CK1delta binds and phosphorylates the microtubule plus-end–binding protein EB1 and promotes centrosome translocation to the immunological synapse in T cells.
Abstract
Although termed central body, the centrosome is located off-center in many polarized cells. T cell receptor (TCR) engagement by antigens induces a polarity switch in T cells. This leads to the recruitment of the centrosome to the immunological synapse (IS), a specialized cell–cell junction. Despite much recent progress, how TCR signaling triggers centrosome repositioning remains poorly understood. In this paper, we uncover a critical requirement for the centrosomal casein kinase I delta (CKIδ) in centrosome translocation to the IS. CKIδ binds and phosphorylates the microtubule plus-end–binding protein EB1. Moreover, a putative EB1-binding motif at the C terminus of CKIδ is required for centrosome translocation to the IS. We find that depletion of CKIδ in T lymphocytes and inhibition of CKI in epithelial cells reduce microtubule growth. Therefore, we propose that CKIδ–EB1 complexes contribute to the increase in microtubule growth speeds observed in polarized T cells, a mechanism that might serve to generate long-stable microtubules necessary for centrosome translocation.
Introduction
The microtubule cytoskeleton is important for a variety of cellular processes such as cell migration, division, or vesicle trafficking. Microtubule organization in interphase animal cells is largely dependent on the centrosome (Doxsey et al., 2005). Although centrosomes nucleate symmetric arrays of microtubules, relocating the centrosome from the geometric center of the cell can generate asymmetries in microtubule organization. The ability to reposition the centrosome is particularly crucial in helper and cytotoxic T cells (Poo et al., 1988; Huse et al., 2008). When a T lymphocyte encounters a cognate antigen-presenting cell, a signaling platform known as the immunological synapse (IS) assembles around the T cell receptor (TCR; Monks et al., 1998; Grakoui et al., 1999; Dustin, 2008). Formation of the IS is accompanied by the remodeling of the actin cytoskeleton and the repositioning (referred to as polarization hereafter) of the centrosome to the IS, where it contacts the plasma membrane (Geiger et al., 1982; Kupfer and Dennert, 1984; Das et al., 2002; Stinchcombe et al., 2006; Billadeau et al., 2007).
Intact microtubule cytoskeleton is essential for centrosome polarization, as microtubule poisons inhibit the process (Kupfer and Dennert, 1984). Proteins so far implicated in centrosome polarization include mediators of TCR signaling such as the tyrosine kinases Lck, ZAP70, and Fyn (Lowin-Kropf et al., 1998; Blanchard et al., 2002; Martín-Cófreces et al., 2006; Tsun et al., 2011). Recent work indicates that in cytotoxic T cells, Fyn controls centrosome polarization, whereas Lck is responsible for its docking at the plasma membrane (Tsun et al., 2011). Important roles for centrosome polarization have also been assigned to several cytoskeletal proteins and their regulators like the formins FMNL1 and DIA1 (Gomez et al., 2007), the tubulin deacetylase HDAC6 (Serrador et al., 2004), and the dynein/dynactin complex (Combs et al., 2006). Dynein and dynactin are involved in centrosome positioning during directed cell migration (Etienne-Manneville and Hall, 2001; Palazzo et al., 2001; Dujardin et al., 2003; Gomes et al., 2005; Manneville et al., 2010). Recruitment of dynein to the IS by diacylglycerol has been shown to promote centrosome translocation (Quann et al., 2009), reinforcing the current model that dynein at the cell cortex pulls on centrosomal microtubules and hence facilitates centrosome movement toward the IS (Combs et al., 2006; Kim and Maly, 2009).
Many aspects of dynein function are modulated by dynactin (Berrueta et al., 1999; Vale, 2003). The dynactin subunit p150glued belongs to a group of proteins called +TIPs that track the growing tips of microtubules. Many +TIPs are recruited to the plus-ends of microtubules by members of the EB protein family, +TIPs themselves (Watson and Stephens, 2006; Slep, 2010). EB1 and EB3 contain a C-terminal α-helical sequence consisting of a dimerization motif, an EB homology domain, and an acidic tail (Honnappa et al., 2005; Slep et al., 2005; Slep, 2010). Some +TIPs like p150glued bind the acidic tail of EB1 via a cytoskeleton-associated protein–glycine-rich (CAP-Gly) domain (Steinmetz and Akhmanova, 2008). Other +TIPs interact with the EB homology domain through a Ser-X-Pro-Ile (SxIP) polypeptide (Honnappa et al., 2005; Slep et al., 2005; Akhmanova and Steinmetz, 2008; Honnappa et al., 2009). Such SxIP-containing factors include the adenomatous polyposis coli protein APC (Honnappa et al., 2009). Collectively, +TIPs are important modulators of microtubule behavior. Their interaction with EB1 brings them close to microtubule plus-ends, organelles, and the cell cortex, where they can locally modulate microtubule behavior and control cell migration, polarity, and differentiation (Tirnauer et al., 1999; Rogers et al., 2002; Green et al., 2005; Mimori-Kiyosue et al., 2005; Vaughan, 2005; Coquelle et al., 2009). The role of +TIPs in T cell centrosome polarization has not yet been investigated in detail, but, IQGAP1, a protein that bridges microtubule plus-ends with actin filaments, participates in the process (Fukata et al., 2002; Stinchcombe et al., 2006).
To gain better understanding into the molecular mechanism responsible for centrosome polarization, a candidate gene approach has led us to the casein kinase 1 (CKI) family of proteins. In yeast, casein kinase plays an important role in chromosome segregation during meiosis and ER-to-Golgi transport (Petronczki et al., 2006; Ishiguro et al., 2010; Katis et al., 2010; Lord et al., 2011). CKIs have been implicated in several cell polarity–dependent processes such as spindle orientation in Caenorhabditis elegans (Walston et al., 2004; Panbianco et al., 2008), Drosophila melanogaster planar polarity establishment (Klein et al., 2006), or vertebrate gastrulation (Tsai et al., 2007). Several members of the CKI family are core centrosome components (Andersen et al., 2003). Indicative of potential roles in microtubule organization, CKIδ mediates neurite outgrowth and the function of brain-specific microtubule-associated proteins (Behrend et al., 2000; Li et al., 2004; Wolff et al., 2005; Flajolet et al., 2007; Greer and Rubin, 2011). In addition, CKIs regulate the circadian clock in mammals (Ning et al., 2004; Eide et al., 2005; Xu et al., 2005).
Here, we report that the centrosomal kinase CKIδ promotes efficient centrosome polarization to the IS. We further show that CKIδ binds and phosphorylates EB1 and promotes microtubule growth. Our findings indicate that CKIδ-dependent regulation of microtubule behavior is important for centrosome translocation to the IS during T cell activation.
Results
Inhibitors of the CKI family of kinases prevent centrosome polarization to the IS
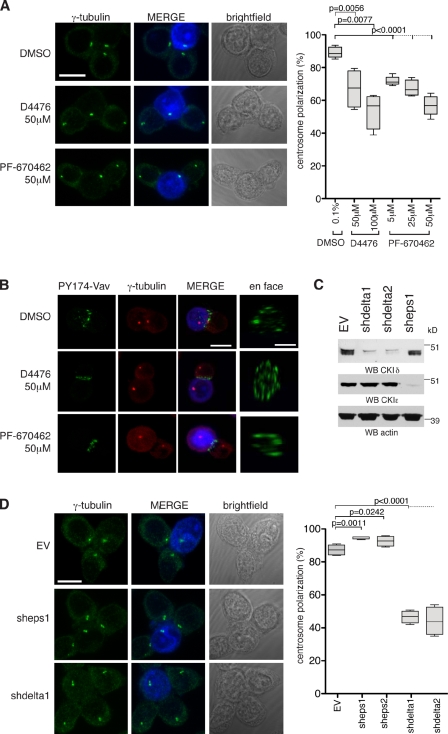
We first investigated the effect of CKI inhibition on centrosome polarization using the CKI-specific inhibitors D4476 (Rena et al., 2004) and PF-670462 (Badura et al., 2007). Jurkat T cells were conjugated with staphylococcal enterotoxin E (SEE)–pulsed Raji B cells to induce IS assembly. Jurkat cells were treated with DMSO, D4476, or PF-670462 for 2 h before conjugate formation and then scored for centrosome polarization to the IS. In DMSO, centrosomes polarized to the IS in 89% ± 3 of cells. 50 and 100 µM D4476 reduced the polarization efficiency to 67% ± 11 and 54% ± 11, respectively, whereas 25 and 50 µM PF-670462 reduced it to 67% ± 5 and 57% ± 6, respectively (Fig. 1 A; for scoring criteria and examples, see Fig. S1 [A and B]). The wide data distribution of D4476-treated cells in the box plot in Fig. 1 A may reflect the poor solubility of D4476 in aqueous media (Rena et al., 2004). D4476 also inhibits the TGF-β type I receptor ALK5 kinase (Rena et al., 2004), but, as the ALK5-inhibitor LY-364947 did not perturb centrosome polarization, the effect of D4476 is likely to be independent of ALK5 (Fig. S1 C).
Figure 1.
CKIδ regulates TCR-mediated centrosome polarization to the IS. (A) Conjugates of Jurkat and SEE-pulsed Raji cells in the presence of the indicated drugs. Centrosomes are stained with anti–γ-tubulin antibody. Raji cells are blue in merge. The box plot shows quantification of centrosome polarization to the IS based on scoring criteria in Fig. S1 A (n = 4 experiments; 200 conjugates/experiment). (B) Conjugates of Jurkat and SEE-pulsed Raji cells in the presence of the indicated drugs. Cells are stained with anti–PY174-Vav and anti–γ-tubulin antibodies. Raji cells are blue in merge. (C) Cytoplasmic cell extracts of Jurkat cells containing stably integrated EV (control), vector encoding CSNK1D shRNA (clones shdelta1 and shdelta2), or CSNK1E shRNA (clone sheps1) are immunoblotted with antibodies against CKIδ or CKIε. Actin serves as a loading control. WB, Western blotting. (D) Conjugates of control (EV), CKIε- or CKIδ-depleted Jurkat cell clones, and SEE-pulsed Raji cells. Centrosomes are stained with anti–γ-tubulin antibody. Raji cells are blue in merge. The box plot shows quantification of centrosome polarization to the IS (n = 4 experiments; 200 conjugates/experiment). In the box plots, whiskers are set at minimum and maximum, and horizontal lines mark the median, whereas boxes indicate the interquartile range (25–75%). Bars:10 µm; (en face) 5 µm.
Impaired centrosome polarization can arise from defective IS formation and/or signaling. To address whether CKI inhibition affected TCR signaling or IS assembly, we assayed the localization of phosphorylated Vav (Tyr174), a ZAP-70 effector (Tybulewicz, 2005). The inhibitors did not preclude either the phosphorylation or the accumulation of Vav at the IS, indicating that inhibition of CKI does not impede either processes (Fig. 1 B).
CKIδ promotes centrosome polarization to the IS
D4476 and PF-670462 are both ATP-competitive inhibitors, and it was therefore crucial to confirm that their effect on centrosome polarization was specific to their action against CKI kinases. As D4476 and PF-670462 inhibit CKIδ and CKIε (Rena et al., 2004; Badura et al., 2007) and CKIδ is highly active in isolated lymphocytes (Maritzen et al., 2003), we first assayed the role of the closely related CKIδ and CKIε in centrosome polarization using a loss-of-function approach. Short hairpin RNAs (shRNAs) targeting either CSNK1D or CSNK1E were embedded into sequences derived from the miR30 microRNA and introduced into Jurkat cells (Chang et al., 2006). The same vector without a hairpin sequence (referred to as empty vector [EV]) was used as a control. Of two CSNK1E and five CSNK1D-specific short hairpin sequences, one CSNK1E (sheps) and two CSNK1D (shdelta and shdeltaB) hairpins achieved substantial depletions (60–70% reduction) in cell populations. Single clones of CKIδ- or CKIε-depleted cells were isolated, and the most efficiently depleted ones were characterized further (88 and 90% reduction in CKIδ levels in clones shdelta1 and shdelta2, respectively; 92% reduction in CKIε levels in clone sheps1; Fig. 1 C). Note that shdelta1 and shdelta2 are independent Jurkat cell clones that contain the same shdelta short hairpin sequence. In humans, the CKIδ gene can be alternatively spliced to produce two mRNAs that differ only in their last coding exon: the canonical 415-aa-long protein and a shorter 409-aa protein (hereafter called CKIδ-2). The shdelta RNA sequence targets the 3′ untranslated region (UTR), and, therefore, shdelta cells are depleted of both isoforms.
EV, sheps1, shdelta1, and shdelta2 cells were viable in long-term cell culture and exhibited no morphological or cell cycle defects (unpublished data). When conjugated to SEE-pulsed Raji cells, only CKIδ-depleted cells displayed an impairment in centrosome polarization (Figs. 1 D and S2 A). Whereas clones shdelta1 and shdelta2 were used for subsequent experiments, centrosome polarization was similarly impaired by a different CSNK1D-specific hairpin sequence (Fig. S2 B). Thus, despite sharing over 80% sequence identity with CKIε, only CKIδ is required for centrosome polarization. CKIα, another centrosomal CKI (Gross et al., 1995), was dispensable for centrosomal polarization (Fig. S2 C).
CKIδ is required for translocation of the centrosome to the IS
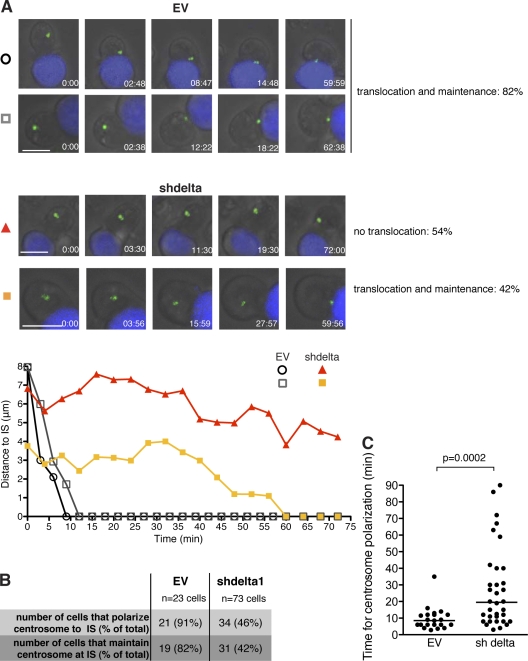
Centrosome polarization involves the translocation of the centrosome to the IS and its subsequent docking to the plasma membrane (Stinchcombe et al., 2006). To identify which of these is abrogated by CKIδ depletion, we followed centrosome movement in real time in Raji-conjugated Jurkat cells transiently transfected with the GFP-tagged centrosome component, centrin-1. 82% of EV cells translocated and maintained their centrosomes at the IS for the duration of filming (Fig. 2 [A and B] and Videos 1 and 2). In contrast, only 46% of CKIδ-depleted cells translocated their centrosomes to the IS. We observed no major defect in centrosome maintenance at the IS in these cells (Fig. 2 [A and B] and Videos 3 and 4). However, in those CKIδ-depleted cells that polarized their centrosome, the centrosome took longer to reach the IS (Fig. 2 C). Therefore, we conclude that CKIδ is required for efficient centrosome translocation to the IS.
Figure 2.
CKIδ is required for centrosome translocation to the IS. (A) Still images from time-lapse videos showing conjugate formation between Jurkat cells transfected with GFP-centrin1 (green) and SEE-pulsed Raji cells (blue). Control (EV; top; Videos 1 and 2) and CKIδ-depleted Jurkat cells (shdelta; bottom; Videos 3 and 4) are shown. Bar, 10 µm. The percentages next to panels indicate the frequency of the particular phenotype. The plots below depict the distance between the centrosome and the IS as a function of time; time 0 is when the conjugate forms. Tracks are symbol- and color-matched to individual cells. The shdelta cell (orange) polarizes its centrosome with a delay. (B) Summary of the time-lapse data collected. (C) Distribution of the time required for centrosome polarization to the IS in control (EV) and CKIδ-depleted (shdelta) cells. Median values are marked by horizontal lines.
CKIδ is dispensable for early TCR signaling and high-order IS assembly
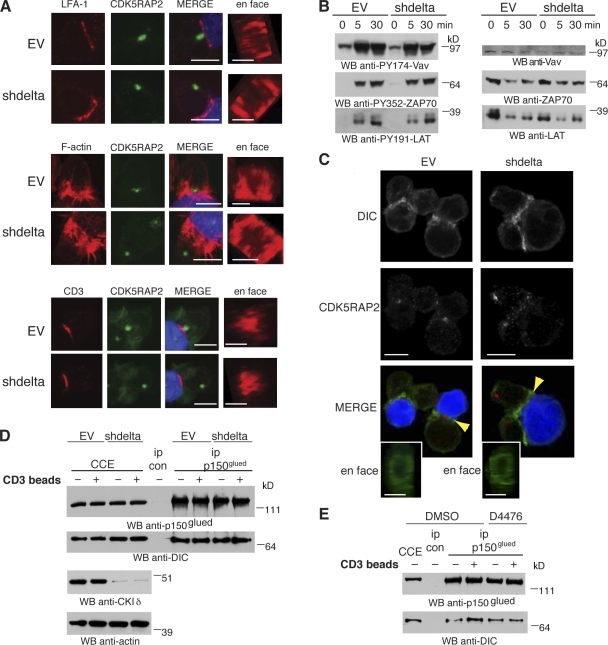
We examined IS assembly in EV and CKIδ-depleted Jurkat cells by studying three key features of the IS (Monks et al., 1998; Billadeau et al., 2007): LFA-1 integrin accumulation, F-actin cap formation, and clustering of the TCR subunit CD3. All three features appeared normal in the absence of CKIδ (Fig. 3 A), indicating that high-order IS assembly can occur without the kinase. Next, we assayed the kinetics of early TCR signaling events in CKIδ-depleted cells. Tyrosine phosphorylations of ZAP70 (Tyr352), LAT (Tyr191), and Vav (Tyr174) were similar in EV and CKIδ-depleted cells over a time course of 30 min after conjugate formation with SEE-pulsed Raji cells (Fig. 3 B). Anti-CD3 antibody–coated beads that cluster TCRs on the surface of T cells can induce centrosome polarization. Unlike the IS of Jurkat–Raji cell conjugates, this IS forms without integrin engagement and thus drives centrosome polarization solely by TCR signaling (Tsun et al., 2011). As anti-CD3–activated CKIδ-depleted cells still displayed a centrosome polarization defect, we conclude that CKIδ is required for centrosome polarization independent of integrin engagement (59 ± 1.6% centrosome polarization in CKIδ-depleted cells compared with 82 ± 2.5% in control cells in two experiments [150 cells/experiment]).
Figure 3.
CKIδ is dispensable for IS formation and early TCR signaling. (A) Conjugates of control (EV) or CKIδ-depleted (shdelta) Jurkat and SEE-pulsed Raji cells are stained with anti–LFA-1 antibody (top), phalloidin (F-actin; middle), or anti-CD3 antibody (bottom). Centrosomes are stained with anti-CDK5RAP2 antibody. Raji cells are in blue in merge. (B) Cytoplasmic cell extracts of control (EV) or CKIδ-depleted (shdelta) Jurkat cells were lysed at different time points after conjugate formation with SEE-pulsed Raji cells and were immunoblotted with the indicated antibodies. WB, Western blotting. (C) Conjugates between control (EV; left) or CKIδ-depleted (shdelta; right) Jurkat and SEE-pulsed Raji cells are stained with anti-DIC (green in merge) and anti-CDK5RAP2 (red in merge) antibodies. Arrowheads highlight DIC accumulation at the IS. (D and E) Cytoplasmic cell extracts (CCE) were prepared from control (EV) and CKIδ-depleted (shdelta; D) or DMSO- and D4476-treated (E) Jurkat cells that were unconjugated (−) or conjugated (+) to anti-CD3 antibody beads. Immunoprecipitations (ip) were performed with random IgG (ip con) or anti-p150glued (ip p150glued) antibodies. Bars: 10 µm; (en face) 5 µm.
Cortical localization of the retrograde microtubule motor dynein has been implicated in centrosome translocation to the IS (Combs et al., 2006; Martín-Cófreces et al., 2008; Quann et al., 2009). Thus, impaired centrosome polarization in the absence of CK1δ could reflect inefficient recruitment of dynein to the IS. This is unlikely, however, as dynein accumulated at the IS of CKIδ-depleted cells (Fig. 3 C). Cytoplasmic dynein acts with dynactin to mediate centrosome translocation to the IS (Martín-Cófreces et al., 2008). Could CKIδ depletion affect the formation of dynein/dynactin complexes in cells? To eliminate molecular contamination from Raji cells, immunoprecipitation was performed on Jurkat cells activated by anti-CD3–coated beads. The interaction between dynein and dynactin is mediated by the dynein intermediate chain (DIC) and the p150glued subunit of dynactin (Vaughan and Vallee, 1995). Binding between DIC and p150glued persisted in both CKIδ-depleted (Fig. 3 D) and in D4476-treated cells (Fig. 3 E), indicating that dynein/dynactin complexes assemble independently of CKIδ. Therefore, CKIδ is dispensable for IS assembly and early TCR signaling.
CKIδ is a microtubule-associated protein in T cells
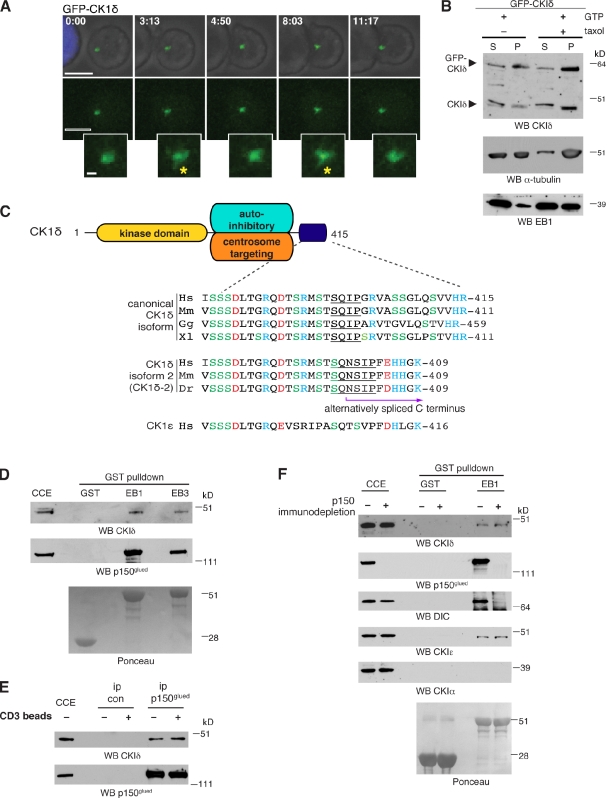
Consistent with previous studies (Andersen et al., 2003; Sillibourne et al., 2002; Greer and Rubin, 2011), CKIδ is an integral centrosomal protein in Jurkat cells (Fig. S2 D). Time-lapse imaging confirmed the presence of GFP-tagged CKIδ at the centrosome of Jurkat cells during activation (Fig. 4 A), but no GFP signal was detected at the IS. GFP-CKIδ also appeared on dynamic fiberlike structures that extended out of the centrosome, reminiscent of microtubule ends (Fig. 4 A and Video 5). To enable better visualization of this behavior, we tried to increase expression levels of GFP-CKIδ, but high levels proved toxic to Jurkat cells. Expression of GFP-CKIδ diminished endogenous kinase levels in Jurkat cells, revealing tight cellular control of kinase levels (Fig. S3 A). Consistent with its dynamic localization pattern, both endogenous and GFP-fused CKIδ copelleted with microtubules (Fig. 4 B). Thus, CKIδ is a microtubule-associated protein in lymphocytes, similar to the brain (Behrend et al., 2000; Li et al., 2004; Wolff et al., 2005; Flajolet et al., 2007).
Figure 4.
CKIδ interacts with the microtubule plus-end–binding proteins EB1 and p150glued. (A) Still images from time-lapse imaging (Video 5) showing conjugate formation between a Jurkat cell expressing GFP-CKIδ (green) and an SEE-pulsed Raji cell (blue). Insets correspond to higher magnifications of the centrosomal region. Asterisks mark fiberlike extrusions near the centrosome. Bars: 10 µm; (inset) 1 µm. (B) In vitro microtubule-pelleting assay. Pure tubulin was incubated in the presence of GTP without (−) or with (+) taxol and then added to cytoplasmic extracts of Jurkat cells transfected with GFP-CKIδ. High-speed supernatants (S) and pellets (P) were collected and immunoblotted with antibodies as indicated. WB, Western blotting. (C) A schematic view of key functional domains of CKIδ: kinase, autoinhibitory (Longenecker et al., 1998), and centrosome-targeting (Greer and Rubin, 2011) domains. Sequence alignments of the extreme C termini of two CKIδ isoforms and CKIε are shown below. Acidic and basic amino acids are in red and blue, respectively. Hs, Homo sapiens; Mm, Mus musculus; Gg, Gallus gallus; Xl, Xenopus laevis; Dr, Danio rerio. (D) Cytoplasmic cell extracts (CCE) of Jurkat cells were subjected to pull-down assays with GST, GST-EB1 (EB1), or GST-EB3 (EB3) and were immunoblotted with antibodies as indicated. Recombinant GST products are shown in Ponceau S staining below. (E) Jurkat cells were unconjugated (−) or conjugated (+) with anti-CD3–coated beads. Cytoplasmic cell extracts were processed for immunoprecipitation (ip) with random IgG (ip con) or anti-p150glued (ip p150glued) antibodies and immunoblotted with the indicated antibodies. (F) Cytoplasmic cell extracts of Jurkat cells were mock depleted with random IgG (−) or immunodepleted of p150glued (+). Depleted extracts were then processed for pull-down assays with GST or GST-EB1 (EB1) and immunoblotted with the indicated antibodies. Recombinant GST products are visible in Ponceau S staining below.
CKIδ interacts with microtubule plus-end proteins EB1 and p150glued
Several +TIPs interact with the microtubule end-binding proteins EB1 and EB3 via an SxIP motif (Honnappa et al., 2009). At the C terminus of CKIδ, we identified a SQIP motif embedded in a sequence rich in serine and basic amino acids (Fig. 4 C). GST pull-down assays revealed a specific interaction between endogenous CKIδ (from Jurkat cell extracts) and recombinant GST-EB1 and -EB3 (Fig. 4 D). Similar to other SxIP-containing +TIPs (Honnappa et al., 2009), interaction between endogenous CKIδ and EB1 seems too weak to be detectable. However, CKIδ coimmunoprecipitated with the dynactin subunit p150glued from Jurkat cell extracts (Fig. 4 E). As p150glued binds EB1 directly (Askham et al., 2002), it could potentially mediate the interaction between EB1 and CK1δ. To address whether this is indeed the case, GST-EB1 pull-downs were performed from cell lysates previously immunodepleted of p150glued. As reported before, GST-EB1 failed to precipitate DIC from p150glued-depleted lysates, consistent with p150glued mediating the binding between DIC and EB1 (Fig. 4 F; Berrueta et al., 1999). In contrast, binding between CK1δ and GST-EB1 persisted in the absence of p150glued, implying that the interaction between CK1δ and GST-EB1 is independent of p150glued.
At least two other members of the CKI family are centrosomal: CKIα and CKIε. A conserved SxIP motif is present in the C terminus of CKIε but not in CKIα (Fig. 4 C). Consistently, CKIε but not CKIα interacted with GST-EB1 (Fig. 4 F). As CKIδ and CKIε exhibit similar substrate specificity, their kinase domains might also contribute to their interaction with EB1 (Dahlberg et al., 2009). In summary, our results point to the existence of a specific interaction between EB1 and the CKI kinases CKIδ and CK1ε.
Identification of a CKIδ phosphorylation site on EB1
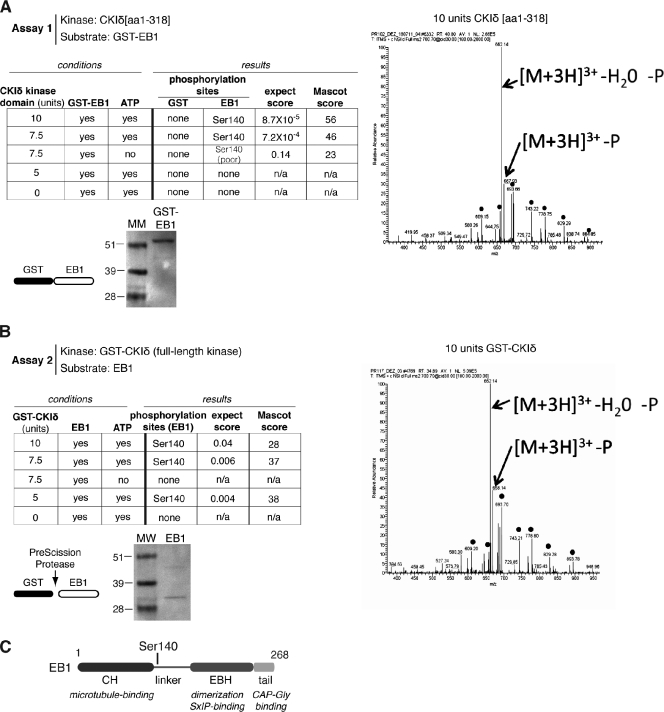
The kinase activity of CKIδ is required for centrosome polarization to the IS (Fig. 1 A). Our finding that CKIδ and EB1 are part of the same protein complex prompted us to investigate whether EB1 is a substrate of CKIδ. We performed two independent in vitro kinase assays and subsequently analyzed the substrates with phosphopeptide mapping using mass spectrometry. The assays were designed with two considerations in mind. First, we wanted to find out whether the kinase required the SQIP domain to phosphorylate EB1, and, therefore, we used the C-terminal–truncated CKIδ[aa 1–318] in assay 1 and the full-length kinase in assay 2. Second, as a GST tag can drive oligomerization, in each assay, only the kinase or the substrate was tagged but not both. In assay 1, untagged CKIδ[aa 1–318] was tested on GST-EB1, whereas in assay 2, GST-CKIδ was tested on untagged EB1 (Fig. 5 A). Mass spectrometry analysis achieved >95% coverage of EB1. The two assays identified the same single phosphopeptide carrying an ATP- and dosage-dependent phosphorylation event on Ser140 (Fig. 5, B and C). Thus, the SQIP motif of CKIδ is dispensable for EB1 phosphorylation in vitro. CKIδ[aa 1–318] did not phosphorylate the GST part of the GST-EB1 fusion protein, reinforcing the specificity of the phosphorylation event on EB1 (Fig. 5 A).
Figure 5.
Identification of a CKIδ phosphorylation site on EB1. (A) A table summarizing the experimental conditions and results for assay 1. Fragmentation spectra were obtained and analyzed for all conditions. The spectrum on the right corresponds to 10 units of kinase. Ponceau S staining shows purified recombinant GST-EB1 used in the assay. Molecular mass (MM) is indicated in kilodaltons. (B) A table summarizing the experimental conditions and results for assay 2. Fragmentation spectra were obtained and analyzed for all conditions. The spectrum on the right corresponds to 10 units of kinase. Ponceau S staining shows the purified recombinant EB1 used in the assay. (A and B) Fragmentation spectra of the triply charged [M + 3H]3+ peptide ion at m/z 700.3676 (A) and m/z 700.367 (B) correspond to the phosphorylated peptide of aa 131–150 with the sequence phospho-QGQETAVAPSLVAPALNKPK of EB1. The predominant y-ion series for both spectra, extending from y11 to y17, confirm the sequence ETAVAP-phosphoS (dots). The fragment ions observed at m/z 667.93 (A) and 668.14 (B) correspond to the neutral loss of H3PO4 from the precursor ions, whereas those observed at m/z 662.14 (A) and 662.14 (B) correspond to the neutral loss of H3PO4 and H2O. GST-CKIδ and CKIδ[1–318] were commercially sourced. (C) A schematic diagram shows the position of the phosphorylation site in EB1. The domains shown are calponin homology (CH) and end binding homology (EBH).
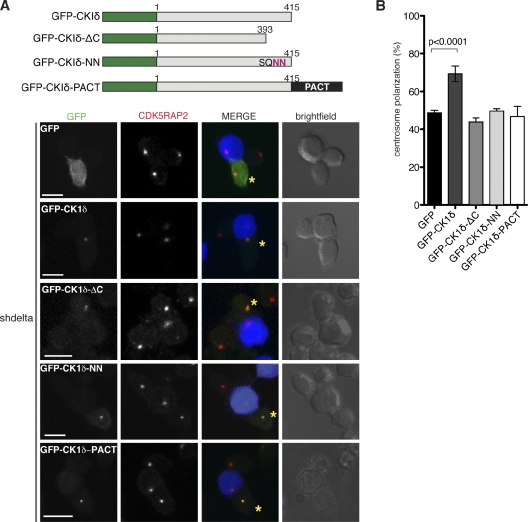
The SQIP motif of CKIδ is essential for centrosome polarization
To address the role of the SQIP motif in CKIδ, we first asked whether the C terminus of CKIδ containing the putative EB1-binding region but lacking the kinase domain (ΔKD) could bind EB1. ΔKD did not bind either EB1 or p150glued (Fig. S3 B), suggesting that the kinase domain is essential for these interactions. Second, we created two CKIδ mutants with disrupted SQIP motifs: CKIδ-ΔC, lacking the last 22 aa, and CKIδ-NN, in which the hydrophobic Ile-Pro dipeptide of the SQIP motif was replaced with Asn residues (Fig. 6 A; Honnappa et al., 2009). GST-EB1 pull-downs were performed as previously described, but, to minimize contribution by endogenous CKIδ, GFP-CKIδ, GFP-CKIδ-ΔC, or GFP-CKIδ-NN was expressed in CKIδ-depleted Jurkat cells. Plasmids encoding these products did not contain CSKN1D 3′ UTR sequences and were therefore resistant to shRNA. Contrary to our expectations, GST-EB1 precipitated both GFP-CKIδ-ΔC and GFP-CKIδ-NN (Fig. S3 C). This was not caused by p150glued, as binding persisted in p150glued-depleted extracts. In addition, GFP-CKIδ-NN copelleted with microtubules (Fig. S3 D). Thus, the SQIP motif of CKIδ is dispensable for interactions with EB1 and microtubules in vitro.
Figure 6.
The SQIP motif of CKIδ is required for centrosome polarization. (A) A schematic view of various GFP-fused CKIδ constructs is shown on top. Examples for conjugates formed between SEE-pulsed Raji cells and CKIδ-depleted Jurkat cells (shdelta) expressing the indicated GFP fusion product (green in merge). Asterisks mark cells with GFP signal. Centrosomes are stained with anti-CDK5RAP2 antibodies (red in merge). Raji cells are blue in merge. Bars, 10 µm. (B) A graph showing quantification of centrosome polarization to the IS in cells expressing the different GFP constructs (n = 4 experiments; at least 50 GFP-positive cells were scored per experiment). Error bars represent SD.
Next, we tested whether the SQIP motif of the kinase has a role in centrosome polarization in vivo. CKIδ-depleted Jurkat cells were transfected with GFP-CKIδ, GFP-CKIδ-ΔC, or GFP-CKIδ-NN and then conjugated to Raji cells. GFP-CKIδ complemented the centrosome polarization defect of CKIδ-depleted cells, whereas GFP-CKIδ-ΔC and GFP-CKIδ-NN did not (Fig. 6 B). Therefore, the SQIP motif in the kinase is dispensable for binding to EB1 in vitro, but this sequence seems important for centrosome polarization in vivo. Interestingly, both mutants were present at the centrosome (Fig. 6 A), implying that centrosomal localization of CKIδ is not sufficient for its function. Dynamic localization of CKIδ is likely to be important for its role because when the kinase is immobilized at the centrosome with a PACT tag (a centrosome-targeting domain derived from the scaffolding protein AKAP450 [Gillingham and Munro, 2000]), it no longer supports centrosome polarization (Fig. 6 B).
The shdelta shRNA targeted both CKIδ isoforms (Fig. S4 A), and, therefore, we asked whether CKIδ-2 could also complement the centrosome polarization defect in CKIδ-depleted cells. Although GFP-CKIδ-2 attenuated the defect, it was less effective than canonical CKIδ (Fig. S4 B). CKIδ-2 also interacted with GST-EB1 despite containing the sequence SQNSIP instead of the consensus SxIP (Fig. S4, C and D). In addition to EB1, GST-fused canonical isoform also precipitated CKIδ-2 (Fig. S4 D). As the two CKIδ isoforms exist in multimeric protein complexes, CKIδ-2 may interact with EB1 via canonical CKIδ, hence explaining the poor performance of GFP-CKIδ-2 in complementing CKIδ-depleted cells (Fig. S4 B).
CKIδ increases microtubule growth speeds in T cells
Centrosome polarization requires an intact microtubule cytoskeleton. Interactions between CKIδ and the microtubule-associated factors EB1 and p150glued suggested that CKIδ could control microtubule behavior. Indeed, microtubules appeared somewhat disorganized in CKIδ-depleted Jurkat cells conjugated to Raji cells (Fig. S5 A). In fixed cells, however, growing microtubule plus-ends were visible in D4476-treated cells (Fig. S5 B).
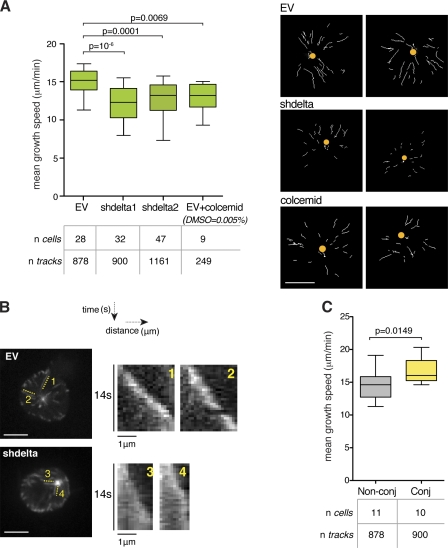
Microtubules switch between phases of growth and shrinkage, a behavior termed dynamic instability (Mitchison and Kirschner, 1984). Transitions from growth to shrinkage or shrinkage to growth are called catastrophes or rescues, respectively. To characterize the effect of CKIδ on microtubule behavior, we first tried to visualize microtubules with GFP-tubulin in live cells, but no microtubule bundles were detectable as a result of the small cytoplasmic volume of interphase T cells. Fluorescently tagged EB1 and EB3 exhibit characteristic cometlike patterns that arise as a result of a transient association between these proteins and growing microtubule plus-ends. The tracking of comets is a widely used method for the analysis of microtubule dynamics (Tirnauer et al., 2002; Goodson et al., 2010). Although this approach seems counterintuitive to study EB-binding proteins, as CKIδ is not required for comet formation by EB1 (Fig. S5 B) or EB3 (Fig. 7 A and Videos 6 and 7), we reasoned that overexpression of EB3 may not mask the effects of the kinase on microtubules (if any). In time-lapse experiments, EB3-GFP signal was filmed for 1 min in a single focal plane containing the centrosome. Data were analyzed with plusTipTracker, an open source software described in Matov et al. (2010). This method identifies +TIP comets using a particle-tracking algorithm (Jaqaman et al., 2008) and selects collinear and sequential growth tracks to reconstruct microtubule trajectories with inferred states such as pause and shrinkage. In Jurkat cells, such trajectories were too short to provide reliable information other than the direct measurement of growth speeds. Nonetheless, CKIδ depletion reduced microtubule growth speeds by 20% in shdelta1 and 16% in shdelta2 cells (Fig. 7 A and Videos 6 and 7). Likewise, manually constructed kymographs of EB3-GFP comets tracked shorter distances in CK1δ-depleted cells (Fig. 7 B), confirming the defective growth observed with plusTipTracker. Next, we asked whether results obtained from CK1δ-depleted cells could be mimicked by treatments with microtubule poisons. We tested different concentrations of colcemid in Jurkat cells and found that 50 nM did not obliterate the microtubule network yet reduced centrosome polarization by 25% and mean microtubule growth speeds by 14% (Figs. 7 A and S5 [C and D]).
Figure 7.
CKIδ promotes microtubule growth in Jurkat cells. (A) Analysis of EB3-GFP comets in control (EV), CKIδ-depleted (shdelta) Jurkat cell clones, and colcemid-treated EV cells. The box plot shows the distribution of mean growth speeds of tracked EB3-GFP comets per cell. On the right, an overlay of EB3 growth tracks collected over 1 min is shown in individual cells (dots mark centrosome position). Bar, 5 μm. (B) Maximum intensity projection of EB3-GFP signal in individual cells over 28 sequential frames (0.5 s per frame). Corresponding kymographs on the right show the growth of individual microtubules in control (EV) and CKIδ-depleted (shdelta) Jurkat cells. Bars, 5 µm. (C) Analysis of EB3-GFP comets in nonconjugated (Non-conj) or Raji-conjugated (Conj) Jurkat cells. The box plot shows the distribution of mean growth speeds of tracked EB3-GFP comets per cell. In box plots, whiskers are set at 5–95 percentiles; horizontal lines mark the median, and boxes indicate interquartile range (25–75%).
As a result of the rapid movement of the centrosome that follows engagement between Jurkat and Raji cells, it proved difficult to assay microtubule behavior in Jurkat cells undergoing centrosome polarization. Instead, we measured microtubule plus-end dynamics in Raji-conjugated Jurkat cells that already polarized their centrosomes. 15–20 min after conjugation, Jurkat cells displayed a 15% increase in mean microtubule growth speeds compared with unconjugated T cells (Fig. 7 C). This raises the possibility of a persistent change in microtubule behavior after activation of T cells.
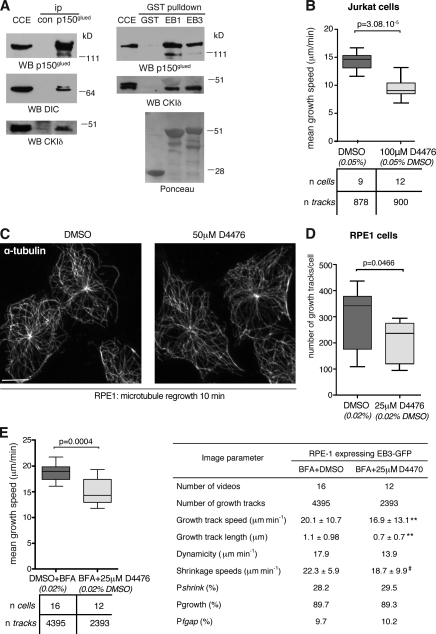
The CKI inhibitor D4476 reduces microtubule growth in epithelial cells
To assess whether CKIδ contributes to microtubule behavior in cell types other than lymphocytes, we first confirmed the interaction between CKIδ, p150glued, and EB1 in retinal pigment epithelial (RPE1) cell extracts (Fig. 8 A). Next, using the CKI inhibitor D4476, we asked whether CKI function is required for microtubule growth in RPE1 cells. Whereas EB3 growth tracks (i.e., comets that persist for a minimum of four frames) were less frequent in Jurkat cells treated with 100 µM D4476 (Fig. 8 B), tracks were significantly reduced in RPE1 cells treated with 50 µM D4476 (not depicted). Yet, this dose did not prevent microtubule formation, as microtubules regrew in 50 µM D4476 after cold-induced depolymerization (Fig. 8 C). 25 µM D4476 also decreased EB3 track numbers but to a lesser extent (Fig. 8 D). The Golgi apparatus nucleates almost half of all microtubules in RPE1 cells (Efimov et al., 2007; Rivero et al., 2009) and is in close proximity with the centrosome. To prevent Golgi-nucleated microtubules from masking an effect on the centrosome, the Golgi apparatus was dispersed by Brefeldin A (BFA), a fungal toxin that disassembles Golgi stacks without affecting microtubule numbers or behavior in RPE1 cells (Efimov et al., 2007). Similar to Jurkat cells, the focal plane containing the centrosome was selected for imaging. The larger size and flat morphology of RPE1 allowed a more in-depth analysis of microtubule behavior. 25 µM D4476 decreased growth and shrinkage speeds and reduced microtubule dynamicity, a parameter describing the distance plus-ends track while shrinking and growing over their lifetime (Fig. 8 E). To address whether this effect was caused by inhibition of CKIδ, single clones of RPE1 cells carrying shdelta shRNA were isolated. The best clones displayed ∼70% depletion, but, when assayed with plusTipTracker, they showed normal plus-end microtubule dynamics (unpublished data). Thus, the extent of CKIδ depletion in RPE1 cells is either suboptimal, or, perhaps, in these cells, CKIδ is redundant with CKIε. Results obtained by D7746 treatment do not distinguish between the respective roles of CKIδ and CKIε in the process.
Figure 8.
Inhibition of CKI suppresses microtubule growth in epithelial cells. (A, left) Cytoplasmic cell extracts (CCE) of RPE1 cells were processed for immunoprecipitation (ip) with random IgG (control [con]) or anti-p150glued (p150glued) antibodies. (right) Cytoplasmic cell extracts of RPE1 cells were subjected to pull-down assays with GST, GST-EB1 (EB1), or GST-EB3 (EB3) and were immunoblotted with antibodies as indicated. Recombinant GST products are visible in Ponceau S staining below. WB, Western blotting. (B) Analysis of EB3-GFP comets in Jurkat cells treated with DMSO or 100 µM D4476. The box plot shows the distribution of mean growth speeds of tracked EB3-GFP comets per cell. (C) Microtubules were depolymerized on ice in DMSO- or 50 µM D447-treated RPE1 cells. Cells were then shifted to 37°C for 10 min to allow microtubule polymerization. Cells are stained with anti–α-tubulin antibodies. Bar, 10 µm. (D) Number of EB3-GFP growth tracks in DMSO- or 25 µM D4476-treated RPE1 cells. The box plot shows the number of EB3-GFP growth tracks per cell. (E) Analysis of EB3-GFP comets in RPE1 cells treated with DMSO or 25 µM D4476. (left) The box plot shows distribution of mean growth speeds of tracked EB3-GFP comets per cell. (right) The table lists microtubule dynamic parameters by plusTipTracker. Pshrink indicates the probability of shrinkage at the end of grouped growth tracks. Pgrowth and Pfgap indicate the percentage of time all microtubules spend in growth or in pause between growth phases, respectively. **, P < 0.0001; #, P = 0.05 (using permutation t tests). In box plots, whiskers are set at 5–95 percentiles; horizontal lines mark the median, and boxes indicate interquartile range (25–75%).
Discussion
Using a candidate gene approach, we identified CKIδ as a crucial kinase required for centrosome polarization to the IS in T cells. CKIδ binds and phosphorylates EB1, and disruption of a putative EB1-binding motif in the kinase perturbs its function in centrosome positioning. We propose that CKIδ-induced changes in microtubule behavior may contribute to centrosome polarization during the immune response.
Insight into the role of centrosome polarization in immune response
As depletion of CKIδ does not preclude IS formation, CKIδ-depleted cells represent a useful tool to dissect the role of centrosome polarization in IS assembly and function. In agreement with previously published observations (Gomez et al., 2007), our results indicate that the absence of centrosome from the IS does not preclude TCR clustering, actin cap formation, and LFA-1 integrin accumulation. The basic bull’s eye pattern of IS components is intact in the absence of a centrosome at the IS (Billadeau et al., 2007). Centrosome translocation to the IS is thought to contribute to sustained TCR signaling, as dynein-depleted cells that did not polarize their centrosomes failed to maintain TCR signaling beyond 15 min (Martín-Cófreces et al., 2008). Despite defects in centrosome polarization, however, CKIδ-depleted cells show normal induction and maintenance of early TCR signaling. This discrepancy may be a result of dynein depletion having a more profound effect on centrosome polarization than depletion of CKIδ (Martín-Cófreces et al., 2008). Alternatively, dynein/dynactin may play additional roles in TCR signaling.
The molecular interplay between CKIδ and EB1
We find that CKIδ binds and phosphorylates EB1 on Ser140 in vitro. Ser140 maps to a flexible linker region that lies between the N- and C-terminal globular domains (Fig. 5 C). The precise role of this linker is unclear, but it is required for full EB1 functionality (Komarova et al., 2009; Buey et al., 2011). Phosphorylation of EB1 by CKIδ may modulate its interactions with CAP-Gly, SxIP-containing proteins, or even microtubules. The kinase might also phosphorylate other EB1-binding proteins. Indeed, the dynactin subunit p150glued coimmunoprecipitated with CKIδ, raising the possibility of a ternary complex between EB1, p150glued, and CKIδ. CKIδ may also be present in a complex with EB1 and APC. APC binds EB1 via a SQIP motif (Su et al., 1995; Honnappa et al., 2009) and is also a CKIδ substrate (Gao et al., 2002).
The conserved SQIP motif is neither necessary nor sufficient for the interaction between EB1 and CKIδ, yet it is required for centrosome polarization in intact cells. How can we explain this discrepancy? One possibility is that CKIδ uses the motif for molecular interactions other than with EB1, and these underlie its role in centrosome polarization. Alternatively, the SQIP motif may contribute to the EB1–CKIδ interaction in intact cells. Indeed, in the case of the highly similar CKIε, there is evidence that the C-terminal tail can influence substrate binding, even though substrate specificity is conferred by the kinase domain (Dahlberg et al., 2009). Therefore, the C-terminal tail of CKIδ could stabilize binding between the kinase and EB1.
The roles of CKIδ in microtubule dynamics and centrosome polarization in T cells
Although our study clearly implicates CKIδ in regulating both microtubule growth and centrosome polarization, a conclusive link between the two roles remains to be established. Centrosome polarization in T cells activated by CD3-coated surfaces does not depend on dynamic microtubules (Baratt et al., 2008). Our results are consistent with this finding, as we had to use 50 nM colcemid to impair centrosome polarization, which is greater than the minimal concentration required for suppressing microtubule dynamics (Jordan and Wilson, 2004; Yang et al., 2010). Instead of dynamic microtubules by itself, centrosome positioning in cells seems sensitive to microtubule length and numbers (Baratt et al., 2008; Kim and Maly, 2009; Maly and Maly, 2010). Indeed, short microtubules cannot polarize centrosomes effectively (Kupfer and Dennert, 1984), whereas overly long microtubules interfere with centrosome polarization to and centrosome positioning within the IS (Baratt et al., 2008). Regulators of microtubule dynamics contribute to steady-state microtubule length (Verde et al., 1992; Tournebize et al., 2000). Therefore, we postulate that when CKIδ function is impaired in cells, lower growth speeds generate microtubule populations of suboptimal length to mediate centrosome translocation. One caveat of using +TIPs to characterize microtubule behavior is the inability to detect catastrophes. However, 50 nM colcemid in CHO cells reduces growth speeds, dynamicity, and shrinkage rates, yet it does not alter catastrophe frequencies (Yang et al., 2010). Thus, the effect of colcemid on centrosome polarization in Jurkat cells is unlikely to be a result of a change in catastrophe rates. Unlike colcemid in CHO cells (Yang et al., 2010), D4476 did not increase pause times in RPE1 cells, indicating that the two drugs have distinct mechanisms of action. The molecular mechanism by which CKIδ promotes microtubule growth remains to be identified but likely targets include EB1 and APC. The clustering of APC at microtubule plus-ends near the leading edge of migrating cells contributes to centrosome reorientation (Etienne-Manneville and Hall, 2003; Etienne-Manneville et al., 2005). The CKI inhibitor D4476 reduced the number of these APC clusters and perturbed directed cell migration (Harris and Nelson, 2010). Therefore, CKI could modulate local microtubule behavior via APC.
EB1 also participates in microtubule anchoring at the centrosome together with dynactin (Askham et al., 2002), CAP350, and FGFR1 oncogene partner (Yan et al., 2006). As an integral centrosomal protein, CKIδ is well placed to contribute to microtubule anchoring at the centrosome (Askham et al., 2002; Ligon et al., 2003; Yan et al., 2006; Vitre et al., 2008). Such a function could be particularly important for centrosome polarization in T cells, as forces generated by dynein at the cell cortex can only translocate the centrosome if these are balanced by microtubule anchoring within the centrosome or other microtubule bundling/cross-linking activity near the centrosome. Colcemid treatment of Jurkat cells partially mimicked effects of CKIδ depletion in terms of lowering microtubule growth speeds and centrosome polarization. Interestingly, similar concentrations of colcemid in CHO cells cause microtubule release from the centrosome (Yang et al., 2010).
Finally, EB1 function has also been implicated in interactions between the cortex and microtubules (Lee et al., 2000; Buttrick et al., 2008). Although we could not observe CKIδ at the IS and our results are more consistent with a role for CKIδ in generating rather than capturing long microtubules, CKIε has been recently reported to promote dynein-dependent trafficking of pigment granules in Xenopus laevis melanophores (Ikeda et al., 2011). Therefore, it is feasible that CKIδ travels to the cortex on microtubule plus-ends with EB1, where it subsequently phosphorylates dynein enriched at the IS. Phosphorylation could enhance the minus-end–directed motility of dynein. Whether or how TCR signaling modulates CKIδ activity remains to be established. Last but not least, the involvement of CKI family members in Wnt and Hedgehog signaling (Price, 2006) raises the question whether these signaling pathways could play a role in modulating cell polarity during immune response.
Materials and methods
Cell lines, drugs, and reagents
The Jurkat E6.1 and Raji cell lines were obtained from Cancer Research UK Cell Services and grown in RPMI1640 containing 5 (Jurkat) or 10% (Raji) FBS supplemented with penicillin and streptomycin. hTERT-RPE1 cells were obtained from the American Type Culture Collection and grown in DMEM:F12 (Life Technologies) containing 10% FBS supplemented with penicillin and streptomycin. For time-lapse imaging, cells were incubated in serum-free Leibovitz medium (Life Technologies). The Phoenix amphotrophic packaging cell line was a gift from M. Narita (Cancer Research UK Cambridge Research Institute, Cambridge, England, UK). D4476 was obtained from Merck Biosciences. PF-670462, LY364947, and colcemid were obtained from Tocris Bioscience. SEE was purchased from Toxin Technology, Inc. BFA was purchased from Enzo Life Sciences. Cell Tracker blue CMAC was obtained from Life Technologies, and Paclitaxel and poly-l-lysine were obtained from Sigma-Aldrich.
Antibodies
The primary antibodies used in this study were mouse monoclonal antibodies against γ-tubulin (GTU88; Sigma-Aldrich); α-tubulin (Dm1α; Sigma-Aldrich); p150glued (BD); DIC (clone 70.1; Sigma-Aldrich); LFA-1 (TS1/22), CD3 (OKT3), and EB1 (Cancer Research UK Clare Hall laboratories); rabbit polyclonal antibodies against CDK5RAP2 (Bethyl Laboratories, Inc.); pericentrin (Covance); ZAP-70, LAT, PY352-ZAP-70, and PY191-LAT (Cell Signaling Technology); and rabbit monoclonal antibodies against Vav and PY174-Vav (Epitomics, Inc.). CKIδ was detected with goat (Abcam) antibodies, CKIδ-2 and CKIα with rabbit antibodies (Bethyl Laboratories, Inc.), and CKIε with a mouse monoclonal antibody (BD). Phalloidin-TRITC and Alexa Fluor–labeled secondary antibodies (Alexa Fluor 488, 555, or 647) were obtained from Life Technologies.
Expression vectors and transfections
Jurkat cells (3 × 106) were transfected with 1.5 µg of plasmid DNA using the cell line nucleofector kit V and the program X-001 (Lonza). Modified MSCV-miR30puro vector was a gift from S. Lowe (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). pEGFP-centrin1 (Piel et al., 2000) was provided by M. Bornens (Institut Curie, Paris, France). EB3-GFP was a gift from A. Akhmanova (Utrecht University, Utrecht, Netherlands). CKIδ and CKIδ-ΔC were subcloned into pEGFP-C1 (Takara Bio Inc.) vector to generate GFP fusion proteins. GFP-CKIδ-NN was generated using the QuikChange site-directed mutagenesis kit (Agilent Technologies). EB1 and CKIδ were inserted in frame into pGex6P2 to create GST fusions. aa 278–415 of CKIδ (ΔKD) was inserted in frame into pMalc4x to create maltose-binding protein (MBP)–ΔKD fusion.
Retrovirus-mediated RNAi
Oligonucleotides containing shRNAs specific for the CSNK1D, CSNK1E, or CSNK1A genes were designed as follows. First, target sequences were selected using the prediction algorithm developed by the Novartis Institutes for BioMedical Research (access to the Software was provided by the Friedrich Miescher Institute). The 19-mer short hairpin sequences used were CSNK1D: shdelta 5′-CGATGAGAACTCTCCTT-3′ (3′ UTR target) and shdeltaB 5′-GGGATGTGAAGCCAGACAA-3′; CSNK1E: sheps 5′-CCTCCGAATTCTCAACATA-3′; and CSNK1A: shalpha 5′-AGAACTTAATTTCAGTATA-3′.
Corresponding 110-bp shRNA oligonucleotides were then designed from the Cold Spring Harbor Hannon laboratory website for cloning into the miR30 context of the modified MSCV-miR30puro vector. XhoI and EcoRI overhangs were added for direct cloning into the modified MSCV-miR30puro vector (a gift from M. Narita and S. Lowe). Empty MSCV-miR30puro vector was used as a control (EV). These constructs were transfected in amphotrophic Phoenix cells by the calcium phosphate method, and viral supernatants were collected 48 h after transfection and were added to Jurkat or RPE1 cells (1 ml of viral supernatant was mixed with 106 cells). Polybrene was added to 5 µg/ml to Jurkat cells, and mixtures were centrifuged at 1,300 g for 45 min. 0.6 µg/ml puromycin was added 48 h after infection to select for stable integration events. Single cell clones were generated by dilution cloning in the presence of puromycin. Depletion of CKI isoforms was assessed by immunoblotting.
Conjugate formation
Raji cells were pulsed with 1 µg/ml SEE for 1 h, stained with 10 µM CMAC for 20 min, washed three times in RPMI 1640 media containing 5% FBS, and mixed with Jurkat cells at a 1:2 ratio (105 Raji and 2.105 Jurkat for one 13-mm-diameter coverslip). Conjugates were allowed to form for 20 min on poly-l-lysine–coated glass coverslips at 37°C before fixation.
Immunofluorescence, image acquisition, and analysis
Cells were fixed in cold methanol (for γ-tubulin, pericentrin, CDK5RAP2, and EB1 staining), 3.7% formaldehyde (for LFA-1, CD3, and phalloidin staining), and 3% PFA (for DIC) or 3% PFA/0.25% glutaraldehyde (for α-tubulin and CDK5RAP2 staining). Permeabilization was achieved with 0.2% saponin or 0.2% Triton X-100 for 2 min. For microtubule regrowth experiments, RPE1 cells were incubated in PBS/0.1% Triton X-100 for 1 min before methanol fixation. Blocking was performed in PBS/5% BSA followed by incubation with the relevant primary and secondary antibodies. Coverslips were mounted in ProLong Gold (Life Technologies) and imaged with a 60× oil/1.40 NA objective on a scanning confocal microscope (Eclipse 90i; Nikon) fitted with a camera (Eclipse C1Si; Nikon). Images were acquired using EZ-C1 software (Nikon). The images presented are 3D projections of z stacks (0.5-µm steps). Images of any individual figure were acquired using the same settings and were imported into Volocity (PerkinElmer) and Photoshop software (Adobe) before being processed in an identical manner. Images were adjusted to use the full range of pixel intensities in Photoshop CS4. The fluorochromes used in this study were Alexa fluorophores (Life Technologies) Alexa Fluor 488, 555, and 637. Raji cells were visualized with CMAC, a cell tracker dye. For time-lapse imaging of Raji/Jurkat conjugates, SEE-pulsed CMAC-stained Raji cells were allowed to adhere onto a poly-l-lysine–coated glass-bottom dish (MatTek Corporation) for 10 min in Leibovitz medium. Dishes were then transferred to an incubation chamber (Tokai Hit) at 37°C. Jurkat cells expressing centrin-GFP were added to the dish to initiate conjugate formation. For microtubule plus-end analysis, EB3-GFP–transfected Jurkat or RPE1 cells were seeded onto poly-l-lysine–coated glass-bottom dishes and were filmed in Leibovitz medium. All live-cell imaging was performed using a spinning-disk confocal system (PerkinElmer), mounted on an inverted microscope (Eclipse TE2000-S; Nikon), and equipped with an electron microscope charge-coupled device digital camera (C9100-13; Hamamatsu Photonics). A 60×/1.40 NA oil immersion objective was used to film conjugate formation (Fig. 2 and Videos 1–4), whereas a 100×/1.40 NA oil immersion objective was used to analyze microtubules (Figs. 4, 7 and 8 and Videos 5–7). Multiple fields were imaged every 3 or 4 min (Fig. 2 and Videos 1–4) or every minute (Fig. 4 and Video 5) as z stacks (1-µm steps) using Volocity (version 5.0). For EB3-GFP signal, a single focal plane containing the centrosome was imaged at a 2-Hz frame rate. 2D volume-rendered image sequences were exported as QuickTime files. For video stills, snapshots of 2D volume-rendered time points were taken in Volocity and were processed as previously described.
Quantitative analysis of microtubule plus-end behavior
The MATLAB-based open source software package plusTipTracker (Matov et al., 2010) was downloaded and used according to the accompanying technical report to analyze videos of Jurkat and RPE1 cells transfected with EB3-GFP and acquired as described under image acquisition. The following settings were used in plusTipTracker: maximum gap length (frames) was set to 14; maximum angle for forward growth was set to 30; maximum angle for backward growth was set to 10; minimum subtrack length (frames) was set to 4; search radius range (pixels) was set to 5–10; and max shrinkage factor (relative to growth speed) was set to 1.5. The cortex of Jurkat cells was excluded from the analysis using the sub-ROI feature of plusTipTracker. This was necessary, as the signal-to-noise ratio was poor in this region as a result of the roundness of the cell. In RPE1 cells, all comets were included in analyses. Kymographs were produced in ImageJ (National Institutes of Health) using the kymograph plugin.
Western blots and immunoprecipitation
Cell pellets were washed in ice-cold PBS and lysed in a buffer containing 50 mM Tris-HCl, pH 8, 1% NP-40, 150 mM NaCl, 10% glycerol, and cocktails of protease and phosphatase inhibitors (Sigma-Aldrich). Postnuclear supernatants were separated by SDS-PAGE. Immunoblotting was performed using the relevant primary and peroxidase-labeled secondary antibodies (Dako) and quantified by densitometry using the ImageJ software. For immunoprecipitation, 107 Jurkat cells were washed in serum-free RPMI, left untreated or mixed with 5 × 106 anti-CD3 antibody–bound MACSi beads (T cell activation and expansion kit; Miltenyi Biotec) at 37°C for 20 min, and treated with 1 µg/ml cytochalasin D and 2 µM nocodazole for 30 min before lysis in 1 mM Hepes, pH 7, 0.5% NP-40, 50 µM MgCl2, and 0.1% β-mercaptoethanol, supplemented with protease inhibitors. Postnuclear supernatants were then incubated with Dynabeads (Life Technologies) coupled to anti-p150glued or random IgG antibodies for 2 h at 4°C. The beads were washed four times and eluted with 0.1 M glycine, pH 2.2. The neutralized eluates (1 M Tris, pH 8) were separated by SDS-PAGE and analyzed by immunoblotting.
Protein purification and GST pull-down assays
Recombinant MBP and GST fusions were expressed in Escherichia coli T7 Express (New England Biolabs, Inc.) and purified using amylose resin (New England Biolabs, Inc.) or glutathione Sepharose beads (GE Healthcare), respectively. For GST pull-downs, Jurkat cells were lysed in a buffer containing 30 mM Hepes, pH 7.4, 150 mM NaCl, and 1% TX-100, supplemented with protease and phosphatase inhibitors. Beads were washed in lysis buffer, added to cytoplasmic cell extract, and incubated for 2 h at 4°C before extensive washing in wash buffer (30 mM Hepes, pH 7.4, 150 mM NaCl, and 0.1% TX-100). Beads were boiled in 2× SDS-PAGE loading buffer. Complexes were separated by SDS-PAGE and analysis by immunoblotting.
Microtubule pelleting and microtubule regrowth
Whole Jurkat cell extracts were made in a lysis buffer containing 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 0.1 mM EGTA, and 0.5% Triton X-100, supplemented with protease and phosphatase inhibitor cocktails, and passed through a 26G gauge needle 10 times. After a first spin at 13,000 g, the cytoplasmic extracts were precleared at 189,000 g for 20 min at 4°C. After addition of 0.5 mM MgGTP and 2 mM MgATP, extracts were warmed to RT before sequential addition of 5 µM taxol (or DMSO for control extracts) and 15 µM taxol (or DMSO for control extracts). Tubulin was obtained from Cytoskeleton. Taxol-stabilized microtutubules (0.2 mg/ml) or nontaxol-treated tubulin (control extracts; 0.2 mg/ml) was added, and extracts were incubated at 30°C for 30 min before layering onto a 1 M sucrose cushion made in BRB80 buffer (80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) with 0.5 mM ATP and with or without 10 µM taxol. Microtubules were pelleted at 69,500 g for 20 min at 22°C. Supernatants were removed and saved for immunoblotting. Pellets were washed twice in BRB80 and resuspended in 1× SDS-PAGE loading buffer to one fourth of the volume of supernatant. For microtubule regrowth experiments, RPE1 cells were treated for 2 h with 5 µg/ml BFA and chilled on ice for 45 min before being transferred to a 37°C incubator for the indicated time period.
Kinase assay
For assay 1, 5 µg GST-EB1 was mixed with CKIδ[aa 1–318] (the number of units is indicated in Fig. 5 A; Promega). For assay 2, to remove the GST tag from GST-EB1, 300 µg GST-EB1 was incubated with 10 µl PreScission protease (GE Healthcare) for 4 h on ice following the manufacturer’s instruction. Uncleaved GST-EB1 was captured by glutathione Sepharose. 5 µg of EB1 was then mixed with GST-CKIδ (the number of units is indicated in Fig. 5 A; Life Technologies). Assays were performed in 25 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 1 mM DTT, 0.01% Triton X-100, and 0.1 mM ATP (apart from no ATP condition). Reactions were incubated for 5 min at 37°C and then boiled in SDS 4× sample buffer and separated by SDS-PAGE.
Peptide separation, mass spectrometry, and database analysis
Digested peptides mixtures were subjected to liquid chromatography tandem mass spectrometry (MS/MS) using a mass spectrometer (LTQ Orbitrap Velos; Thermo Fisher Scientific) coupled to a rapid separation liquid chromatography system (UltiMate Nano LC; Dionex Corporation) fitted with an Acclaim PepMap100 column (C18, 3 µm, and 100 Å; Dionex Corporation) with an internal diameter of 75 µm and a capillary length of 25 cm. A flow rate of 350 nL/min was used with a solvent gradient of 5 to 50% solvent B in 57 min. Solvent A was 0.1% (volume/volume) formic acid, and aqueous 80% (volume/volume) acetonitrile in 0.1% (volume/volume) formic acid was used as solvent B. The mass spectrometer was operated using an Nth order double play method to automatically switch between Orbitrap mass spectrometry and LTQ Velos MS/MS acquisition. Survey full-scan Mass spectra (from m/z 400 to 1,600) were acquired in the Orbitrap with a resolution of 60,000 at m/z 400 (after accumulation to a target of 1,000,000 charges in the LTQ). The method used allowed sequential isolation of the 20 most intense ions for fragmentation in the linear ion trap, depending on signal intensity, using collision-induced dissociation at a target value of 5,000 charges. For accurate mass measurements, the lock mass option was enabled in mass spectrometry mode, and the polydimethylcyclosiloxane ions generated in the electrospray process from ambient air were used for internal recalibration during the analysis. Target ions already selected for MS/MS were dynamically excluded for 60 s. General mass spectrometry conditions were as follows: an electrospray voltage at 1.76 kV with no sheath or auxiliary gas flow and an ion selection threshold of 1,000 counts for MS/MS. An activation Q value of 0.25, an activation time of 30 ms, a capillary temperature of 250°C, and an S-Lens radio frequency level of 60% were also applied for MS/MS. Raw files were processed using Proteome Discoverer software (version 1.3; Thermo Fisher Scientific). Processed files were searched against the Swiss-Prot human database using the Mascot search engine (version 2.3.0; Matrix Science). The possible structure modifications allowed were carbamidomethyl cysteine as a fixed modification. Oxidized methionine, deamidation of asparagine and glutamine, phosphorylated serine, threonine, and tyrosine were searched as variable modifications. Searches were performed with tryptic specificity, allowing up to two miscleavages and a tolerance on mass measurement of 10 parts per million in mass spectrometry mode and 0.6 D for MS/MS ions. Phosphorylation sites assigned by Mascot were cross-checked against the fragmentation data in Proteome Discoverer.
Centrosome purification
Centrosomes were purified as described in Bornens and Moudjou (1999). In brief, Jurkat cytoplasmic cell lysates were spun onto 2 ml of 60% sucrose cushion in an SW40 rotor at 10,000 g for 30 min. Centrosome-containing supernatant was then loaded onto a discontinuous sucrose gradient consisting of 1 ml of 70% sucrose, 600 µl of 50% sucrose, and 600 µl of 40% sucrose. Samples were centrifuged for 2 h at 120,000 g in an SW55 Ti rotor. Fractions were collected by punching a small hole in the bottom of the centrifuge tube. After addition of 10 mM Pipes-KOH, pH 7.2, centrosomes in each fraction were pelleted at 115,000 g in the MLA-55 rotor in an ultracentrifuge (Beckman Coulter).
Statistical analyses
Prism software (GraphPad Software) was used for statistical data analysis. Error bars are mean ± SD. Normality was tested with a D’Agostino and Pearson omnibus test, and equality of variance among datasets was tested with an F-test. Apart from microtubule growth parameters (in the table of Fig. 8 E) that were analyzed using permutation t tests, p-values have been derived by two-tailed unpaired Student’s t test or t test with Welch correction when variances were unequal.
Online supplemental material
Fig. S1 shows criteria for scoring centrosome polarization in Fig. 1 (A and D). Fig. S2 illustrates that depleting CKIδ with a different CSNK1D-specific shRNA affects centrosome polarization and that CKIα is not required for centrosome polarization. Fig. S3 demonstrates that CKIδ lacking the SQIP domain still interacts with EB1. Fig. S4 shows that CKIδ isoform-2 promotes centrosome polarization and binds EB1. Fig. S5 shows the microtubule network in CKIδ-depleted and colcemid-treated cells. Videos 1–4, 5, and 6 and 7 correspond to Figs. 2 A, 4 A, and 7 B, respectively. Conjugate formation between SEE-pulsed Raji cells and control (EV) or CKIδ-depleted Jurkat cells expressing GFP-centrin is depicted in Videos 1 and 2 or Videos 3 and 4, respectively. Video 5 corresponds to Fig. 4 A and shows conjugate formation between SEE-pulsed Raji cells and Jurkat cells transfected with GFP-CKIδ. Videos 6 and 7 show EB3-GFP comets in control (EV) and CKIδ-depleted Jurkat cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106025/DC1.
Acknowledgments
We would like to thank K.J. Patel, R. Rios, and the Griffiths and Gergely laboratories for useful discussions. We would also like to thank Drs. Yang and D’Santos at the Cambridge Research Institute Proteomics Core Facility for their expert help.
F. Gergely is supported by a Royal Society University Research Fellowship. Research at F. Gergely’s laboratory is funded by Cancer Research UK.
The authors have no other conflicting financial interests.
Footnotes
Abbreviations used in this paper:
- BFA
- Brefeldin A
- DIC
- dynein intermediate chain
- EV
- empty vector
- IS
- immunological synapse
- MBP
- maltose-binding protein
- SEE
- staphylococcal enterotoxin E
- shRNA
- short hairpin RNA
- TCR
- T cell receptor
- UTR
- untranslated region
References
- Akhmanova A., Steinmetz M.O. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 426:570–574 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- Askham J.M., Vaughan K.T., Goodson H.V., Morrison E.E. 2002. Evidence that an interaction between EB1 and p150(Glued) is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol. Biol. Cell. 13:3627–3645 10.1091/mbc.E02-01-0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badura L., Swanson T., Adamowicz W., Adams J., Cianfrogna J., Fisher K., Holland J., Kleiman R., Nelson F., Reynolds L., et al. 2007. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J. Pharmacol. Exp. Ther. 322:730–738 10.1124/jpet.107.122846 [DOI] [PubMed] [Google Scholar]
- Baratt A., Arkhipov S.N., Maly I.V. 2008. An experimental and computational study of effects of microtubule stabilization on T-cell polarity. PLoS ONE. 3:e3861 10.1371/journal.pone.0003861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrend L., Stöter M., Kurth M., Rutter G., Heukeshoven J., Deppert W., Knippschild U. 2000. Interaction of casein kinase 1 delta (CK1delta) with post-Golgi structures, microtubules and the spindle apparatus. Eur. J. Cell Biol. 79:240–251 10.1078/S0171-9335(04)70027-8 [DOI] [PubMed] [Google Scholar]
- Berrueta L., Tirnauer J.S., Schuyler S.C., Pellman D., Bierer B.E. 1999. The APC-associated protein EB1 associates with components of the dynactin complex and cytoplasmic dynein intermediate chain. Curr. Biol. 9:425–428 10.1016/S0960-9822(99)80190-0 [DOI] [PubMed] [Google Scholar]
- Billadeau D.D., Nolz J.C., Gomez T.S. 2007. Regulation of T-cell activation by the cytoskeleton. Nat. Rev. Immunol. 7:131–143 10.1038/nri2021 [DOI] [PubMed] [Google Scholar]
- Blanchard N., Di Bartolo V., Hivroz C. 2002. In the immune synapse, ZAP-70 controls T cell polarization and recruitment of signaling proteins but not formation of the synaptic pattern. Immunity. 17:389–399 10.1016/S1074-7613(02)00421-1 [DOI] [PubMed] [Google Scholar]
- Bornens M., Moudjou M. 1999. Studying the composition and function of centrosomes in vertebrates. Methods Cell Biol. 61:13–34 10.1016/S0091-679X(08)61973-1 [DOI] [PubMed] [Google Scholar]
- Buey R.M., Mohan R., Leslie K., Walzthoeni T., Missimer J.H., Menzel A., Bjelic S., Bargsten K., Grigoriev I., Smal I., et al. 2011. Insights into EB1 structure and the role of its C-terminal domain for discriminating microtubule tips from the lattice. Mol. Biol. Cell. 22:2912–2923 10.1091/mbc.E11-01-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttrick G.J., Beaumont L.M., Leitch J., Yau C., Hughes J.R., Wakefield J.G. 2008. Akt regulates centrosome migration and spindle orientation in the early Drosophila melanogaster embryo. J. Cell Biol. 180:537–548 10.1083/jcb.200705085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K., Elledge S.J., Hannon G.J. 2006. Lessons from Nature: microRNA-based shRNA libraries. Nat. Methods. 3:707–714 10.1038/nmeth923 [DOI] [PubMed] [Google Scholar]
- Combs J., Kim S.J., Tan S., Ligon L.A., Holzbaur E.L., Kuhn J., Poenie M. 2006. Recruitment of dynein to the Jurkat immunological synapse. Proc. Natl. Acad. Sci. USA. 103:14883–14888 10.1073/pnas.0600914103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle F.M., Vitre B., Arnal I. 2009. Structural basis of EB1 effects on microtubule dynamics. Biochem. Soc. Trans. 37:997–1001 10.1042/BST0370997 [DOI] [PubMed] [Google Scholar]
- Dahlberg C.L., Nguyen E.Z., Goodlett D., Kimelman D. 2009. Interactions between Casein kinase Iepsilon (CKIepsilon) and two substrates from disparate signaling pathways reveal mechanisms for substrate-kinase specificity. PLoS ONE. 4:e4766 10.1371/journal.pone.0004766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das V., Nal B., Roumier A., Meas-Yedid V., Zimmer C., Olivo-Marin J.C., Roux P., Ferrier P., Dautry-Varsat A., Alcover A. 2002. Membrane-cytoskeleton interactions during the formation of the immunological synapse and subsequent T-cell activation. Immunol. Rev. 189:123–135 10.1034/j.1600-065X.2002.18911.x [DOI] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., Theurkauf W. 2005. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21:411–434 10.1146/annurev.cellbio.21.122303.120418 [DOI] [PubMed] [Google Scholar]
- Dujardin D.L., Barnhart L.E., Stehman S.A., Gomes E.R., Gundersen G.G., Vallee R.B. 2003. A role for cytoplasmic dynein and LIS1 in directed cell movement. J. Cell Biol. 163:1205–1211 10.1083/jcb.200310097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin M.L. 2008. T-cell activation through immunological synapses and kinapses. Immunol. Rev. 221:77–89 10.1111/j.1600-065X.2008.00589.x [DOI] [PubMed] [Google Scholar]
- Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P.M., Andreyeva N., Gleeson P., Galjart N., Maia A.R., McLeod I.X., et al. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 12:917–930 10.1016/j.devcel.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide E.J., Kang H., Crapo S., Gallego M., Virshup D.M. 2005. Casein kinase I in the mammalian circadian clock. Methods Enzymol. 393:408–418 10.1016/S0076-6879(05)93019-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 106:489–498 10.1016/S0092-8674(01)00471-8 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. 2003. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 421:753–756 10.1038/nature01423 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Manneville J.B., Nicholls S., Ferenczi M.A., Hall A. 2005. Cdc42 and Par6-PKCzeta regulate the spatially localized association of Dlg1 and APC to control cell polarization. J. Cell Biol. 170:895–901 10.1083/jcb.200412172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajolet M., He G., Heiman M., Lin A., Nairn A.C., Greengard P. 2007. Regulation of Alzheimer’s disease amyloid-beta formation by casein kinase I. Proc. Natl. Acad. Sci. USA. 104:4159–4164 10.1073/pnas.0611236104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M., Watanabe T., Noritake J., Nakagawa M., Yamaga M., Kuroda S., Matsuura Y., Iwamatsu A., Perez F., Kaibuchi K. 2002. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 109:873–885 10.1016/S0092-8674(02)00800-0 [DOI] [PubMed] [Google Scholar]
- Gao Z.H., Seeling J.M., Hill V., Yochum A., Virshup D.M. 2002. Casein kinase I phosphorylates and destabilizes the beta-catenin degradation complex. Proc. Natl. Acad. Sci. USA. 99:1182–1187 10.1073/pnas.032468199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B., Rosen D., Berke G. 1982. Spatial relationships of microtubule-organizing centers and the contact area of cytotoxic T lymphocytes and target cells. J. Cell Biol. 95:137–143 10.1083/jcb.95.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., Munro S. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1:524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., Gundersen G.G. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Gomez T.S., Kumar K., Medeiros R.B., Shimizu Y., Leibson P.J., Billadeau D.D. 2007. Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity. 26:177–190 10.1016/j.immuni.2007.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson H.V., Dzurisin J.S., Wadsworth P. 2010. Methods for expressing and analyzing GFP-tubulin and GFP-microtubule-associated proteins. Cold Spring Harb. Protoc. 2010:pdb top85 10.1101/pdb.top85 [DOI] [PubMed]
- Grakoui A., Bromley S.K., Sumen C., Davis M.M., Shaw A.S., Allen P.M., Dustin M.L. 1999. The immunological synapse: a molecular machine controlling T cell activation. Science. 285:221–227 10.1126/science.285.5425.221 [DOI] [PubMed] [Google Scholar]
- Green R.A., Wollman R., Kaplan K.B. 2005. APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol. Biol. Cell. 16:4609–4622 10.1091/mbc.E05-03-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer Y.E., Rubin J.S. 2011. Casein kinase 1 delta functions at the centrosome to mediate Wnt-3a-dependent neurite outgrowth. J. Cell Biol. 192:993–1004 10.1083/jcb.201011111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.D., Hoffman D.P., Fisette P.L., Baas P., Anderson R.A. 1995. A phosphatidylinositol 4,5-bisphosphate-sensitive casein kinase I alpha associates with synaptic vesicles and phosphorylates a subset of vesicle proteins. J. Cell Biol. 130:711–724 10.1083/jcb.130.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.S., Nelson W.J. 2010. Adenomatous polyposis coli regulates endothelial cell migration independent of roles in beta-catenin signaling and cell-cell adhesion. Mol. Biol. Cell. 21:2611–2623 10.1091/mbc.E10-03-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., John C.M., Kostrewa D., Winkler F.K., Steinmetz M.O. 2005. Structural insights into the EB1-APC interaction. EMBO J. 24:261–269 10.1038/sj.emboj.7600529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J., Buey R.M., Lawera A., et al. 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 138:366–376 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Huse M., Quann E.J., Davis M.M. 2008. Shouts, whispers and the kiss of death: directional secretion in T cells. Nat. Immunol. 9:1105–1111 10.1038/ni.f.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Zhapparova O., Brodsky I., Semenova I., Tirnauer J.S., Zaliapin I., Rodionov V. 2011. CK1 activates minus-end-directed transport of membrane organelles along microtubules. Mol. Biol. Cell. 22:1321–1329 10.1091/mbc.E10-09-0741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro T., Tanaka K., Sakuno T., Watanabe Y. 2010. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat. Cell Biol. 12:500–506 10.1038/ncb2052 [DOI] [PubMed] [Google Scholar]
- Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., Danuser G. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M.A., Wilson L. 2004. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer. 4:253–265 10.1038/nrc1317 [DOI] [PubMed] [Google Scholar]
- Katis V.L., Lipp J.J., Imre R., Bogdanova A., Okaz E., Habermann B., Mechtler K., Nasmyth K., Zachariae W. 2010. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev. Cell. 18:397–409 10.1016/j.devcel.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.J., Maly I.V. 2009. Deterministic mechanical model of T-killer cell polarization reproduces the wandering of aim between simultaneously engaged targets. PLOS Comput. Biol. 5:e1000260 10.1371/journal.pcbi.1000260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T.J., Jenny A., Djiane A., Mlodzik M. 2006. CKIepsilon/discs overgrown promotes both Wnt-Fz/beta-catenin and Fz/PCP signaling in Drosophila. Curr. Biol. 16:1337–1343 10.1016/j.cub.2006.06.030 [DOI] [PubMed] [Google Scholar]
- Komarova Y., De Groot C.O., Grigoriev I., Gouveia S.M., Munteanu E.L., Schober J.M., Honnappa S., Buey R.M., Hoogenraad C.C., Dogterom M., et al. 2009. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 184:691–706 10.1083/jcb.200807179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer A., Dennert G. 1984. Reorientation of the microtubule-organizing center and the Golgi apparatus in cloned cytotoxic lymphocytes triggered by binding to lysable target cells. J. Immunol. 133:2762–2766 [PubMed] [Google Scholar]
- Lee L., Tirnauer J.S., Li J., Schuyler S.C., Liu J.Y., Pellman D. 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 287:2260–2262 10.1126/science.287.5461.2260 [DOI] [PubMed] [Google Scholar]
- Li G., Yin H., Kuret J. 2004. Casein kinase 1 delta phosphorylates tau and disrupts its binding to microtubules. J. Biol. Chem. 279:15938–15945 10.1074/jbc.M314116200 [DOI] [PubMed] [Google Scholar]
- Ligon L.A., Shelly S.S., Tokito M., Holzbaur E.L. 2003. The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol. Biol. Cell. 14:1405–1417 10.1091/mbc.E02-03-0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longenecker K.L., Roach P.J., Hurley T.D. 1998. Crystallographic studies of casein kinase I delta toward a structural understanding of auto-inhibition. Acta Crystallogr. D Biol. Crystallogr. 54:473–475 10.1107/S0907444997011724 [DOI] [PubMed] [Google Scholar]
- Lord C., Bhandari D., Menon S., Ghassemian M., Nycz D., Hay J., Ghosh P., Ferro-Novick S. 2011. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 473:181–186 10.1038/nature09969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowin-Kropf B., Shapiro V.S., Weiss A. 1998. Cytoskeletal polarization of T cells is regulated by an immunoreceptor tyrosine-based activation motif-dependent mechanism. J. Cell Biol. 140:861–871 10.1083/jcb.140.4.861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly V.I., Maly I.V. 2010. Symmetry, stability, and reversibility properties of idealized confined microtubule cytoskeletons. Biophys. J. 99:2831–2840 10.1016/j.bpj.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manneville J.B., Jehanno M., Etienne-Manneville S. 2010. Dlg1 binds GKAP to control dynein association with microtubules, centrosome positioning, and cell polarity. J. Cell Biol. 191:585–598 10.1083/jcb.201002151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritzen T., Löhler J., Deppert W., Knippschild U. 2003. Casein kinase I delta (CKIdelta) is involved in lymphocyte physiology. Eur. J. Cell Biol. 82:369–378 10.1078/0171-9335-00323 [DOI] [PubMed] [Google Scholar]
- Martín-Cófreces N.B., Sancho D., Fernández E., Vicente-Manzanares M., Gordón-Alonso M., Montoya M.C., Michel F., Acuto O., Alarcón B., Sánchez-Madrid F. 2006. Role of Fyn in the rearrangement of tubulin cytoskeleton induced through TCR. J. Immunol. 176:4201–4207 [DOI] [PubMed] [Google Scholar]
- Martín-Cófreces N.B., Robles-Valero J., Cabrero J.R., Mittelbrunn M., Gordón-Alonso M., Sung C.H., Alarcón B., Vázquez J., Sánchez-Madrid F. 2008. MTOC translocation modulates IS formation and controls sustained T cell signaling. J. Cell Biol. 182:951–962 10.1083/jcb.200801014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matov A., Applegate K., Kumar P., Thoma C., Krek W., Danuser G., Wittmann T. 2010. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods. 7:761–768 10.1038/nmeth.1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., Akhmanova A. 2005. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168:141–153 10.1083/jcb.200405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- Monks C.R., Freiberg B.A., Kupfer H., Sciaky N., Kupfer A. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 395:82–86 10.1038/25764 [DOI] [PubMed] [Google Scholar]
- Ning K., Li L., Liao M., Liu B., Mielke J.G., Chen Y., Duan Y., El-Hayek Y.H., Wan Q. 2004. Circadian regulation of GABAA receptor function by CKI epsilon-CKI delta in the rat suprachiasmatic nuclei. Nat. Neurosci. 7:489–490 10.1038/nn1236 [DOI] [PubMed] [Google Scholar]
- Palazzo A.F., Joseph H.L., Chen Y.J., Dujardin D.L., Alberts A.S., Pfister K.K., Vallee R.B., Gundersen G.G. 2001. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr. Biol. 11:1536–1541 10.1016/S0960-9822(01)00475-4 [DOI] [PubMed] [Google Scholar]
- Panbianco C., Weinkove D., Zanin E., Jones D., Divecha N., Gotta M., Ahringer J. 2008. A casein kinase 1 and PAR proteins regulate asymmetry of a PIP(2) synthesis enzyme for asymmetric spindle positioning. Dev. Cell. 15:198–208 10.1016/j.devcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Matos J., Mori S., Gregan J., Bogdanova A., Schwickart M., Mechtler K., Shirahige K., Zachariae W., Nasmyth K. 2006. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 126:1049–1064 10.1016/j.cell.2006.07.029 [DOI] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C.L., Bornens M. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo W.J., Conrad L., Janeway C.A., Jr 1988. Receptor-directed focusing of lymphokine release by helper T cells. Nature. 332:378–380 10.1038/332378a0 [DOI] [PubMed] [Google Scholar]
- Price M.A. 2006. CKI, there’s more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 20:399–410 10.1101/gad.1394306 [DOI] [PubMed] [Google Scholar]
- Quann E.J., Merino E., Furuta T., Huse M. 2009. Localized diacylglycerol drives the polarization of the microtubule-organizing center in T cells. Nat. Immunol. 10:627–635 10.1038/ni.1734 [DOI] [PubMed] [Google Scholar]
- Rena G., Bain J., Elliott M., Cohen P. 2004. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 5:60–65 10.1038/sj.embor.7400048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero S., Cardenas J., Bornens M., Rios R.M. 2009. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 28:1016–1028 10.1038/emboj.2009.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., Rogers G.C., Sharp D.J., Vale R.D. 2002. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158:873–884 10.1083/jcb.200202032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrador J.M., Cabrero J.R., Sancho D., Mittelbrunn M., Urzainqui A., Sánchez-Madrid F. 2004. HDAC6 deacetylase activity links the tubulin cytoskeleton with immune synapse organization. Immunity. 20:417–428 10.1016/S1074-7613(04)00078-0 [DOI] [PubMed] [Google Scholar]
- Sillibourne J.E., Milne D.M., Takahashi M., Ono Y., Meek D.W. 2002. Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J. Mol. Biol. 322:785–797 10.1016/S0022-2836(02)00857-4 [DOI] [PubMed] [Google Scholar]
- Slep K.C. 2010. Structural and mechanistic insights into microtubule end-binding proteins. Curr. Opin. Cell Biol. 22:88–95 10.1016/j.ceb.2009.10.009 [DOI] [PubMed] [Google Scholar]
- Slep K.C., Rogers S.L., Elliott S.L., Ohkura H., Kolodziej P.A., Vale R.D. 2005. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J. Cell Biol. 168:587–598 10.1083/jcb.200410114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M.O., Akhmanova A. 2008. Capturing protein tails by CAP-Gly domains. Trends Biochem. Sci. 33:535–545 10.1016/j.tibs.2008.08.006 --> [DOI] [PubMed] [Google Scholar]
- Stinchcombe J.C., Majorovits E., Bossi G., Fuller S., Griffiths G.M. 2006. Centrosome polarization delivers secretory granules to the immunological synapse. Nature. 443:462–465 10.1038/nature05071 [DOI] [PubMed] [Google Scholar]
- Su L.K., Burrell M., Hill D.E., Gyuris J., Brent R., Wiltshire R., Trent J., Vogelstein B., Kinzler K.W. 1995. APC binds to the novel protein EB1. Cancer Res. 55:2972–2977 [PubMed] [Google Scholar]
- Tirnauer J.S., O’Toole E., Berrueta L., Bierer B.E., Pellman D. 1999. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145:993–1007 10.1083/jcb.145.5.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., Grego S., Salmon E.D., Mitchison T.J. 2002. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell. 13:3614–3626 10.1091/mbc.02-04-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R., Popov A., Kinoshita K., Ashford A.J., Rybina S., Pozniakovsky A., Mayer T.U., Walczak C.E., Karsenti E., Hyman A.A. 2000. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2:13–19 10.1038/71330 [DOI] [PubMed] [Google Scholar]
- Tsai I.C., Amack J.D., Gao Z.H., Band V., Yost H.J., Virshup D.M. 2007. A Wnt-CKIvarepsilon-Rap1 pathway regulates gastrulation by modulating SIPA1L1, a Rap GTPase activating protein. Dev. Cell. 12:335–347 10.1016/j.devcel.2007.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun A., Qureshi I., Stinchcombe J.C., Jenkins M.R., de la Roche M., Kleczkowska J., Zamoyska R., Griffiths G.M. 2011. Centrosome docking at the immunological synapse is controlled by Lck signaling. J. Cell Biol. 192:663–674 10.1083/jcb.201008140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tybulewicz V.L. 2005. Vav-family proteins in T-cell signalling. Curr. Opin. Immunol. 17:267–274 10.1016/j.coi.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Vale R.D. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480 10.1016/S0092-8674(03)00111-9 [DOI] [PubMed] [Google Scholar]
- Vaughan K.T. 2005. TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J. Cell Biol. 171:197–200 10.1083/jcb.200509150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K.T., Vallee R.B. 1995. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol. 131:1507–1516 10.1083/jcb.131.6.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Dogterom M., Stelzer E., Karsenti E., Leibler S. 1992. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol. 118:1097–1108 10.1083/jcb.118.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B., Coquelle F.M., Heichette C., Garnier C., Chrétien D., Arnal I. 2008. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 10:415–421 10.1038/ncb1703 [DOI] [PubMed] [Google Scholar]
- Walston T., Tuskey C., Edgar L., Hawkins N., Ellis G., Bowerman B., Wood W., Hardin J. 2004. Multiple Wnt signaling pathways converge to orient the mitotic spindle in early C. elegans embryos. Dev. Cell. 7:831–841 10.1016/j.devcel.2004.10.008 [DOI] [PubMed] [Google Scholar]
- Watson P., Stephens D.J. 2006. Microtubule plus-end loading of p150(Glued) is mediated by EB1 and CLIP-170 but is not required for intracellular membrane traffic in mammalian cells. J. Cell Sci. 119:2758–2767 10.1242/jcs.02999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S., Xiao Z., Wittau M., Süssner N., Stöter M., Knippschild U. 2005. Interaction of casein kinase 1 delta (CK1 delta) with the light chain LC2 of microtubule associated protein 1A (MAP1A). Biochim. Biophys. Acta. 1745:196–206 10.1016/j.bbamcr.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Xu Y., Padiath Q.S., Shapiro R.E., Jones C.R., Wu S.C., Saigoh N., Saigoh K., Ptácek L.J., Fu Y.H. 2005. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 434:640–644 10.1038/nature03453 [DOI] [PubMed] [Google Scholar]
- Yan X., Habedanck R., Nigg E.A. 2006. A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol. Biol. Cell. 17:634–644 10.1091/mbc.E05-08-0810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Ganguly A., Cabral F. 2010. Inhibition of cell migration and cell division correlates with distinct effects of microtubule inhibiting drugs. J. Biol. Chem. 285:32242–32250 10.1074/jbc.M110.160820 [DOI] [PMC free article] [PubMed] [Google Scholar]