Abstract
Protein phosphorylation is a reversible post-translational modification controlling many biological processes. Most phosphorylation occurs on serine and threonine, and to a less extend on tyrosine (Tyr). In animals, Tyr phosphorylation is crucial for the regulation of many responses such as growth or differentiation. Only recently with the development of mass spectrometry, it has been reported that Tyr phosphorylation is as important in plants as in animals. The genes encoding protein Tyr kinases and protein Tyr phosphatases have been identified in the Arabidopsis thaliana genome. Putative substrates of these enzymes, and thus Tyr-phosphorylated proteins have been reported by proteomic studies based on accurate mass spectrometry analysis of the phosphopeptides and phosphoproteins. Biochemical approaches, pharmacology and genetic manipulations have indicated that responses to stress and developmental processes involve changes in protein Tyr phosphorylation. The aim of this review is to present an update on Tyr phosphorylation in plants in order to better assess the role of this post-translational modification in plant physiology.
Key words: protein tyrosine phosphorylation, kinases, phosphatases, proteomics, mass spectrometry, signaling
Introduction
Proteins can undergo various post-translational modifications (PTMs) that affect their conformation, their activity, their stability and their localization. Thus, PTMs are recognized as regulators of many cellular processes. A few hundred types of PTMs have been described so far such as acetylation, thiolation, adenylation, ribosylation and phosphorylation.1 Protein phosphorylation is one of the most studied PTMs. One-third of all eukaryotic proteins are presumed to be phosphorylated.2,3 Phosphorylation is the result of the antagonistic action of protein kinases (PKs) and protein phosphatases (PPs) that allow to add or remove respectively a phosphate group in a protein. It has been shown that the human genome encodes at least 518 PKs4 and approximately 148 PPs5 indicating the importance of protein phosphorylation. In Arabidopsis thaliana, more than 800 PKs6 and 150 PPs5 have been identified. Phosphorylation is mostly found on hydroxylated amino acids such as serine (Ser), threonine (Thr) and tyrosine (Tyr). The classification of PKs and PPs has been established following the amino acids they are specifically targeting.7 The protein Ser/Thr kinases and protein Tyr kinases (PTKs) strictly phosphorylate Ser/Thr and Tyr residues respectively, whereas dual-specificity PTKs (DsPTKs) act on both Ser, Thr and Tyr. Similarly, protein Ser/Thr phosphatases and protein Tyr phosphatases (PTPs) dephosphorylate phospho-Ser/Thr or phospho-Tyr respectively and dual-specificity PTPs (DsPTPs) remove the phosphate group from Ser, Thr and Tyr. First experiments have reported that the majority of phosphorylation occurs on Ser and Thr residues whereas Tyr phosphorylation accounts only for 0.05%.8 Recently, phosphoproteomics studies based on more accurate detection of phosphopeptides have shown that phosphorylation on Ser, Thr and Tyr residues occurs at a ratio of 88:11:1.3 It clearly appears that Tyr phosphorylation is minor compared to Ser and Thr. Nevertheless, Tyr phosphorylation plays a crucial role as it regulates many cellular processes in animal cells like division, growth and differentiation.9 Until recently, very few studies have tried to elucidate the involvement of Tyr phosphorylation in plant cells. This is due to the lack of a typical PTK gene in plants and also to the fact that few PTP genes have been identified in the genome of Arabidopsis.10 Sensitive proteomic approaches have however, confirmed the existence of protein Tyr phosphorylation in plants. The combination of pep-tides enrichment methods with sensitive mass spectrometry (MS) instrumentation has allowed the identification of low abundance phosphopeptides, in particular Tyr-phosphorylated peptides. This review first presents the early studies on Tyr phosphorylation in plants and the different PTKs and PTPs which have been identified. Then, the latest proteomic studies identifying the Tyr-phosphorylated plant proteins and the physiological processes regulated by Tyr phosphorylation in plants are discussed.
Discovery of Protein Tyrosine Phosphorylation and First Studies in Plants
Detection by radioactive labeling.
In 1980, Ser and Thr were the only known phosphorylated amino acids in proteins. Hunter and Eckart11 were studying polyomavirus middle T and Rous sarcoma virus associated kinase activities and discovered that a third amino acid, identified as Tyr, could also be phosphorylated. The impact of this observation proved to be fundamental for the study of regulation of many physiological processes. Indeed, 30 years later, this PTM has emerged to be one of the most crucial mechanisms of regulation in animal cells.9 First experiments were based on radioactive labeling by 32P-ortho-phosphate of proteins in the cells, separation of proteins by electrophoresis and partial acid hydrolysis. Following thin-layer chromatography of the hydrolyzates and autoradiography, phospho-Ser, phospho-Thr and phospho-Tyr could be detected. The phospho-Tyr residues had escaped detection previously as they were 3,000-fold less abundant than the phosphorylated-Ser and -Thr, and because phospho-Thr and phospho-Tyr were difficult to separate by traditional electrophoretic procedures.11 In plants, protein Tyr phosphorylation was initially demonstrated by the ability of kinases to autophosphorylate on Tyr, Ser and Thr in vitro.12–18 The kinase STY13 autophosphorylates on Tyr, Ser and Thr in vitro.19 Using this radioactive labeling approach, it was also shown that mitogen-activated protein kinases (MAPKs) and somatic embryogenesis receptor-like kinase can transphosphorylate in vitro artificial substrates on Tyr.16,18,20,21
Immunochemical detection.
Developments in the immunochemical field have provided monoclonal antibodies that recognize phosphorylated-residues regardless of the surrounding sequences.22 Therefore, antibodies raised specifically against phospho-Tyr, enabled to detect the phospho-Tyr residues in proteins by western blot test. Tyr phosphorylation was first detected in Pisum sativum using these specific antibodies.23 Immunoblotting with anti-phospho-Tyr antibodies have permitted detection of plant proteins phosphorylated on Tyr in several other species.17,19,24–37
Involvement of PTKs and PTPs by pharmacological approaches.
One way to probe specifically elements of signal transduction pathways is by perturbating these pathways through the loss of function of one of these elements. Pharmacological approaches enable a fast, reversible and dose-dependent inactivation of single components in intact cells organisms. Inhibitors that have been designed to specifically block PTKs and PTPs, have been used in plants. PTKs inhibitors such as genistein, lavendustin A,38,39 erbstatin and tyrphostins,40 and PTPs inhibitors such as phenylarsine oxide (PAO),41 dephostatin42 and orthovanadate43 have permitted to identify plant PTKs and PTPs and to specify the physiological processes in which they are involved. Studies based on genistein have reported PTK activity in Catharanthus roseus44 and in Pisum sativum.45 Genistein has been used to show involvement of PTKs in the ABA signaling pathways leading to stomatal closure and to RAB18 gene expression in Arabidopsis thaliana.33 Genistein and tyrphostin AG18 altered root hair growth and development suggesting a role for PTKs in the organization of cortical microtubules.46 In the pathway leading to the production of inositol-1,4,5-trisphosphate in Citrus limon, implication of PTKs has been shown using genistein, lavendustin A and erbstatin.47 Lavendustin A also inhibits a DsPTK activity in Zea mays.48 Tyrphostins inhibit the activity of a DsPTK in Pisum sativum.45 PTP activities sensitive to PAO have been reported in Lycopersicon esculentum,49 in Mimosa pudica36 and in Vicia faba.50 PAO was used to show the implication of PTPs in the ABA-induced expression of RAB16 in barley aleurone cells51 and of RAB18 expression in A. thaliana suspension cells.33 The same inhibitor allowed to implicate PTPs in ABA-activated stomatal closure in Arabidopsis.33,50,52 A PTP activity inhibited by dephostatin has been detected in soybean.53 Using sodium ortho-vanadate, it has been shown that a PTP is implicated in the copper-dependent transduction pathway leading to cell death34 and in the dynamics and organization of microtubules.46 Once involved by these pharmacological approaches in several plant physiological processes, the genes encoding these enzymes were identified by analysis of the plant genomes using bioinformatics.
Identification of PTK and PTP Genes in Plant Genomes
PTK genes.
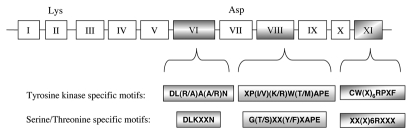
Although Tyr kinases are separated into two broad classes, PTKs and DsPTKs, they all share a common catalytic domain of approximately 250 amino acids19 (Fig. 1). Eleven major conserved subdomains are evident, out of which they are residues specifically conserved in either Ser/Thr or Tyr kinases and may play a role in correct recognition of hydroxylated amino acid.19,54 In the N-terminal end of the catalytic region, the subdomain II contains an invariant lysine residue which has been recognized as essential for maximal enzyme activity as it helps to anchor ATP (Fig. 1). In the middle of the catalytic region, the subdomain VII has an aspartic acid residue believed to be important in the catalytic activity of the kinase (Fig. 1). The subdomain VI confers Ser/Thr specificity but also Tyr specificity. Subdomain VIII appears to play a major role in recognition of peptide substrates and subdomain XI has a specific consensus motif CW(X)6RPXF (Fig. 1). These last subdomains both confer PTK specificity. A first genomic survey with CW(X)6RPXF sequence motif of sub-domain XI revealed the existence of 57 Arabidopsis Tyr kinases possessing 11 kinase subdomains.19 The catalytic domains of all the identified Arabidopsis kinases have motifs for Ser/Thr in subdomains VI and PTK motifs in subdomain XI. These datas suggested that all the kinases belong to a DsPTK family. This survey could not detect Tyr kinase specificity alone in the complete genome of Arabidopsis.19 This is similar to yeast which has DsPTKs but seems to lack PTKs.55 More recently, another survey has shown that three plant species seemed to encode putative Tyr-specific kinases.56 Two A. thaliana putative Tyr-specific kinases lack the conserved lysine residue of subdomain II and the conserved aspartate of the activation loop in subdomain VII.56 This suggests that these two enzymes are catalytically inactive. Two rice species were found to encode six and seven putative Tyr-specific kinases. Out of the six predicted PTK-strict of Oryza sativa ssp. Indica, only three of them were found to include all the residues known to be essential for catalytic activity. Out of the seven putative Tyrspecific kinases of Oryza sativa ssp. Japonica, four were found to have all residues known to be required for catalytic activity.56 Other possible candidates would be Tyr-specific protein kinase-like kinases (TKLs), which are especially abundant in plants. There are 776 TKLs in Arabidopsis and nearly 1,000 in Oryza sativa, compared to 55 in humans.56 Plant TKLs' function remains unknown, but the large number of TKLs in plants may suggest that they carry out important and diverse plant-specific functions. Plant receptor Tyr kinases have not been reported. In animals, they all share the same basic topology: an extracellular ligand-binding domain, a single transmembrane domain and a cytoplasmic domain which contains the kinase activity.57
Figure 1.
Catalytic domain of the serine/threonine and tyrosine protein kinases (adapted from Rudrabhatla et al.19). The catalytic kinase domain of around 250 amino acids contains 11 conserved subdomains (I to XI). At the N-terminal extremity, the subdomain II has a conserved lysine residue believed to be involved in ATP binding. In the central part, an aspartic acid residue in subdomain VII is important for the catalytic activity of the kinase. Subdomains VI and VIII are specifically conserved in either serine/threonine or tyrosine kinases. Consensus DL(R/A)A(A/R)N is specific of tyrosine kinases whereas the consensus DLKXXN is serine/threonine-specific. Subdomain VIII has a tyrosine-specific consensus XP(I/V)(K/R)W(T/M)APE and a poorly conserved serine/threonine-specific consensus G(T/S)XX(Y/F)XAPE. In subdomain XI, the consensus CW(X)6RPXF in subdomain XI is tyrosine-specific.
PTP genes.
Notwithstanding that the identification of plant PTKs is poorly documented, studies on PTP genes are more abundant. The overall protein sequence of Tyr-specific PTPases and DsPTPases share little homology but these phosphatases all contain the CX5R motif in their catalytic domain: (V/I)HCXAGXGR(S/T).58 This catalytic core has a conserved cysteine residue, which acts as a nucleophile, to displace the phosphate group from the substrate and form the phosphoenzyme intermediate59 (Fig. 2). Several genomic surveys with the CX5R motif have identified one PTP-strict and 24 DsPTPs in Arabidopsis.5 In humans, more than 100 members of PTP superfamily which includes 60 DsPTPs have been reported in reference 5. Among these 100 human PTPs, 11 are catalytically inactive and 16 dephosphorylate either glycogen,60 mRNA61 or phosphoinositides58 and not proteins.
Figure 2.
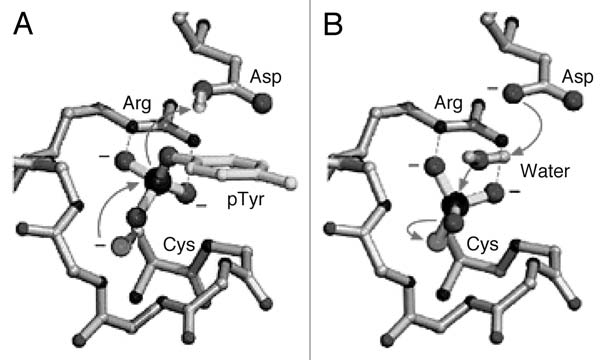
Catalytic mechanisms of protein tyrosine phosphatases (adapted from Denu et al.59). The model is derived from Yersinia protein tyrosine phosphatase and the backbone atoms of the active site loop from cysteine to arginine are shown as a ball-and-stick model. (A) Enzyme-Substrate complex: The dianion of the phosphoryl group is coordinated by the nitrogens of the arginine. The catalytic cysteine thiolate does a nucleophilic attack on the phospho-tyrosine substrate and forms the phosphoenzyme intermediate. (B) Hydrolysis of the phosphoenzyme intermediate: The phosphate is covalently bound to the cysteine. The aspartic acid activates a water molecule which hydrolyzes the phosphoenzyme intermediate. The arginine allows to position the substrate for the nucleophilic attack. Reprinted with permission from Denu et al., Cell 1996; 87:361–364 from Elsevier.59
Some Arabidopsis PTPs have also been shown to be inactive. Inactive PTPs display PTP-like domain structures but possess mutations in the cysteine or arginine residue of the CX5R motif, in an upstream conserved aspartate residue or in combinations of the three. Such alterations are thought to disrupt the enzymatic activity of the phosphatases yet potentially maintain interaction with phosphorylated substrates.62 For example in plants, PASTICCINO2 (PAS2) is a PTP-like member with an inactive phosphatase catalytic site. It functions as an anti-phosphatase by binding a kinase and preventing its dephosphorylation by other activating phosphatases.63 It has been shown finally that PAS2 is a dehydratase which functions in the elongase complex required for the production of very-long-chain fatty acids.64 Another plant PTP has been reported to act on starch. PTPKIS1/SEX4/DSP4 is a DsPTP interacting with plant SNF1-related kinase AKIN11,49 that has a carbohydrate-binding domain allowing it to bind starch granules.65–67 A genetic screen for plants producing excess starch has permitted the identification of a mutant impaired in this DsPTP gene. In the chloroplast, SEX4 dephosphorylates starch. SEX4 resembles to animal laforin62 that regulate glycogen accumulation indicating striking parallels in the regulation of starch metabolism in plants and glycogen metabolism in animals.
The remainder of the PTPs in humans are protein-specific and dephosphorylate phospho-Tyr, and in some cases, phospho-Ser and phospho-Thr. They have been grouped in three classes. Class I includes the classic and dual-specificity enzymes and have a common PTP domain structural fold. They are by far the largest group of PTPs and are further divided into subfamilies. The classic enzymes (receptor and non-receptor) are given the name of PTPs as they all dephosphorylate Tyr residues only. The receptor-like PTPases all contain an extracellular domain of variable length, a single transmembrane region, and one or two cytoplasmic PTPase catalytic domains. No typical PTP receptor has been identified in plants. In A. thaliana, there is only one Tyr-strict phosphatase which is non-receptor protein. This only Tyr-strict phosphatase, AtPTP1, was characterized using a systematic PCR approach and sequence alignment.68 It contains a typical signature motif present in all other PTPases. In Class I, animal DsPTP are divided into the MAPK phosphatases (MKPs), slingshots, phosphatases of regenereting liver (PRL), atypical DsPTPs, cell division cycle 14 (CDC14) phosphatases, tensin homologue (PTEN) and myotubularins (MTMs).69 In A. thaliana, no slingshots, PRLs or CDC14 have been reported. Five MKPs have been identified, three DsPTPs have been classified as atypical and nine have not been classified.5 Four PTEN containing SH2 (Src homology 2) domain and PTB (phospho-Tyr-binding) domain have been identified as a tumor suppressor. This subclass contains the catalytic core of PTPs and has a structural domain with high similarity to a cytoskeleton protein called tensin. PTEN proteins have a low DsPTP activity towards proteins whereas they have a high activity against phosphatidyl inositol 3,4,5-triphosphate.70 In Arabidopsis, two MTMs phosphoinositides phosphatases have been reported in reference 5 and 69. Class II and III are represented by the lowmolecular-mass PTP isoforms and CDC25 phosphatases respectively. One Class II PTP has been reported in A. thaliana but Class III PTPs are not represented.69
Recent development in MS has allowed the efficient identification of the PTKs' and PTPs' protein substrates.
Identification of Phosphorylation Sites by Mass Spectrometry
Estimation of plant Tyr phosphorylation.
Phosphoproteome analysis by large-scale MS-based studies have shown that Tyr phosphorylation in animal cells ranges from 2% to 3%.3,71,72 The proportion of phospho-Tyr residues in human cells is estimated between 1.8 and 6.0%, depending on the analyzed samples.3,72,73 This PTM concerns less than 1% of proteins in yeast.55,74 Phosphorylation estimations by MS approaches have to be considered with caution as they are established on the basis of the phosphopeptides which were detected by MS and thus, they only represent a part of all the phosphopeptides existing within the cell.
First MS studies performed in plants predicted a low occurrence, between 0% and 0.7%, of Tyr phosphorylation as in yeast.75–77 Improvement of both technology based on MS and phosphopeptide enrichment methods have allowed to explore more efficiently the plant phosphoproteome. Out of 2,172 phosphorylation sites analyzed in A. thaliana, 94 have been shown to occur on Tyr residues.35,78 In Arabidopsis suspension cells, the relative abundances of phospho-Ser, phospho-Thr and phospho-Tyr were estimated to be 85%, 10.7% and 4.3% respectively.35 This study reported an unexpected high proportion of Tyr phosphorylation in plants compared to the previous studies, close to the proportion found in human phosphoproteome. A phosphoproteome analysis developed by Reiland et al.79 of Arabidopsis seedlings identified a similar number of phosphorylation sites. The phosphoproteome of A. thaliana shoots and rosette leafs was studied using a LTQ-Orbitrap mass spectrometer for phosphopeptide detection combined with immobilized metal affinity chromatography (IMAC) and titanium dioxide (TiO2) phosphopeptide enrichment. This survey79 identified 2,349 phosphopeptides and reported that 88% of all phosphorylation events occur on Ser and 11% on Thr similarly to the observations made by Sugiyama et al.35 Nevertheless, the amount of Tyr phosphorylation was estimated to only 0.3% of the phosphopeptides detected. This discrepancy in the rate of phospho-Tyr peptides could be explained by the differences between the Arabidopsis suspension cells and Arabidopsis seedlings. Also, the recovery rate of phospho-Tyr peptides could be different because of the enrichment techniques and the detection limitations of liquid chromatography coupled to MS. However, even if some of the putative Tyr phosphorylation sites reported by Sugiyama et al.35 may need to be reconsidered,78 numerous other studies have confirmed their estimations. Indeed, Nakagami et al.80 have reported that the contributions of phospho-Ser, phospho-Thr and phospho-Tyr sites were estimated to be respectively 82.7%, 13.1% and 4.2% in Arabidopsis and to be 84.8%, 12.3% and 2.9% in Oryza sativa. All the phosphopeptides for Oryza sativa and Arabidopsis are available on web-based RIKEN database called Plant Phosphoproteome Database (phosphoproteome.psc.database.riken.jp).35,80 In A. thaliana, a proteomic study mapping the phosphorylation sites of soluble proteins identified 122 phospho-Ser, 44 phospho-Thr and 15 phospho-Tyr which results in a contribution of 67.4% to 24.3% to 8.3% for phospho-Ser:phospho-Thr:phospho-Tyr.81 There are 14 phospho-Tyr sites that have been identified among 105 phosphosites in proteins from the moss Physcomitrella patens which indicates a Tyr phosphorylation rate of 13%.82 A phosphoproteome study83 has shown that the proportion of phospho-Tyr in Medicago trunculata is 1.3% similar to the estimations from Nakagami et al.80 Phosphorylation sites have been characterized from M. trunculata root proteins including both cell lysates and membrane-enriched fractions and using metal affinity chromatography and tandem MS. The Medicago phosphoproteomic database contains more than 3,457 phosphopeptides (phospho.medicago.wisc.edu).
Finally, a global survey has analyzed all the datas from Arabidopsis. A phosphorylation sites prediction was obtained for 500,000 residues in A. thaliana (203,622 Ser, 174,301 Thr, 120,983 Tyr). Experimental evidence from biochemical approaches or MS was obtained for 12,000 residues (9,406 Ser, 2,352 Thr, 699 Tyr) covering about 5,000 proteins in Arabidopsis.84 A Phosphoproteomic database was created from all these compiled datas to form the largest database called PhosPhAT (phosphat. mpimgolm.mpg.de). This database contains 5,170 Arabidopsis phosphoproteins and 32,601 phosphosites. This global survey conclusion was that the percentage of validated unique phos-pho-Ser, phospho-Thr and phospho-Tyr is 76%, 17% and 1.3% respectively.
Enrichment of phosphopeptides.
Large-scale studies of phosphoproteomes were first based on protein separation by 2D-gel electrophoresis and detection of the phosphoproteins using radioactive labeling with 32P-ortho-phosphate,85 antibodies raised specifically against the phosphoresidues22 or a phosphoprotein staining dye called ProQ diamond.86 These very sensitive techniques allowed to detect phosphoproteins but not to identify them. Protein identification was permitted by MS analysis of the protein spots detected in gel. Comparatively to biochemical approaches, MS allows a more specific detection of phosphoproteins as it maps the phosphorylation sites and thus identifies precisely the residues which are phosphorylated in the protein. On the other hand, MS detection is less sensitive than classical biochemical techniques. Indeed, western blot test appears more sensitive for detection of peptides compared to MS by one or two orders of magnitude. Standard peptides are detected in a range of 10–100 fmoles by MS whereas detection by immunochemical method coupled to chemiluminescence is achieved for less than 0.1 fmoles.33 Numerous limitations have been reported for the efficient identification of phosphoproteins by this combined approach of 2D-gel electrophoresis separation with MS analysis. A first limitation is due to the biochemical technique used for protein separation itself. Indeed, the number of proteins that could be analyzed at one glance is limited as 2D-gel electrophoresis could not separate on a single gel more than 1,000 proteins. A second limitation is linked to the preparation of the samples for their analysis by MS. Trypsin is the commonly used enzyme for protein digestion in gel and generates tryptic peptides which have to be eluted from the 2D-gel in order to be analyzed in the mass spectrometer. Selective suppression of phosphopeptides seems enhanced when peptides mixture are extracted from gel probably due to the presence of contaminants from the gel. This leads to a low abundance of the phosphopeptides that can not be detected efficiently. A third limitation is inherent to the physico-chemical properties of the phosphopeptides themselves. Protein phosphorylation is a transient state which is low represented within the cell and thus phosphopeptides are naturally low represented. Furthermore, ionization of phosphopeptides in mixtures with non-phosphorylated peptides is suppressed which hampers their analysis by MS.
To circumvent these difficulties, especially in large scale proteomic studies, specific purification strategies have been elaborated. They allow the enrichment of a small proportion of peptide fragments containing the phosphorylated residues once the phosphoproteins have been proteolytically cleaved. A first approach based on immunoaffinity has been developed. The antibodies recognizing phospho-Tyr can be immobilized on a solid support to allow the enrichment of Tyr-phosphorylated proteins from complex cell extracts. This approach was used successfully in a large-scale phosphoproteomic study intending to establish the profiling of phospho-Tyr peptides from tissu extracts.87 Another approach called IMAC achieves enrichment of phosphopeptides using Fe(III) immobilized on nitrilotriacetate support.74 This enrichment was used to analyze Arabidopsis suspension cell cultures treated with the bacterial pathogen flagellin.88 Metal Oxide Affinity Chromatography (MOAC) is based on a separation using TiO2 that has amphoteric ion-exchange properties which make it suitable for phosphopeptides enrichment.77,89,90 Different reversible covalent binding techniques have been proposed to convert phosphorylated groups into properties amenable to affinity purification. One method involves the activation of the phosphate group to a reactive phosphoramidate that can be subsequently coupled to glass beads or to a dendrimer.91,92 Several groups have been taken advantage of the negatively charged phosphate moiety of phosphopeptides to enrich them via interaction chromatography methods such as strong anion exchange or strong cation exchange. These methods can be combined with IMAC or TiO2 to improve recovery yields of phosphopeptides.77,88
Phosphopeptide identification by MS.
Downstream of the enrichment step, the improvement of the mass spectrometers has allowed a more efficient identification of the phosphopeptides. Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS) permits estimation of peptide mass with very high accuracy. It is now possible to differentiate between nonmodified and phosphorylated peptides directly by their mass as they are determined with an accuracy better than ±0.1 ppm.93 Soft ion fragmentation methods have been developed in order to optimize the analysis of phosphopeptides.72 Liquid chromatography tandem MS based approaches including collision-induced dissociation (CID) as fragmentation method were first employed for deciphering large scale phosphoproteome. CID is based upon collision of selected digest peptide as multiprotonated molecules with gas (Nitrogen, Argon, Helium, etc.) at low energy (10–100 eV).94 Upon collision, peptide ions undergo isomerization reaction involving the mobility of one or more protons to sites that are thermodynamically less stable. As a result, amide bonds are cleaved forming b and y ions series.95,96 For phosphopeptides, a potential difficulty arises from the labile nature of this modification and the amide bond, undergoing a predominant neutral loss of H3PO4(∼98 Da) and to a lesser extend of HPO3 (∼80 Da).97,98 Today, the sensitivity and the reliability are significantly improved when the information of MS/MS and MS/MS/MS are combined for the phosphopeptide identifications.99,100 More recently new fragmentation modes have been developed based on the interaction of ion with electron (electron capture dissociation, ECD) or with radical anion (electron transfer dissociation, ETD).101–103 Both interactions are fast and cleave randomly along the peptide backbone (so called c and z ions) while side chains and modifications such as phosphorylation are left intact. The combination of these new fragmentation modes with mass analyzers offering high resolution and mass accuracy such as FTICR and Orbitrap offers valuable tools for deciphering phosphoproteome. The ability to fragment peptide ions while simultaneously preserving the labile phospho-ester bonds provides ETD with an inherent advantage in large-scale phosphoproteome analyses compared to currently most used CID method. All previous large-scale plant phosphoproteomic studies have relied solely on CID during tandem mass spectrometry (MS/MS).102,103 Medicago phosphoproteome analysis utilized the ETD technology and confirmed that ETD is more efficient than CID in phosphorylation-site localization.83
Physiological Processes Regulated by Tyrosine Phosphorylation in Plants
In animal cells, many physiological roles of Tyr phosphorylation such as regulation of growth, differentiation or oncogenesis, have been reviewed in reference 9. On the other hand, evidence for regulation by Tyr phosphorylation in plant physiological processes is just starting to emerge. This section presents an overview of the signaling pathways and the biological responses shown to be under the control of Tyr phosphorylation in plant cells.
Developmental processes and adaptation to environmental signals.
Tyr phosphorylation has been involved in the development of the male and female gametophytes in higher plants. The AtDsPTP1 cDNA has been isolated from an Arabidopsis cDNA library following homology with mammalian PTPs and its expression is restricted to stamens and pollen.104,105 Another DsPTP, which gene AtPTEN1 is specifically expressed in pollen grains, is essential for pollen development. Indeed, suppression of AtPTEN1 gene expression by RNA interference causes pollen cell death after mitosis.70 Some DsPTK genes are expressed specifically in late flowering stage and have been reported to be highly expressed in stamens.19 Finally, PAO and genistein arrest pollen germination and pollen tube growth confirming involvement of PTKs and PTPs in these processes.106
Embryo development and seed germination are also modulated by Tyr phosphorylation. Changes in the protein Tyr phosphorylation occurs during embryogenesis in Coco nucifera32 and in Daucus carota.24 A PTK has been involved in the establishment of zygotic polarity in Fucus as genistein provokes an alteration of the embryo pattern.107 A group of DsPTK genes is expressed specifically during germination and early seedling stages.19 Reyes et al.108 have shown that PAO enhances the inhibition of germination of Arabidopsis seeds. PTKs and PTPs have been implicated in the control of germination as ABA modulates the Tyr phosphorylation of seed proteins during this stage.33 Also, the mutant phs1–3 impaired in a DsPTP gene, is hypersensitive to ABA during germination.109
Some developmental aspects regulated by phytohormones involve Tyr phosphorylation. Brassinotseroids (BRs) regulate many mechanisms of plant growth and development. Brassinolide induces phosphorylation of many proteins on Tyr residues in Pisum sativum.31 BRI1 (Brassinosteroid Insensitive 1), the receptor of BRs, is a leucine-rich repeat kinase located in the plasma membrane that interacts with the cytosolic protein BAK1 (BRI1-associated kinase 1). Oh et al.110 reported that BRI1 and BAK1 autophosphorylate on Tyr residues and thus are DsPTKs. The autophosphorylation of BRI and BAK1 are necessary for root growth inhibition induced by BRs.110 It was also shown that autophosphorylation/dephosphorylation of the kinase Brassinosteroid Insensitive 2 (BIN2) on Tyr-200 is a critical switch in downstream regulation of BR signaling.111 Furthermore, upon BR perception, BRI1 phosphorylates the BRI1 kinase inhibitor 1 (BKI1) on Tyr-211 to displace the kinase inhibitor into the cytosol where it is active.112 Another phytohormone, auxin, also influences many processes such as vascular development, apical dominance, lateral root formation, gravitropism and phototropism. The ibr5 Arabidopsis mutant which is impaired in a DsPTP gene, is less sensitive to auxin and exhibits the characteristics of some auxin mutants such as a long root and a short hypocotyl when grown in light, fewer lateral roots, aberrant vascular patterning and an increased leaf serration.113 In a proteomic study, Tyr phosphorylation of the auxin receptor, TIR1, has been reported in reference 35. The double mutant ibr5/tir1 enhances auxin resistance compound compared to that of either parent.114 All these datas clearly suggest the involvement of Tyr phosphorylation in the developmental aspects controlled by BRs and by auxin.
Response to stresses.
Water deficit occurs as a result of drought, salinity, low temperature or heat. Plants respond to these stresses with similar signaling mechanisms, biochemical and metabolic responses. Tyr phosphorylation has been implicated in the abiotic stress responses. In Arachis hypogaea, a PTK gene expression and its activity are induced by cold and salt.17 Cold, salt and heat induce high levels of gene expression of 14 DsPTK genes and downregulate three DsPTK genes.19 The only member of classical PTP in Arabidopsis, AtPTP1, was upregulated by salt and downregulated by cold treatment.53,68 MAPK phosphatase 1 (MKP1) is a DsPTP that participates to the response to salt stress. This was shown by expression profiling of wild-type vs. mkp1 mutant lines and increased resistance to salinity of mkp1 mutant plants.115
During water stress, transpirational loss of water is reduced as stomata close in response to ABA. Using specific inhibitors, PTKs and PTPs have been involved in the signaling pathway leading to stomatal closure.33,50,52 The phs1-3 mutant affected in a DsPTP gene, presented a deregulation of the ABA-dependent stomatal closure.109 Perception of water deficit gives rise to increases in internal ABA concentrations. Involvement of Tyr phosphorylation have been observed in ABA signaling as the DsPTP PHS1 is a negative regulator of ABA transduction pathway whereas IBR5, another DsPTP, has been shown to regulate positively this pathway.109,113
Moreover, Tyr phosphorylation controls both oxidative stress tolerance and the response to genotoxic stress. Reactive oxygen species are produced during the normal operation of respiratory and photosynthetic electron transport. They are toxic for the plant as they induce the production of highly destructive species. Plants have elaborated mechanisms to counter the action of these compounds. AtDsPTP2 also called MKP2 has been shown to regulate positively the physiological responses to oxidative stress generated during ozone treatment.116 The mkp2 mutant plants inhibit hypersensitivity to oxidative stress induced by methyl viologen during germination, confirming the role of this DsPTP as a positive regulator.117 Besides the stress of increased light intensity, plants are subjected to stress from the ultraviolet (UV) wavelengths in incident irradiation. UV-C is the most potentially damaging component as it modifies DNA and proteins. Plants use both restorative and repair mechanisms to counter this stress. Screens for UV-sensitive mutants in Arabidopsis led to the identification of the DsPTP MKP1 as essential for UV resistance, that interacts with stress activated MAPK3, MAPK4 and MAPK6.118,119 The Arabidopsis mkp1 mutant which was shown to be resistant to elevated salinity, is also hypersensitive to UV radiation.115,118
Defense, cytoskeleton organization and response to hormones.
The DsPTK BAK1 is the positive regulator of the signaling pathways induced by the BRs but also of the transduction cascade induced by the bacterial pathogen flagellin. This suggested that Tyr phosphorylation might regulate some plant defense aspects. Indeed, phosphorylation on Tyr-610 of BAK1 is critical for BR-signaling and expression of a large number of defense-related genes.120 Immunoblots and immunoprecipitation analysis with anti-phospho-Tyr antibody showed that a fungal elicitor induces Tyr phosphorylation of a kinase in Nicotiana tabacum.121 MKP1 is implicated in the response to pathogens as an mkp1 null mutation Columbia (Col) accession, mkp1 (Col), showed constitutive defense responses including pathogenesis related (PR) gene expression, accumulation of salicylic acid and resistance to the bacterial pathogen Pseudomonas syringae. A genetic study implicated PTP1 in the negative regulation of plant defense. Indeed, the mutant phenotype mkp1 (Col) is strongly enhanced by the ptp1 null mutation indicating redundant function of MKP1 and PTP1.122 Finally, salicylic acid upregulates eight DsPTK genes and suppresses gene expression of three DsPTKs.19
Genetic studies and pharmacology have reported the involvement of Tyr phosphorylation in the organization of the cytoskeleton. The DsPTP PHS1 controls microtubule organization in A. thaliana. A semi-dominant phs1-1 allele was identified in a screen for mutants with increased sensitivity to a microtubule-destabilizing drug.123 It has also been shown that both α- and β-subunits of plant tubulin undergo phosphorylation on Tyr residues.124 Involvement of PTKs and PTPs in the organization of microtubules in primary root cells was shown using specific inhibitors.46,125 Other components of cytoskeleton are also modulated by Tyr phosphorylation. For example, in Mimosa pudica, Tyr phosphorylation of actin is induced by stimulation of the pulvinus and its stimulation is abolished by PAO.36 Tyr phosphorylation affects pollen germination and polarized pollen tubes via the actin cytoskeleton.106 Phaseolus vulgaris profilin can be phosphorylated on Tyr residues in vivo.25
Finally, in a recent large scale proteomic study, very few proteins have been shown to be differentially phosphorylated on Tyr in response to several hormones.126 Nevertheless, it appears that one protein, the G protein α subunit 1 (GPA1) has a Tyr residue which is phosphorylated in response to all hormones (ABA, auxin, gibberellic acid, jasmonic acid and kinetin). Tyr phosphorylation of Gα subunits is known to activate G protein-coupled receptors.127 It was suggested that phosphorylation of Gα subunit on Tyr-166 was a common response for multiple hormones and thus constitutes a key switch in the cross-talk between several signaling pathways.126
All the studies mentioned in this last chapter have permitted to have a better view of the roles of Tyr phosphorylation in plants. However, they need to be completed in order to establish more precisely the implication of Tyr phosphorylation in these regulatory processes.
Conclusions and Future Prospects
Knowledge about plant PTKs, PTPs and their protein substrates has emerged this last decade. The contribution of protein Tyr phosphorylation to the regulation of a vast array of physiological processes in plant is just starting to be elucidated. However, biochemical studies on PTPs and PTKs will have to confirm their specificity towards Tyr residues. Similarly to PTPs, the functions of the plant PTKs will have to be explored by a reverse genetic approach based on insertional T-DNA mutants. Identification based on enhanced MS technology of more plant Tyr-phosphorylated proteins will allow to complete the existing databases of Tyr-phosphorylated peptides. The next important challenge will be to identify the targets of the Tyr-phosphorylated proteins detected by MS. Lim and Pawson128 have suggested that Tyr phosphorylation is due to a three part tool kit which includes the PTK, the PTP and the proteins recognizing Tyr-phosphorylated substrates. These last proteins bind to the substrates of the couple PTK/PTP via an SH2 domain that interacts specifically with phospho-Tyr containing motifs.129 In human cells, there are around 100 of these proteins whereas only four SH2 domain proteins have been reported in Arabidopsis.78,130 Plants might be similar to yeast that has only one SH2 protein and which is supposed to completely lack Tyr phosphorylation. But, recent proteomic studies have clearly established that Tyr phosphorylation in plants is more abundant than expected. This tends to show that plants probably developed a different strategy from mammalians to read the phospho-Tyr signals. PTB domains are known to bind phospho-Tyr131 but such domain has not yet been identified in plants. Recently, another type of phospho-Tyr-binding domain has been characterized in animals, the C2 domain of human protein kinase C delta.132 A. thaliana was found to possess 85 proteins containing C2 domain.56 These questions should be addressed in the near future. The complexity of the signaling networks involving Tyr phosphorylation in plants will also have to be unravelled by positioning each PTK, each PTP and their substrates within each cascade and further precise the functions controlled by these pathways.
Acknowledgments
I would like to thank Prof. Anthony J. Trewavas (University of Edinburgh, UK) for helpful discussions and critical reading of the manuscript. I am grateful to Dr. Gérard Bolbach (Université UPMC Paris 06, Plateforme de protéomique et spectrométrie de masse, CNRS UMR7613, IFR83, France) for stimulating suggestions and discussions on the MS aspects of the review. I finally would like to thank Prof. Arnould Savouré (Université UPMC Paris 06, PCMP, EAC 7180 CNRS, France) for his constant support concerning the exploration of the role of post-translational modifications in proteins.
Abbreviations
- PTM
post-translational modification
- Ser
serine
- Thr
threonine
- Tyr
tyrosine
- PTK
protein tyrosine kinase
- Ds
dual-specificity
- PTP
protein tyrosine phosphatase
- MAPK
mitogen-activated protein kinase
- PAO
phenylarsine oxide
- TLK
tyrosine-specific protein kinase-like kinase
- MS
mass spectrometry
- CID
collision-induced dissociation
- ECD
electron capture dissociation
- ETD
electron transfer dissociation
- BR
brassinosteroid
- ABA
abscisic acid
- SH2
Src homology 2
References
- 1.Wold F. In vivo chemical modification of proteins (post-translational modification) Annu Rev Biochem. 1981;50:783–814. doi: 10.1146/annurev.bi.50.070181.004031. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Aleosi DR, et al. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 3.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 4.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 5.Kerk D, Templeton G, Moorhead GB. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae and higher plants. Plant Physiol. 2008;146:351–367. doi: 10.1104/pp.107.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 7.Luan S. Protein phosphatases in plants. Ann Rev Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- 8.Hunter T, Shefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci USA. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luan S. Tyrosine phosphorylation in plant cell signaling. Proc Natl Acad Sci USA. 2002;99:11567–11569. doi: 10.1073/pnas.182417599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter T, Eckart W. The discovery of tyrosine phosphorylation: it's all in the buffer! Cell. 2004;116:35–39. doi: 10.1016/s0092-8674(04)00049-2. [DOI] [PubMed] [Google Scholar]
- 12.Hirayama T, Oka A. Novel protein kinase of Arabidopsis thaliana (APK1) that phosphorylates tyrosine, serine and threonine. Plant Mol Biol. 1992;20:653–662. doi: 10.1007/BF00046450. [DOI] [PubMed] [Google Scholar]
- 13.Ali N, Halfter U, Chua NH. Cloning and biochemical characterization of a plant protein kinase that phosphorylates serine, threonine and tyrosine. J Biol Chem. 1994;269:31626–31629. [PubMed] [Google Scholar]
- 14.Mu JH, Lee HC, Kao TH. Characterization of a pollen-expressed receptor-like kinase gene of Petunia inflata and the activity of its encoded kinase. Plant Cell. 1994;6:709–721. doi: 10.1105/tpc.6.5.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sessa G, Raz V, Savaldi S, Fluhr R. PK12, a plant dual-specificity protein kinase of the LAMMER family, is regulated by the hormone ethylene. Plant Cell. 1996;8:2223–2234. doi: 10.1105/tpc.8.12.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Li H, Gupta R, Morris PC, Luan S, Kieber JJ. ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol. 2000;122:1301–1310. doi: 10.1104/pp.122.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudrabhatla P, Rajasekharan R. Developmentally regulated dual-specificity kinase from peanut that is induced by abiotic stresses. Plant Physiol. 2002;130:380–390. doi: 10.1104/pp.005173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayrose M, Bonshtein A, Sessa G. LeMPK3 is a mitogen-activated protein kinase with dual specificity induced during tomato defense and wounding responses. J Biol Chem. 2004;279:14819–14827. doi: 10.1074/jbc.M313388200. [DOI] [PubMed] [Google Scholar]
- 19.Rudrabhatla P, Reddy MM, Rajasekharan R. Genome-wide analysis and experimentation of plant serine/threonine/tyrosine-specific protein kinases. Plant Mol Biol. 2006;60:293–319. doi: 10.1007/s11103-005-4109-7. [DOI] [PubMed] [Google Scholar]
- 20.Shah K, Vervoot J, de Vries SC. Role of threonines in the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J Biol Chem. 2001;276:41263–41269. doi: 10.1074/jbc.M102381200. [DOI] [PubMed] [Google Scholar]
- 21.Stulemeijer IJ, Stratmann JW, Joosten MH. Tomato mitogen-activated protein kinases LeMPK1, LeMPK2 and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol. 2007;144:1481–1494. doi: 10.1104/pp.107.101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaydes JP, Vojtesek B, Bloomberg GB, Hupp TR. The development and use of phospho-specific antibodies to study protein phosphorylation. Methods Mol Biol. 2000;99:177–189. doi: 10.1385/1-59259-054-3:177. [DOI] [PubMed] [Google Scholar]
- 23.Torruella M, Casano LM, Vallejos RH. Evidence of the activity of tyrosine kinase(s) and of the presence of phosphotyrosine proteins in pea plantlets. J Biol Chem. 1986;261:6651–6653. [PubMed] [Google Scholar]
- 24.Barizza E, Sciavo FL, Terzi M, Filippini F. Evidence suggesting protein tyrosine phosphorylation in plants depends on the developmental conditions. FEBS Lett. 1999;447:191–194. doi: 10.1016/s0014-5793(99)00272-0. [DOI] [PubMed] [Google Scholar]
- 25.Guillén G, Valdés-López V, Noguez R, Olivaries J, Rodriguez-Zapata LC, Pérez H, et al. Profilin in Phaseolus vulgaris is encoded by two genes (only one expressed in root nodules) but multiple isoforms are generated in vivo by phosphorylation on tyrosine residues. Plant J. 1999;19:497–508. doi: 10.1046/j.1365-313x.1999.00542.x. [DOI] [PubMed] [Google Scholar]
- 26.Nühse TS, Peck SC, Hirt H, Boller T. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK6. J Biol Chem. 2000;275:7521–7526. doi: 10.1074/jbc.275.11.7521. [DOI] [PubMed] [Google Scholar]
- 27.Carpi A, Di Maria G, Vedovato M, Rossi V, Naccari T, Floriduz M, et al. Comparative proteome bioinformatics: identification of a whole complement of putative protein tyrosine kinases in the model flowering plant Arabidopsis thaliana. Proteomics. 2002;2:1494–1503. doi: 10.1002/1615-9861(200211)2:11<1494::AID-PROT1494>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 28.Ndimba BK, Chivasa S, Hamilton JM, Simon WJ, Slabas AR. Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics. 2003;3:1047–1059. doi: 10.1002/pmic.200300413. [DOI] [PubMed] [Google Scholar]
- 29.Boudolf V, Inzé D, De Veylder L. What if higher plants lack a CDC25 phosphatase? Trends Plant Sci. 2006;11:474–479. doi: 10.1016/j.tplants.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Wan L, Ross AR, Yang J, Hegedus DD, Kermode AR. Phosphorylation of the 12 S globulin cruciferin in wild-type and abi1-1 mutant Arabidopsis thaliana (thale cress) seeds. Biochem J. 2007;404:247–256. doi: 10.1042/BJ20061569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedina EO, Karimova FG, Tarchevsky IA. Effect of brassinolide on tyrosine phosphorylation of pea leaf proteins. Biochemistry (Mosc) 2006;71:423–429. doi: 10.1134/s0006297906040109. [DOI] [PubMed] [Google Scholar]
- 32.Islas-Flores I, Oropeza C, Hernandez-Sotomayor SM. Protein phosphorylation during coconut zygotic embryo development. Plant Physiol. 1998;118:257–263. doi: 10.1104/pp.118.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghelis T, Bolbach G, Clodic G, Habricot Y, Miginiac E, Sotta B, et al. Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol. 2008;148:1668–1680. doi: 10.1104/pp.108.124594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hung WC, Huang DD, Chien PS, Yeh CM, Chen PY, Chi WC, et al. Protein tyrosine dephosphorylation during copper-induced cell death in rice roots. Chemosphere. 2007;69:55–62. doi: 10.1016/j.chemosphere.2007.04.073. [DOI] [PubMed] [Google Scholar]
- 35.Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, et al. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol. 2008;4:1–7. doi: 10.1038/msb.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kameyama K, Kishi Y, Yoshimura M, Kanzawa N, Sameshima M, Tsuchiya T. Tyrosine phosphorylation in plant bending. Nature. 2000;407:37. doi: 10.1038/35024149. [DOI] [PubMed] [Google Scholar]
- 37.Huang HJ, Lin YM, Huang DD, Takahashi T, Sugiyama M. Protein tyrosine phosphorylation during phytohormone-stimulated cell proliferation in Arabidopsis hypocotyls. Plant Cell Physiol. 2003;44:770–775. doi: 10.1093/pcp/pcg082. [DOI] [PubMed] [Google Scholar]
- 38.Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe SI, Itoh N, et al. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem. 1987;262:5592–5595. [PubMed] [Google Scholar]
- 39.Onoda T, Iinuma H, Sasaki Y, Hamada M, Isshiki K, Naganawa H, et al. Isolation of a novel tyrosine kinase inhibitor, lavendustin A, from Streptomyces griseolavendus. J Nat Prod. 1989;52:1252–1257. doi: 10.1021/np50066a009. [DOI] [PubMed] [Google Scholar]
- 40.Levitzki A, Mishani E. Tyrphostins and other tyrosine kinase inhibitors. Annu Rev Biochem. 2006;75:93–109. doi: 10.1146/annurev.biochem.75.103004.142657. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Morales P, Minami Y, Luong EL, Klausner RD, Samuelson LR. Tyrosine phosphorylation in T-cells is regulated by phosphatase activity: studies with phenylarsine oxide. Proc Natl Acad Sci USA. 1990;87:9255–9259. doi: 10.1073/pnas.87.23.9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umezawa K, Kawakami M, Watanabe T. Molecular design and biological activities of protein-tyrosine phosphatase inhibitors. Pharmacol Ther. 2003;99:15–24. doi: 10.1016/s0163-7258(03)00050-0. [DOI] [PubMed] [Google Scholar]
- 43.Swarup G, Cohen S, Gabers DL. Inhibition of membrane phosphotyrosil-protein phosphatase activity by vanadate. Biochem Biophys Res. 1982;107:1104–1109. doi: 10.1016/0006-291x(82)90635-0. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez-Zapata LC, Hernandez-Sotomayor SMT. Evidence of protein-tyrosine activity in Catharanthus roseus roots transformed by Agrobacterium rhizogenes. Planta. 1998;204:70–77. doi: 10.1007/s004250050231. [DOI] [PubMed] [Google Scholar]
- 45.Rudrabhatla P, Rajasekharan R. Functional characterization of peanut serine/threonine/tyrosine protein kinase: molecular docking and inhibition kinetics with tyrosine kinase inhibitors. Biochemistry. 2004;43:12123–12132. doi: 10.1021/bi0497042. [DOI] [PubMed] [Google Scholar]
- 46.Yemets A, Sheremet Y, Vissenberg K, Van Orden J, Verbelen JP, Blume YB. Effects of tyrosine kinase and phosphatase inhibitors on microtubules in Arabidopsis root cells. Cell Biol Int. 2008;32:630–637. doi: 10.1016/j.cellbi.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Ortega X, Velasquez JC, Perez LM. IP3 production in the hypersensitive response of lemon seedlings against Alternaria alternata involves active protein tyrosine kinases but not G-protein. Biol Res. 2005;38:89–99. doi: 10.4067/s0716-97602005000100011. [DOI] [PubMed] [Google Scholar]
- 48.Trojanek JB, Klimecka MM, Fraser A, Dobrowolska G, Muszynska G. Characterization of dual specificity protein kinase from maize seedlings. Acta Biochim Pol. 2004;51:635–647. [PubMed] [Google Scholar]
- 49.Fordham-Skelton AP, Chilley P, Lumbreras V, Reignoux S, Fenton TR, Dahm CC, et al. A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J. 2002;29:705–715. doi: 10.1046/j.1365-313x.2002.01250.x. [DOI] [PubMed] [Google Scholar]
- 50.Shi WL, Liu X, Jia WS, Zhang SQ. Protein tyrosine phosphatases mediate the signalling pathway of stomatal closure in Vicia faba L. J Int Plant Biol. 2005;47:319–326. [Google Scholar]
- 51.Knetsch MLW, Wang M, Snaar-Jagalska BE, Heimovaara-Dijkstra S. Abscisic acid induces mitogen-activated protein kinase activation in barley aleurone protoplasts. Plant Cell. 1996;8:1061–1067. doi: 10.1105/tpc.8.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacRobbie E. Evidence for a role for protein tyrosine phosphatase in the control of ion release from the guard cell vacuole in stomatal closure. Proc Natl Acad Sci USA. 2002;99:11963–11968. doi: 10.1073/pnas.172360399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fordham-Skelton AP, Skipsey M, Eveans IM, Edwards R, Gatehouse JA. Higher plant tyrosine-specific protein phosphatases (PTPs) contain novel amino-terminal domain: expression during embryogenesis. Plant Mol Biol. 1999;39:593–605. doi: 10.1023/a:1006170902271. [DOI] [PubMed] [Google Scholar]
- 54.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 55.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JEP, Bai DL, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miranda-Saavedra D, Barton GJ. Classification and functional annotation of eukaryotic protein kinases. Proteins. 2007;68:893–914. doi: 10.1002/prot.21444. [DOI] [PubMed] [Google Scholar]
- 57.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1143. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 59.Denu JM, Stuckey JA, Saper MA, Dixon JE. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 60.Cohen PT. Protein phosphatase 1: targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 61.Kennelly PJ. Archaeal protein kinases and protein phosphatses: insights from genomics and biochemistry. Biochem J. 2003;370:373–389. doi: 10.1042/BJ20021547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 63.Da Costa M, Bach L, Landrieu I, Bellec Y, Catrice O, Brown S, et al. Arabidopsis PASTICCINO2 is an antphosphatse involved in regulation of cyclin-dependent kinase A. Plant Cell. 2006;18:1426–1437. doi: 10.1105/tpc.105.040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bach L, Michaelson LV, Halsam R, Bellec Y, Gissot L, Marion J, et al. The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA. 2008;105:14727–14731. doi: 10.1073/pnas.0805089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niittylä T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MD, et al. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem. 2006;281:11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 66.Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, et al. A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J. 2006;46:400–413. doi: 10.1111/j.1365-313X.2006.02704.x. [DOI] [PubMed] [Google Scholar]
- 67.Sokolov LN, Dominguez-Soliz JR, Allary AL, Buchanan BB, Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA. 2006;103:9732–9737. doi: 10.1073/pnas.0603329103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu Q, Fu HH, Gupta R, Luan S. Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell. 1998;10:849–857. doi: 10.1105/tpc.10.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moorhead GBG, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 70.Gupta R, Ting JT, Sokolov LN, Johnson SA, Luan S. A tumor suppressor homology, AtPEN1, is essential for pollen development in Arabidopsis. Plant Cell. 2002;14:2495–2507. doi: 10.1105/tpc.005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bodenmiller B, Malmstrom J, Gerrits B, Campbell D, Lam H, Schmidt A, et al. PhosphoPep-a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007;6:1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- 74.Ficarro SB, McClealand ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 75.Nühse TS, Stensballe A, Jensen ON, Peck SC. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell. 2004;16:2394–2405. doi: 10.1105/tpc.104.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de la Fuente van Bentem S, Anrather D, Dohnal I, Roitinger E, Csaszar E, Joore J, et al. Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J Proteome Res. 2008;7:2458–2470. doi: 10.1021/pr8000173. [DOI] [PubMed] [Google Scholar]
- 77.Benschop JJ, Mohammed S, O'Flaherty M, Heck AJR, Slijper M, Menke FLH. Quantitative Phosphoproteomics of Early Elicitor Signaling in Arabidopsis. Mol Cell Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 78.de la Fuente van Bentem S, Hirt H. Protein tyrosine phosphorylation in plants: More abundant than expected? Trends Plant Sci. 2009;14:71–76. doi: 10.1016/j.tplants.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Reiland S, Messerli G, Baerenfäller K, Gerrits B, Endler A, Grossmann J, et al. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, et al. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010;153:1161–1174. doi: 10.1104/pp.110.157347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lohrig K, Müller B, Davydova J, Leister D, Wolters DA. Phosphorylation site mapping of soluble proteins: bioinformatical filtering reveals potential plastidic phosphoproteins in Arabidopsis thaliana. Planta. 2010;229:1123–1134. doi: 10.1007/s00425-009-0901-y. [DOI] [PubMed] [Google Scholar]
- 82.Heintz D, Erxleben A, High AA, Wurtz V, Reski R, Van Dorsselaer A, et al. Rapid alteration of the phosphoproteome in the moss Physcomitrella patens after cytokinin treatment. J Proteome Res. 2006;5:2283–2293. doi: 10.1021/pr060152e. [DOI] [PubMed] [Google Scholar]
- 83.Grimsrud PA, den Os D, Wenger CD, Swaney DL, Schwartz D, Sussman MR, et al. Large-scale phospho-protein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 2010;152:19–28. doi: 10.1104/pp.109.149625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, et al. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010;38:828–834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peck SC, Nuhse TS, Iglesias A, Hess D, Meins F, Boller T. Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell. 2001;13:1467–1475. doi: 10.1105/tpc.13.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.El-Khatib RT, Good AG, Muench DG. Analysis of the Arabidopsis cell suspension phosphoproteome in response to short-term low temperature and abscisic acid treatment. Physiol Plant. 2007;129:687–697. [Google Scholar]
- 87.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci USA. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nühse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 89.Ikeguchi Y, Nakamura H. Determination of organic phosphates by column-switching high performance anion-exchange chromatography using on-line preconcentration on Titania. Anal Sci. 1997;13:479–483. [Google Scholar]
- 90.Wolschin F, Wienkoop S, Wekcwerth W. Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC) Proteomics. 2005:4389–4397. doi: 10.1002/pmic.200402049. [DOI] [PubMed] [Google Scholar]
- 91.Zhou H, Tian R, Ye M, Xu S, Feng S, Pan C, et al. Highly specific enrichment of phosphopeptides by zirconium dioxide nanoparticles for phosphoproteome analysis. Electrophoresis. 2007;28:2201–2215. doi: 10.1002/elps.200600718. [DOI] [PubMed] [Google Scholar]
- 92.Tao WA, Wollscheid B, O'Brien R, Eng JK, Li XJ, Bodenmiller B, et al. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 93.Spengler B, Hester A. Mass-based classification (MBC) of peptides: highly accurate precursor ion mass values can be used to directly recognize peptide phosphorylation. J Am Soc Mass Spectrom. 2008;19:1808–1812. doi: 10.1016/j.jasms.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 94.Wells JM, McLuckey SA. Collision-induced dissociation (CID) of peptides and proteins. Methods Enzymol. 2005;402:148–185. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 95.Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 96.Medzihradszky KF. Peptide sequence analysis. Methods Enzymol. 2005;402:209–244. doi: 10.1016/S0076-6879(05)02007-0. [DOI] [PubMed] [Google Scholar]
- 97.DeGnore JP, Qin J. Fragmentation of phosphopeptides in an ion trap mass spectrometer. J Am Soc Mass Spectrom. 1998;9:1175–1188. doi: 10.1016/S1044-0305(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 98.Lehmann WD, Kruger R, Salek M, Hung CW, Wolschin F, Weckwerth W. Neutral loss-based phosphopeptide recognition: A collection of caveats. J Proteome Res. 2007;6:2866–2873. doi: 10.1021/pr060573w. [DOI] [PubMed] [Google Scholar]
- 99.Ulintz PJ, Bodenmiller B, Andrews PC, Aebersold R, Nesvizhskii AI. Investigating MS2/MS3 matching statistics. Mol Cell Proteomics. 2008;7:71–87. doi: 10.1074/mcp.M700128-MCP200. [DOI] [PubMed] [Google Scholar]
- 100.Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Nat Acad USA. 2004;101:13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. J Am Chem Soc. 1998;120:3265–3266. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 102.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kelleher NL, Zubarev RA, Bush K, Furie B, Furie BC, McLafferty FW, et al. Localization of labile posttranslational modifications by electron capture dissociationn: the case of gamma-carboxyglutamic acid. Anal Chem. 1999:4250–4253. doi: 10.1021/ac990684x. [DOI] [PubMed] [Google Scholar]
- 104.Gupta R, Huang Y, Kieber J, Luan S. Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 1998;16:581–589. doi: 10.1046/j.1365-313x.1998.00327.x. [DOI] [PubMed] [Google Scholar]
- 105.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 106.Zi H, Xiang Y, Li M, Wang T, Ren H. Reversible protein tyrosine phosphorylation affects pollen germination and pollen tube growth via the actin cytoskeleton. Protoplasma. 2007;230:183–191. doi: 10.1007/s00709-006-0232-9. [DOI] [PubMed] [Google Scholar]
- 107.Correllou F, Potin P, Brownlee C, Kloareg B, Bouget FY. Inhibition of the establishment of zygotic polarity by protein tyrosine inhibitors leads to an alteration of embryo pattern in Fucus. Dev Biol. 2000;219:165–182. doi: 10.1006/dbio.1999.9603. [DOI] [PubMed] [Google Scholar]
- 108.Reyes D, Rodriguez D, Nicolas G, Nicolas C. Evidence of a role for tyrosine dephosphorylation in the control of postgermination arrest of development by abscisic acid in Arabidopsis thaliana L. Planta. 2006;223:381–385. doi: 10.1007/s00425-005-0135-6. [DOI] [PubMed] [Google Scholar]
- 109.Quettier AL, Bertrand C, Habricot Y, Miginiac E, Agnes C, Jeannette E, et al. The phs1-3 mutation in a putative dual-specificity protein tyrosine phosphatase gene provokes hypersensitive responses to abscisic acid in Arabidopsis thaliana. Plant J. 2006;47:711–719. doi: 10.1111/j.1365-313X.2006.02823.x. [DOI] [PubMed] [Google Scholar]
- 110.Oh MH, Wang X, Kota U, Goshe MB, Clouse SD, Huber SC. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:658–663. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25:232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Monroe-Augustus M, Zolman BK, Bartel B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell. 2003;15:2979–2991. doi: 10.1105/tpc.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Strader LC, Monroe-Augustus M, Bartel B. The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 2008;8:41. doi: 10.1186/1471-2229-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ulm R, Revenkova E, di Sansebastiano GP, Bechtold N, Paszkowski J. Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev. 2001;15:699–709. doi: 10.1101/gad.192601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee JS, Ellis BE. Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J Biol Chem. 2007;282:25020–25029. doi: 10.1074/jbc.M701888200. [DOI] [PubMed] [Google Scholar]
- 117.Lumbreras V, Vilela B, Irar S, Solé M, Capellades M, Valls M, et al. MAPK phosphatse MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J. 2010;63:1017–1030. doi: 10.1111/j.1365-313X.2010.04297.x. [DOI] [PubMed] [Google Scholar]
- 118.Ulm R, Ichimura K, Mizoguchi T, Peck SC, Zhu T, Wang X, et al. Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 2002;21:6483–6493. doi: 10.1093/emboj/cdf646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seo S, Katou S, Seto H, Gomi K, Ohashi Y. The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J. 2007;49:899–909. doi: 10.1111/j.1365-313X.2006.03003.x. [DOI] [PubMed] [Google Scholar]
- 120.Oh MH, Wang X, Wu X, Zhao Y, Clouse SD, Huber SC. Autophosphorylation of Tyr-610 in the recptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc Natl Acad Sci USA. 2010;107:17827–17832. doi: 10.1073/pnas.0915064107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121.Suzuki K, Shinshi H. Transient activation and tyrosine phosphorylation of a protein kinase in tobacco cells treated with fungal elicitor. Plant Cell. 1995;7:639–647. doi: 10.1105/tpc.7.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, et al. MAP KINASE PHOSPHATASE1 and PROTEIN TYROSINE PHOSPHATASE1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 2009;21:2884–2897. doi: 10.1105/tpc.109.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Naoi K, Hashimoto T. A semidominant mutation in an Arabidopsis mitogen-activated protein kinase phosphatase-like gene compromises cortical microtubule organization. Plant Cell. 2004;16:1841–1853. doi: 10.1105/tpc.021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Blume Y, Yemets A, Sulimenko V, Sulimenko T, Chan J, Lloyd C, et al. Tyrosine phosphorylation of plant tubulin. Planta. 2008;229:143–150. doi: 10.1007/s00425-008-0816-z. [DOI] [PubMed] [Google Scholar]
- 125.Blume Y, Yemets A, Sheremet Y, Nyporko A, Sulimenko V, et al. Exposure of beta-tubulin regions defined by antibodies on an Arabidopsis thaliana microtubule protofilament model and in the cells. BMC Plant Biology. 2010;10:29. doi: 10.1186/1471-2229-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen Y, Hoehenwarter W, Weckwerth W. Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J. 2010;63:1–17. doi: 10.1111/j.1365-313X.2010.04218.x. [DOI] [PubMed] [Google Scholar]
- 127.Umemori H, Inoue T, Kume S, Sekiyama N, Nagao M, Itoh H, et al. Activation of the G protein Gq/11 through tyrosine phosphorylation of the α subunit. Science. 1997;276:1878–1881. doi: 10.1126/science.276.5320.1878. [DOI] [PubMed] [Google Scholar]
- 128.Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell. 2010;142:661–667. doi: 10.1016/j.cell.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 130.Williams JG, Zvelebil M. SH2 domains in plants imply new signalling scenarios. Trends Plant Sci. 2004;9:161–163. doi: 10.1016/j.tplants.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 131.Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE. 2003:12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- 132.Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCd is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]