Abstract
The eukaryotic hetrohexameric mini-chromosome maintenance (MCM2-7) proteins complex provides DNA unwinding function during the DNA replication. The complex also functions as DNA replication licensing factor which ensures that the DNA in genome is replicated only once per cell division cycle. Recently, a single subunit MCM6 from pea has been shown to contain helicase and ATPase activities in vitro. Recently, the transcript of a single subunit was reported to be upregulated in pea plant in response to high salinity and cold stress and not with ABA, drought and heat stress. The first direct evidence that overexpression of single subunit MCM6 confers salinity stress tolerance without yield loss has also been reported. Here we report the promoter of the pea MCM6 single subunit that contains stress responsive elements which may be responsible for regulating the MCM6 under abiotic stress conditions.
Key words: cell cycle, DNA replication, MCM6 promoter, salinity stress, stress responsive cis-elements
The initiation of DNA replication in eukaryotes starts from origins where many protein factors including MCMs proteins bind to start and control the process.1 In yeast and animal, all the six MCM proteins are required for DNA replication initiation and DNA synthesis.2,3 When the MCM complex is loaded on the chromatin, the replication origin is formally defined as being licensed for replication, therefore MCMs are also known as licensing factors. The MCMs protein complex functions as essential replicative helicase for DNA replication. The complexes of MCM4/7,4 MCM4/6/7,5,6 or MCM2-77 have been shown to contain helicase and ATPase activities in vitro. Recently, a first direct evidence of pea MCM6 single subunit functions as DNA helicase has been reported in reference 8. This report is unique to plant MCM6 protein, as this activity was only reported for heteromultimers of MCM proteins in animal system. The pea MCM6 is reported to contain all the known canonical MCM motifs including zinc finger, MCM specific Walker A and Walker B and arginine finger. Recently, the first direct evidence that overexpression of pea MCM6 gene in tobacco plant confers salinity stress tolerance without affecting yield has also been reported in reference 9. This report suggested a previously undescribed pathway for manipulating stress tolerance in crop plants.
A full-length cDNA (2.89 kbp) encoding MCM6 single subunit was cloned (Accession number AY169793) from pea cDNA library as described earlier.8 The cDNA is consisted of an open reading frame (ORF) of 2.48 kbp, a 5′ untranslated region (UTR) of 99 bp and a 3′ UTR of 312 bp including a 13 bp poly(A) tail. A genomic fragment (2.48 kb) corresponding to the ORF was also cloned from the pea genomic DNA. The same size and sequence of the genomic fragment of PsMCM6 and the cDNA showed that PsMCM6 is an intron-less gene.8 However, the Arabidopsis MCM6 gene contains 17 introns (TAIR Accession number: 504952844, gene model: AT5G44635.1) and rice MCM6 also contains 17 introns (LOC_Os05g14590). The MCM6 transcript was reported to be upregulated in response to NaCl (300 mM) and cold (4°C) stress. The other stresses such as dehydration, heat, ABA and NAA could not show any induction in the PsMCM6 mRNA level.9 MCM6 overexpression driven by a constitutive cauliflower mosaic virus-35S promoter in tobacco plants has been shown to confer salinity tolerance. T1-transgenic plants grow normally and set viable seeds without yield penalty under salinity stress.9 The exact mechanism of PsMCM6-mediated tolerance of salinity stress is not understood.
To check whether the stress-regulated cis-elements are present in the promoter of MCM6 gene, the promoter has also been isolated. For this first the pea genomic DNA library was prepared and then the MCM6 promoter isolated as described below:
The genomic “DNA library” is a pool of specially prepared DNA fragments from which specific pieces of DNA can be identified, isolated and cloned. Construction of BD Genome Walker Libraries (BD Bioscience Clontech) begins with isolation of very clean genomic DNA of considerably higher quality. Four separate aliquots were thoroughly digested with four different restriction enzymes (EcoRV, DraI, PvuII and SspI) that recognize a 6-base site, leaving blunt ends. Following digestion, each pool of DNA fragment was ligated to the BD Genome Walker Adaptor. The adaptor forward primers (Ap1: 5′-GTA ATA CGA CTC ACT ATA GGG C-3′, nested AP2: 5′-ACT ATA GGG CAC GCG TGG T-3′) are based on adaptor region and MCM6 gene specific reverse primers (GSP1: 5′-CGT AAT ATA GTT CAT TCC GTT GTC CG-3′) was designed from first exon near start translation site and the second (nested) primer from 5′-UTR region (nested GSP2: 5′-CAG TTT ATG CTT CTG AGT ATT GAG TAC-3′). The amplified PCR product(s) using first set of primers (GSP1 and AP1) was used as a template for the nested PCR using second set of primers (nested GSP2 and AP2). The resulting specific fragment (MCM6 promoter) was cloned into pGEMT-T easy vector followed by sequencing analysis for identification of cis-elements by using PLACE and PlantCARE database.
The complete sequence of MCM6 promoter along with all the cis-elements is shown in Figure 1. Many of cis-acting elements are present in the MCM6 promoter region which is probably bound by different transcription factors to either enhance or inhibit the expression of MCM6 gene. The results show that MCM6 promoter contains cis-elements which are related to salt, drought, ABA cold and wound stresses, and many more (Fig. 1A). The putative functions of the cis-acting elements present in the MCL6 promoter are described below:
Figure 1.
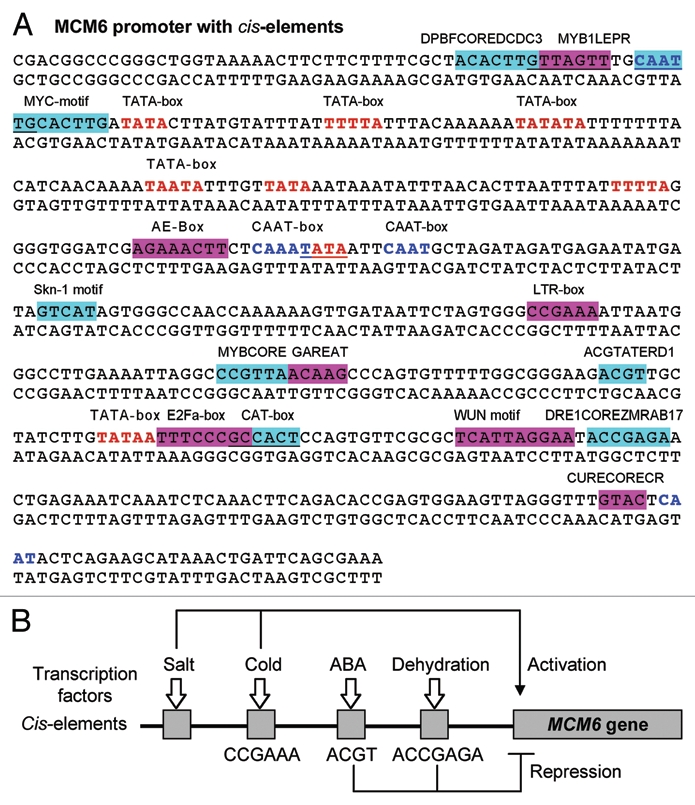
(A) Pea PCM promoter sequence showing various cis-elements as determined by PLACE and PlantCARE program. Blue and red letters are TATA-box and CAAT sequences, respectively. Highlight boxes are different cis-acting elements. (B) A hypothetical model representing the regulation of expression of stress-induced pea MCM6 gene under stress by cis-elements in the promoter region.
ACGTATERD1: required for etiolation-induced expression of erd1 (early responsive to dehydration) in Arabidopsis. Seq: ACGT.
AE-box: part of a module for light response: Seq: 178: AGAAACTT.
CAAT-box: common cis-acting element in promoter and enhancer regions. Seq CAATT, CAAT, CAAAT.
CAT-box: cis-acting regulatory element related to meristem expression. Seq: 354: GCCACT.
CURECORECR: is the core of a CuRE (copper-response element) found in Cyc6 and Cpx1 genes in Chlamydomonas; Also involved in oxygen-response of these genes; Seq: GTAC.
DPBFCOREDCDC3: A novel class of bZIP transcription factors, DPBF-1 and 2 (Dc3 promoter-binding factor-1 and 2) binding core sequence; Found in the carrot (D.c.) Dc3 gene promoter; Dc3 expression is normally embryo-specific, and also can be induced by ABA; The Arabidopsis abscisic acid response gene ABI5 encodes a bZIP transcription factor; abi5 mutant have a pleiotropic defects in ABA response; ABI5 regulates a subset of late embryogenesis-abundant genes; GIA1 (growth-insensitivity to ABA) is identical to ABI5. Seq: ACACNNG.
DRE1COREZMRAB17: “DRE1” core found in maize (Z.M.) rab17 gene promoter; “DRE1” was protected, in in vivo footprinting, by a protein in embryos specifically, but in leaves, was protected when was treated with ABA and drought; rab17 is expressed during late embryo-genesis, and is induced by ABA. Seq: ACCGAGA.
E2Fa-box: Related to regulation during cell cycle, Seq: TTTCCCGC.
GAREAT: GARE (GA-responsive element); Occurrence of GARE in GA-inducible, GA-responsible and GA-nonresponsive genes found in Arabidopsis seed germination was 20, 18 and 12%, respectively. Seq: TAACAAR.
LTR-box: cis-acting element involved in low-temperature responsiveness. Seq: 267: CCGAAA
MYB1LEPR: Tomato Pti4 (ERF) regulates defence-related gene expression via GCC box and non-GCC box cis elements [Myb1 (GTTAGTT), G box (CACGTG)]. Seq: GTTAGTT.
MYBCORE: Binding site for all animal MYB and at least two plant MYB proteins ATMYB1 and ATMYB2, both isolated from Arabidopsis; ATMYB2 is involved in regulation of genes that are responsive to water stress in Arabidopsis; A petunia MYB protein (MYB.Ph3) is involved in regulation of flavonoid biosynthesis. Seq: CNGTTR.
MYCCONSENSUSAT: MYC recognition site found in the promoters of the dehydration-responsive gene rd22 and many other genes in Arabidopsis; Binding site of ATMYC2 (previously known as rd22BP1); see S000144 (E-box; CANNTG), S000174 (MYCATRD22); N = A/T/G/C; MYC recognition sequence in CBF3 promoter; Binding site of ICE1 (inducer of CBF expression 1) that regulates the transcription of CBF/DREB1 genes in the cold in Arabidopsis; ICE1; This sequence is also known as RRE (R response element). Seq: CANNTG.
Skn-1 Motif: cis-acting regulatory element required for endosperm expression. Seq: 226, GTCAT.
TATA-box: core promoter element around -30 of transcription start. Seq: TTTTA, TATA, TAATA, TATAA, TATATA.
WUN Motif: wound-responsive element. Seq: 373, TCATTACGAA.
The schematic model representing the regulation of expression stress-induced pea MCM6 gene with some cis-elements responsible for salt, cold, ABA and dehydration is shown in Figure 1B. Stress responsive genes can be expressed either through an ABA-dependent or ABA-independent pathway.10 The earlier results showed that MCM6 followed ABA-independent pathways in abiotic stress.9 The MCM6 transcript results indicated that the stress response was specific to salinity and cold stress related pathways.9 Earlier, an upregulation of DEAD-box helicases (PDH45 and PDH47) was also reported in salinity and cold stresses.11,12 To check whether the stress related cis-elements were present in the gene, the promoter of MCM6 was analyzed. The sequence analysis showed the presence of various stress related cis-elements including salt, cold ABA and dehydration. The hypothetical model in Figure 1B shows that probably some transcription factors bind to the salt and cold related cis-elements and activate the PsMCM6 gene transcription. In contrast some transcription factors bind to the ABA and dehydration related cis-elements and repress the PsMCM6 gene transcription. Further work is needed to understand the presence of many cis-elements in the promoter region of MCM6.
Acknowledgments
Work on DNA replication and plant abiotic stress tolerance in N.T.'s laboratory is partially supported by Department of Biotechnology (DBT), Government of India.
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 3.Hyrien O, Marheineke K, Goldar A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays. 2003;25:116–125. doi: 10.1002/bies.10208. [DOI] [PubMed] [Google Scholar]
- 4.Kanter DM, Bruck I, Kaplan DL. Mcm subunits can assemble into two different active unwinding complexes. J Biol Chem. 2008;283:31172–31182. doi: 10.1074/jbc.M804686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 6.You Z, Komamura Y, Ishimi Y. Biochemical analysis of the intrinsic Mcm4-Mcm6-mcm7 DNA helicase activity. Mol Cell Biol. 1999;19:8003–8015. doi: 10.1128/mcb.19.12.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Tran NQ, Dang HQ, Tuteja R, Tuteja N. A single subunit MCM6 from pea forms homohexamer and functions as DNA helicase. Plant Mol Biol. 2010;74:327–336. doi: 10.1007/s11103-010-9675-7. [DOI] [PubMed] [Google Scholar]
- 9.Dang HQ, Tran NQ, Gill SS, Tuteja R, Tuteja N. A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol Biol. 2011;76:19–34. doi: 10.1007/s11103-011-9758-0. [DOI] [PubMed] [Google Scholar]
- 10.Tuteja N. Abscisic acid and abiotic stress signalling. Plant Signal Behav. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA. 2005;102:509–514. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vashisht AA, Tuteja N. Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J Photochem Photobiol Biology. 2006;84:150–160. doi: 10.1016/j.jphotobiol.2006.02.010. [DOI] [PubMed] [Google Scholar]


