Abstract
Ca2+-dependent protein kinases (CDPKs) are encoded by a multigene family and are thought to play central roles in Ca2+ signaling in plants. Although the primary structures of CDPK isoforms are highly conserved, several studies suggested a distinct physiological function for each CDPK isoform in plants. Hence, there should be mechanisms by which individual CDPK specifically recognizes its substrate. Recently, the variable N-terminal domain of NtCDPK1 was shown to play an essential role in the specific recognition of the substrate. Because the variable N-terminal domain of other CDPKs may also be involved in the substrate recognition, the search for interacting proteins of the variable N-terminal domain would provide important clues to identify the physiological substrates of each CDPK. Additionally, manipulation of the variable N-terminal domain may enable us to engineer the substrate specificity of CDPK, leading a rational rewiring of cellular signaling pathways.
Key words: Ca2+-dependent protein kinase (CDPK), substrate specificity, phosphorylation, signal transduction, calcium
Ca2+ ions play a vital role as second messengers in plant cells during various developmental processes and in response to environmental stimuli.1 Ca2+-dependent protein kinases (CDPKs) are thought to play central roles in the Ca2+ signaling pathway in plants and some protozoans.2–5 There are 34 genes encoding CDPKs in Arabidopsis6 and 29 genes in rice.7 CDPK proteins are Ser/Thr protein kinases that are composed of a variable N-terminal domain, a catalytic domain, a junction domain and a calmodulin-like domain.2 Recently, structural analysis of CDPK showed that Ca2+ binding triggers the reorganization of the CAD (CDPK activation domain), which is composed of the junction domain and the calmodulin-like domain, into a highly intricate fold, leading to the relocation of the CAD around the base of the catalytic domain to a site remote from the substrate binding site.8
A tobacco transcription factor RSG (REPRESSION OF SHOOT GROWTH), which is involved in the gibberellin homeostasis, is negatively regulated by 14-3-3 signaling proteins.9–11 The 14-3-3 proteins bind to RSG depending on the RSG phosphorylation of Ser-114 and thereby sequester RSG in the cytoplasm so that it is unable to regulate its target genes in the nucleus.12 NtCDPK1 was identified as an RSG kinase that promotes 14-3-3 binding to RSG by phosphorylation of Ser-114 of RSG. NtCDPK1 interacts with RSG in vivo and in vitro and specifically phosphorylates Ser-114 of RSG in vitro. Knockdown of NtCDPK1 by RNAi repressed the GA-induced phosphorylation of Ser-114 of RSG, while overexpression of NtCDPK1 in transgenic plants promoted phosphorylation of Ser-114.13 These results showed that RSG is a direct target of NtCDPK1.
Cells must somehow maintain the specificity of distinct Ca2+ signaling and avoid unwanted crosstalk. The functional specificity of individual CDPKs may be determined by Ca2+ and lipid sensitivity, expression pattern, posttranslational regulation, targeted subcellular compartmentalization and substrate recognition.14,15 The first level of substrate specificity arises from the interaction between the active site of the kinase and the amino acid sequences surrounding the phosphorylation site of the substrate.16 Additional conserved docking motifs on the substrate that interacts with specific regions of the catalytic domain may increase the selectivity of the kinase substrate.17 However, because the sequences of phosphorylation sites and docking motifs are rather simple and ambiguous, they are insufficient to account for the substrate specificity. Other molecular mechanisms are required to select the functional targets among potential phosphorylation sites. Scaffold proteins or targeting subunits, which help to enhance substrate specificity, were not found for CDPKs.
The primary structures of CDPK isoforms are highly conserved, especially within their catalytic domains. Thus, it had been considered unlikely that CDPKs would have distinguishable substrate specificities. However, several studies using loss-of-function mutants and knockdown plants suggested that each CDPK isoform in plants possesses a distinct physiological function, such as stress response,18,19 oxidative burst,20 GA response,13 pollen tube growth,21 and stomatal regulation.22 There should be a mechanism by which the substrates are specifically recognized by CDPKs.
We found that the variable N-terminal domain of NtCDPK1 plays an essential role in the specific recognition of substrate RSG.23 The recognition by the variable N-terminal domain of NtCDPK1 may strictly determine the substrate specificity, in concert with the interaction between the catalytic domain of NtCDPK1 and the phosphorylation site of RSG. The variable N-terminal domain of other CDPKs may be involved in the substrate recognition. Yeast two-hybrid analysis suggested that the variable N-terminal domain of an Arabidopsis CDPK, AtCPK32, participates in the interaction with transcription factor ABF4.24 AtCPK11 and a Mesembryanthemum crystallinum CDPK, McCDPK1, also interact with their substrate.25,26 They may need the variable N-terminal domain for the substrate recognition. Although CDPKs have been reported to be involved in diverse physiological processes, very limited information is available about the direct substrates in vivo. The search for interacting proteins of the variable N-terminal domain by yeast two-hybrid screen or the tandem affinity purification technique would provide important clues to identify the physiological substrates of each CDPK. Such comprehensive studies would improve our understanding of both the physiological roles of each CDPK and the complicated network of Ca2+ signaling in plants.
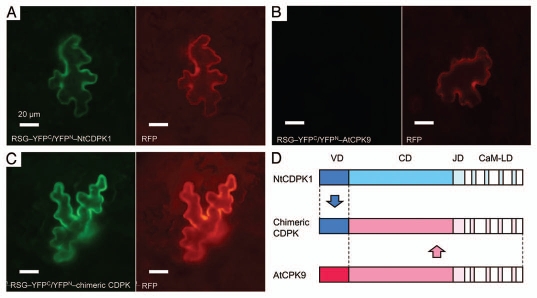
The redesign of the substrate specificity of kinases is a major challenge of protein engineering.27 The variable N-terminal domain of NtCDPK1 conferred sufficient activities as RSG kinase to an Arabidopsis CDPK, AtCPK9, that only poorly phosphorylates RSG (Fig. 1).23 Substitutions of amino acids near the active center in the catalytic domain might affect the substrate specificity but often simultaneously decrease the kinase activity.28 This is major obstacle in the specificity engineering of kinases. The finding that the substrate recognition of CDPK is separable from the catalytic activity opens the possibility of engineering the substrate specificity of CDPK by manipulations of the variable N-terminal domain, which would provide an approach for the rewiring the signaling pathways.
Figure 1.
Visualization of the interactions between RSG and CDPKs in plant cells using BiFC (bimolecular fluorescence complementation) analysis. (A) NtCDPK1 interacts with RSG. YFPN and YFPC represent N-terminal and C-terminal portion of a yellow fluorescent protein, respectively. RFP (red fluorescent protein) was used as a control for transfection efficiency. (B and C) The chimeric CDPK in which the variable N-terminal domain of AtCPK9 was substituted with that of NtCDPK1 interacts with RSG, although AtCPK9 does not. (D) Schematic diagram of the chimeric CDPK. VD, variable N-terminal domain; CD, catalytic domain; JD, junction domain; CaM-LD, calmodulin-like domain.
Abbreviations
- CDPK
Ca2+-dependent protein kinase
- RSG
repression of shoot growth
- CAD
CDPK activation domain
References
- 1.Kudla J, Batistic O, Hashimoto K. Calcium Signals: The lead currency of plant information processing. Plant Cell. 2010;22:541–546. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harper JF, Breton G, Harmon A. Decoding Ca2+ signals through plant protein kinases. Annu Rev Plant Biol. 2004;55:263–288. doi: 10.1146/annurev.arplant.55.031903.141627. [DOI] [PubMed] [Google Scholar]
- 4.Harper JF, Harmon A. Plants, symbiosis and parasites: a calcium signaling connection. Nat Rev Mol Cell Biol. 2005;6:555–566. doi: 10.1038/nrm1679. [DOI] [PubMed] [Google Scholar]
- 5.DeFalco TA, Bender KW, Snedden WA. Breaking the code: Ca2+ sensors in plant signaling. Biochem J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- 6.Arabidopsis Genome Initiative, author. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 7.Asano T, Tanaka N, Yang G, Hayashi N, Komatsu S. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 2005;46:356–366. doi: 10.1093/pcp/pci035. [DOI] [PubMed] [Google Scholar]
- 8.Wernimont AK, Artz JD, Finerty P, Jr, Lin YH, Amani M, Allali-Hassani A, et al. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol. 2010;17:596–601. doi: 10.1038/nsmb.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukazawa J, Nakata M, Ito T, Yamaguchi S, Takahashi Y. The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J. 2010;62:1035–1045. doi: 10.1111/j.1365-313X.2010.04215.x. [DOI] [PubMed] [Google Scholar]
- 10.Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell. 2000;12:901–915. doi: 10.1105/tpc.12.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Igarashi D, Ishida S, Fukazawa J, Takahashi Y. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell. 2001;13:2483–2497. doi: 10.1105/tpc.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishida S, Fukazawa J, Yuasa T, Takahashi Y. Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell. 2004;16:2641–2651. doi: 10.1105/tpc.104.024604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishida S, Yuasa T, Nakata M, Takahashi Y. A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell. 2008;20:3273–3288. doi: 10.1105/tpc.107.057489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng SH, Willmann MR, Chen HC, Sheen J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002;129:469–485. doi: 10.1104/pp.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klimecka M, Muszyńska G. Structure and functions of plant calcium-dependent protein kinases. Acta biochim Pol. 2007;54:219–233. [PubMed] [Google Scholar]
- 16.Ubersax JA, Ferrell JE. Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 17.Sharrocks AD, Yang SH, Galanis A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 18.Ma SY, Wu WH. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol Biol. 2007;65:511–518. doi: 10.1007/s11103-007-9187-2. [DOI] [PubMed] [Google Scholar]
- 19.Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19:3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, et al. Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell. 2007;19:1065–1080. doi: 10.1105/tpc.106.048884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers C, Romanowsky SM, Barron YD, Garg S, Azuse CL, Curran A, et al. Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 2009;59:528–539. doi: 10.1111/j.1365-313X.2009.03894.x. [DOI] [PubMed] [Google Scholar]
- 22.Zou JJ, Wei FJ, Wang C, Wu JJ, Ratnasekera D, Liu WX, et al. Arabidopsis calcium-dependent protein kinase AtCPK10 functions in ABA and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 2010;154:1232–1243. doi: 10.1104/pp.110.157545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y. Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell. 2010;22:1592–1604. doi: 10.1105/tpc.109.073577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi HI, Park HJ, Park JH, Kim S, Im MY, Seo HH, et al. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression and modulates its activity. Plant Physiol. 2005;139:1750–1761. doi: 10.1104/pp.105.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patharkar OR, Cushman JC. A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J. 2000;24:679–691. doi: 10.1046/j.1365-313x.2000.00912.x. [DOI] [PubMed] [Google Scholar]
- 26.Milla MAR, Uno Y, Chang IF, Townsend J, Maher EA, Quilici D, et al. A novel yeast two-hybrid approach to identify CDPK substrates: characterization of the interaction between AtCPK11 and AtDi19, a nuclear zinc finger protein. FEBS Lett. 2006;580:904–911. doi: 10.1016/j.febslet.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Benner SA, Sismour AM. Synthetic biology. Nat Rev Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]