SUMMARY
Leptotrichia buccalis ATCC 14201 is a Gram-negative, anaerobic rod-shaped bacterium resident in oral biofilm at the tooth surface. The sequenced genome of this organism reveals three contiguous genes at loci: Lebu_1525,1526 and 1527. The translation products of these genes exhibit significant homology with phospho-α-glucosidase (Pagl), a regulatory protein (GntR) and a phosphoenol pyruvate-dependent sugar transport protein (EIICB), respectively. In non-oral bacterial species, these genes comprise the sim operon that facilitates sucrose isomer metabolism. Growth studies showed that L. buccalis fermented a wide variety of carbohydrates, including four of the five isomers of sucrose. Growth on the isomeric disaccharides elicited expression of a 50kDa polypeptide comparable in size to that encoded by Lebu_1525. The latter gene was cloned, and the expressed protein was purified to homogeneity from Escherichia coli TOP 10 cells. In the presence of two cofactors, NAD+ and Mn2+ ion, the enzyme readily hydrolyzed p-nitrophenyl-α-glucopyranoside 6-phosphate (pNPαG6P), a chromogenic analog of the phosphorylated isomers of sucrose. By comparative sequence alignment, immuno-reactivity and signature motifs, the enzyme can be assigned to the phospho-α-glucosidase (Pagl) clade of Family 4 of the glycosyl hydrolase super family. We suggest that the products of Lebu_1527 and 1525, catalyze the phosphorylative translocation and hydrolysis of sucrose isomers in L. buccalis, respectively. Four genetically diverse, but 16S rDNA related species of Leptotrichia have recently been described: L. goodfellowii, L. hofstadii, L. shahii and L. wadei. The phenotypic traits of these new species, with respect to carbohydrate utilization, have also been determined.
Keywords: Leptotrichia, sucrose isomers, dental caries, phospho-α-glucosidase, oral microbiology, glycosyl hydrolase Family 4
INTRODUCTION
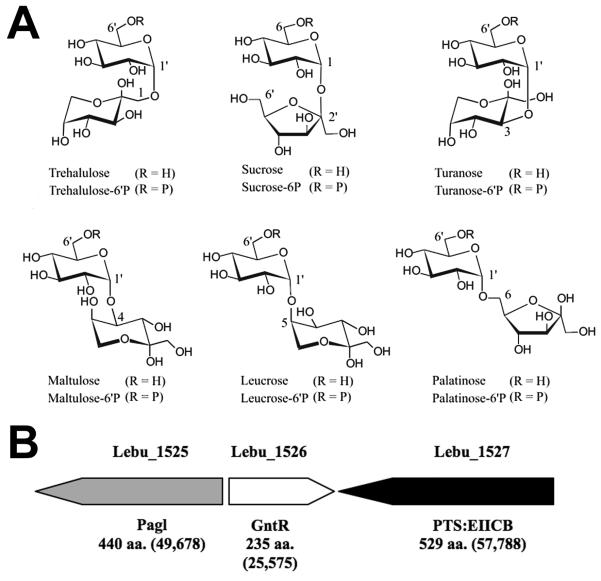
Of the many microbial species resident in oral biofilms (plaque) at the tooth surface (Becker et al., 2002; Jenkinson & Lamont, 2005; Paster et al., 2006; Dewhirst et al., 2010), mutans streptococci have long been recognized as a primary contributor in the etiology of dental caries (Loesche, 1986; van Houte, 1994; Banas, 2004). The pathogenicity of organisms such as Streptococcus mutans and S. sobrinus, is attributable in part to: (i) the capacity of these species to produce, from dietary sucrose, extracellular glycan(s) that facilitate microbial adherence to the tooth surface, and (ii) the transport and fermentation of the disaccharide to lactic acid, the causative agent in the demineralization of tooth enamel (Slee & Tanzer, 1979; St. Martin & Wittenberger, 1979). Remarkably, mutans streptococci are unable to metabolize the five structural isomers of sucrose designated: trehalulose, turanose, maltulose, leucrose and palatinose (Fig. 1A, Ziesenitz et al., 1989; Ooshima et al., 1983, 1991; Matsuyama et al., 2003). Consequently, these comparatively sweet, and ostensibly non-fermentable disaccharides are increasingly used as “non-cariogenic” substitutes for dietary sucrose in Japan, Europe and the United States (Hamada, 2002; Matsukubo & Takazoe, 2006). In contrast with mutans streptococci, numerous non-oral species including Bacillus subtilis, Klebsiella pneumoniae, Clostridium acetobutylicum, Lactobacillus casei (Thompson et al., 1998, 2001a,b, 2004, 2008) and Fusobacterium mortiferum (Thompson et al., 1995; Pikis et al., 2002) readily utilize all five isomers of sucrose as energy sources for growth. In these bacteria, the isomers are translocated via an α-glucoside-specific phosphoenolpyruvate-dependent: phosphotransferase system (PEP-PTS, Postma et al., 1993) and the phosphorylated derivatives are hydrolyzed intracellularly to glucose-6P and fructose by an NAD+ and Mn2+-dependent phospho-α-glucosidase (Thompson et al., 1998; Rajan et al., 2004; Yip et al., 2007; Vocadlo & Davies, 2008). This catalytically unique enzyme has been assigned to Family GH4 of the glycosyl hydrolase superfamily (Henrissat & Davies, 1997; Cantarel et al., 2009; Hall et al., 2009).
Figure 1.
A. Structural representations of sucrose and its five linkage- isomeric α-D-glucosyl-D-fructoses. In the free form R at C6′= H, and in the phosphorylated derivatives R= phosphate. B. Genetic composition and organization of the putative sucrose isomer metabolic (sim) operon of L. buccalis 14201.
The inability of mutans streptococci to ferment sucro-disaccharides is well established, but whether this deficiency is characteristic of other oral microorganisms is the subject of debate (Peltroche-Llacsahuanga et al., 2001; Matsuyama et al., 2003). Among the ~ 700 bacterial species presently catalogued in the Human Oral Microbiome Database (HOMD) (Dewhirst et al., 2010; Chen et al., 2010), Leptotrichia buccalis (Human Oral Taxon 563) occurs as a minor constituent of the total flora of plaque (Hofstad, 1992; Hofstad & Olsen, 2007; Eribe & Olsen, 2008). However, L. buccalis is highly saccharolytic (Kasai, 1965; Thompson, 2002) and, because it ferments a wide variety of mono- and disaccharides to lactic acid, the bacterium has evident cariogenic potential (Brown & Gross, 1981; Hofstad, 1992). For many years, L. buccalis was believed to be the sole member of the genus Leptotrichia, but four novel oral species (L. goodfellowii, L. hofstadii, L. shahii and L. wadei) have recently been assigned to the genus (Eribe, et al., 2004). Equally significant was the discovery, in the recently sequenced genome of the type strain L. buccalis ATCC 14201, of a locus of three genes (Fig.1B) with extensive similarity to those previously shown to encode the requisite proteins for dissimilation of sucro-disaccharides by non-oral bacteria, including Escherichia coli (Pikis et al., 2006). These in silico findings, prompted us to hypothesize that L. buccalis 14201 (and related species) might also metabolize the isomers of sucrose. We have tested this hypothesis by growth studies, enzymatic assays and proteomic analyses. Our findings are presented in this communication.
METHODS
Reagents and materials
High purity sugars including glucose, sucrose, maltose and 1-O-methyl-α-D-glucopyranoside were purchased from Pfanstiehl Laboratories, Inc, Waukegan, IL. The five isomers of sucrose were obtained from the following sources: trehalulose was a generous gift from Südzucker, Germany; turanose was obtained from Pfanstiehl; maltulose from TCI America; leucrose from Fluka, and palatinose was purchased from Wako Chemicals. Phosphorylated chromogenic derivatives including, p-nitrophenyl-α-D-glucopyranoside -6-phosphate (pNPαG6P) were synthesized in this laboratory (Thompson et al; 1995). Tris-Acryl M-DEAE, Ultrogel AcA-44 chromatography media and other materials, were obtained from Sigma-Aldrich. Polyclonal rabbit antibody to phospho-α-glucosidase (MalH) from F. mortiferum ATCC 25557 was prepared by Covance Research Laboratories, Denver, PA (Thompson et al; 1995). Glucose-6-phosphate dehydrogenase (G6PDH, baker’s yeast, E.C. 1.1.1.49) and phosphoglucose isomerase (PGI, baker’s yeast, E.C. 5.3.1.9) were obtained from Boehringer.
Bacterial strains
Leptotrichia buccalis ATCC 14201 (DSM 1135/C-1013-bT) was purchased from the American Type Culture Collection (Manassas, VA). The four novel Leptotrichia species: L. goodfellowii CCUG 32286T (DSM 19756/LB 57T), L. shahii CCUG 47503T(LB 37T), L. hofstadii CCUG 47504T (LB 23T) and L. wadei CCUG 47505T (LB 16T), were obtained from the Culture Collection University of Göteborg, Sweden. Leptotrichia goodfellowii FO264 was kindly provided by Dr. F. Dewhirst from the Forsyth Institute Culture Collection (FICC), Boston, MA. Chromosomal DNA from L. goodfellowii CCUG 32286T / DSM 19756 was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ), Braunschweig, Germany.
Maintenance of Leptotrichia species
Organisms were grown under anaerobic conditions (BD GasPak) at 37°C in 10 ml of Reinforced Clostridial Medium (Difco). At mid-log stage, the cells were collected by centrifugation and supernatants were decanted. The white cell pellets were resuspended in 0.5 ml of filter sterilized Clostridial Medium containing 20% glycerol and dispensed into sterile 1 ml tubes (Sarstedt). The samples were immediately frozen in dry ice, and maintained at −80°C until required.
Growth studies and large - scale preparation of cells
To determine their carbohydrate specificities, Leptotrichia species were grown anaerobically (24-48 h) at 37°C in 10 ml of modified salts/vitamins-supplemented Todd Hewitt broth (MTH, Robrish & Thompson, 1990) containing 0.4% (wt/vol) of desired sugar. For large-scale preparations, the organisms were grown anaerobically in 800 ml volumes of appropriate MTH. Stationary phase cells were harvested by centrifugation (13,000 x g for 20 min at 5°C), and washed by resuspension and centrifugation from 25 mM Tris-HCl buffer (pH 7.5) containing 0.1 mM NAD+, 1 mM Mn2+ and 1mM dithiothreitol (TMND buffer). The yield of cells was ~ 1.1 g wet weight l −1.
Cloning and expression of L. buccalis ATCC 14201 gene Lebu_1525 in E. coli TOP10
From the complete genome sequence of L. buccalis ATCC 14201 (Ivanova et al. 2009; GenBank Accession no. NC_013192) the following primers were designed to amplify the Lebu_1525 gene. Forward primer: 5′-GTGCACGCCATGGGTATGAAGAAATTTTCAATTGTAGTAGC-3′ (Lebu_1525 is boldfaced and the NcoI restriction site is underlined). Reverse primer: 5′-GCGGTCCCGAATTCCTACTATTTCAACACTGGCCAGAAGTC-3′ (the sequence complimentary to the downstream region of Lebu_1525 is boldfaced and the EcoRI restriction site is underlined). PCR amplification was carried out with a thermal cycler (GeneAmp 9700; PE Applied Biosystems) using Pfu high fidelity DNA polymerase from Stratagene. The components of the amplification mixture (100 μl) were as follows: 5 U of Pfu high fidelity DNA polymerase, 1X reaction buffer provided by the manufacturer, 20 mM each of the four deoxynucleotide triphosphates, 250 ng of each primer and 100 ng of L. buccalis ATCC 14201 chromosomal DNA. After an initial 2 min incubation at 95°C, the mixture was subjected to 30 cycles of amplification under the following conditions: denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min/kb of insert. This was followed by a 10 min runoff at 72°C. The amplicon was digested with restriction endonucleases NcoI and EcoRI, electrophoresed in 1% agarose gel, and purified using QIAquick gel extraction kit. The PCR product (~1.3 kbp) was ligated to similarly digested and purified pTrcHis2B expression vector (Invitrogen) to form the recombinant plasmid pLBpagl. In this construct, the Lebu_1525 gene is under control of the trc promoter, which is also regulated by the lacO operator and the product of the laclq gene. Therefore, expression of the Lebu_1525 gene is fully induced in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG). Plasmid pLBpagl was transformed into E. coli TOP10 competent cells (Invitrogen), and colonies were selected on Luria-Bertani agar plates containing 150 μg mL−1 ampicillin. It should be noted that in the pTrcHis2B vector the fusion peptide is located at the C terminus. However, the stop codon included in the reverse primer prevents the expression of the fusion peptide.
Purification of Pagl from E. coli TOP 10 (pLBpagl)
The plasmid-containing cells were grown in 800 ml Luria-Bertani broth containing ampicillin (150 μg ml −1) in 2-liter baffled flasks at 37°C on a rotary shaker. At A600nm ~ 0.6, IPTG (0.5 mM) was added, and growth was continued for 3-4 h. The cells were harvested by centrifugation (13,000 x g for 10 min at 5°C) and washed by centrifugation from TMND buffer. Washed cells (16 g wet weight) of E. coli TOP 10 (pLBpagl) were resuspended in 40 ml of TMND buffer, and the organisms were disrupted at 0°C by sonic oscillation. The extract was clarified by high -speed centrifugation (180,000 x g for 2 h at 5°C), and the supernatant was dialyzed for 16 h in a cold room against 4 liters of TMND buffer. The purification procedure for LB Pagl, was essentially that used previously for the purification of Pagl from L. casei ATCC 334 (Thompson et al; 2008). This two-stage process, involving Tris-Acryl M-DEAE (anion exchange) and Ultrogel AcA-44 (gel fltration) chromatography, yielded 9.8 mg of highly purified enzyme.
Enzyme Assays
1. Phospho-α-glucosidase (Pagl) activity
Enzyme activity was determined using a discontinuous assay and a 2 ml mixture that contained Tris-HCl buffer (50 mM, pH 7.5), 1 mM MnCl2, 0.1 mM NAD+, 25 mM DTT and requisite amount of extract or purified Pagl. After 5 min of incubation at 37°C, reactions were initiated by addition of the chromogenic substrate (0.5mM pNPαG6P). Samples of 0.25 ml were removed at 20 s intervals, and immediately injected into 0.75 ml of a solution of 0.5 M Na2CO3 containing 0.1 M EDTA to stop the reaction. The absorbance at 400 nm was measured, and the rates of p-nitrophenol (pNP) formation were calculated from progress plots, assuming a molar extinction coefficient at pH 10.2 for the yellow p-nitrophenolate anion ε= 18,000 M −1 cm −1. Pagl activity was expressed in nmol or μmol of pNPαG6P hydrolyzed min−1 mg −1 of protein.
2. Fructokinase activity
In this assay fructose is first phosphorylated to fructose-6P by ATP-dependendent fructokinase present in the cell extract, and subsequently is converted to G6P by PGI. Oxidation of G6P is coupled to the reduction of NADP+ by G6PDH, and the increase in A340nm was followed in a Beckman DU 640 recording spectrophotometer. The standard 1-ml assay contained: 0.1M potassium phosphate buffer (pH 7.0), 5 mM ATP, 10 mM Mg2+, 1 mM NADP+, 10 mM fructose, 5 U G6PDH, 5 U PGI and the reaction was initiated by addition of 50 μl of cell extract. The initial rates of NADPH formation (equivalent to fructose-6P) were determined using the kinetics program installed in the instrument. In all calculations, a molar extinction coefficient ε = 6220 M −1 cm −1 was assumed for NADPH.
Amplification and characterization of a region encoding the Pagl and EII(CB) genes of L. goodfellowii CCUG 32286 / DSM 19756
Based on the incomplete genome sequence of L. goodfellowii CCUG 32286 / DSM 19756, the following two pairs of primers were designed to amplify and characterize the DNA fragment encoding the putative Pagl and EII(CB) genes (LepgoDRAFT_1708 and LepgoDRAFT_1710, respectively). The two primer sets were constructed as follows: F1 5′-CCGGTGTAAATGTAC TTCCTCCTC-3′ and R1 5′-ATACCCTCCTAAAAAGCGAAAAA-3′ for the amplification of Pagl; F2 5′-GCCGAGACTGAGAAGTTAGTATTAATAAAGGG-3′ and R2 5′-GAATAACTGTGCGACAAGACTTAGAGTAAGTG-3′ for the amplification of the EII(CB) gene. Amplifications were carried out with a thermal cycler (GeneAmp 9700; PE Applied Biosystems) using Phusion high-fidelity DNA polymerase (New England Biolabs, Inc.). For the amplification of LepgoDRAFT_1708 the following PCR settings were used: after an initial 1 min incubation at 98°C, the mixture was subjected to the following cycling parameters: 2 cycles of amplification under the following conditions: denaturation at 98°C for 20 sec, annealing at 65°C for 20 sec, and extension at 72°C for 2 min; 2 cycles with denaturation at 98°C for 20 sec, annealing at 63°C for 20 sec, and extension at 72°C for 2 min; and 2 cycles with denaturation at 98°C for 20 sec, annealing at 61°C for 20 sec, and extension at 72°C for 2 min. These cycles were followed by 29 cycles with the following conditions: denaturation at 98°C for 20 sec, annealing at 59°C for 20 sec, and extension at 72°C for 2 min. This procedure was followed by a 10-min runoff at 72°C. For the amplification of the region encoding LepgoDRAFT_1710, the PCR parameters used were as follows: after an initial 1 min of incubation at 95°C, the mixture was subjected to 32 cycles of amplification. Each cycle consisted of 20 sec denaturation at 95°C, 30 sec annealing at 60°C, and extension at 72°C for 2 min. These were followed by a 10-min runoff at 72°C. All sequencing was performed by OriGene Technologies Inc. (Rockville, MD). The MacVector sequence analysis package (Version 11.0, Genetics Computer Group, Madison, WI) was used to assemble, edit, and analyze the results.
Analytical procedures
The molecular weight of denatured Pagl was determined by sodium dodecyi sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) using the Novex XCell mini-cell system (Invitrogen). Novex NUPage (4-12% acrylamide) bis-Tris gels and morpholineethanesulfonic acid (MES)-SDS running buffer (pH 7.3) were used together with All Blue Precision Plus Protein Standards (BioRad). Polypeptides were visualized by staining with Coomassie blue R-250. For Western blots, proteins and See-Blue prestained standards were transferred to nitrocellulose membranes using NuPage transfer buffer. Immuno-detection of Pagl was performed by incubation of the membrane with polyclonal antibody to MalH from F. mortiferum, and subsequently with goat anti-rabbit horseradish peroxidase-conjugated antibody as described previously (Thompson et al; 1995). The native Mr of Pagl from L. buccalis was estimated chromatographically in an Ultrogel AcA-44 filtration column (1.6 by 94 cm) previously calibrated with TMND buffer containing the standards: alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (29 kDa) and Blue Dextran. Protein concentrations of cell extracts were determined with the BCA assay kit (Pierce), and the concentration of purified Pagl was determined spectrophotometrically (A280nm) assuming a theoretical molar absorption coefficient of 62,340 M −1 cm −1. The N-terminal sequence of Pagl was determined with an ABI 477A protein sequencer (Applied Biosystems Inc.) connected online to an ABI 120A phenylthiohydantoin analyzer.
RESULTS
Growth of L. buccalis 14201 on sucrose isomers
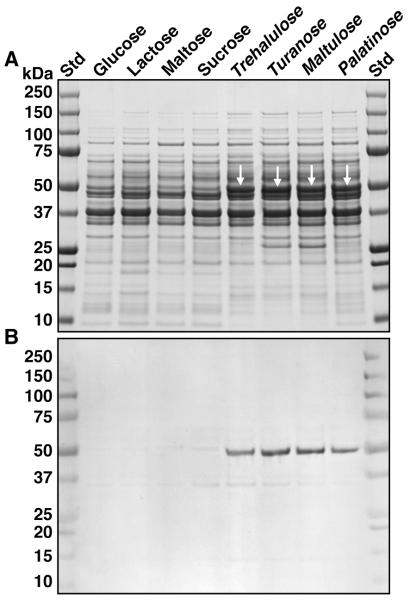
Leptotrichia buccalis 14201 showed excellent growth on all isomers of sucrose with the exception of leucrose (Table 1). SDS-PAGE analyses of cell extracts, revealed expression of a unique polypeptide during growth of L. buccalis on trehalulose, turanose, maltulose and palatinose (Fig. 2A, white arrows). The Mr of this polypeptide (~ 50 kDa), was of the size expected for the product encoded by Lebu_1525. Importantly, the results of a Western blot (Fig. 2B) showed that the induced polypeptide cross-reacted strongly with polyclonal antibody prepared against the NAD+ and Mn2+ -dependent phospho-α-glucosidase (MalH) from Fusobacterium mortiferum (Thompson et al; 1995). Data from enzymatic analyses (Table 2) provided convincing evidence that the ~ 50 kDa protein induced by growth on sucrose isomers also exhibited NAD+ and Mn2+ - dependent phospho-α-glucosidase (Pagl) activity. The latter cytoplasmic enzyme catalyzes the hydrolysis of phosphorylated sucrose isomers to yield G6P and fructose. It seemed likely that in order to facilitate the fermentation of the ketohexose, an ATP-dependent fructokinase might also be induced by growth on the sucro disaccharides. Surprisingly, enzymatic analyses (Table 2) revealed constitutively high levels of fructokinase in all cell extracts. Inspection of the L. buccalis genome suggests that this ATP-dependent kinase may reside at locus Lebu_0629, whose translation product predicts a polypeptide of 295 residues and Mr = 32,626 (GenBanK Accession no. YP_003163528).
Table 1.
Growth of Leptotrichia species on various sugars a
| Sugar Added b |
L. buccalis ATCC 14201 |
L. goodfellowii CCUG 32286 |
L. goodfellowii FICC F0264 |
L. shahii CCUG 47503 |
L. hofstadii CCUG 47504 |
L. wadei CCUG 47505 |
|---|---|---|---|---|---|---|
| None | NG | 0.33 c | 0.30 c | NG | NG | NG |
| Glucose | 1.24 | 1.04 | 1.05 | 1.02 | 0.90 | 0.82 |
| α-MGlc d | 0.19 | 0.13 | 1.00 | 0.61 | NG | NG |
| Maltitol | NG | 0.24 | 0.77 | 0.32 | NG | NG |
| Maltose | 1.21 | 0.75 | 1.02 | 0.96 | 0.86 | 0.91 |
| Lactose | 1.29 | 0.40 | 0.76 | 0.99 | 0.71 | NG |
| Sucrose | 1.26 | 0.93 | 1.12 | 1.00 | 0.87 | NG |
| Trehalulose e | 1.08 | 0.30 (NG) f | 1.09 | 1.20 | 0.77 | 0.94 |
| Turanose | 0.95 | 0.23 (NG) | 1.08 | 1.15 | 0.91 | 0.96 |
| Maltulose | 0.92 | 0.26 (NG) | 1.05 | 0.95 | 0.82 | (NG) |
| Leucrose | (NG) | 0.23 (NG) | 0.83 | 1.04 | 0.22 | (NG) |
| Palatinose | 0.91 | 0.29 (NG) | 1.01 | 1.09 | 0.89 | 0.81 |
Organisms were grown anaerobically at 37 °C, and after 48h, growth was recorded (A600nm) using a Beckman DU 640 spectrophotometer. Results are mean values from two experiments.
All sugars were present at a final concentration of 0.4 % (w/v).
Note growth of L. goodfellowii strains in absence of sugar.
α-MGlc: methyl-α-D-glucopyranoside.
Sugars in bold face are sucrose isomers.
(NG), no growth on designated sucrose isomer.
Figure 2.
Proteomic analysis by SDS- PAGE, and Western blot assay of extracts from cells of L. buccalis 14201 grown on various sugars. A. SDS-PAGE and visualization of proteins by staining with Coomassie blue R-250. Approx. 30μg of protein was applied per lane. Note the expression (white arrowheads) of a 50 kDa protein induced by growth of the organism on the four isomers of sucrose. B. Western blot of a duplicate gel showing the immuno-cross reactivity of the induced 50 kDa protein with polyclonal antibody raised against the NAD+ and Mn2+ -dependent phospho-α-glucosidase (MalH) from F. mortiferum.
Table 2.
Enzyme activities in extracts from cells of L. buccalis 14201 grown on various sugars
| Growth sugar | P-α-glucosidase a. (Pagl) |
ATP-dependent b. Fructokinase |
|---|---|---|
| Glucose | NDA c. | 36 |
| Maltose (α, 1g – 4g) | 0.52 | 116 |
| Lactose (β, 1gal – 4g) | NDA | 102 |
| Trehalulose d. (α, 1g –1f) | 8.41 | 185 |
| Sucrose (α, 1g – 2f) | NDA | 159 |
| Turanose (α, 1g – 3f) | 7.72 | 187 |
| Maltulose (α, 1g – 4f) | 3.28 | 75 |
| Palatinose (α, 1g – 6f) | 5.26 | 212 |
Specific activity: nmol pNPαG6P hydrolyzed min −1 mg protein −1.
Specific activity: nmol F6P formed min −1 mg protein −1.
NDA: no detectable activity: all values are average rates from three separate assays.
Sugars in bold face are sucrose isomers.
Purification and properties of Pagl from L. buccalis 14201
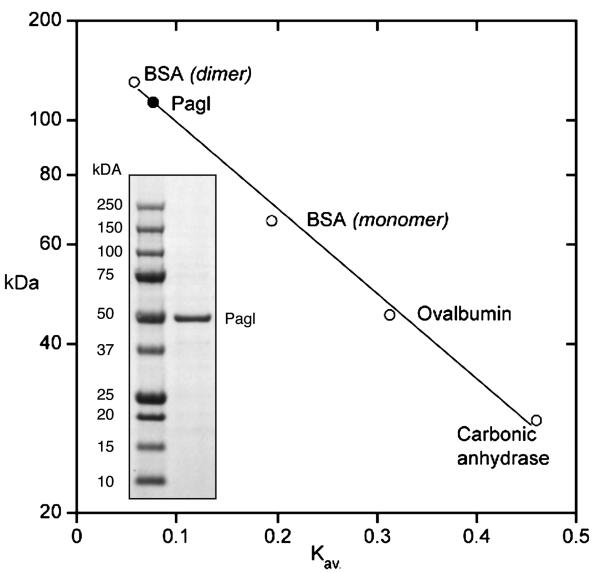
Lebu_1525 from L. buccalis 14201 was cloned, and the phospho-α-glucosidase (Pagl) was expressed in E. coli TOP 10. The enzyme was purified by two-stage chromatography (anion exchange and gel filtration), in 25 mM Tris/HCl buffer (pH 7.5) containing 0.1 mM NAD+ and I mM MnCl2. During purification, enzyme activity was monitored by hydrolysis of pNPαG6P, a chromogenic analog of the phosphorylated O-α-linked isomers of sucrose. The specific activity of purified Pagl was 0.74 μmol of pNPαG6P hydrolyzed min −1 mg protein −1. The MW of the denatured enzyme determined by SDS-PAGE (~ 50 kDa, Fig. 3, inset), was in excellent agreement with the theoretical Mr of 49,678. However, the molecular weight of the native enzyme (110 ± 10 kDa) estimated by calibrated gel filtration chromatography (Fig. 3), suggests that in the catalytically- active solution state, Pagl assumes a homodimeric conformation. The homogeneity of the preparation was confirmed by the unambiguous micro-sequence determination of the first 26 residues from the N-terminus: MKKFSIVVAGGGSTFTPGIVLMLLE. Optimum conditions for Pagl activity were found in the pH range 6.9-7.3, at an assay temperature of 36-38 °C. The enzyme was stable during prolonged incubation (> 0.5 h) at 37 °C in the presence of NAD+ and Mn2+ ion, but was irreversibly inactivated within 8 min when the two cofactors were omitted from the assay. Whereas pNPαG6P was readily hydrolyzed by Pagl, neither the C2 nor C4 chromogenic stereomers (pNPα-mannopyranoside-6P and pNPα-galactopyranoside-6P, respectively) were substrates for the enzyme.
Figure 3.
Estimation of the solution – state molecular weight of NAD+ and Mn2+ -dependent phospho-α-glucosidase (Pagl) of L. buccalis 14201 by gel filtration chromatography. Inset : Demonstration of the purity, and estimated Mr ~ 50 kDa of the denatured Pagl monomer.
Sugar utilization and metabolic diversity of Leptotrichia species
Data presented in Table 1, show considerable variation in the range of carbohydrates able to support growth of Leptotrichia species. In the absence of sugar(s), there is no growth of L. buccalis, L. shahii, L. hostadii and L. wadei. However, there is significant growth of both strains of L. goodfellowii in sugar-free MTH medium. Arginine dihydrolase activity (GenBank accession no. ZP_06011002) has been reported in this organism (Eribe et al., 2004), and it is likely that arginine catabolism provides the requisite ATP for growth of L. goodfellowii in this medium. By extrapolation, we may assume that the enzymes comprising the arginine fermentation pathway are either absent or non-functional in other species. All species grew readily on glucose, maltose (and, with the exception of L. wadei) on lactose and sucrose also. Examination of the published DNA sequence of L. buccalis 14201 (Ivanova et al; 2009) reveals genetic loci for the phosphoenolpyruvate-dependent phosphotransferase systems (PEP-PTS) for glucose (Lebu_1946), sucrose (Lebu_1906) and lactose (Lebu_0588). Similar PEP-PTS operons are likely present in the genomes of L. goodfellowii, L. shahii and L. hofstadii, but presumably are absent in L. wadei. The results from growth experiments involving sucrose isomers revealed considerable diversity in those compounds able to support growth of the Leptotrichia species. For example, L. shahii and L. hofstadii, showed excellent growth on all five isomers, whereas L. buccalis 14201 failed to grow on leucrose, and L. wadei was unable to metabolize either maltulose or leucrose.
Metabolism of sucrose isomers by L. goodfellowii strains FO264 and 32286 / DSM 19756
Two strains have been assigned to the species L. goodfellowii. The complete chromosome of L. goodfellowii FO264 has been sequenced by the J. Craig Venter Institute as part of the Human Microbiome Project (HMP). A draft (incomplete) sequence of the chromosome of L. goodfellowii CCUG 32286/ DSM 19756 was released January 10th, 2010 by the US Department of Energy, Joint Genome Institute (http://www.jgi.doe.gov). As reported in Table 1, L. goodfellowii FO264 showed excellent growth on all five sucrose isomers whereas, unexpectedly, strain CCUG 32286 failed to grow on any of the sucro-disaccharides. To explain the latter findings, we hypothesized that L. goodfellowii 32286 might lack (or had incurred mutational inactivations in) the requisite genes for metabolism of the isomers. To obtain support for this hypothesis, an in silico analysis was undertaken of the contigs available from the incomplete sequence of the L. goodfellowii CCUG 32286/ DSM 19756 genome. Contig 00026 encoded a gene, LepgoDRAFT_1708, whose translation product was of similar size (440 residues, Mr = 49,672) and with 90% identity to Pagl from L. buccalis 14201 (Fig.1). Contig 00026 also contained a presumptive pseudogene, LepgoDRAFT_1710, whose predicted polypeptide (302 amino acids, Mr = 33,020) was considerably smaller, but with 67% identity to the α-glucoside-specific EII(CB) PTS transporter encoded by Lebu_1527 in L. buccalis 14201 (Fig. 1). We have confirmed these results by re-sequencing of the appropriate region of chromosomal DNA from L. goodfellowii CCUG 32286 / DSM 19756 containing contig 00026 (see, Methods). Importantly, we have found that a glutamate residue at position 302 in LepgoDRAFT_1710 is immediately followed by a tag stop codon, The presence of this codon presumably causes premature termination, and production of a functionally inactive EII(CB) transporter of the sucrose isomers.
DISCUSSION
Growth of non-oral bacteria on the five isomers of sucrose has been well documented for species from Gram-negative and Gram-positive genera. However, to our knowledge, this report is the first to document the genetic organization and enzymatic basis for growth of a common resident of the oral microflora on sucro-disaccharides. Our findings show that for both Leptotrichia and non-oral species, the phosphorylative transport and hydrolysis of the isomeric derivatives are mediated by the products of chromosomal genes encoding an α-glucoside-specific PTS transport system and a unique NAD+ and Mn2+-dependent phospho-α-glucosidase. Notably, Pagl is only the second phospho-glycosidase to have been isolated and characterized from L. buccalis 14201. Previously, a phospho-β-galactosidase (Pbgal; EC. 3.2.1.85; gene locus Lebu_ 0590) was purified to electrophoretic homogeneity from cells of L. buccalis grown on lactose or lactulose (Thompson, 2002). Phospho-β-galactosidase (UniProtKB/Swiss-Prot C7N8L9; 467 residues, Mr =54,271) hydrolyzes lactose 6′-phosphate to yield galactose-6P and glucose, but in contrast with Pagl, Pbgal has no cofactor requirements for catalytic activity. Importantly, by the sequence-based CAZy classification (Henrissat & Davies, 1997; Cantarel et al, 2009) Pbgal from L. buccalis 14201 is included in Family GH 1, whilst Pagl is assigned to GH 4 of the glycosyl hydrolase superfamily.
Non-oral species, including B. subtilis, Lb. casei, C. acetobutylicum, F. mortiferum and K. pneumoniae, invariably grow on all five sucrose isomers. However, in the present study we find considerable variation in the number(s) of isomers that support the growth of a particular species of Leptotrichia. For example, L. shahii metabolizes all five isomers, L. buccalis utilizes four of the sucro-disaccharides whereas, under the same conditions, L. wadei fails to grow on maltulose or leucrose. At the molecular level, such isomeric discrimination presumably reflects subtle species-specific mutations or conformational changes in either the transport or intracellular Pagl proteins. The genetic basis for the variability of metabolic traits may be revealed by comparative analyses when the genomes of the different species of Leptotrichia have been sequenced in their entirety. Earlier surveys have reported the generation of acid from selected isomers of sucrose by some (but not all) oral species of Actinomyces and lactobacillae (Peltroche-Llacsahuanga et al, 2001; Matsuyama et al, 2003). Interestingly, A. israelii ATCC12102 and A. gerencseriae ATCC 23860 produced acid from all five sucrose isomers, but other species including A. odontolyticus ATCC 17929, were unable to ferment any of the disaccharides. The recently sequenced genomes of A. odontolyticus strains ATCC 17982 and F0309 encode a gene whose predicted polypeptide (460 residues, Mr ~‘49,430) shows 28% identity with Pagl from L. buccalis 14201, but a comparable gene is not present in the genomes of A. viscosus C505 or A. oris K20. Of the oral lactobacillae investigated, Lb. crispatus strains JV-V01, ST1 and 214-1 encode a gene whose translation product (445 amino acids, Mr = 50,639) exhibits 57% identity with Pagl from L. buccalis 14201, but this gene is absent from the genomes of Lb. salivarius strains ATCC 11741 and GJ-24. The metabolic pathway(s) involved in the fermentation of sucrose isomers by Actinomyces and lactobacillae have yet to be elucidated. However, as genetic information accumulates in the Human Oral Microbiome Database, it highly likely that genetic loci similar to the sim operon in Leptotrichia and Lb. casei ATCC 334 will be discovered in currently unsequenced genomes of oral bacteria. In this context, the fermentation of presumed “non-cariogenic” disaccharides by oral bacteria may be more widespread than presently envisaged. It remains to be seen whether continued commercial scale use of the various sucrose isomers will impact upon the species composition or ecological distribution of the oral microflora (Hamada, 2002; Matsukubo & Takazoe, 2006).
ACKNOWLEDGEMENTS
We thank Rick Dreyfuss for assistance with photography and computer graphics and Nga Nguyen for microsequence analyses. This work was supported by the Intramural Research Program of the NIDCR, National Institutes of Health, Bethesda, MD.
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas JA. Virulence properties of Streptococcus mutans. Front Biosci. 2004;9:1267–1277. doi: 10.2741/1305. [DOI] [PubMed] [Google Scholar]
- Becker MR, Paster BJ, Leys EJ, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AT, Gross ML. Regulation of lactic acid production by Leptotrichia buccalis. Caries Res. 1981;15:21–25. doi: 10.1159/000260495. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [PMID: 18838391] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu W-Han., Izard J, et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;Vol. 2010 doi: 10.1093/database/baq013. Article ID baq013, doi: 10. 1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eribe ERK, Olsen I. Leptotrichia species in human infections. Anaerobe. 2008;14:131–137. doi: 10.1016/j.anaerobe.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Eribe ERK, Paster BJ, Caugant DA, et al. Genetic diversity of Leptotrichia and description of Leptotrichia goodfellowii sp.nov., Leptotrichia hofstadii sp.nov., Leptotrichia shahii sp. nov. and Leptotrichia wadei sp. nov. Int J Syst Evol Microbiol. 2004;54:583–592. doi: 10.1099/ijs.0.02819-0. [DOI] [PubMed] [Google Scholar]
- Hall BG, Pikis A, Thompson J. Evolution and biochemistry of Family 4 glycosidases: Implications for assigning enzyme function in sequence annotations. Mol Biol Evol. 2009;26:2487–2497. doi: 10.1093/molbev/msp162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H. Role of sweeteners in the etiology and prevention of dental caries. Pure Appl Chem. 2002;74:1293–1300. [Google Scholar]
- Henrissat B, Davies GJ. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- Hofstad T. The Genus Leptotrichia. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H, editors. The Prokaryotes. 2nd edn Vol. IV. Springer-Verlag; New York: 1992. pp. 3983–3986. [Google Scholar]
- Hofstad T, Olsen I. Fusobacterium and Leptotrichia. In: Boriello SP, Murray PR, Funke G, editors. Topley and Wilson’s Microbiology and Microbial Infections: Bacteriology. 10th edn Vol 1 and 2. Hodder Arnold; London: 2007. [Google Scholar]
- Ivanova N, Gronow S, Lapidus A, et al. Complete genome sequence of Leptotrichia buccalis type strain (C· 1013-bT) Stand Genomic Sci. 2009;1:126–132. doi: 10.4056/sigs.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Kasai GJ. A study of Leptotrichia buccalis. II. Biochemical and physiological observations. J Dent Res. 1965;44:1015–1022. doi: 10.1177/00220345650440050301. [DOI] [PubMed] [Google Scholar]
- Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukubo T, Takazoe I. Sucrose substitutes and their role in caries prevention. Int Dent J. 2006;56:119–130. doi: 10.1111/j.1875-595x.2006.tb00083.x. [DOI] [PubMed] [Google Scholar]
- Matsuyama J, Sato T, Hoshino E, et al. Fermentation of five sucrose isomers by human dental plaque bacteria. Caries Res. 2003;37:410–415. doi: 10.1159/000073392. [DOI] [PubMed] [Google Scholar]
- Ooshima T, Izumitani A, Minami T, et al. Trehalulose does not induce dental caries in rats infected with mutans streptococci. Caries Res. 1991;25:277–282. doi: 10.1159/000261376. [DOI] [PubMed] [Google Scholar]
- Ooshima T, Izumitani A, Sobue S, et al. Non - cariogenicity of the disaccharide palatinose in experimental dental caries of rats. Infect Immun. 1983;39:43–49. doi: 10.1128/iai.39.1.43-49.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Olsen I, Aas. JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Peltroche-Llacsahuanga H, Hauk CJ, Kock R, et al. Assessment of acid production by various human oral micro-organisms when palatinose or leucrose is utilized. J Dent Res. 2001;80:378–384. doi: 10.1177/00220345010800011401. [DOI] [PubMed] [Google Scholar]
- Pikis A, Hess S, Arnold I, Erni B, Thompson J. Genetic requirements for growth of Escherichia coli K12 on methyl-α-D-glucopyranoside and the five α-D-glucosyl-D-fructose isomers of sucrose. J Biol Chem. 2006;281:17900–17908. doi: 10.1074/jbc.M601183200. [DOI] [PubMed] [Google Scholar]
- Pikis A, Immel S, Robrish SA, Thompson J. Metabolism of sucrose and its five isomers by Fusobacterium mortiferum. Microbiology. 2002;148:843–852. doi: 10.1099/00221287-148-3-843. [DOI] [PubMed] [Google Scholar]
- Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan SS, Yang X, Collart F, et al. Novel catalytic mechanism of glycoside hydrolysis based on the structure of an NAD+/Mn2+ -dependent phospho-α-glucosidase from Bacillus subtilis. Structure. 2004;12:1619–1629. doi: 10.1016/j.str.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Robrish SA, Thompson J. Regulation of fructose metabolism and polymer synthesis by Fusobacterium nucleatum ATCC 10953. J Bacteriol. 1990;172:5714–5723. doi: 10.1128/jb.172.10.5714-5723.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee AM, Tanzer JM. Phosphoenolpyruvate – dependent sucrose phosphotransferase activity in Streptococcus mutans NCTC 10449. Infect Immun. 1979;24:821–828. doi: 10.1128/iai.24.3.821-828.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Martin EJ, Wittenberger CL. Characterization of a phosphoenolpyruvate - dependent sucrose phosphotransferase system in Streptococcus mutans. Infect Immun. 1979;24:865–868. doi: 10.1128/iai.24.3.865-868.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. Purification and some properties of phospho-β-galactosidase from the Gram- negative oral bacterium Leptotrichia buccalis ATCC 14201. FEMS Microbiol Lett. 2002;214:183–188. doi: 10.1111/j.1574-6968.2002.tb11344.x. [DOI] [PubMed] [Google Scholar]
- Thompson J, Gentry-Weeks CR, Nguyen NY, et al. Purification from Fusobacterium mortiferum ATCC 25557 of a 6-phosphoryl-O-α-D-glucopyranosyl: 6-phosphoglucohydrolase that hydrolyzes maltose 6-phosphate and related phospho-α-D-glucosides. J Bacteriol. 1995;177:2505–2512. doi: 10.1128/jb.177.9.2505-2512.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Hess S, Pikis A. Genes malH and pagL of Clostridium acetobutylicum ATCC 824 encode NAD+ and Mn2+- dependent phospho-α-glucosidase(s) J Biol Chem. 2004;279:1553–1561. doi: 10.1074/jbc.M310733200. [DOI] [PubMed] [Google Scholar]
- Thompson J, Jakubovics N, Abraham B, et al. The sim operon facilitates the transport and metabolism of sucrose isomers in Lactobacillus casei ATCC 334. J Bacteriol. 2008;190:3362–3373. doi: 10.1128/JB.02008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Pikis A, Ruvinov SB, et al. The gene glvA of Bacillus subtilis 168 encodes a metal-requiring, NAD(H)-dependent 6-phospho-α-glucosidase. Assignment to family 4 of the glycosyl hydrolase superfamily. J Biol Chem. 1998;273:27347–27356. doi: 10.1074/jbc.273.42.27347. [DOI] [PubMed] [Google Scholar]
- Thompson J, Robrish SA, Immel S, et al. Metabolism of sucrose and its five linkage-isomeric α-D-glucosyl-D-fructoses by Klebsiella pneumoniae. Participation and properties of sucrose-6-phosphate hydrolase and phospho-α-glucosidase. J Biol Chem. 2001;276:37415–37425. doi: 10.1074/jbc.M106504200. [DOI] [PubMed] [Google Scholar]
- Thompson J, Robrish SA, Pikis A, et al. Phosphorylation and metabolism of sucrose and its five linkage-isomeric α-D-glucosyl-D-fructoses by Klebsiella pneumoniae. Carbohyd Res. 2001;331:149–161. doi: 10.1016/s0008-6215(01)00028-3. [DOI] [PubMed] [Google Scholar]
- van Houte J. Role of micro-organisms in caries etiology. J Dent Res. 1994;73:672–681. doi: 10.1177/00220345940730031301. [DOI] [PubMed] [Google Scholar]
- Vocadlo DJ, Davies GJ. Mechanistic insights into glycosidase chemistry. Curr Opin Chem Biol. 2008;12:539–555. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Yip VLY, Thompson J, Withers SG. Mechanism of GlvA from Bacillus subtilis : A detailed kinetic analysis of a 6-phospho-α-glucosidase from glycoside hydrolase family 4. Biochemistry. 2007;46:9840–9852. doi: 10.1021/bi700536p. [DOI] [PubMed] [Google Scholar]
- Ziesenitz SC, Siebert G, Imfeld T. Cariological assessment of leucrose [D-glucopyranosyl- α(1→5) -D-fructopyranose] as a sugar substitute. Caries Res. 1989;23:351–357. doi: 10.1159/000261206. [DOI] [PubMed] [Google Scholar]