Abstract
Phosphate and sulfate are two macro-elements essential for plant growth and development. Both elements play a central role in numerous aspects of plant metabolism and their deficiencies have profound effects on the transcriptome as well as on numerous metabolic pathways. The research emphasis so far has been on elucidating the molecular physiology of these individual nutritive elements. Recent data proved the existence of complex connections between the various regulatory layers of the homeostasis of these elements, but the molecular bases and biological significance of such interconnections remains poorly understood. This review provides an update on recent advances to identify the components involved in phosphate and sulfate homeostasis crosstalk. In light of this case study, developing a comprehensive understanding of the coordination of the ion homeostasis and identifying genes which can be used as good molecular markers for monitoring the “integrative ionic status” of plants is not only of great scientific interest, but also crucial for biotechnological and agronomic applications.
Key words: plant, mineral nutrition, ion homeostasis, signaling, phosphate, sulfate, cross-talks
Phosphate (Pi) and sulfate (SO42-) are involved in numerous aspect of plant metabolism and their deficiencies have profound effects on numerous metabolic pathways resulting in limitations in plant growth, development and productivity. Shoot growth is effected by both phosphorus (P) or sulfur (S) deficiencies, which manifests itself as a reduction in the total biomass and accumulation of anthocyanins. Root development is also influenced by P or S availability, where deficiencies in either macro-nutrient cause striking changes in the root architecture.1 When challenged with a SO42- deficiency, the primary root continues to grow, but the lateral roots form closer to the root tip with an increase in density.1 In contrast, when plants are grown in very low Pi concentrations, the primary roots stop growing and lateral root density increases.1 Besides the aforementioned phenotypic differences of Pi- or SO42--deficient plants, an interconnection of responses to the internal concentration of these two elements has been reported at the metabolic level. One such response is the rapid replacement of sulfolipids by phospholipids under S deficiency and a replacement of phospholipids by sulfolipids during P deficiency.2,3 Transcriptome analysis of plants grown with different Pi or SO42- concentrations reveals extensive reprogramming of the gene expression in both the shoots and the roots.4,5 Interestingly, a deficiency or surplus of only one of the elements often involves modifications in the expression of genes encoding for proteins specific to the homeostasis of others elements, including proteins that control the flux of these elements and their assimilation or regulatory molecules acting in the signaling pathways. These observations strongly indicate the existence of multi-level coordination in the regulation of homeostasis of the two ions in which currently unidentified key elements are actively cross talking between these signaling pathways. Such connections are likely to provide flexibility and plasticity for the adaptation to ever-fluctuating environmental conditions, which is vital for sessile organisms, such as plants, to complete their life cycle. How these connections work at molecular level is the challenge for future research.
In the last decade, research efforts have provided us with a comprehensive view on the adaptive strategies employed by plants to cope with deficiencies of these elements. In Arabidopsis and other plant species, many key genes/proteins involved in the Pi or SO42- uptake and inter-organ transport have been cloned, and functionally characterized by the complementation of mutants or by expression in heterologous systems, including Saccharomyces and/or Xenopus Oocyte.6–9 The number of signaling molecules and cis- and trans-regulatory elements that play an important role in the regulation of either Pi or SO42- homeostases have been also identified and extensively described in references 10 and 11.
It is interesting to note that in addition to the similarities between the topology of the SO42- and the Pi transporters12 there are comparable molecular mechanisms that exist which regulate the expression these transporters, including metabolites such as carbohydrate, phytohormones and regulatory miRNAs. It is likely that molecular mechanisms involved in photosynthetic activity and carbon status also influence the regulation of Pi and SO42- homeostasis in order to support the metabolic demands of the plant.13,14 Remarkably, the expression profiles of the two major up-take transporters (PHT2 for Pi and SULTR1;2 for SO42-) appeared to follow a similar diurnal rhythm13,14 and are sensitive to the carbon status, either upstream or downstream, of the hexokinase (HXK) activity in glycolysis.15
We have learned that the Pi and the SO42- homeostasis are also coordinated by the status of diverse phytohormones.16,17 The similarity in homeostatic regulation of these two elements by phytohormones is best illustrated by viewing their relationships to the plant's response to cytokinins.16,17 The frequency of crosstalk between either Pi or SO42-, and the cytokinin signal transduction pathways is extensive beginning with the uptake of these elements from the media into the plants. Indeed, both uptake systems for Pi and the SO42- acquisition are similarly regulated by the plant hormone cytokinin. Exogenous application of cytokinin was shown to repress the expression of transporters involved in Pi and SO42- uptake.17,18 We now know that cytokinins and the cytokinin receptor CYTOKININ RESPONSE 1/WOODEN LEG/ARABIDOPSIS HISTIDINE KINASE 4 (CRE1/WOL/AHK4) pathways not only play an important role in suppressing the upregulation of several genes following Pi deficiency,16 but also the downregulation of the two higher affinity SO42- transporters in Arabidopsis.17
As mentioned above, an additional important similarity is revealed by the discovery of the molecular mechanism employing regulatory microRNAs (miRNAs) for both Pi and SO42- homeostasis.19,21 In Arabidopsis, our knowledge on the role of two miRNA molecules is particularly advanced on the miRNA395 and the miRNA399, which are characterized for their involvement in SO42- and Pi starvation signaling pathways, respectively.19,21 The expression of miRNA399 is controlled by the MYB transcription factor, PHR1.22 The mature miRNA399 plays a key role in the shoot-to-root Pi-deficiency signaling pathway by targeting the transcript of an E2-conjugase (PHO2), which results in Pi transport in throughout plant starting with Pi-uptake in the roots to its translocation into the shoots.19,20 Likewise, regulation of the SO42- homeostasis involves miR395,22 which acts downstream of the transcription factor SLIM1,23 and contributes to the regulation of SULTR2; 1 transcript,24 which is involved in the SO42- root-to-shoot transfer.
In addition to the aforementioned mechanistic similarities in the regulation of the Pi and SO42- transport systems, the presence of cross talk between the signaling pathways of these two elements are starting to surface in the face of new data. Recently, it has been shown that miRNAs and the transcription factor PHR1 (Phosphate Response 1) play an important role in the cross talk between the SO42- and Pi signaling pathways. Also, Hsieh et al. (2009),25 showed that the miRNA (miRNA395), which is known to be upregulated by SO42- starvation and to trigger SULTR2;1, is suppressed in Pi-deficient plants. An attractive explanation for this phenomenon could be that this acts as a means to increase the expression of SULTR2;1, which would serve to increase SO42- translocation and increase sulfolipid biosynthesis to replace phospholipids under Pi-deficient conditions. Although the physiological impact of the regulation of miR395 on sulfate metabolism in Pi-deficient plants is currently unknown, these observations strengthen the hypothesis that there is a genetic program allowing the plant to respond to the starvation of one of the elements, while enhancing the availability and utilization of both by orchestrating the compensation of one for the other.
As already mentioned, the PHR1 (phosphate starvation response 1) transcription factor plays a role in the regulation of phosphate homeostasis in plants. The universal PHR1-binding sequence (PIBS) has been defined as an 8 bp imperfect palindrome in the promoter of many genes.22 In the context of cross talk between the SO42- and Pi response pathways, the PHR1 appear to play an important role. For example, two genes involved in the replacement of phospholipids by sulfolipids in Pi-deficient plants, SQD1 and SQD2, contain a PHR1 binding site in their promoter and are upregulated by Pi deficiency in a PHR1-dependant manner.22,26–28 The earlier observation, in which the MYB transcription factor PHR1 is an active molecular component linking the sulfate homeostasis and the phosphate signaling, has recently been strengthened by Rouached et al. (2011).26 The authors showed that upon Pi deficiency, the expression of SULTR1.3, which encodes a transporter involved in shoot-to-root SO42- distribution,29 appeared to be dependent on PHR1. In agreement with this finding, structural analysis revealed that SULTR1.3 possesses in its promoter a PHR1-binding sequence.26 In line with this observation, the flux of sulfate from shoots to roots is compromised in the Pi-deficient phr1 mutant.26 It is also important to note that in addition to its positive regulatory role on SULTR1;3 expression, PHR1 inhibits the expression of a distinct group of SULTR genes known to be upregulated by sulfate starvation (e.g., SULTR2;1).26 Interestingly, a similar repressive effect of the Chlamydomonas reinhardtii PHR1 orthologue PSR1 on the expression of genes involved in SO42- scavenging and acquisition has recently been reported in reference 30. These results on PSR1 in C. reinhardtii and PHR1 in Arabidopsis reveal an unexpected level of complexity and interconnection in the regulation of sulfate and Pi homeostasis and highlight the conservation of the importance of the PSR1/PHR1 transcription factor in these processes through evolution.
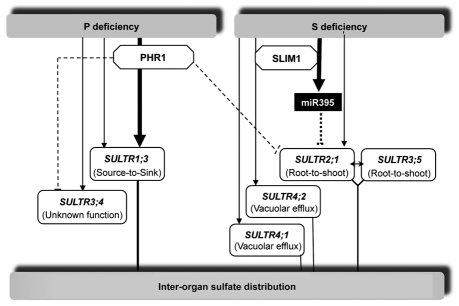
Taken together, we have reviewed the evidence for the genetic basis of the multilevel regulation that mediates the adaptation of the plant to the available phosphate and sulfate. Based on this evidence, obtained mainly from studies in Arabidopsis thaliana, it is becoming possible to propose an integrative model for the regulation of genes involved in intracellular and inter-organ sulfate homoeostasis upon Pi deficiency. It therefore can be proposed that SLIM1 and miR395 are major regulators of gene expression upon sulfate deficiency, while PHR1 acts both positively to regulate SULTR1;3 and shoot-to-root sulfate transfer, and negatively to suppress the induction of other sulfate transporters upon Pi deficiency (Fig. 1). The future challenge will be to develop a comprehensive understanding of the coordination of the homeostasis of these two elements by discovering new signaling and regulatory networks.31
Figure 1.
Schematic representation for the regulation of the expression of the SULTR genes involved in the internal sulfate distribution in response to sulfate and Pi nutritional status. In this model, the expression of SULTR1;3 and SULTR3;4 are controlled by Pi starvation.26 The upregulation of SULTR1;3 expression upon Pi limitation is partially controlled by the transcription factor PHR1, while PHR1 plays a repressive role on SULTR3;4 overexpression under Pi-deficiency in shoots.26 Limitation in sulfate external availability mainly results in the activation of SULTR2;1 (in roots), SULTR4;1 and SULTR4;2 expression. The repression of SULTR2;1 in shoots is downstream of the trans-acting factor SLIM1 and targeted by miR395.21 SLIM1 also controls the expression of SULTR4;2 in response to S deficiency.23 Double black arrow between SULTR3;5 and SULTR2;1 indicates the synergy between these proteins in mediating sulfate uptake.32 Solid lines with arrows and dotted lines symbolize positive and negative effects, respectively. In this scheme, the thick and thin arrows indicate major and minor signaling networks, respectively.
References
- 1.López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol. 2003;6:280–287. doi: 10.1016/s1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- 2.Essigmann B, Guler S, Narang RA, Linke D, Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugimoto K, Sato N, Tsuzuki M. Utilization of a chloroplast membrane sulfolipid as a major internal sulfur source for protein synthesis in the early phase of sulfur starvation in Chlamydomonas reinhardtii. FEBS Lett. 2007;581:4519–4522. doi: 10.1016/j.febslet.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 4.Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005;42:305–314. doi: 10.1111/j.1365-313X.2005.02363.x. [DOI] [PubMed] [Google Scholar]
- 6.Poirier Y, Bucher M. Phosphate transport and homeostasis in Arabidopsis. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. Available at: www.bioone.org/doi/full/10.1199/tab.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Ribot C, Rezzonico E, Poirier Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 2004;135:400–411. doi: 10.1104/pp.103.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouached H, Secco D, Arpat AB. Getting the most sulfate from soil: Regulation of sulfate uptake transporters in Arabidopsis. J Plant Physiol. 2009;166:893–902. doi: 10.1016/j.jplph.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi H. Regulation of sulfate transport and assimilation in plants. Int Rev Cell Mol Biol. 2010;281:129–159. doi: 10.1016/S1937-6448(10)81004-4. [DOI] [PubMed] [Google Scholar]
- 10.Rouached H, Arpat AB, Poirier Y. Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant. 2010;3:288–299. doi: 10.1093/mp/ssp120. [DOI] [PubMed] [Google Scholar]
- 11.Gojon A, Nacry P, Davidian JC. Root uptake regulation: a central process for NPS homeostasis in plants. Curr Opin Plant Biol. 2009;12:328–338. doi: 10.1016/j.pbi.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Smith FW, Rae AL, Hawkesford MJ. Molecular mechanisms of phosphate and sulphate transport in plants. Biochim Biophys Acta. 2000;1465:236–245. doi: 10.1016/s0005-2736(00)00141-3. [DOI] [PubMed] [Google Scholar]
- 13.Lejay L, Gansel X, Cerezo M, Tillard P, Muller C, Krapp A, et al. Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. Plant Cell. 2003;15:2218–2232. doi: 10.1105/tpc.013516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouached H, Wirtz M, Alary R, Hell R, Arpat AB, Davidian JC, et al. Differential regulation of the expression of two high-affinity sulfate transporters, SULTR1.1 and SULTR1.2, in Arabidopsis. Plant Physiol. 2008;147:897–911. doi: 10.1104/pp.108.118612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lejay L, Wirth J, Pervent M, Cross JM, Tillard P, Gojon A. Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol. 2008;146:2036–2053. doi: 10.1104/pp.107.114710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Pena A, et al. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000;24:559–567. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- 17.Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H. A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 2004;38:779–789. doi: 10.1111/j.1365-313X.2004.02079.x. [DOI] [PubMed] [Google Scholar]
- 18.Franco-Zorrilla JM, Martin AC, Leyva A, Paz-Ares J. Interaction between phosphate-starvation, sugar and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 2005;138:847–857. doi: 10.1104/pp.105.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bari RP, Pant BD, Stitt M, Scheible WR. PHO2, micro RNA399 and PHR1 define a phosphate signalling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. Regulation of phoshate homeostasis by microRNA in Arabidopsis. Plant Cell. 2006;18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, et al. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 2009;57:313–321. doi: 10.1111/j.1365-313X.2008.03690.x. [DOI] [PubMed] [Google Scholar]
- 22.Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 2000;23:171–182. doi: 10.1046/j.1365-313x.2000.00768.x. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh L, Lin S, Shih A, Chen J, Lin W, Tseng C, et al. Uncovering small RNA-mediated responses to phosphate-deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouached H, Secco D, Arpat B, Poirier Y. The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 2011;11:19. doi: 10.1186/1471-2229-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franco-Zorrilla JM, Gonzalez E, Bustos R, Linhares F, Leyva A, Paz-Ares J. The transcriptional control of plant responses to phosphate limitation. J Exp Bot. 2004;55:285–293. doi: 10.1093/jxb/erh009. [DOI] [PubMed] [Google Scholar]
- 28.Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, et al. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 2007;50:982–994. doi: 10.1111/j.1365-313X.2007.03108.x. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H. Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 2003;131:1511–1517. doi: 10.1104/pp.014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moseley JL, Gonzalez-Ballester D, Pootakham W, Bailey S, Grossman AR. Genetic interactions between regulators of Chlamydomonas phosphorus and sulfur deprivation responses. Genetics. 2009;181:889–905. doi: 10.1534/genetics.108.099382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouached H, Secco D, Arpat BA. Regulation of ion homeostasis in plants: Current approaches and future challenges. Plant Signal Behav. 2010;5:501–502. doi: 10.4161/psb.11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kataoka T, Hayashi N, Yamaya T, Takahashi H. Rootto-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 2004;136:4198–4204. doi: 10.1104/pp.104.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]