Abstract
Lower vertebrates develop a unique set of primary sensory neurons located in the dorsal spinal cord. These cells, known as Rohon-Beard (RB) sensory neurons, innervate the skin and mediate the response to touch during larval stages. Here we report the expression and function of the transcription factor Xaml1/Runx1 during RB sensory neurons formation. In Xenopus embryos Runx1 is specifically expressed in RB progenitors at the end of gastrulation. Runx1 expression is positively regulated by Fgf and canonical Wnt signaling and negatively regulated by Notch signaling, the same set of factors that control the development of other neural plate border cell types, i.e. the neural crest and cranial placodes. Embryos lacking Runx1 function fail to differentiate RB sensory neurons and lose the mechanosensory response to touch. At early stages Runx1 knockdown results in a RB progenitor-specific loss of expression of Pak3, a p21-activated kinase that promotes cell cycle withdrawal, and of N-tub, a neuronal-specific tubulin. Interestingly, the pro-neural gene Ngnr1, an upstream regulator of Pak3 and N-tub, is either unaffected or expanded in these embryos, suggesting the existence of two distinct regulatory pathways controlling sensory neuron formation in Xenopus. Consistent with this possibility Ngnr1 is not sufficient to activate Runx1 expression in the ectoderm. We propose that Runx1 function is critically required for the generation of RB sensory neurons, an activity reminiscent of that of Runx1 in the development of the mammalian dorsal root ganglion nociceptive sensory neurons.
Keywords: Rohon-Beard, Sensory neurons, Runx1, Ngnr1, N-Tub, Pak3, Xenopus
Introduction
The ectoderm of the vertebrate embryos can be divided into three regions at the end of gastrulation: the neural plate, which is the precursor of the central nervous system, the non-neural ectoderm forming the epidermis, and the neural plate border (NPB) that arises at the boundary between the neural plate and the non-neural ectoderm. The NPB is the source of two important cell populations: the neural crest (NC) and the pre-placodal ectoderm (PE). The NC is located lateral to the neural plate but is excluded from its most anterior region. NC cells will migrate in the periphery and give rise to a broad array of derivatives including craniofacial structures, the pigment cell lineage and peripheral nervous system (LeDouarin et al., 2004). The PE is restricted to the anterior-most region of the neural plate and lateral to the NC. The PE will eventually segregate into individual cranial placodes to give rise to the sensory organs in the head (Schlosser, 2010; Park and Saint-Jeannet, 2010a).
In anamniotes such as the frog Xenopus laevis the NPB gives rise to two additional cell populations: the hatching gland (HG) cells and a group of primary neurons known as Rohon-Beard (RB) sensory neurons. The HG is located in the outer layer of the ectoderm of the anterior neural folds, medial to the prospective NC. The HG produces proteolytic enzymes, which digest the vitelline envelope and jelly coat to release the tadpole into the environment (Drysdale and Elinson, 1991). The RB sensory neurons arise from the posterior-most region of the NPB. At the end of neurulation, these neurons are located in the dorsal spinal cord and innervate the skin to mediate the escape response to touch at the larval stages (Roberts and Smyth, 1974). Later in development RB neurons will undergo apoptosis (Lamborghini, 1987) and their function will be assumed by the NC-derived dorsal root ganglia neurons (reviewed in Roberts, 2000). Genes typically expressed in RB sensory neuron progenitors are also detected in two additional primary neuron subpopulations confined to a more medial region of the neural plate, the primary interneurons and motoneurons. This is the case, for example, of the basic helix loop helix (b-HLH) gene neurogenin-related-1 (Ngnr1; Ma et al., 1996), and the neural-specific tubulin gene, N-Tubulin (N-Tub; Chitnis et al., 1995). The absence of molecular markers restricted to RB sensory neuron progenitors has made it difficult to analyze the more unique requirements of this population of primary neurons in terms of specification and differentiation.
The gene Runx1 encodes a runt domain transcription factor with a critical role in hematopoietic stem cell formation and definitive hematopoiesis in mammals (reviewed in Swiers et al., 2010). Runx1 is also expressed in a subpopulation of dorsal root ganglia (DRG) sensory neurons involved in pain transduction and regulates aspects of the differentiation of this group of nociceptive neurons (reviewed in Stifani and Ma, 2009). In Xenopus in addition to its expression and function in blood progenitors, Xaml1/Runx1 is expressed in RB neuron precursors at the end of gastrulation (Tracey et al., 1998), and therefore represents a unique tool to analyze the development of this population of primary sensory neurons. Moreover little is known of the role Xaml1/Runx1 in the formation of these mechanosensory neurons.
Here we describe the detailed expression pattern of Runx1 in RB progenitors as compared to other primary neuron-specific genes. We characterize the regulatory inputs controlling Runx1 expression at the NPB and analyze the consequences of Runx1 knockdown on the sensory function of Xenopus tadpoles. We also analyze the position of Runx1 in the regulatory cascade leading to RB sensory neurons specification. Our findings indicate that Runx1 function is critically required in RB progenitors to promote cell cycle exit and neuronal differentiation, and that Runx1 is acting in parallel with Ngnr1 to regulate sensory neuron formation.
Materials and Methods
Plasmid constructs
Vertebrate Runx1 genes are expressed from two alternative promoters, a distal (P1) and a proximal (P2), that encode isoforms with distinct amino-terminal sequences (Supplementary Fig S1), here referred to as Runx1 (accession # BC057739.1) and Xaml1 (accession # AF035446), respectively. Xenopus laevis Runx1 (pCMV-Sport6) was obtained from Open Biosystems. Xaml1 was amplified by PCR from stage 30 cDNA using primers based on the published sequence (Tracey et al., 1998), and subcloned into pGEM-T Easy (Promega). Both ORFs including 9bp (Runx1) and 14bp (Xaml1) upstream of the ATG were amplified by PCR and subcloned into pCS2+ expression plasmid digested with ClaI and XbaI. These two constructs were used to test the specificity of the translation blocking morpholino antisense oligonucleotides (Supplementary Fig S1). We generated a hormone-inducible version of Ngnr1 (Ma et al., 1996) by sub-cloning the coding region of Ngnr1 into pCS2+GR (Ngnr1-GR). All constructs were sequenced and the corresponding proteins monitored using an in vitro transcription/translation assay.
In vitro transcription/translation
The in vitro transcription/translation coupled rabbit reticulocyte lysate system (SP6-TNT, Promega) was performed following the manufacturer recommendations (Promega), in the presence of 35S-methionine. The reaction was resolved on a NuPAGE BIS-Tris gel (Invitrogen). The gel was dried using GelAir Drying System (Bio-Rad) and the product of the TNT reaction was detected by exposure onto a BioMax film (Kodak).
Morpholino antisense oligonucleotides
β-Catenin (βcatMO; 25 ng; Heasman et al., 2000), Wnt8 (Wnt8MO; 30 ng; Park and Saint-Jeannet, 2008), Fgf8a (Fgf8aMO; 50 ng; Fletcher et al., 2006), Pax3 (Pax3MO; 50 ng; Monsoro-Burq et al., 2005; Hong and Saint-Jeannet, 2007), Zic1 (Zic1MO; 45 ng; Sato et al., 2005; Hong and Saint-Jeannet, 2007) and control (CoMO; 60 ng) morpholino antisense oligonucleotides were purchased from Gene-Tools LLC (Philomath, OR). To interfere with Runx1 function we used two translation blocking and a splice blocking morpholinos. Runx1MO (CACTATGTGAGGCCATTGCGTTTCC) and Aml1MO (GGGATACGCATCACAACAAGCCTGG) specifically block translation of Runx1 (P1 promoter) and Xaml1 (P2 promoter) mRNA, respectively. The specificity of Runx1MO and Aml1MO was tested in an in vitro transcription/translation coupled rabbit reticulocyte lysate assay (Supplementary Fig S1). Based on Xenopus tropicalis genome information (Ensembl Gene ID: ENSXETG00000014140), several intronic regions within Runx1 were selected as candidate sites for a splice-inhibitory morpholino. Primers flanking these introns in the Xenopus laevis Xaml1/Runx1 mRNA sequence were used to amplify Xenopus laevis genomic DNA fragments, which were cloned into pGEM-T (Promega) and sequenced. We designed a splice-inhibitory morpholino (Runx1SMO: AAACAGAGCCAGGGTCTTACCTTGA) targeting the Exon1-Intron1 junction (Supplementary Fig S1).
Embryo injections and in situ hybridization
Embryos were staged according to Nieuwkoop and Faber (1967). Fgf8a (2 pg; Christen and Slack, 1997), Notch-ICD (0.5 ng; Chitnis and Kintner, 1996) and Ngnr1- GR (0.5 ng; Perron et al., 1999) mRNAs were synthesized in vitro using the Message Machine kit (Ambion, Austin, TX). Synthetic mRNA and morpholino antisense oligonucleotides were injected in the animal pole of 2-cell stage embryos. All embryos were co-injected with the lineage tracer β-gal mRNA (β-gal; 0.5 ng) to identify the injected side. Ngnr1-GR (0.5 ng) injected embryos were treated with 10 µM dexamethasone (Sigma) in NAM 0.1X at stage 10.5 or stage 12.5, as described (Gammill and Sive, 1997). Untreated sibling embryos were used as a control (not shown). For in situ hybridization embryos were fixed in MEMFA and were successively processed for Red-Gal (Research Organics) staining to detect β-gal activity, and in situ hybridization. Antisense DIG-labeled probes (Genius kit, Roche) were synthesized using template cDNA encoding Xaml1/Runx1 (Tracey et al., 1998), Xhe (Katagiri et al., 1997), N-Tub (Chitnis et al., 1995), Pak3 (Souopgui et al., 2002), Ngnr1 (Ma et al., 1996), Islet1 (Brade et al., 2007), Krox20 (Bradley et al., 1993), Snail2 (Mayor et al., 1995), XK81 (Jonas et al., 1989), Kv1.1 (Burger and Ribera, 1996) and Ccndx (Chen et al., 2007). Whole-mount in situ hybridization was performed as previously described (Harland, 1991). For in situ hybridization on sections, embryos were fixed in MEMFA for 1 hour, embedded in Paraplast+, 12 µm sections cut on a rotary microtome and hybridized with the appropriate probes as described (Henry et al. 1996). Sections were then briefly counter stained with Eosin.
Proliferation assay
For phosphohistone H3 detection (Saka and Smith, 2001), Sox9MO-injected albinos embryos were fixed in MEMFA. Embryos were incubated successively in α-phosphohistone H3 antibody (Upstate Biotechnology; 1 µg/ml) and anti-rabbit IgG conjugated to alkaline phosphatase (Jackson ImmunoResearch; 1:1000). Alkaline phosphatase activity was revealed using NBT/BCIP (Roche). Fluorescein lysine dextran (FLDX; MW 10,000, Molecular Probes) was used as a lineage tracer to identify the injected side.
Touch response assay
During embryogenesis Xenopus embryos develop a dual touch sensory system largely mediated by RB sensory neurons (Roberts and Smyth, 1974). The touch response assay and quantification was performed as previously described (Fein et al., 2008). Briefly, the dorsal trunk of stage 32 embryos was gently touched or stroked with a metal probe. After a period of approximately 3 seconds the probe was reapplied for a total of 10 trials. Responses were scored as follows: 0, no response; 0.5, non-swimming response (restricted trunk bend); 1.0, normal swimming response. The scores of each of the 10 trials were summed to yield a final touch response score between 0 and 10.
Results
Xaml1/Runx1 is expressed in Rohon-Beard sensory neurons
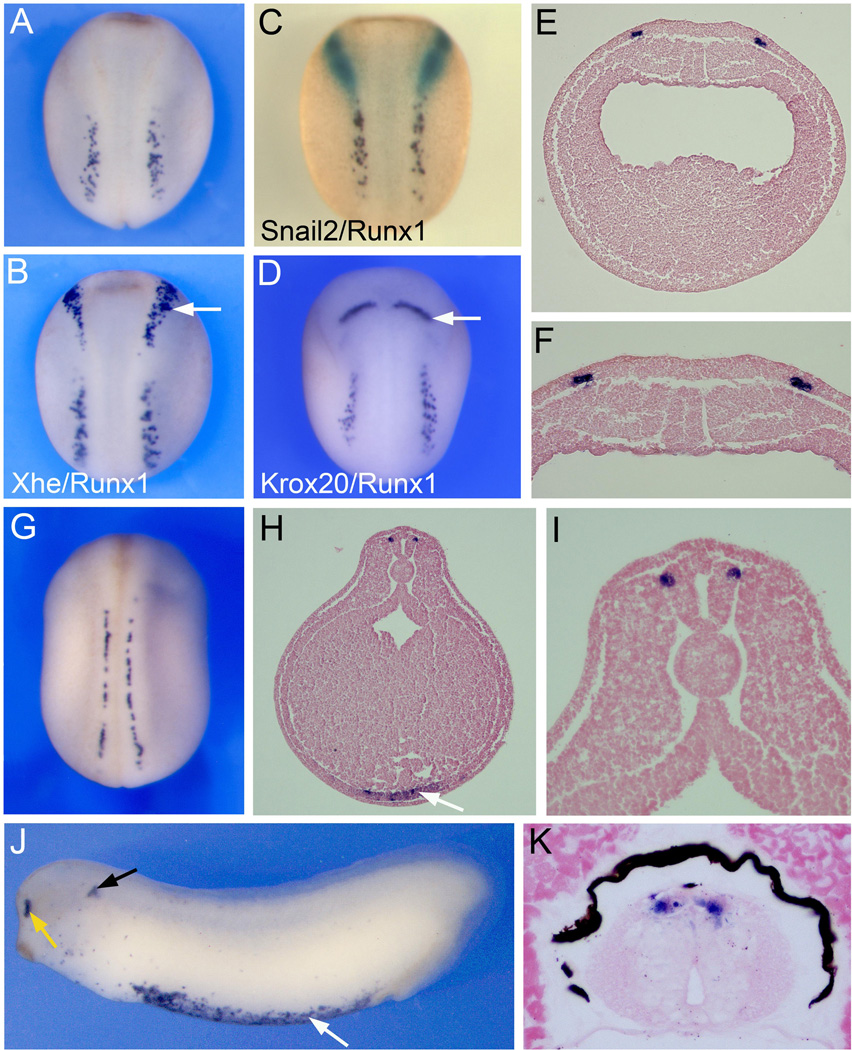
The cloning and expression of Xaml1/Runx1 has been previously reported (Tracey et al., 1998), however this initial study was primarily focused on Runx1 expression in the hematopoietic lineage. To further evaluate the developmental expression of Runx1 in the ectoderm, we performed whole-mount in situ hybridization on embryos at various stages. At the neurula stage (stage 15) the Runx1 expression domain in the ectoderm is located posterior to the prospective NC and HG, as seen by Snail2 and Xhe expression, respectively (Fig 1A–C). Runx1 expression is restricted to a region of the NPB that is also posterior to the hindbrain marker Krox20 (Fig 1D). Transverse sections indicate that Runx1 is expressed in the deep layer of the ectoderm, lateral to the neural plate (Fig 1E–F), which anatomically corresponds to the position of the prospective RB sensory neurons (Roberts, 2000). By stage 23, the neural plate has folded into a tube, resulting in the repositioning of Runx1-expressing cells to the dorsolateral region of the spinal cord (Fig 1G–I). At this stage Runx1 is also detected in blood progenitors in the ventral mesoderm (Fig 1H), as previously reported (Tracey et al., 1998). Around stage 28, additional domains of expression of Runx1 include the olfactory epithelium and the developing statoaccoustic ganglia associated with the otic vesicle (Fig 1J; Park and Saint-Jeannet, 2010b). Runx1 expression persists in RB sensory neurons at least up to stage 45 (Fig 1K).
Figure 1. Expression of Runx1/Xaml1 in Rohon-Beard sensory neurons by whole-mount in situ hybridization.
(A–F) Runx1 expression in stage 15 embryos. (A) Runx1 is detected at the posterior portion of the NPB. (B) Double in situ hybridization for Runx1 and the HG marker Xhe. Runx1 positive cells are located posterior to HG cells (arrow). (C) Double in situ hybridization for Runx1 and the neural crest marker Snail2. Runx1 expression (purple) is distinct from and posterior to the neural crest forming region (Snail2, green staining). (D) Double in situ hybridization for Runx1 and the hindbrain marker Krox20 (arrow). Panels (A–D), dorsal view, anterior to top. (E) Transverse section, dorsal to top, showing that Runx1 is restricted to two discrete domains at the neural plate border. (F) Higher magnification of the neural plate region of the embryo shown in (E). (G) At stage 22 as the neural tube closes, Runx1 expression is restricted to the dorsal aspect of the spinal cord. Dorsal view, anterior to top. (H–I) Transverse section through a stage 23 embryo, dorsal to top. Runx1 is confined to the dorso-lateral region of the spinal cord. Runx1 is also detected ventrally in the lateral plate mesoderm, precursor of the hematopoietic lineage (arrow). (I) Higher magnification of the neural tube region of the embryo shown in (H). (J) Runx1 expression in a stage 28 embryo. Runx1 is detected in the olfactory epithelium (yellow arrow), periotic mesenchyme (black arrow), and blood precursors (white arrow). Lateral view, anterior to left. (K) Transverse section through the spinal cord of a stage 45 embryo shows Runx1 expression in RB sensory neurons in the dorsal spinal cord. Dorsal to top.
Runx1 is co-expressed with other primary neuron-specific genes
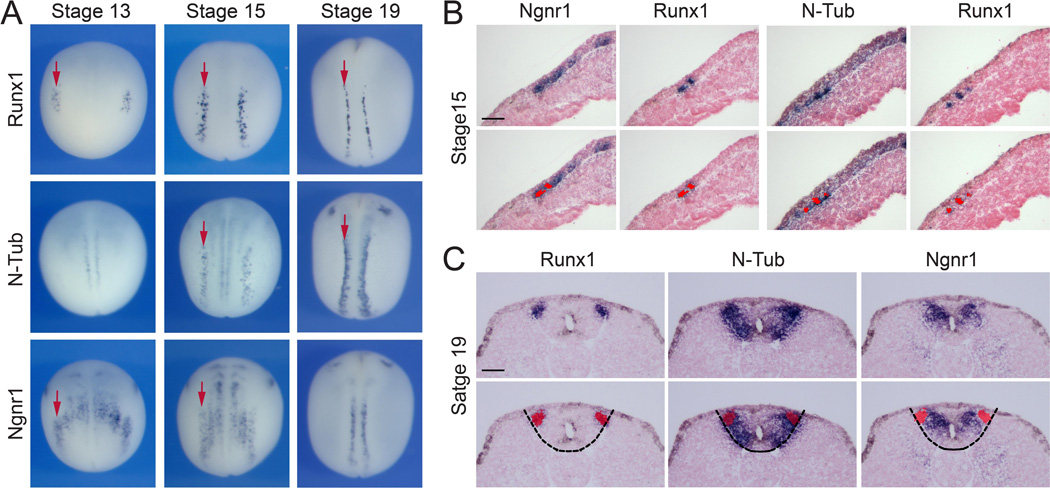
We next performed a comparative analysis of Runx1 expression to that of classic markers for primary neurons such as N-Tub and Ngnr1. Runx1 is first detected at stage 13 in RB progenitors, where it colocalizes with Ngnr1, which is also expressed in progenitors of primary interneurons and motoneurons within the neural plate (Chitnis et al., 1995; Ma et al., 1996; Fig 2A). The onset of Runx1 expression is slightly later than the stage at which RB progenitors have been birthdated in Xenopus, around stage 11.5 to 12 (Lamborghini, 1980; Jacobson and Moody, 1984). N-Tub expression in RB progenitors is initiated around stage 15 as well as in the other two primary neuron subpopulations (Fig 2A). At this stage, N-Tub, Ngnr1, and Runx1 are all co-expressed in RB progenitors. This co-localization was confirmed by in situ hybridization on adjacent sections of the same embryo (Fig 2B). By stage 19, as the neural plate folds to form a tube, Runx1 and N-Tub are still co-expressed in RB progenitors, however Runx1 expression is more sparse than that of N-Tub suggesting that Runx1 is only expressed in a subset of RB cells (Fig 2A). At this stage Ngnr1 expression appears to be more medial than that of N-Tub and Runx1 as Ngnr1 is now downregulated in RB progenitors (Fig 2A). In situ hybridization on adjacent sections of stage 19 embryos using all three probes confirmed the loss of Ngnr1 expression in RB progenitors (Fig 2C), while the Runx1 expression domain largely overlaps with that of N-Tub in the dorsolateral region of the spinal cord containing RB progenitors (Fig 2C). In other regions of the spinal cord Ngnr1 overlaps with N-Tub in the prospective primary interneurons and motoneurons. These results show that Runx1, Ngnr1 and N-Tub are initially co-expressed in RB progenitors, however by neural tube closure Ngnr1 expression is lost in this population of primary neurons.
Figure 2. Comparative expression of Runx1, N-Tub and Ngnr1 in primary neurons.
(A) Developmental expression of Runx1, N-Tub, and Ngnr1 at the neural plate border in stage-matched embryos. The arrows indicate the position of the row of RB sensory neurons. Dorsal views, anterior to top. (B–C) In situ hybridization on adjacent transverse sections of stage 15 and stage 19 embryos, dorsal to top. (B) At stage 15 Runx1 is co-expressed with N-Tub and Ngnr1, as indicated by the red overlay in the lower panels. (C) At stage 19 while Runx1 and N-Tub are still co-expressed, Ngnr1 expression is lost in the RB neurons population, as indicated by the red overlay in the lower panels. The dotted lines demarcate the position of the spinal cord.
Fgf, Wnt and Notch signaling regulate Runx1 expression at the NPB
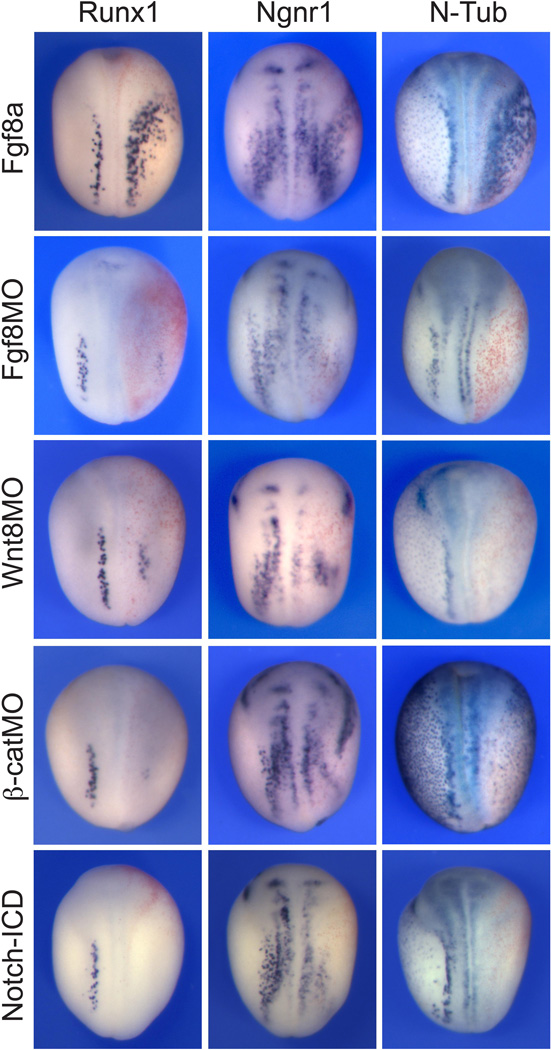
The formation of NPB cells requires attenuation of Bmp signaling in the ectoderm through the activity of Bmp antagonists produced by the axial mesoderm. However, changes in Bmp signaling in the ectoderm are not sufficient to specify the NPB and other signaling pathways have been implicated in this process including Fgf, canonical Wnt and Notch signaling (reviewed in Knecht and Bronner-Fraser, 2002; Huang and Saint-Jeannet, 2004). A recent study indicates that like other NPB cell types RB sensory neuron formation requires Bmp activity (Rossi et al., 2008). We decided to determine whether other signaling pathways were also implicated in the generation of RB neurons. We specifically analyzed the role of Fgf8a and Wnt8, two ligands implicated in NPB induction in Xenopus (Hong and Saint-Jeannet, 2007; Hong et al., 2008). Overexpression of Fgf8a by injection of Fgf8a mRNA resulted in a dramatic ventrolateral expansion of the Runx1 expression domain in all embryos examined (Table 1; Fig 3). N-Tub and Ngnr1 expression domains were also expanded in a majority of embryos. The expansion of these genes was associated with a loss of epidermal keratin (Supplementary Fig S2). Conversely, knockdown of Fgf8a using a splice-blocking morpholino antisense oligonucleotide (Fgf8aMO) caused a severe reduction of all three genes in most embryos examined (Table 1; Fig 3). Interference with canonical Wnt signaling by injection of Wnt8 or β-catenin morpholino antisense oligonucleotides (Wnt8MO and β-catMO) had a similar outcome: a reduction of Runx1 expression domain at a high frequency, as well as a reduction of N-Tub and Ngnr1 in all three populations of primary neurons (Table 1; Fig 3). Because the formation of primary neurons is negatively regulated by Notch signaling (Chitnis et al., 1995), we next asked whether Notch signaling had an inhibitory effect on Runx1 expression. We found that expression of mRNAs encoding an activated form of Notch (Notch-ICD; Chitnis and Kintner, 1996) also represses Runx1 expression (Table 1; Fig 3), consistent with a previous study (Perron et al., 1999). Taken together, these results indicate that Runx1 expression in RB progenitors is negatively regulated by Notch signaling, and under the positive influence of Fgf and canonical Wnt signaling.
Table 1.
Regulation of Runx1 expression by Fgf, Wnt and Notch signaling
| Injection | Probe | N | Normal | Reduced/Lost | Expanded/Ectopic |
|---|---|---|---|---|---|
| Fgf8a | Runx1 Ngnr1 N-Tub |
52 57 45 |
0% 2% 9% |
0% 2% 7% |
100% 96% 84% |
| Fgf8MO | Runx1 Ngnr1 N-Tub |
76 47 43 |
15% 34% 10% |
84% 66% 88% |
1% 0% 2% |
| Wnt8MO | Runx1 Ngnr1 N-Tub |
51 53 52 |
4% 11% 2% |
96% 89% 98% |
0% 0% 0% |
| β-CatMO | Runx1 Ngnr1 N-Tub |
32 26 29 |
0% 0% 0% |
100% 100% 100% |
0% 0% 0% |
| Notch ICD | Runx1 Ngnr1 N-Tub |
37 33 42 |
0% 6% 0% |
100% 73% 100% |
0% 21% 0% |
Figure 3. Wnt, Fgf and Notch signaling pathways regulate Runx1.
Overexpression of Fgf8a by injection of Fgf8a mRNA in one blastomere at the 2-cell stage results in an expansion of the Runx1 expression domain. The expression domain of N-Tub and Ngnr1 is also expanded in these embryos. Knockdown of Fgf8a (Fgf8aMO) causes a severe reduction of all three genes. Similarly, interference with canonical Wnt signaling, by injection of Wnt8MO or β-catMO, reduces Runx1, N-Tub and Ngnr1 expression in all three populations of primary neurons. Expression of an activated form of Notch (Notch-ICD) also represses Runx1 expression. In all panels embryos are viewed from the dorsal side, anterior to top. The injected side is on the right.
The NPB specifiers Pax3 and Zic1 regulate Runx1 expression
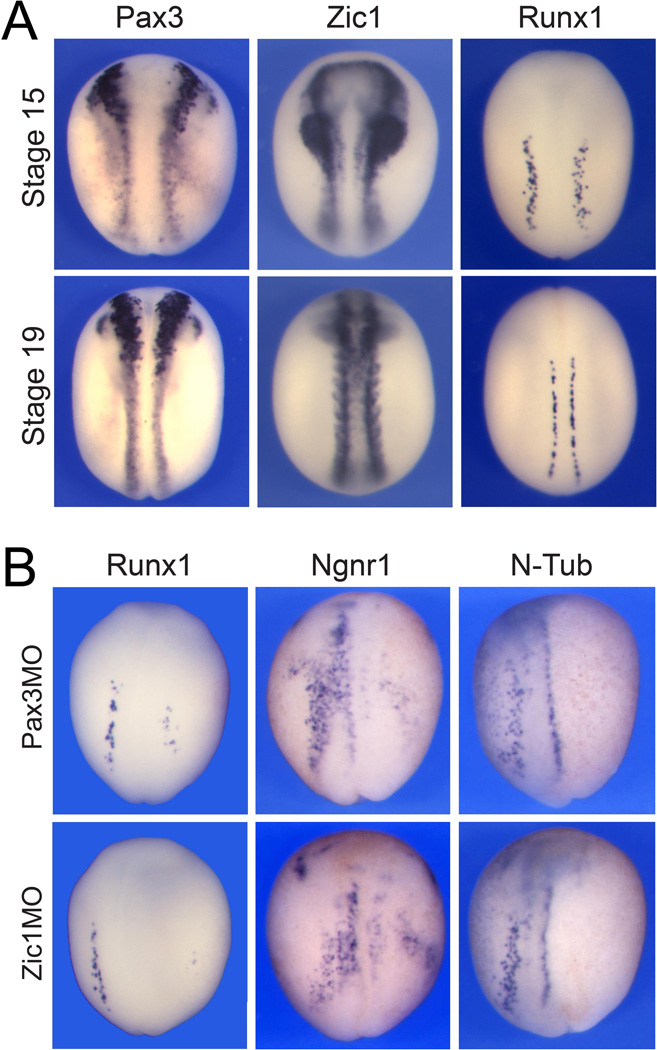
Downstream of these signaling events Pax3 and Zic1 are two genes activated early at the NPB and required for the development of three distinct lineages: the NC, PE and HG (Monsoro-Burq et al., 2005; Sato et al., 2005; Hong and Saint-Jeannet, 2007). At early neurula stages Pax3 and Zic1 expression overlaps with the Runx1 expression domain in RB progenitors (Fig 4A) suggesting that these factors may also regulate Runx1 expression and RB sensory neuron formation. To test this possibility, we used Pax3- and Zic1-specific morpholino antisense oligonucleotides (Pax3MO and Zic1MO), which block Pax3 and Zic1 translation, respectively. Unilateral injection of either Pax3MO or Zic1MO at the 2-cell stage inhibited Runx1 expression in a vast majority of the embryos (Table 2; Fig 4B). Pax3 or Zic1 knockdown also prevented Ngnr1 and N-Tub expression in RB progenitors as well as in the other two primary neuron populations (Table 2; Fig 4B), consistent with the expression of Pax3 and Zic1 in the lateral neural plate (Hong and Saint-Jeannet, 2007). These results demonstrate that the NPB specifiers Pax3 and Zic1 are functioning upstream of Runx1, Ngnr1 and N-Tub, and suggest that Pax3 and Zic1 are required for the formation of all four NPB cell types: the NC, PE, HG and RB progenitors.
Figure 4. Runx1 expression in RB sensory neurons depends on Pax3 and Zic1.
(A) Comparative expression of Pax3, Zic1 and Runx1 in stage-matched embryos indicate that the Runx1 expression domain overlaps with that of these two NPB specifiers. (B) Embryos injected with Pax3MO (50 ng) or Zic1MO (45 ng) in one blastomere at the 2-cell stage exhibit a strong reduction of Runx1 as well as N-Tub and Ngnr1 expression in RB progenitors. The injected side is on the right. In all panels embryos are viewed from the dorsal side, anterior to top.
Table 2.
Pax3 and Zic1 regulates Runx1 expression at the NPB
| Injection | Probe | N | Normal | Reduced/Lost | Expanded/Ectopic |
|---|---|---|---|---|---|
| Pax3MO | Runx1 Ngnr1 N-Tub |
48 47 44 |
6% 21% 7% |
94% 79% 93% |
0% 0% 0% |
| Zic1MO | Runx1 Ngnr1 N-Tub |
56 52 39 |
4% 16% 5% |
96% 67% 95% |
0% 17% 0% |
Embryos lacking Runx1 function fail to differentiate RB sensory neurons and lose response to touch
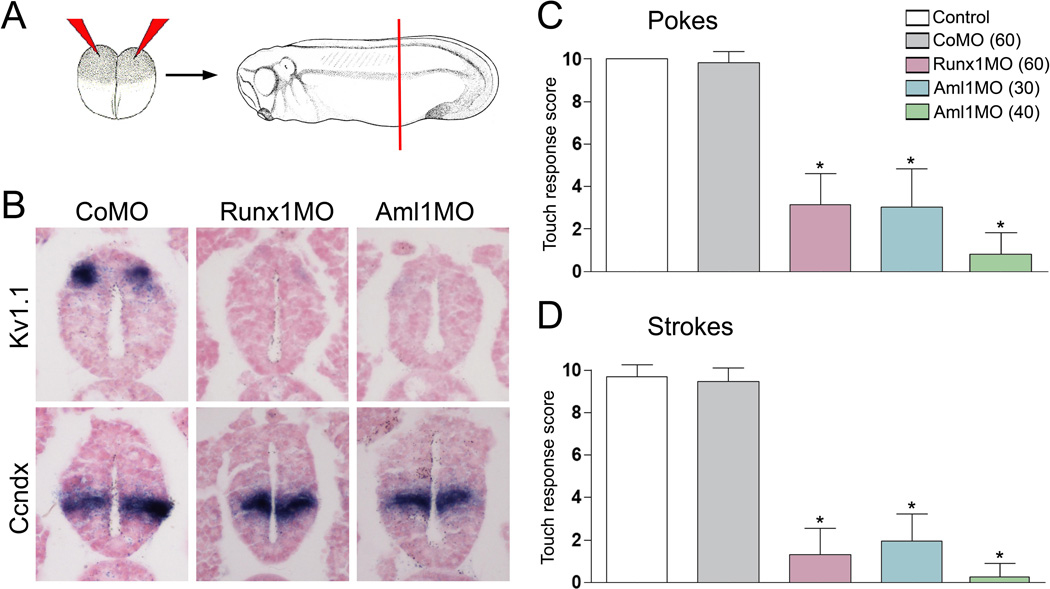
To determine whether Runx1 function is required for RB neuron development we performed Runx1 knockdown in developing embryos using morpholino antisense oligonucleotides. A conserved feature of the vertebrate Runx genes is their expression from two adjacent promoters (distal, P1 and proximal, P2) encoding two isoforms with distinct amino-terminal sequences (reviewed in Blyth et al., 2005). We designed two morpholinos to specifically interfere with the translation of each isoform, Runx1MO (P1 promoter) and Aml1MO (P2 promoter) (Supplementary Fig S1). We also designed a third morpholino (Runx1SMO) that blocks Runx1 splicing at the Exon1-Intron-1 junction (Supplementary Fig S1). To evaluate the formation of RB sensory neurons in morphant embryos we analyzed the expression of the potassium-gated channel Kv1.1, which is normally expressed in differentiated RB neurons (Burger and Ribera, 1996). Bilateral injection of either morpholino in the animal region of 2-cell stage embryos resulted in a severe reduction or loss of Kv1.1 expression in the dorsal spinal cord at stage 32 (Fig. 5A–B). In these embryos the development of other neuronal subpopulations in the spinal cord was largely unaffected (Fig. 5B), as exemplified by unperturbed expression of Ccndx, a gene expressed in motoneurons (Chen et al., 2007).
Figure 5. Runx1-deficient tadpoles lack Rohon-Beard sensory neurons and lose response to touch.
(A) Two-cell stage embryos received a bilateral injection of control (CoMO), Runx1 (Runx1MO) or Aml1 (Aml1MO) morpholino antisense oligonucleotides. At stage 28 the corresponding embryos were sectioned in the trunk region (red line) and analyzed by in situ hybridization. (B) Expression of Rohon-Beard (Kv1.1) and motor neuron (Ccndx) marker genes in the spinal cord of morphant embryos. Runx1MO and Aml1MO show a loss of Kv1.1 expression, while the ventral motor neurons are largely unaffected. (C–D) At stage 32 Runx1MO and Aml1MO injected embryos have a severely reduced response to touch (pokes and strokes) as compared to control uninjected or control morpholino (CoMO) injected embryos. (C) Pokes: Control (uninjected), 10.00±0.00 (n=30); CoMO (60 ng), 9.83±0.53 (n=30); Runx1MO (60 ng), 3.14±1.45 (n=28); Aml1MO (30 ng), 3.03±1.82 (n=20); Aml1MO (40 ng), 1.00±1.19 (n=15). (D) Strokes: Control (uninjected), 9.68±0.56 (n=30); CoMO (60 ng), 9.47±0.63 (n=30); Runx1 MO (60 ng), 1.30±1.26 (n=28); Aml1MO (30 ng), 1.95±1.28 (n=20); Aml1MO (40 ng), 0.33±0.75 (n=15). Statistical significance was determined using one-way ANOVA. The values are presented as mean SEM; * = P<0.0001, versus Control and CoMO).
Because RB neurons serve as primary mechanosensory neurons and are involved in the response to touch in the developing embryo, we next analyzed the behavior of morphant embryos to determine the functional consequences of the loss of Runx1. Bilateral Injection of any one of the three morpholinos led to a significant reduction in touch sensitivity in a dose dependent manner (Fig 5C–D; Supplemental movies). The fact that two translation blocking morpholinos and a splice-inhibitory morpholino give identical results establishes the strong specificity of the phenotype. These results indicate that Runx1 function is critically required for the formation of RB sensory neurons and is essential for the development of a functional mechanosensory system at larval stages.
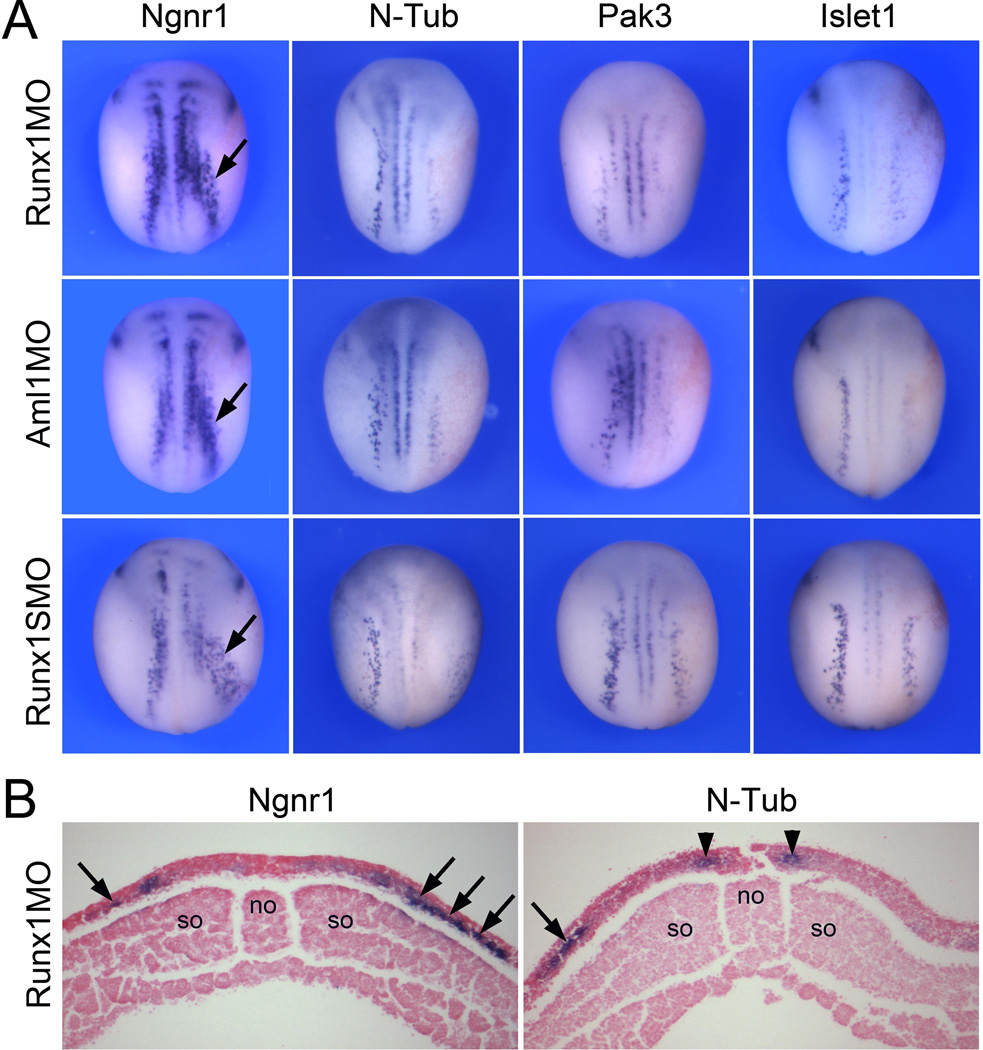
Runx1 regulates Pak3, Islet1 and N-Tub expression in RB progenitors
In order to gain insights into the mechanisms by which Runx1 regulates RB sensory neuron formation, we analyzed the influence of Runx1 function on the expression of several components of the proneural gene network regulating the emergence of primary neurons at the neurula stage. These genes include: Ngnr1, a b-HLH proneural factor, and an upstream regulator of the pathway (Ma et al., 1996); Islet 1, a LIM homeodomain transcription factor involved in neuronal specification (Brade et al., 2007); Pak3, a p21-activated kinase 3 essential for cell cycle withdraw (Souopgui et al., 2002); and N-Tub, a marker of neuronal differentiation (Chitnis et al., 1995). Runx1 knockdown by injection of Runx1MO, Aml1MO or Runx1SMO resulted in a similar phenotype characterized by a reduction of Islet1, Pak3 and N-Tub expression in RB progenitors at the neurula stage (Table 3; Fig 6A–B). In contrast Ngnr1 was either unaffected or expanded in morphant embryos (Table 3; Fig 6A–B). Phosphohistone H3 staining did not show any significant change in the rate of cell proliferation at the neural plate border of morphant embryos (Supplementary Fig S2). Altogether these experiments indicate that Runx1 is functioning upstream of Pak3, Islet1 and N-Tub in the regulatory cascade leading to RB progenitors formation, and suggest that Runx1 may regulate the differentiation of these cells by promoting cell cycle withdrawal.
Table 3.
Summary of Runx1 knockdown phenotype at the neurula stage
| Injection | Probe | N | Normal | Reduced/Lost | Expanded/Ectopic |
|---|---|---|---|---|---|
| Aml1MO | N-Tub Pak3 Islet1 Ngnr1 |
78 80 37 81 |
13% 29% 22% 42% |
87% 70% 78% 14% |
0% 1% 0% 44% |
| Runx1MO | N-Tub Pak3 Islet1 Ngnr1 |
70 40 35 73 |
20% 33% 31% 42% |
79% 65% 69% 0% |
1% 2% 0% 57% |
| Runx1SMO | N-Tub Pak3 Islet1 Ngnr1 |
41 46 45 45 |
17% 24% 20% 58% |
83% 76% 80% 9% |
0% 0% 0% 33% |
Figure 6. Runx1-deficient embryos downregulate N-Tub, Pak3 and Islet1.
(A) Embryos were injected in one blastomere at the 2-cell stage with Runx1MO, Aml1MO or Runx1SMO and analyzed at stage 15 for the expression of Ngnr1, N-Tub, Pax3 and Islet1. The RB expression domain of N-Tub, Pak3 and Islet1 is reduced while Ngnr1 expression is expanded (arrows). In all panels the injected side is on the right. Dorsal view anterior to top. (B) Transverse sections of representative Runx1MO-injected embryos. The expression of N-Tub is lost in RB progenitors while N-Tub expression in primary motor neurons precursors is unaffected (arrow heads). Ngnr1 expression is expanded (arrows). The injected side is on the right. Dorsal to top. no, notochord; so, somites.
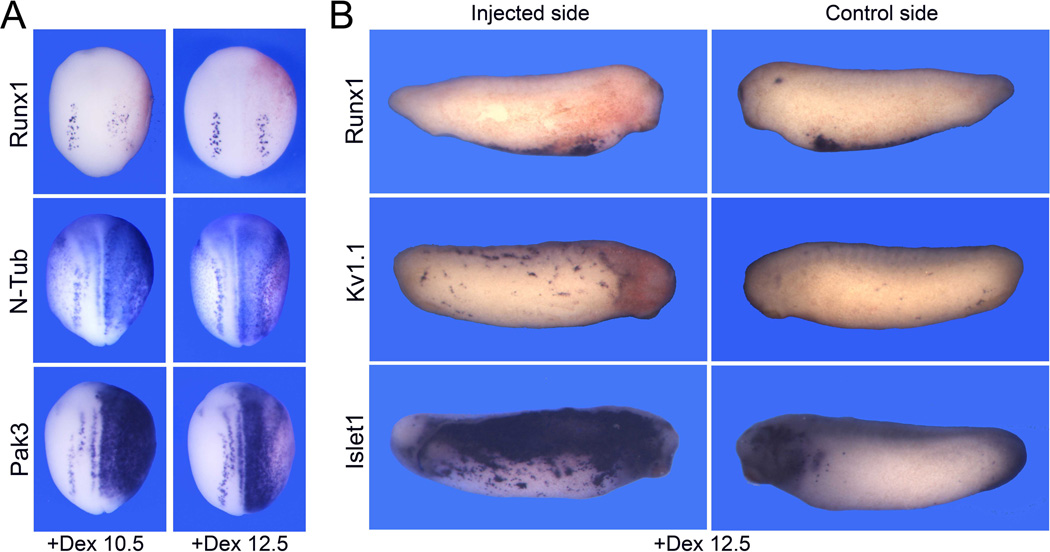
Ngnr1 is not sufficient to activate Runx1 expression
To further evaluate the role of Ngnr1 in the regulation of Runx1 expression in RB sensory neurons we expressed an hormone inducible version of Ngnr1 in which Ngnr1 was fused to the hormone-binding domain of human glucocorticoid receptor (Ngnr1-GR). The activity of this fusion protein can be regulated by addition of dexamethasone to the embryo culture medium (Kolm and Sive, 1995). Embryos injected with 0.5 ng of Ngnr1-GR mRNA were treated with dexamethasone at the early (stage 10.5) or late (stage 12.5) gastrula stages and analyzed for gene expression at the neurula (stage 15) or tailbud (stage 27) stages. Activation of Ngnr1-GR at the gastrula stages resulted in a reduction of the NC-specific gene Snail2, and a dramatic expansion of N-Tub and Pak3 expression domain at the neurula stage, converting the entire ectoderm into primary neuron progenitors (Table 4; Fig 7A), as previously reported (Ma et al., 1996; Olson et al., 1998; Perron et al., 1999). For both genes the lateral expansion was more pronounced for a treatment with dexamethasone at stage 10.5 (Fig 7A). In contrast, Runx1 expression appeared more diffuse or partially inhibited after dexamethasome treatment at stage 10.5, while for a later treatment (stage 12.5) a majority of the embryos were unaffected (Table 4; Fig 7A), suggesting that Ngnr1 expression is not sufficient to induce Runx1, consistent with a previous study (Perron et al., 1999). Interestingly, at later stage these embryos showed massive ectopic Islet1 and Kv1.1 expression in the ectoderm without any upregulation of Runx1 expression (Table 4; Fig 7B). These results indicate that Ngnr1 has the ability to promote the induction of neuronal sensory characteristics in the ectoderm, and this is occurring in a Runx1 independent manner, suggesting the existence of two distinct and parallel regulatory pathways controlling sensory neurons formation in Xenopus.
Table 4.
Summary of the phenotype of Ngnr1 misexpression
| Injection | Probe | N | Normal | Reduced/Lost | Expanded/Ectopic |
|---|---|---|---|---|---|
|
Ngnr1-GR +Dex 10.5 (Stage 15) |
Snail2 Runx1 N-Tub Pak3 |
32 69 88 65 |
6% 7% 0% 0% |
94% 86% 0% 0% |
0% 7% 100% 100% |
|
Ngnr1-GR +Dex 12.5 (Stage 15) |
Snail2 Runx1 N-Tub Pak3 |
27 55 62 62 |
0% 66% 0% 0% |
100% 7% 3% 0% |
0% 27% 97% 100% |
|
Ngnr1-GR +Dex 12.5 (Stage 27) |
Runx1 Kv1.1 Islet1 |
38 44 36 |
100% 9% 0% |
0% 0% 0% |
0% 91% 100% |
Figure 7. Ngnr1 expression is not sufficient to activate Runx1 expression.
(A) Embryos at the 2-cell stage were injected in one blastomere with 0.5 ng of Ngnr1-GR mRNA. Embryos were subsequently incubated with dexamethasone at early (+Dex 10.5) or late (+Dex 12.5) gastrula stages, and fixed at stage 15 for detection of Runx1, N-Tub or Pak3 by whole mount in situ hybridization. N-Tub and Pak3 are dramatically upregulated while the Runx1 expression domain is only marginally affected. The injected side is on the right. Dorsal view anterior to top. (B) At the tailbud stage (stage 27) these embryos show ectopic Kv1.1 and Islet1 expression in the ectoderm, independently of any upregulation of Runx1. Lateral views dorsal to top. Control and injected sides of the same embryo are shown for comparison.
Discussion
In this study we provide novel information on the regulatory mechanisms controlling the specification and differentiation of RB sensory neurons in Xenopus, focusing on the activity of the transcription factor Xaml1/Runx1. Runx1 is detected in RB progenitors at the end of gastrulation (stage 13) and its expression is regulated by the same factors that control the development of other NPB cell types. Runx1 is specifically required for the expression of the p21-activated serine/threonine kinase 3 (Pak3), allowing RB progenitors to exit the cell cycle and initiate differentiation. In the absence of Runx1, RB sensory neurons failed to form resulting in embryos with impaired mechanosensory function. These results indicate that Runx1 is critically required for RB sensory neurons formation in Xenopus, reminiscent of Runx1 function in the development of the mammalian DRG nociceptive sensory neurons (Chen et al., 2006; Marmigere et al., 2006; Kramer et al., 2006).
Recent transplantation experiments have shown that RB neurons form as the result of an inductive interaction between the neural and non-neural ectoderm in Xenopus (Rossi et al., 2008). In these experiments Bmp4 protein was reported to induce RB neurons ectopically, as assayed by XHox11L2 expression,and was shown to be required for RB induction at sites of newly formed neural plate-epidermal boundaries (Rossi et al., 2008). Other studies using N-Tub as a marker have proposed a role for Fgf, Wnt and Notch signaling in RB neuron induction (Bang et al., 1999; Pera et al., 2003; Fletcher et al., 2006; Garcia-Morales et al, 2009). However, because N-Tub is expressed in all three subpopulations of primary neurons, it is difficult in some of these studies to determine whether the effects observed reflect changes in RB progenitor fate or a broader effect on the development of other populations of primary neurons. Our work using Runx1 as an early marker for RB progenitors demonstrates unambiguously that in addition to Bmp, canonical Wnt, Fgf and Notch signaling are all implicated at some level in the induction of RB progenitors. It is interesting that the same set of signals regulating the formation of the NC, PE and HG at the NPB are also involved in the induction of RB progenitors (McGrew et al., 1999; Brugmann et al., 2004; Glavic et al., 2004a; Glavic et al., 2004b; Ahrens and Schlosser, 2005; Hong and Saint-Jeannet, 2007; Hong et al., 2008). How these signals are integrated at the NPB to generate distinct fates is an important and still unresolved question.
Downstream of these signaling molecules a number of genes are broadly activated at the NPB. These genes are referred as NPB specifiers and include members of the Zic, Pax, Dlx and Msx families of transcriptional regulators. In turn these genes are responsible for the activation of a subset of genes with more restricted expression domains, known as NC specifiers or PE specifiers (Meulemans and Bronner-Fraser, 2004; Litsiou et al., 2005). Pax3 and Zic1 are two NPB specifiers that have the ability, alone or in combination, to regulate the formation of three distinct fates at the NPB. Pax3 and Zic1 are critical for HG and PE fate, respectively, while in combination they synergize to specify the NC (Monsoro-Burq et al., 2005; Sato et al., 2005; Hong and Saint-Jeannet, 2007). Our observations indicate that Runx1 is also under the control of Pax3 and Zic1, suggesting that the regulatory network underlying the emergence of the NC and PE (Meulemans and Bronner-Fraser, 2004; Litsiou et al., 2005) can be extended to RB sensory neurons, and we propose that Runx1 represents a bona fide RB specifier downstream of the NPB specifiers Pax3 and Zic1 (Fig 8). Recent work in zebrafish indicates that the transcription factor prdm1a is an important upstream regulator of NPB cell fates, through the selective activation of two target genes, sox10 in the NC, and islet1 in RB neurons (Olesnicki et al., 2010), and the repression of the basic helix-loop-helix gene olig4 (Hernandez-Lagunas et al., 2011). In light of these results it would be of particular importance to also evaluate the role of Prdm1 in the regulation of cell fate at the NPB in Xenopus.
Figure 8. Model of the gene regulatory network regulating cell fate at the neural plate border.
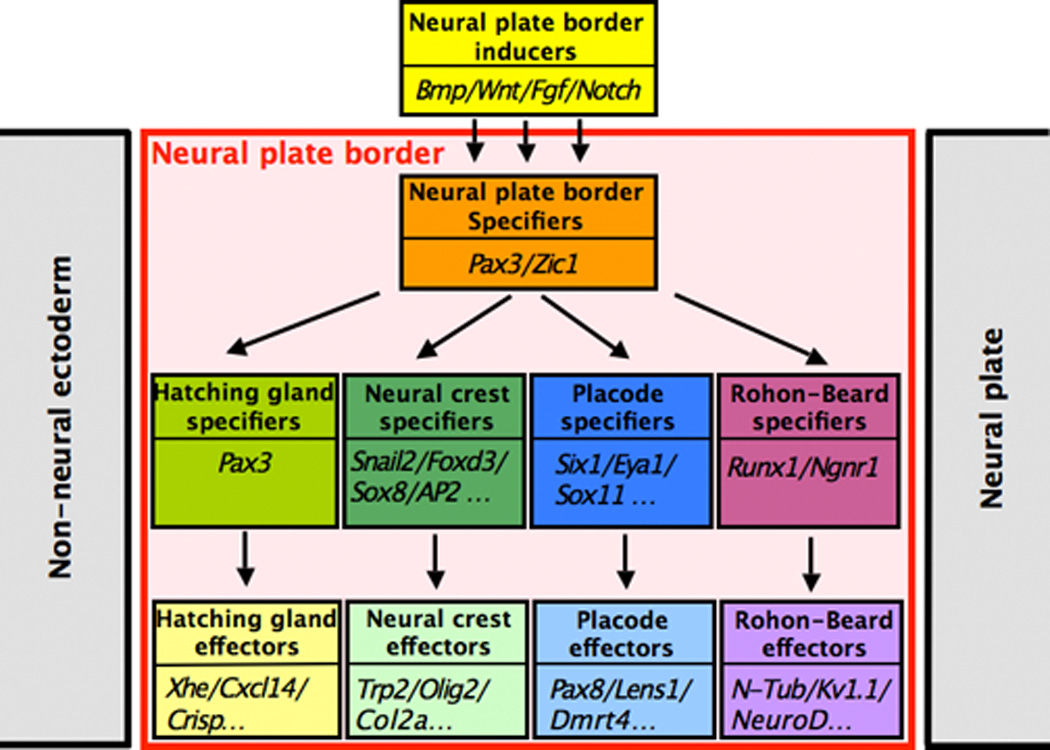
This model is an extension of the model proposed by Meulemans and Bronner-Fraser (2004) and Litsiou et al., (2005) for NC and PE specification. Based on our current observations and other studies (Hong and Saint-Jeannet, 2007) this regulatory cascade has been expanded to include two additional NPB cell types, RB neurons and HG cells.
Loss of Runx1 function using 3 distinct morpholino antisense oligonucleotides interfering either with Runx1 translation or splicing show an extremely consistent phenotype at the tadpole stage, characterized by the failure to form RB neurons in the dorsal spinal cord. Later, morphant tadpoles have a defective escape response to touch, consistent with the loss of RB neurons. These observations demonstrate that Runx1 is critically required for the development of RB sensory neurons and the establishment of the larval mechanosensory system. Vertebrate Runx genes are expressed from two alternative promoters, the distal P1 and proximal P2 encoding isoforms with distinct amino-terminal sequences (reviewed in Blyth et al., 2005). This is also the case for Xenopus Runx1, which has two isoforms that differ by a few amino acids. Interestingly in Xenopus elimination of one of the isoforms using Runx1MO or Aml1MO appears to be sufficient to abrogate all mechanosensory functions. Moreover, the splice blocking MO (Runx1SMO) that is believed to interfere with both isoforms does not produce a phenotype that is any stronger than each translation blocking morpholino individually. This suggests that the formation of RB progenitors is extremely sensitive to Runx1 dosage. This dosage sensitivity has also been described in the context of Runx1 function during hematopoiesis. For example loss of P1-Runx1 in the mouse embryo is reminiscent of the Runx1 heterozygote phenotype with sufficient definitive hematopoietic cells to permit embryonic survival. With loss of P2-Runx1, in contrast, definitive hematopoiesis is dramatically affected, resulting in a phenotype resembling the Runx1 null (Bee et al., 2009; 2010).
RB progenitors constitute one of the three groups of primary neurons specified at the end of gastrulation in anamniotes. The other two, the primary motoneurons and interneurons, are confined to the medial neural plate. The differentiation of the primary neurons is driven by proneural transcription factors, which promote the activation of a number of factors required for cell fate determination, cell cycle exit and terminal differentiation. In Xenopus most of these factors have fairly similar expression patterns in all three primary neuron populations, suggesting that the formation of primary neurons is regulated by the same mechanisms (reviewed in Henningfeld et al., 2007). The b-HLH gene Ngnr1 is at the top of this regulatory cascade (Ma et al., 1996). Ngnr1 indirectly activates Pak3, a p21-activated serine/threonine kinase 3, which promotes cell cycle withdrawal, thereby allowing neuronal differentiation to proceed (Souopgui et al., 2002). Our results using morpholinos interfering with Runx1 function demonstrate that Runx1 is also required for Pak3 and N-Tub expression in RB progenitors. In these embryos Ngnr1 expression was unperturbed or expanded. A similar phenotype was observed using a dominant negative form of Runx1 (Tracey et al., 1998; not shown). These results indicate that Runx1 can regulate the generation of RB cells independently of Ngnr1. Interestingly, while Runx1 and Ngnr1 are initially coexpressed in RB progenitors (stage 13), a few hours later (stage 19) Ngnr1 is no longer detected in RB cells (Fig 2). Transient Ngnr1 expression in RB progenitors distinguishes this cell type from the other two populations of primary neurons, and suggests that the formation of this primary neuron subtype is regulated by distinct mechanisms. One interpretation of our results is that Runx1 is required in RB progenitors to downregulate Ngnr1. In the absence of Runx1 function, Ngnr1 is maintained in RB progenitors preventing their differentiation.
Misexpression of Ngnr1 is known to repress NC fate by converting the entire ectoderm into primary neuron progenitors (Ma et al., 1996; Olson et al., 1998; Perron et al., 1999). Interestingly, unlike N-Tub and Pak3, we found that Runx1 was not ectopically induced upon Ngnr1 misexpression, while Islet-1 and Kv1.1 were dramatically upregulated throughout the ectoderm at the tailbud stage. One interpretation is that Ngnr1 can bypass the need for Runx1 to induce sensory neuron characteristics in the ectoderm, suggesting the existence of two distinct pathways regulating the emergence of sensory neurons. Alternatively, it is also possible that Runx1 and Ngnr1 are involved in the differentiation of distinct classes of sensory neurons. Additional studies will be needed to fully evaluate these possibilities, define the interplay between these factors, and identify the downstream targets they regulate to establish the identity of this cell population. Interestingly, a recent study is also pointing to the existence of Ngnr1-dependent and Ngnr1-independent pathways in the specification of cranial sensory neurons in Xenopus (Schlosser et al., 2008).
In the mouse DRG, Runx1 is expressed in a subpopulation of sensory neurons involved in pain transduction, the nociceptive neurons. Runx1 is required for the generation of nociceptive neurons during embryonic and early postnatal phases of DRG development (Chen et al., 2006; Marmigere et al., 2006; Kramer et al., 2006). There is an interesting parallel between Runx1 function in these two anatomically distinct groups of sensory neurons across species. It has been proposed that the NC may have evolved from a subset of RB progenitors that had delaminated from the dorsal spinal cord and migrated in the periphery, to eventually give rise to the modern DRG sensory neurons (Fritzsch and Northcutt, 1993; reviewed in Donohue et al., 2008). Consistent with the idea of a common origin to both RB and NC cells, in zebrafish the segregation of the two fates is tightly linked and depends on Notch/Delta signaling (Cornell and Eisen, 2000; 2002). Moreover, the transcription factor prdm1a is required for specification of both RB and NC cells (Artinger et al., 1999; Roy and Ng, 2004; Rossi et al., 2009; Olesnicky et al., 2010). The conserved function of Runx1 in anamniote RB sensory neurons and in the NC-derived DRG sensory neurons of higher vertebrates may represent additional evidence in support of the evolutionary derivation of these cells.
Highlights.
-
➢
Runx1 is expressed in Rohon-Beard (RB) progenitors at the end of gastrulation.
-
➢
In the absence of Runx1 function RB sensory neurons failed to form.
-
➢
The resulting embryos lose the mechanosensory response to touch.
-
➢
Runx1 is critically required for RB sensory neurons formation in Xenopus.
Supplementary Material
Supplemental Figure S1: Analysis of the specificity of Runx1 morpholino antisense oligonucleotides. (A) Structure of Runx1 protein. Runx1 genes are expressed from two promoters (P1 or distal, and P2 or proximal), which give rise to isoforms with distinct amino-terminal sequences, here referred to as Runx1 (P1) and Xaml1 (P2). The Runt domain and the transactivation domain are indicated. (B) Sequence of the Runx1 translation blocking morpholino, Runx1MO. (C) Sequence of the Xaml1 translation blocking morpholino, Aml1MO. (D) Increasing amounts of Runx1MO blocks translation directed by Runx1 mRNA. Similarly increasing amounts of Aml1MO blocks translation directed by Xaml1 mRNA. Runx1MO and Aml1MO did not interfere with translation directed by Xaml1 and Runx1 mRNAs, respectively. (E) Sequence of the Runx1 splice blocking morpholino (Runx1SMO) targeting the Exon1-Intron1 junction. (F) Schematic representation Runx1 Exon1-Intron1 junction with the position of the 2 sets of primers (F1-R1 and F2-R2) used to analyze splicing upon morpholino injection. (G) Two-cell-stage embryos were injected into both blastomeres with 60 ng of each Runx1SMO or Runx1MO and analyzed at stage 17 by RT-PCR. Runx1SMO blocked splicing almost completely as compared to uninjected or Runx1MO-injected embryos. EF-1a primers are used as control.
Supplemental Figure S2: Fgf8a expands Runx1 expression domain at the expanse of epidermal keratin. Overexpression of Fgf8a by injection of 2 pg of Fgf8a mRNA in one blastomere at the 2-cell stage results in an expansion of the Runx1 expression domain (100% embryos; n=44) and a reduction of epidermal keratin (XK81) expression in the ectoderm (100% of embryos; n=37).
Supplemental Figure S3: Proliferation is not affected in Xaml1/Runx1 morphants. Whole-mount phosphohistone H3 staining at stage 15, after unilateral injection of Aml1MO, Runx1MO or Runx1SMO at the 2-cell stage. The red lines indicate the position of the midline. The injected side is on the right (arrows). Dorsal views, anterior to top.
Supplemental Movie 1: Normal response to touch in a CoMO (60 ng) injected embryo at stage 32.
Supplemental Movie 2: Lack of response to touch in a Runx1MO (40 ng) injected embryo at stage 32.
Supplemental Movie 3: Lack of response to touch in an Aml1MO (30 ng) injected embryo at stage 32.
Supplemental Movie 4: Lack of response to touch in a Runx1SMO (40 ng) injected embryo at stage 32.
Acknowledgements
We would like to thank Drs. Enrique Amaya, Peter Klein, Tomas Pieler, Angela Ribera, Thomas Sargent and Jonathan Slack for plasmids. We are grateful to members of the Saint-Jeannet lab for discussion. This work was supported by research funds from Chonbuk National University (2010) to B-Y P, by a grant from the Korean Research Foundation (KRF-2008-331-C00207) to C-S H, and by a grant from the National Institutes of Health (RO1-DE014212) to J-P S-J.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahrens K, Schlosser G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev. Biol. 2005;288:40–59. doi: 10.1016/j.ydbio.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Artinger KB, Chitnis AB, Mercola M, Driever W. Zebrafish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons. Development. 1999;126:3969–3979. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang AG, Papalopulu N, Goulding MD, Kintner C. Expression of Pax-3 in the lateral neural plate is dependent on a Wnt-mediated signal from posterior Nonaxial mesoderm. Dev. Biol. 1999;212:366–380. doi: 10.1006/dbio.1999.9319. [DOI] [PubMed] [Google Scholar]
- Bee T, Liddiard K, Swiers G, Bickley SRB, Vink CS, Jarratt A, Hughes JR, Medvinsky A, de Bruijn MFTR. Alternative Runx1 promoter usage in mouse developmental hematopoiesis. Blood Cell Mol. Dis. 2009;43:35–42. doi: 10.1016/j.bcmd.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Bee T, Swiers G, Muroi S, Pozner A, Nottingham W, Santos AC, Li P-S, Taniuchi I, de Bruijn MFTR. Nonredundant roles of Runx1 alternative promoters reflects their activity at discreyte stages of developmental hematopoiesis. Blood. 2010;115:3042–3050. doi: 10.1182/blood-2009-08-238626. [DOI] [PubMed] [Google Scholar]
- Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat. Rev. Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Brade T, Gessert S, Kühl M, Pandur P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev. Biol. 2007;311:297–310. doi: 10.1016/j.ydbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Bradley LC, Snape A, Bhatt S, Wilkinson DG. The structure and expression of the Xenopus Krox-20 gene: conserved and divergent patterns of expression in rhombomeres and neural crest. Mech. Dev. 1993;40:73–84. doi: 10.1016/0925-4773(93)90089-g. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–5881. doi: 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- Burger C, Ribera AB. Xenopus spinal neurons express Kv2 potassium channel transcripts during embryonic development. J. Neurosci. 1996;16:1412–1421. doi: 10.1523/JNEUROSCI.16-04-01412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Broom DC, Liu Y, de Nooij JC, Li Z, Cen C, Samad OA, Jessell TM, Woolf CJ, Ma Q. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Chen J-A, Chu S-T, Amaya E. Maintenance of motor neuron progenitors in Xenopus requires a novel cyclin. EMBO Reports. 2007;8:287–292. doi: 10.1038/sj.embor.7400903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A, Kintner CR. Sensitivity of proneural genes to lateral inhibition affects the pattern of primary neurons in Xenopus embryos. Development. 1996;122:2295–2301. doi: 10.1242/dev.122.7.2295. [DOI] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis L, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Christen B, Slack RM. FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev. Biol. 1997;192:455–466. doi: 10.1006/dbio.1997.8732. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000;127:2873–2882. doi: 10.1242/dev.127.13.2873. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta/Notch signaling promotes of zebrafish neural crest by repressing neurogenin1 function. Development. 2002;129:2639–2648. doi: 10.1242/dev.129.11.2639. [DOI] [PubMed] [Google Scholar]
- Donoghue PCJ, Graham A, Kelsh RN. The origin and evolution of the neural crest. BioEssays. 2008;30:530–541. doi: 10.1002/bies.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale TA, Elinson RP. Development of the Xenopus laevis hatching gland and its relationship to surface ectoderm patterning. Development. 1991;111:469–478. doi: 10.1242/dev.111.2.469. [DOI] [PubMed] [Google Scholar]
- Fein AJ, Wright MA, Slat EA, Ribera AB, Isom LL. scn1bb, a Zebrafish Ortholog of SCN1B Expressed in Excitable and Nonexcitable Cells, Affects Motor Neuron Axon Morphology and Touch Sensitivity. J. Neurosci. 2008;28:12510–12522. doi: 10.1523/JNEUROSCI.4329-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Baker JC, Harland RM. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development. 2006;133:1703–1714. doi: 10.1242/dev.02342. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Northcutt RG. Cranial and spinal nerve organizations in amphioxus and lampreys: evidence for an ancestral craniate pattern. Acta Anat. 1993;148:96–109. doi: 10.1159/000147529. [DOI] [PubMed] [Google Scholar]
- Glavic A, Honore SM, Feijoo CG, Bastidas F, Allende ML, Mayor R. Role of BMP signaling and the homeoprotein Iroquois in the specification of the cranial placodal field. Dev. Biol. 2004a;272:89–103. doi: 10.1016/j.ydbio.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Glavic A, Silva F, Aybar MJ, Bastidas F, Mayor R. Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development. 2004b;131:347–359. doi: 10.1242/dev.00945. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Sive H. Identification of Otx2 target genes and restrictions in ectodermal competence during Xenopus cement gland formation. Development. 1997;124:471–481. doi: 10.1242/dev.124.2.471. [DOI] [PubMed] [Google Scholar]
- Garcia-Morales C, Liu C-H, Abu-Elmagb M, Hajihosseini MK, Wheeler GN. Frizzled-10 promotes sensory neuron development in Xenopus embryos. Dev. Biol. 2009;335:143–155. doi: 10.1016/j.ydbio.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Meth Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Henningfeld KA, Locker M, Perron M. Xenopus primary neurogenesis and retinogenesis. Funct. Dev. Embryol. 2007;1:26–36. [Google Scholar]
- Henry GL, Brivanlou IH, Kessler DS, Hemmati-Brivanlou A, Melton DA. TGF-beta signals and a pattern in Xenopus laevis endodermal development. Development. 1996;122:1007–1015. doi: 10.1242/dev.122.3.1007. [DOI] [PubMed] [Google Scholar]
- Hernandez-Lagunas L, Powell D, Law J, Grant K, Artinger KB. prdm1a and olig4 act downstream of Notch signaling to regulate cell fate at the neural plate border. Dev. Biol. 2011;356:496–505. doi: 10.1016/j.ydbio.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C-S, Saint-Jeannet J-P. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol. Biol. Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C-S, Park B-Y, Saint-Jeannet J-P. Fgf8a induces neural crest indirectly through the activation of Wnt8 in the paraxial mesoderm. Development. 2008;135:3903–3910. doi: 10.1242/dev.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Saint-Jeannet JP. Induction of the neural crest and the opportunities of life on the edge. Dev. Biol. 2004;275:1–11. doi: 10.1016/j.ydbio.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Jacobson M, Moody SA. Quantitative lineage analysis of the frog’s nervous system. I. Lineages of Rohon-Beard neurons and primary motoneurons. J. Neurosci. 1984;4:1361–1369. doi: 10.1523/JNEUROSCI.04-05-01361.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas EA, Snape AM, Sargent TD. Transcriptional regulation of a Xenopus embryonic epidermal keratin gene. Development. 1989;106:399–405. doi: 10.1242/dev.106.2.399. [DOI] [PubMed] [Google Scholar]
- Katagiri C, Maeda R, Yamashikia C, Mita K, Sargent TD, Yasamasu S. Molecular cloning of Xenopus hatching enzyme and its specific expression in hatching gland cells. Int. J. Dev. Biol. 1997;41:19–25. [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat. Rev. Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev. Biol. 1995;171:267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, Arber S. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–393. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Lamborghini JE. Rohon-beard cells and other large neurons in Xenopus embryos originate during gastrulation. J. Comp. Neurol. 1980;189:323–333. doi: 10.1002/cne.901890208. [DOI] [PubMed] [Google Scholar]
- Lamborghini JE. Disappearance of Rohon-Beard neurons from the spinal cord of larval Xenopus laevis. J. Comp. Neurol. 1987;264:47–55. doi: 10.1002/cne.902640105. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Creuzet S, Couly G, Dupin E. Neural crest cell plasticity and its limits. Development. 2004;131:4637–4650. doi: 10.1242/dev.01350. [DOI] [PubMed] [Google Scholar]
- Litsiou A, Hanson S, Streit A. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development. 2005;132:4051–4062. doi: 10.1242/dev.01964. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Marmigere F, Montelius A, Wegner M, Groner Y, Reichardt LF, Ernfors P. The Runx1/AML1 transcription factor selectively regulates development and survival of TrkA nociceptive sensory neurons. Nat. Neurosci. 2006;9:180–187. doi: 10.1038/nn1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R, Morgan R, Sargent M. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- McGrew LL, Takemaru K, Bates R, Moon RT. Direct regulation of the Xenopus engrailed-2 promoter by the Wnt signaling pathway, and a molecular screen for Wnt-responsive genes, confirm a role for Wnt signaling during neural patterning in Xenopus. Mech. Dev. 1999;87:21–32. doi: 10.1016/s0925-4773(99)00136-7. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev. Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) Amsterdam: North Holland Publishing Company; 1967. [Google Scholar]
- Olesnicky E, Hernandez-Lagunas L, Artinger KB. Prdm1a regulates sox10 and islet1 in the development of neural crest and Rohon-Beard sensory neurons. Genesis. 2010;48:656–66. doi: 10.1002/dvg.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EC, Schinder AF, Dantzker JL, Marcus EA, Spitzer NC, Harris WA. Properties of ectopic neurons induced by Xenopus Neurogenin1 expression> . Mol. Cell. Neurosc. 1998;12:281–299. doi: 10.1006/mcne.1998.0712. [DOI] [PubMed] [Google Scholar]
- Park B-Y, Saint-Jeannet J-P. Hindbrain-derived Wnt and Fgf signals cooperate to specify the otic placode in Xenopus. Dev. Biol. 2008;324:108–121. doi: 10.1016/j.ydbio.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B-Y, Saint-Jeannet J-P. Induction and segregation of the vertebrate cranial placodes. Colloquium Series on Developmental Biology. 2010a;1 No. 3. Morgan & Claypool Publishers. [PubMed] [Google Scholar]
- Park B-Y, Saint-Jeannet J-P. Expression analysis of Runx3 and other Runx family members during Xenopus development. Gene Exp. Patterns. 2010b;19:157–166. doi: 10.1016/j.gep.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes & Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron M, Opdecamp K, Butler K, Harris WA, Bellefroid EJ. X-ngnr-1 and Xath3 promote ectopic expression of sensory neuron markers in the neurula ectoderm and have distinct inducing properties in the retina. Proc. Natl. Acad. Sci. (USA) 1999;96:14996–15001. doi: 10.1073/pnas.96.26.14996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. Early functional organization of spinal neurons in developing lower vertebrates. Brain Res. Bull. 2000;53:585–593. doi: 10.1016/s0361-9230(00)00392-0. [DOI] [PubMed] [Google Scholar]
- Roberts A, Smyth D. The development of a dual touch sensory system in embryos of the amphibian Xenopus laevis. J. Comp. Phyisol. 1974;88:31–42. [Google Scholar]
- Rossi CC, Hernandez-Lagunas L, Zhang C, Choi IR, Kwok L, Klymkowsky M, Artigner KB. Rohon-Beard sensory neurons are induced by BMP4 expressing non-neural ectoderm inXenopus laevis. Dev Biol. 2008;314:351–361. doi: 10.1016/j.ydbio.2007.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi CC, Kaji T, Artinger KB. Transcriptional control of Rohon-Beard sensory neuron development at the neural plate border. Dev. Dyn. 2009;238:931–943. doi: 10.1002/dvdy.21915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ng T. Blimp-1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo. Curr. Biol. 2004;14:1772–1777. doi: 10.1016/j.cub.2004.09.046. [DOI] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Making senses: development of vertebrate cranial placodes. Int. Rev. Cell Mol. Biol. 2010;283:129–234. doi: 10.1016/S1937-6448(10)83004-7. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson K, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, Klymkowsky MW, Moody SA. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev. Biol. 2008;320:199–124. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifani S, Ma Q. ‘Runxs and regulations’ of sensory and motor neuron subtype differentiation: implications ofr hematopoietic development. Blood, Cell, Mol. Dis. 2009;43:20–26. doi: 10.1016/j.bcmd.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souopgui J, Sotler M, Pieler T. XPak3 promotes cell cycle withdrawal during primary neurogenesis in Xenopus laevis. EMBO J. 2002;21:6429–6439. doi: 10.1093/emboj/cdf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiers G, de Bruijn M, Speck NA. Hematopoietic stem cell emergence in the conceptus and the role of Runx1. Int. J. Dev Biol. 2010;54:1151–1163. doi: 10.1387/ijdb.103106gs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD, Pepling ME, Marko EH, Thomsen GH, Gerben JP. A Xenopus homologue of aml-1 reveals unexpected patterning mechanisms leading to the formation of embryonic blood. Development. 1998;125:1371–1380. doi: 10.1242/dev.125.8.1371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: Analysis of the specificity of Runx1 morpholino antisense oligonucleotides. (A) Structure of Runx1 protein. Runx1 genes are expressed from two promoters (P1 or distal, and P2 or proximal), which give rise to isoforms with distinct amino-terminal sequences, here referred to as Runx1 (P1) and Xaml1 (P2). The Runt domain and the transactivation domain are indicated. (B) Sequence of the Runx1 translation blocking morpholino, Runx1MO. (C) Sequence of the Xaml1 translation blocking morpholino, Aml1MO. (D) Increasing amounts of Runx1MO blocks translation directed by Runx1 mRNA. Similarly increasing amounts of Aml1MO blocks translation directed by Xaml1 mRNA. Runx1MO and Aml1MO did not interfere with translation directed by Xaml1 and Runx1 mRNAs, respectively. (E) Sequence of the Runx1 splice blocking morpholino (Runx1SMO) targeting the Exon1-Intron1 junction. (F) Schematic representation Runx1 Exon1-Intron1 junction with the position of the 2 sets of primers (F1-R1 and F2-R2) used to analyze splicing upon morpholino injection. (G) Two-cell-stage embryos were injected into both blastomeres with 60 ng of each Runx1SMO or Runx1MO and analyzed at stage 17 by RT-PCR. Runx1SMO blocked splicing almost completely as compared to uninjected or Runx1MO-injected embryos. EF-1a primers are used as control.
Supplemental Figure S2: Fgf8a expands Runx1 expression domain at the expanse of epidermal keratin. Overexpression of Fgf8a by injection of 2 pg of Fgf8a mRNA in one blastomere at the 2-cell stage results in an expansion of the Runx1 expression domain (100% embryos; n=44) and a reduction of epidermal keratin (XK81) expression in the ectoderm (100% of embryos; n=37).
Supplemental Figure S3: Proliferation is not affected in Xaml1/Runx1 morphants. Whole-mount phosphohistone H3 staining at stage 15, after unilateral injection of Aml1MO, Runx1MO or Runx1SMO at the 2-cell stage. The red lines indicate the position of the midline. The injected side is on the right (arrows). Dorsal views, anterior to top.
Supplemental Movie 1: Normal response to touch in a CoMO (60 ng) injected embryo at stage 32.
Supplemental Movie 2: Lack of response to touch in a Runx1MO (40 ng) injected embryo at stage 32.
Supplemental Movie 3: Lack of response to touch in an Aml1MO (30 ng) injected embryo at stage 32.
Supplemental Movie 4: Lack of response to touch in a Runx1SMO (40 ng) injected embryo at stage 32.