Kruk I, Giambini H, Veisaga ML, Barbieri MA. Rab5-activating protein 6, a novel endosomal protein with a role in endocytosis. Biochem Biophys Res Commun. 2006;340:967–975. doi: 10.1016/j.bbrc.2005.12.099. [DOI] [PubMed] [Google Scholar]
- 31.Lodhi IJ, Chiang SH, Chang L, Vollenweider D, Watson RT, Inoue M, Pessin JE, Saltiel AR. Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab. 2007;5:59–72. doi: 10.1016/j.cmet.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, He X, Fu XY, Chang Z. Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J Cell Sci. 2006;119:1053–1062. doi: 10.1242/jcs.02810. [DOI] [PubMed] [Google Scholar]
- 33.Dhaka A, Costa RM, Hu H, Irvin DK, Patel A, Kornblum HI, Silva AJ, O’Dell TJ, Colicelli J. The RAS effector RIN1 modulates the formation of aversive memories. J Neurosci. 2003;23:748–757. doi: 10.1523/JNEUROSCI.23-03-00748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam SY, Tsai M, Snouwaert JN, Kalesnikoff J, Scherrer D, Nakae S, Chatterjea D, Bouley DM, Galli SJ. RabGEF1 is a negative regulator of mast cell activation and skin inflammation. Nat Immunol. 2004;5:844–852. doi: 10.1038/ni1093. [DOI] [PubMed] [Google Scholar]
- 35.Delprato A, Merithew E, Lambright DG. Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell. 2004;118:607–617. doi: 10.1016/j.cell.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Gournier H, Stenmark H, Rybin V, Lippe R, Zerial M. Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. Embo J. 1998;17:1930–1940. doi: 10.1093/emboj/17.7.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lippe R, Miaczynska M, Rybin V, Runge A, Zerial M. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 39.Mattera R, Tsai YC, Weissman AM, Bonifacino JS. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub- interacting motif and a zinc finger domain. J Biol Chem. 2006;281:6874–6883. doi: 10.1074/jbc.M509939200. [DOI] [PubMed] [Google Scholar]
- 40.Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 41.Barbieri MA, Fernandez-Pol S, Hunker C, Horazdovsky BH, Stahl PD. Role of rab5 in EGF receptor-mediated signal transduction. Eur J Cell Biol. 2004;83:305–314. doi: 10.1078/0171-9335-00381. [DOI] [PubMed] [Google Scholar]
- 42.Simon I, Zerial M, Goody RS. Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J Biol Chem. 1996;271:20470–20478. doi: 10.1074/jbc.271.34.20470. [DOI] [PubMed] [Google Scholar]
- 43.Diaz R, Mayrga L, Stahl P. In vitro fusion of endosomes following receptor-mediated endocytosis. J Biol Chem. 1988;263:6093–6100. [PubMed] [Google Scholar]
- 44.Zhu Z, Dumas JJ, Lietzke SE, Lambright DG. A helical turn motif in Mss4 is a critical determinant of Rab binding and nucleotide release. Biochemistry. 2001;40:3027–3036. doi: 10.1021/bi002680o. [DOI] [PubMed] [Google Scholar]
- 45.Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. Embo J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts RL, Barbieri MA, Pryse KM, Chua M, Morisaki JH, Stahl PD. Endosome fusion in living cells overexpressing GFP-rab5. J Cell Sci. 1999;112(Pt 21):3667–3675. doi: 10.1242/jcs.112.21.3667. [DOI] [PubMed] [Google Scholar]
- 47.Bucci C, Parton RG, Mather IH, Stunnerberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pahtway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 48.Devon RS, Orban PC, Gerrow K, Barbieri MA, Schwab C, Cao LP, Helm JR, Bissada N, Cruz-Aguado R, Davidson TL, Witmer J, Metzler M, Lam CK, Tetzlaff W, Simpson EM, McCaffery JM, El-Husseini AE, Leavitt BR, Hayden MR. Als2-deficient mice exhibit disturbances in endosome trafficking associated with motor behavioral abnormalities. Proc Natl Acad Sci U S A. 2006;103:9595–600. doi: 10.1073/pnas.0510197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalesnikoff J, Rios EJ, Chen CC, Barbieri MA, Tsai M, Tam SY, Galli SJ. Roles of RabGEF1/Rabex-5 domains in regulating Fc epsilon RI surface expression and Fc epsilon RI- dependent responses in mast cells. Blood. 2007;109:5308–17. doi: 10.1182/blood-2007-01-067363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waksman G, Kominos D, Robertson S, Pant N, Baltimore D, Birge R, Cowburn D, Hanafusa H, Mayer B, Overduin M, Resh M, Rios C, Silverman L, Kuriyan J. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine- phosphorylated peptides. Nature. 1992;358:646–653. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- 51.Truckses DM, Bloomekatz JE, Thorner J. The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:912–28. doi: 10.1128/MCB.26.3.912-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffenberg S, Sanford JC, Liu S, Daniel DS, Tuvin M, Knoll BJ, Wessling-Resnick M, Dickey BF. Biochemical and functional characterization of a recombinant GTPase, Rab5, and two of its mutants. J Biol Chem. 1995;270:5048–56. doi: 10.1074/jbc.270.10.5048. [DOI] [PubMed] [Google Scholar]