Abstract
Periodontal disease is a chronic oral inflammatory disease that is triggered by bacteria such as Porphyromonas gingivalis. P. gingivalis strains exhibit great heterogeneity, with some strains being encapsulated while others are nonencapsulated. Although the encapsulated strains have been shown to be more virulent in a mouse abscess model, so far the role of the capsule in P. gingivalis interactions with host cells is not well understood and its role in virulence has not been defined. Here, we investigated the contribution of the capsule to triggering a host response following microbial infection, as well as its protective role following bacterial internalization by host phagocytic cells with subsequent killing, using the encapsulated P. gingivalis strain W50 and its isogenic nonencapsulated mutant, PgC. Our study shows significant time-dependent upregulation of the expression of various groups of genes in macrophages challenged with both the encapsulated and nonencapsulated P. gingivalis strains. However, cells infected with the nonencapsulated strain showed significantly higher upregulation of 9 and 29 genes at 1 h and 8 h postinfection, respectively, than cells infected with the encapsulated strain. Among the genes highly upregulated by the nonencapsulated PgC strain were ones coding for cytokines and chemokines. Maturation markers were induced at a 2-fold higher rate in dendritic cells challenged with the nonencapsulated strain for 4 h than in dendritic cells challenged with the encapsulated strain. The rates of phagocytosis of the nonencapsulated P. gingivalis strain by both macrophages and dendritic cells were 4.5-fold and 7-fold higher, respectively, than the rates of phagocytosis of the encapsulated strain. On the contrary, the survival of the nonencapsulated P. gingivalis strain was drastically reduced compared to the survival of the encapsulated strain. Finally, the encapsulated strain exhibited greater virulence in a mouse abscess model. Our results indicate that the P. gingivalis capsule plays an important role in aiding evasion of host immune system activation, promoting survival of the bacterium within host cells, and increasing virulence. As such, it is a major virulence determinant of P. gingivalis.
INTRODUCTION
Microbial encapsulation has long been recognized as a way to protect bacteria from clearance by host immune defenses. This is thought to be due to reduction of the ability of the host effectors to gain access to the bacteria. However, encapsulation also has a direct effect on the microbial structures capable of eliciting a host response. By shielding microbial surface components, encapsulation may also have an effect on promoting virulence of a microorganism through reduction of the host's response. This results in a reduction of the clearance potential of the host, thus leading to prolonged survival of the bacterium and resulting, ultimately, in a long-term inflammatory response. Such a mechanism is especially important in the case of infectious chronic inflammatory diseases in which the pathogen may persist for a long time and elicit long-term, low-grade inflammation. Indeed, studies have shown that encapsulation plays a role in immune evasion in several bacteria (41, 42). Other examples also have shown that nonencapsulated mutants are outcompeted by encapsulated parental strains in animal models (10).
Periodontal diseases are chronic inflammatory conditions that destroy tissues supporting the teeth. The etiology of periodontitis is complex and involves both bacteria and the host. The onset of periodontal disease is triggered by pathogenic bacterial communities that exploit immunological susceptibilities of the host and induce the release of proinflammatory mediators that ultimately destroy host tissues. Although bacterial pathogens are required to initiate periodontal disease, their presence alone is not sufficient to cause the severe tissue destruction seen clinically (7, 8, 23). Indeed, it is the host inflammatory response to a bacterial challenge that is crucial for determining the disease outcome. Monocytes and macrophages are critical effectors and regulators of both inflammation and the innate immune response, which is the immediate response arm of the immune system (21, 22, 31). Furthermore, dendritic cells (DCs) initiate and regulate the highly pathogen-specific adaptive immune responses and are central to the development of immunological memory and tolerance (11, 20). Therefore, both cell types are important mediators of the immune response.
Several bacteria have been implicated in the initiation and progression of periodontal diseases, including Porphyromonas gingivalis, Tannerella forsythia, Prevotella intermedia, and Treponema denticola (18). P. gingivalis, an anaerobic Gram-negative member of the Cytophaga-Bacteroidetes family, belongs to a “red complex” of species associated with chronic periodontal infections (25, 43, 50). P. gingivalis is a highly heterologous species, and both the most virulent and the least virulent strains have been identified using clinical, animal, and in vitro studies (3, 4, 15, 16, 24, 36, 37, 48). It is equipped with multiple virulence factors, including fimbriae, proteases, outer membrane proteins, lipopolysaccharides (LPS), and a capsule. However, the expression of these factors in various P. gingivalis strains, as well as their role during the various stages of infection and their contribution to the activation of host cells, still remains controversial (39). Ultimately, the basis for the variation in strain virulence remains unknown.
The higher virulence potentials of encapsulated strains than of nonencapsulated ones, evaluated using a mouse abscess model (15, 29), suggest that the capsule plays a significant role in the virulence of the bacterium. However, as most of the evidence regarding the role of the capsule in virulence/adhesion has been derived from sources that used different P. gingivalis strains that are inevitably pleiotropic with respect to other pathogenic properties, the precise contribution of the capsule to pathogenesis has yet to be established.
Since periodontal disease is an inflammatory disease, the ability of a pathogen to withstand as well as elicit such a response is critical. Recent work has shown that encapsulated P. gingivalis strains trigger different host responses than nonencapsulated mutant strains (27, 47), thus indicating that the presence of a capsule indeed has immunomodulating properties. However, these studies were also done using P. gingivalis strains with different genetic backgrounds. The capsular polysaccharide locus has been identified by Aduse-Opoku et al. (1), which allowed for the generation of isogenic capsule-deficient mutants. The analysis of such a mutant by Brunner et al. has shown that encapsulation reduced the production of cytokines interleukin-1 (IL-1), IL-6, and IL-8 by fibroblasts in response to P. gingivalis infection (9), thus further demonstrating that the capsule modulates the host response to bacteria.
We hypothesize that encapsulation is a hallmark of virulence heterogeneity of P. gingivalis and that the mechanism altering the host's response is the presence of a capsule. Although simple, this concept has not been extensively investigated in the initiation and progression of periodontal disease. The availability of defined mutants deficient in capsule production sets the stage for further examination of its role in host-pathogen interactions. Here, we investigated the role of the capsule in the response of immune cells to P. gingivalis infection. In addition, we determined the contribution of the capsule to phagocytosis and microbial survival with immune cells. Finally, the contribution of the capsule to in vivo virulence was investigated using parental and capsule-deficient mutant strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis strains W50 (wild type) and PgC (capsule-deficient mutant strain) were used in this study. The PgC strain had the sequences coding for the capsular polysaccharide (PG0117 to PG0120) replaced by the ermF-ermAM cassette. Since the transcriptional unit coding for the capsular polysaccharide is distinct from the downstream transcripts, no polar effects were present. The absence of capsule in the mutant strain was confirmed by India ink and fuchsin staining. Another set of strains, W83 and its noncapsulated isogenic mutant (EpsC) were also included in the mouse abscess studies. The EPS strain had an ermF insertion in the PG0120 (epsC) sequence coding for UDP-N-acetylglucosamine (GlcNAc) 2-epimerase. Similar to the PgC strain, the loss of capsule was confirmed microscopically using bacterial cells stained with the combination of India ink and fuchsine. The strains were maintained on blood agar plates (TSA II, 5% sheep blood; BBL, Cockeysville, MD) in an anaerobic atmosphere composed of 10% H2, 10% CO2, and 80% N2 at 37°C. Liquid cultures were prepared using brain heart infusion broth (BHI; Difco Laboratories, Detroit, MI) supplemented with hemin (5 μg/ml) (Sigma, St. Louis, MO), yeast extract (5 mg/ml), cysteine (1 mg/ml) (Sigma, St. Louis, MO), and vitamin K3 (1 μg/ml) (Sigma, St. Louis, MO). For all experiments, bacterial cells were prepared from mid-log phase cultures. Thus, overnight actively growing culture was diluted and grown until the optical density at 660 nm (OD660) reached about 0.5.
Mice.
C57BL/6 mice and BALB/c mice were purchased from Harlan Laboratories and were housed in an accredited and pathogen-free animal facility. All mouse protocols were approved by the Virginia Commonwealth University Institutional Animal Care and Use committee.
Eukaryotic cells. (i) Cell media.
The medium (bone marrow medium [BMM]) for primary mouse macrophages was prepared as follows: 60% Dulbecco's modified Eagle's medium (DMEM; Invitrogen), 30% L929-conditioned medium, 10% heat-inactivated fetal bovine serum (FBS; Gemini Bio-Products), 0.2 M l-glutamine (Invitrogen), 100 U/ml penicillin-streptomycin (HyClone-ThermoScientific), and 2.5 μg/ml amphotericin B (J. R. Scientific) in 10% CO2 at 37°C. The L929-conditioned medium was prepared by growing L929 cells (also called the CCL-1 cell line, ATCC; these are mouse fibroblast cells which secrete colony-stimulating factor [CSF]) until 75% confluence was reached; the medium was then collected and sterile filtered for use as a conditioned medium containing macrophage CSF (M-CSF) to grow macrophages.
The murine dendritic cells were cultured in R10, which is composed of RPMI 1640 (Invitrogen) supplemented with 200 U/ml (20 ng/ml) recombinant murine granulocyte-macrophage CSF (rmGM-CSF) (Peprotech/Tebu, Frankfurt, Germany), 10% heat-inactivated FBS, 0.2 M l-glutamine, 50 μM beta-mercaptoethanol, 100 U/ml penicillin-streptomycin, and 2.5 μg/ml amphotericin B in 10% CO2 at 37°C.
(ii) Bone marrow-derived macrophages.
The macrophages used in this study were derived from male C57BL/6 mice using a protocol adapted from McCaffrey et al. (34). Following euthanasia, femurs and tibias (4 bones/mouse) were isolated, cut at the joint, and transferred into a sterile 0.6-ml tube with a tiny hole in the bottom. This was placed into a sterile 1.5 ml-Eppendorf tube containing 30 μl of BMM. Bone marrow was collected by centrifugation (10,000 rpm for 10 min) and suspended in 10 ml of red blood cell (RBC) lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH 7.4). Following incubation at room temperature for 5 min and centrifugation at 800 rpm for 5 min at room temperature, the pelleted cells were suspended in medium and counted in a hemacytometer. Cells were then plated at 1.25 × 107 cells/plate on day 0 in culture medium. On day 3, cells were fed with half of the original volume by adding 4 ml of fresh (prewarmed to 37°C) BMM. The cells were then used on day 7 (when the cell number reached 5 × 106 cells/plate).
(iii) Bone marrow-derived DCs.
Isolation of DCs was performed similarly to macrophage isolation (as described above) except that the RBCs were not lysed. Cells were plated at 2 × 106 cells/10-cm dish in 10 ml of R10 medium (day 0). At day 3, another 10 ml of R10 medium containing 200 U/ml of rmGM-CSF was added. At day 6, half of the culture supernatant was collected and centrifuged, and the cell pellet was suspended in 10 ml of fresh R10 containing 200 U/ml of rmGM-CSF and put back into the original plate. Day 8 cells were considered to be immature and phagocytosis-ready cells (this protocol is based on a paper and advice from Nikolaus Romani [46]).
Macrophage infection and RNA isolation.
On day 7, the macrophage cell medium was changed to antibiotic-free BMM. The cells were then infected with bacteria (prepared as described above) at a multiplicity of infection (MOI) of 100:1 (100 bacteria to 1 eukaryotic cell). Following incubation for two time periods (1 h and 8 h), supernatant was collected and the cells were washed in phosphate-buffered saline (PBS) and stored at −80°C. RNA was isolated as described previously (5). The concentration was measured using a NanoDrop ND-1000 spectrophotometer. RNA was then frozen at −20°C for short-term and −80°C for long-term storage. The experiment was repeated three times on different days to verify the biological significance of the results.
qRT-PCR analysis of macrophage response to bacterial infection.
The expression of 69 genes was examined using quantitative reverse transcription-PCR (qRT-PCR) with the SYBR green-based detection system. RNA samples from at least two separate experiments were converted to cDNA using the AffinityScript multitemperature cDNA synthesis kit (Stratagene). qPCR was carried out using primers synthesized by Integrated DNA Technologies (Coralville, IA). The primer sequences were obtained from the primer database qPrimerDepot (http://www.rtprimerdb.org/) (30, 40) or the PrimerBank (http://pga.mgh.harvard.edu/primerbank/index.html) (44). Our PCR arrays were generated by deposition of primer pairs in triplicate in 96-well plates. To select the best endogenous control gene, both β-actin and RPL13α were run in the qRT-PCR study; comparison showed that β-actin was more stably expressed over the range of samples. Thus, β-actin was chosen as the endogenous control for the comparative threshold cycle (CT) study; all samples were normalized to β-actin gene expression. Furthermore, the data were normalized to the results for uninfected cells in the same experiment. All PCRs for each gene were run in triplicate in each 96-well plate and had at least two biological replicates. Reverse transcriptase was omitted in samples serving as negative controls.
Amplification was performed in an ABI 7500 fast real-time system. Each PCR mixture contained 20 ng of cDNA (1 μl), 1 μl of a primer pair at a concentration of 2 μm, 3 μl distilled water, and 5 μl SYBR green reagent (SuperArray Bioscience Corporation, Frederick, MD) in a total volume of 10 μl. The PCR thermal profile was as follows: one cycle of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C, and one cycle at 95°C for 15 min, 60°C for 1 min, and 95°C for 15 min. Melting curves, to verify the specificity of the PCR products, were detected by sequentially annealing the PCR products from 60°C to 95°C for 45 s. The data were analyzed using Applied Biosystems sequence detection software, version 1.4 (7500 fast system SDS software). The comparative 2−ΔΔCT method for real-time quantitative PCR was the approach used to analyze the relative gene expression (32). Normalized time-matched values for each gene from infected cells were compared to the values from the uninfected controls, thus giving a ratio value for the treated cells versus the nontreated cells (RQ, or relative quantity; RQ is equal to fold change for values >1.0; for values <1.0, RQ may be converted to fold change by the formula −1/x, where x is the RQ). Triplicates were averaged prior to the calculation of RQ.
For the final values shown in Table 1, the expression ratios were compared between W50- and PgC-infected cells, thus giving the PgC/W50 ratio of induction of gene expression in infected macrophages.
Table 1.
Role of capsule in response to murine bone marrow-derived macrophage infection with P. gingivalis
| Gene product | Expression ata: |
|||||
|---|---|---|---|---|---|---|
| 1 h |
8 h |
|||||
| RQ |
PgC/W50 ratio | RQ |
PgC/W50 ratio | |||
| PgC | W50 | PgC | W50 | |||
| MARCKS | 1.196 | 1.027 | 10.257 | 4.492 | 2.28 | |
| CCL2 | 1.328 | 1.383 | 7.581 | 1.799 | 4.21 | |
| IL-8RB | 0.75 | 0.791 | 0.358 | 0.284 | ||
| SOCS4 | 0.858 | 1.183 | 0.73 | 1.267 | 1.105 | |
| STAT1 | 1.068 | 1.418 | 0.75 | 2.242 | 0.471 | 4.76 |
| IFNG | 3.562 | 1.313 | 2.71 | 83.791 | 3.999 | 20.95 |
| MYD88 | 1.186 | 1.398 | 3.708 | 1.586 | 2.34 | |
| SOCS5 | 1.07 | 1.179 | 1.159 | 0.928 | ||
| TIMP3 | 1.935 | 1.515 | 1.68 | 0.988 | 1.70 | |
| IL-10 | 8.113 | 5.979 | 25.923 | 9.26 | 2.80 | |
| NFKBIA | 2.179 | 1.44 | 1.51 | 5.85 | 4.711 | |
| STAT3 | 1.014 | 1.308 | 1.754 | 0.61 | 2.88 | |
| CCL5 | 1.574 | 1.571 | 25.162 | 3.016 | 8.34 | |
| IL-12A | 1.623 | 3.439 | 0.47 | 14.062 | 13.433 | |
| PIAS1 | 0.981 | 1.29 | 1.288 | 0.94 | ||
| TLR2 | 1.741 | 1.899 | 4.165 | 1.812 | 2.30 | |
| BCL6 | 1.087 | 1.23 | 1.974 | 1.245 | 1.59 | |
| IL-17 | 1.716 | 0.59 | 2.91 | 15.782 | ||
| RELA | 1.144 | 1.14 | 2.816 | 2.429 | ||
| TLR4 | 0.727 | 1.11 | 0.65 | 1.026 | 0.785 | |
| CD46 | 2.24 | 2.226 | 7.846 | 3.488 | 2.25 | |
| IL-18RAP | 1.224 | 1.174 | 1.252 | 1.274 | ||
| RELB | 1.653 | 1.781 | 6.134 | 4.161 | ||
| TRAF4 | 0.876 | 1.449 | 0.60 | 3.747 | 1.437 | 2.61 |
| CHUK | 1.083 | 1.28 | 1.752 | 1.462 | ||
| IL-19 | 4.268 | 1.323 | 3.23 | 720.93 | 32.503 | 22.18 |
| SOCS1 | 0.99 | 1.069 | 27.301 | 4.306 | 6.34 | |
| CD44 | 1.008 | 1.022 | 2.189 | 1.348 | 1.62 | |
| CRP | 0.268 | 0.317 | 6.309 | 4.659 | ||
| IL1A | 1.736 | 1.651 | 90.948 | 44.1 | 2.06 | |
| SOCS3 | 2.436 | 2.247 | 32.075 | 16.991 | 1.89 | |
| CSF1 | 1.282 | 0.992 | 1.492 | 1.326 | ||
| TNFRSF10B | 1.789 | 2.407 | 0.74 | 1.565 | 0.9 | 1.74 |
| AGT | 0.282 | 0.607 | 0.46 | 0.217 | 0.293 | 0.74 |
| CXCL10 | 0.806 | 0.813 | 167.98 | 8.38 | 20.05 | |
| FN1 | 1.348 | 1.237 | 1.08 | 1.072 | ||
| TNFSF10 | 53.175 | |||||
| AKT1 | 0.703 | 0.871 | 1.405 | 1.641 | ||
| CXCR4 | 0.977 | 0.745 | 0.172 | 0.311 | 0.55 | |
| ICAM1 | 0.735 | 0.936 | 4.977 | 5.531 | ||
| IL-13 | 8.434 | 1.537 | 5.49 | 1.759 | 0.831 | 2.12 |
| ATF1 | 0.734 | 0.935 | 1.094 | 1.288 | ||
| IFNB1 | 1.406 | 1.141 | 13.223 | 4.038 | 3.27 | |
| MMP3 | 1.799 | 1.231 | 5.82 | 3.928 | ||
| CXCL2 | 3.002 | 2.078 | 39.119 | 49.203 | ||
| BCL10 | 0.796 | 0.875 | 1.646 | 2.323 | 0.71 | |
| IL-6 | 0.998 | 1.054 | 98.96 | 21.716 | 4.56 | |
| PAX6 | 1.236 | |||||
| BCL2L1 | 0.838 | 0.855 | 0.776 | 0.892 | ||
| JUN | 1.04 | 0.769 | 0.629 | 1.163 | 0.54 | |
| PPARg | 5.521 | 0.952 | 5.80 | 1.527 | 1.535 | |
| CCR10 | 0.155 | |||||
| CASP1 | 0.937 | 0.96 | 2.278 | 1.23 | 1.85 | |
| TNF | 1.868 | 0.821 | 2.28 | 9.016 | 10.186 | |
| SERPINE1 | 1.502 | 1.188 | 1.062 | 1.182 | ||
| CCL3 | 1.084 | 0.958 | 20.72 | 15.084 | ||
| CASP8 | 0.894 | 1.051 | 1.339 | 0.866 | 1.55 | |
| CSF1R | 0.854 | 0.96 | 0.851 | 1.311 | 0.65 | |
| TGFa | 2.929 | 0.348 | 8.42 | 3.551 | 4.74 | 0.75 |
| CCL20 | 36.678 | 21.318 | 1.72 | |||
| CCND2 | 1.157 | 0.975 | 0.988 | 0.47 | 2.10 | |
| CD14 | 1.504 | 1.238 | 2.913 | 2.579 | ||
| CEBPB | 1.13 | 0.941 | ||||
| CFB | 1.156 | 1.311 | 3.737 | 1.948 | 1.92 | |
| CLCN2 | 0.456 | 0.422 | ||||
| CREBBP | 1.267 | 1.089 | 1.248 | 1.016 | ||
| CSF2 | 1.952 | 1.307 | 1.49 | |||
| CSF3 | 1.925 | 2.083 | 160.28 | 130.7 | ||
| CXCL16 | 1.431 | 1.29 | 1.687 | 1.352 | ||
Bone marrow-derived macrophages were infected with P. gingivalis strains (encapsulated W50 and nonencapsulated PgC) at an MOI of 100:1 (ratio of macrophages to bacterial cells) and incubated for 1 h and 8 h under cell culture conditions. The cellular response was determined by qRT-PCR analysis. RQ data are the ratios of expression in infected cells versus expression in noninfected controls. The PgC/W50 ratio is indicative of the difference between the nonencapsulated PgC and the wild-type encapsulated W50 strain. Ratios above 1.5 and below 0.75 are considered to be significantly differentially regulated genes. Results from an experiment done in triplicate are shown.
ELISA.
The amounts of four cytokines present in macrophage culture supernatants were quantified using enzyme-linked immunosorbent assay (ELISA). The cytokines IL-6 and tumor necrosis factor alpha (TNF-α) were measured using the ELISA Max set standard (BioLegend, San Diego CA). The cytokines IL-10 and gamma interferon (IFN-γ) were measured with the mouse ELISA Ready-SET-Go! kit (eBioscience, San Diego, CA). Each ELISA was performed according to the manufacturer's instructions.
Phagocytosis.
The extent of internalization by macrophages and DCs of fluorescently labeled bacteria was determined by flow cytometry. Thus, 2 ml of bacterial culture in log phase (OD660 of 0.4 to 0.7) were collected by centrifugation at 7,000 rpm for 4 min. Pellets were washed twice in PBS and resuspended in 1 ml of PBS. Following the addition of 3.5 μl of FITC (fluorescein-5-EX, succinimidyl ester; Invitrogen), the bacteria were mixed using a tube rotator at 4°C for 1 h 40 min (the time was adjusted from the 1 h suggested by the protocol to 1 h 40 min based on our previous work indicating that this is the optimal time for labeling P. gingivalis cells). The bacteria were then washed with twice with PBS and resuspended in antibiotic-free cell medium (appropriate for the eukaryotic cells used in the study). The labeling efficiency was determined for both the W50 and PgC strain to ensure equal labeling of both bacterial strains using a FluoStar Galaxy fluorescence plate reader (BMG Lab Technologies). The parameters were set to measure the FITC emission by setting the excitation to 485 nm and the peak emission to 520 nm. Infections of eukaryotic cells were done immediately following the labeling. Macrophages or DCs were infected with labeled bacteria at an MOI of 100:1 (100 bacteria to 1 eukaryotic cell) and incubated for various time points (5, 10, and 30 min).
(i) For determination of phagocytosis by dendritic cells, after the addition of bacteria for the specified time points, the infected cells were washed with PBS and incubated with trypan blue (1:500 dilution; Sigma) to quench fluorescence from extracellular bacteria. Internalized bacteria were quantified by flow cytometry using a BD Canto II in the VCU Flow Cytometry Core laboratory. All flow cytometric analyses were performed on live cells, based on incorporation of propidium iodide and/or forward and side scatter, with the BD FACSDiva software.
(ii) Phagocytosis by macrophages was done by exposing the eukaryotic cells to the FITC-labeled bacteria for the specified time periods. The infected macrophages were then washed, removed from the plates and resuspended in PBS. The cells were treated with propidium iodide prior to running on the flow cytometer as described above.
Maturation of DCs.
Immature DCs from day 8 were infected with bacteria at an MOI of 100:1 (100 bacteria to 1 cell). Escherichia coli LPS (Sigma) was used as a positive control, and no treatment served as a negative control. Plates were incubated for various time periods (4, 24, and 48 h). Following incubation, the cells were collected, centrifuged, and suspended in PBS. Aliquots were prepared and labeled as follows: isotype control (eBioscience), CD11c-phycoerythrin (PE) (BD Bioscience) plus major histocompatibility complex class II-allophycocyanin (MHC-II–APC) (eBioscience), CD40-PE (eBioscience) plus MHC-II–APC, and CD86-PE (eBioscience) plus MHC II-APC for each of the samples, as well as a no-stain tube. Blocking antibody, purified anti-mouse CD16/32 (BioLegend), was then added to each tube and incubated on ice for 15 min. Following washing with PBS, the samples were double-stained with the specific fluorescent antibodies as named above. Samples were then washed again, suspended in PBS, and analyzed by flow cytometry (BD Canto II). Cells were first gated on the live cells using forward and side scatter. The quadrant was selected so that >99.0% of the negative cells were in the lower left-hand quadrant. All other populations were single or double positives. The upper box for the upper right quadrant was selected based on where the cell populations showed the greatest concentration of double positives at 48 h with LPS treatment (positive control).
Bacterial survival with host immune cells.
The ability of bacteria to survive was determined by plating. Thus, DCs from day 8 and macrophages from day 7 were transferred into an anaerobic chamber and placed in the appropriate antibiotic-free cell medium. The cells were then infected with P. gingivalis strains at an MOI of 100:1. Plates (macrophages) or tubes (DCs) were incubated anaerobically at 37°C for 30 min. The cells were then washed with PBS three times to wash away extracellular bacteria. Anaerobic BHI–1% saponin (1 ml/well) was added and the mixture incubated in the anaerobic chamber at 37°C for 15 min to lyse the eukaryotic cells. An additional 1 ml of BHI/well was added and the cell mixture was removed using a cell scraper to ensure that all of the cells were lysed and had released their bacterial load. Finally, dilutions (1:4, 1:100, and 1:1,000) were plated (200 μl/plate) on blood agar plates and the number of viable bacteria was determined by the number of colonies formed after 7 to 11 days of incubation under anaerobic conditions.
Virulence study using a mouse abscess model.
The mouse abscess model was done as described previously (19). Bacterial cells (W50 parental and capsule-deficient PgC mutant strain) from 18-h cultures were harvested by centrifugation and washed in sterile PBS. Cell suspensions at the desired concentration in sterile PBS were then prepared. Groups of 8 week-old BALB/c mice (6 to 7 mice per group) were inoculated by subcutaneous injection of three doses, 3 × 109, 1 × 1010, and 3 × 1010, of P. gingivalis cells on the midpoint of the dorsal side. For 4 days, the animals were monitored twice daily for their general health status (mortality, the presence and location of lesions, mobility, and weight loss). Moribund animals (as described in the university's guidelines for the moribund condition as an endpoint) and animals with lesions that measured more than 15 mm were euthanized immediately and counted as nonsurviving.
The numbers of bacteria in the infected animals were also monitored. Although the animals were inoculated on the dorsal side, the bacteria disseminated to various parts of the infected animals. This is based on published data and our data indicating the presence of lesions on the ventral side of the mice and the presence of bacteria in the abscesses, as well as in the bloodstream (37). Thus, to determine the ability of the host to clear the bacteria, the numbers of bacteria recovered from sera, exudate from a lesion or an abscess, lungs, and spleen were assessed. For the quantification of bacteria, serial dilutions of serum and tissue (ground with a homogenizer and suspended in BHI, done under anaerobic conditions) samples were plated onto blood agar plates and cultivated anaerobically at 37°C for at least 7 days. For relative comparison, the values were standardized per gram of tissue (for tissues) or per ml of fluid (blood samples).
RESULTS
Encapsulation reduces host response.
Our 69-gene qRT-PCR array contained probes for cytokines, chemokines, and signal transduction genes. Infection of macrophages with both strains, W50 and PgC, resulted in altered gene expression (Table 1). However, time-dependent activation of gene expression in infected macrophages was observed. At 1 h postinfection, most genes had unaltered expression, while at 8 h postinfection, most genes showed a change in expression compared to their expression in the uninfected controls (Table 1).
The capsule-deficient strain downregulated the expression of only eight genes by 1.5-fold at 1 h postinfection, including those coding for CRP, AGT, and CLCN2 (Table 1). At the same time postinfection, 26 genes were upregulated 1.5-fold; the cytokines IL-13, IL-10, and IL-19 were the most upregulated, with ratios of 8.4, 8.1, and 4.3, respectively. For the macrophages, challenges with the encapsulated W50 strain showed that six genes (those coding for CLCN2, CRP, TGFα, CXCR4, IL-17, and AGT) had 1.5-fold-reduced expression at 1 h postinfection (Table 1). Among the 14 genes upregulated at 1 h postinfection were those coding for cytokines IL-10 and IL-12, with expression ratios of 5.9 and 3.4, respectively. Infection with both strains resulted in upregulation of chemokines and cytokines at 1 h postinfection. Comparison of the PgC-/W50-elicited gene expression ratios showed that only 16 genes showed significant differences, with 9 genes having higher expression in macrophages challenged with PgC than in macrophages infected with W50, while 7 genes had reduced expression (with genes encoding IL-12 and AGT having lower than 2-fold expression). The genes most highly upregulated by the nonencapsulated PgC strain were those coding for IL-13, IL-19, peroxisome proliferator-activated receptor gamma (PPARγ), and transforming growth factor α (TGF-α), with PgC/W50 ratios of 5.5, 3.2, 5.8, and 8.4, respectively (Table 1).
At 8 h postinfection, four genes had 1.5-fold-reduced expression in cells infected with the nonencapsulated PgC strain, including those encoding CXCR4, IL-8RB, AGT, and JUN. At the same time, as many as 43 genes were upregulated by 1.5-fold, with the genes coding for the cytokines and chemokines IL-19, IL-6, IL-1α, IL-10, CXCL10, CXCL2, CCL20, CCL5, and CCL3, as well as the genes coding for SOCS1, SOCS3, IFNG, and CSF3 (fold change over 20), being the most highly upregulated. Challenge of macrophages with the encapsulated W50 strain also elicited a drastic change in gene expression, with downregulation of 6 genes (coding for IL-8-RB, AGT, CXCR4, CCND2, STAT1, and STAT3) and upregulation of 33 genes, 6 of which (coding for CCL20, IL-6, IL-19, IL-1A, CXCL2, and CSF3) had greater than 20-fold-elevated expression.
Comparison of the PgC-/W50-elicited gene expression ratios at 8 h postinfection showed that 28 genes had at least a 1.5 ratio, demonstrating higher upregulation in cells challenged with the PgC strain than in cells challenged with the W50 strain, while only 6 genes had a ratio of less than 0.75, meaning they had higher expression in cells challenged with the W50 strain (Table 1). Among the genes more highly upregulated by the PgC strain were ones coding for cytokines and chemokines. JUN and CXCR4 were more highly upregulated by the W50 strain.
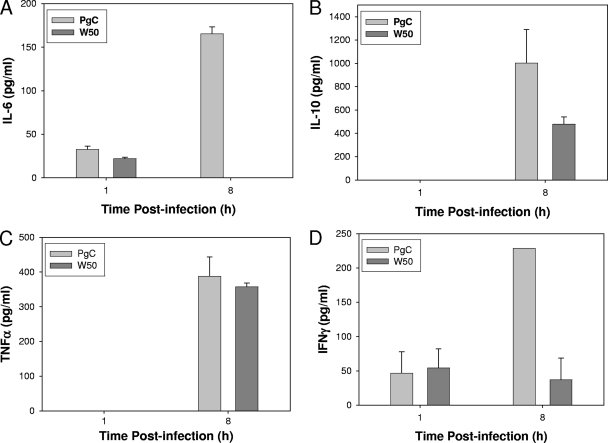
Next, the effect of bacterial infection on protein expression was evaluated. Four cytokines, IL-6, IL-10, TNF-α, and IFN-γ, were examined by ELISA. Supernatants from noninfected macrophages were used as controls. As shown in Fig. 1 A, the amounts of IL-6 produced at 1 h postinfection were similar between PgC- and W50-infected macrophages (with PgC having only a slightly higher production of the cytokine). However, at 8 h postinfection, IL-6 production was approximately 4-fold higher in macrophages challenged with the PgC strain than in cells infected with W50. IL-10 production (Fig. 1B) was elicited by the PgC strain more strongly than by W50 at 8 h, while no protein expression was detectable at 1 h. No TNF-α (Fig. 1C) was detectable at 1 h, but at 8 h both PgC and W50 induced similar amounts of protein. Lastly, the level of IFN-γ (Fig. 1D) induced by PgC at 1 h was similar to that induced by W50, while at 8 h, PgC induced much higher levels of IFN-γ than did W50. All of these data are consistent with the qRT-PCR results shown in Table 1, confirming that the qRT-PCR results are valid and the findings extend to the protein level.
Fig. 1.
IL-6 protein expression levels. ELISA results for IL-6 (A), IL-10 (B), TNF-α (C), and IFN-γ (D) cytokine expression in supernatants from bone marrow-derived macrophages infected with parental (W50) and nonencapsulated mutant (PgC) P. gingivalis strains for 1 h and 8 h at an MOI of 100:1.
Overall, the higher upregulation of genes by the nonencapsulated PgC strain than by the encapsulated W50 strain demonstrates that encapsulation alters the host response toward infection with P. gingivalis.
Phagocytosis of encapsulated P. gingivalis is reduced compared to that of the nonencapsulated strain.
The phagocytosis rates of both macrophages and DCs were compared using fluorescently labeled bacteria. The labeling efficiency has shown that the encapsulated W50 strain was labeled with 80% efficiency compared to the labeling of the nonencapsulated PgC strain (see Table S1 in the supplemental material), thus demonstrating that both strains can be efficiently labeled. DCs challenged with the capsule-deficient mutant, PgC, showed the presence of bacteria after incubation for only 5 min. It is noteworthy that nearly 50% of the cells were bacterium positive (Table 2; also see Fig. S1A in the supplemental material). Cells infected with the encapsulated W50 strain also internalized bacteria following 5 min of incubation; however, only approximately 10% of the cells were bacterium positive. This trend continued through the other incubation periods (10 min and 30 min); thus, nonencapsulated bacteria are phagocytosed by DCs at a 4-fold higher rate than encapsulated bacteria. The phagocytosis results were similar in three independent experiments, thus demonstrating that the results are consistent and biologically relevant. Overall, these results confirm that the presence of a capsule significantly reduces P. gingivalis phagocytosis by DCs.
Table 2.
Role of capsule in phagocytosis of p. gingivalis by immune cells
| Cell type | Incubation time (min) | % of cells positive for bacteria of indicated straina |
|||
|---|---|---|---|---|---|
| PgC |
W50 |
||||
| Avg | SD | Avg | SD | ||
| Dendritic cells | 5 | 46.9 | 1.2 | 9.0 | 0.6 |
| 10 | 45.6 | 0.2 | 9.6 | 0.6 | |
| 30 | 49.1 | 4.6 | 11.0 | 3.4 | |
| Macrophages | 5 | 7.6 | 1.2 | 3.8 | 0.3 |
| 10 | 15.1 | 0.9 | 4.2 | 0.8 | |
| 30 | 31.3 | 4.5 | 14.6 | 1.5 | |
Phagocytosis was assessed by flow cytometric analysis of immune cells incubated with fluorescently labeled bacteria. These results have been corrected for bacterial cell labeling efficiency (see Table S1 in the supplemental material). These results are from one representative experiment that was performed three separate times, with similar results each time.
To further examine the effect of a capsule on phagocytosis, the ability of macrophages to internalize bacteria was analyzed. Again, at 5 min postinfection, approximately 45% of macrophages were bacterium positive when infected with the capsule-deficient PgC strain (Table 2; also see Fig. S1B in the supplemental material). These numbers increased and reached 62% at 30 min postinfection. On the contrary, only 0.5% of macrophages infected with the encapsulated strain W50 were bacterium positive at 5 min postinfection and only 2.6% of the cells were positive for W50 at 30 min postinfection. Thus, overall, there was an approximately 30-fold reduction in phagocytosis of the encapsulated strain compared to the phagocytosis of the nonencapsulated strain by macrophages.
Encapsulation enhances bacterial survival in the presence of host cells.
The capsule forms a thick layer on the bacterial surface and may affect the host's capability to kill microorganisms. To determine if the P. gingivalis capsule plays a role, the survival of the encapsulated W50 and nonencapsulated PgC strains was examined in the presence of two types of immune cells: DCs and macrophages. The survival of bacteria with DCs, determined by plating, differed greatly for the W50 and PgC strains (Table 3; also see Fig. S2A in the supplemental material). Much higher survival rates were recorded for the W50 strain than for the PgC strain. While approximately 200 colonies were found at a 1:10 dilution on plates containing the W50 strain, no colonies were found on plates inoculated with cell mixtures containing PgC. This observation was true even for plates at the lowest dilution (1:4). There were approximately 20 colonies found on the W50 plates at a 1:100 dilution and 3 to 5 colonies on W50 plates at a 1:1,000 dilution.
Table 3.
Role of capsule in survival of P. gingivalis with immune cellsa
| Cell type | Dilution | No. of colonies |
|
|---|---|---|---|
| W50 | PgC | ||
| Dendritic cells | 1:4 | TMTC | 0 |
| 1:10 | 200 | 0 | |
| 1:100 | 20 | 0 | |
| Macrophages | 1:4 | TMTC | 0 |
| 1:10 | 125 | 0 | |
| 1:100 | 5–10 | 0 | |
The P. gingivalis parental strain (W50) and the capsule-deficient mutant (PgC) were used to infect dendritic cells and macrophages. Following washing, different dilutions of lysed host cells containing attached/internalized bacteria were plated, and counts were obtained for each dilution. The table shows quantitative results for one representative experiment that was performed three times on different days with similar results. Plate images are in Fig. S2A and S2B in the supplemental material. TMTC, too many to count.
Similar results were obtained when bacterial strains were incubated with macrophages (Table 3; also see Fig. S2B in the supplemental material). Again, no colonies were observed at any dilution on plates inoculated with the nonencapsulated PgC strain, while at a 1:4 dilution, there were too many colonies to count for the W50 strain, and at 1:10 and 1:100 dilutions, there were 125 and 5 to 10 colonies, respectively, for the W50 plates.
The bacterial survival results with DCs and macrophages were similar in three experiments done on different days, thus demonstrating that the data are biologically consistent. These results show that the P. gingivalis capsule is indispensable for survival of the bacterium with immune cells.
Capsule reduces DC maturation induced by P. gingivalis.
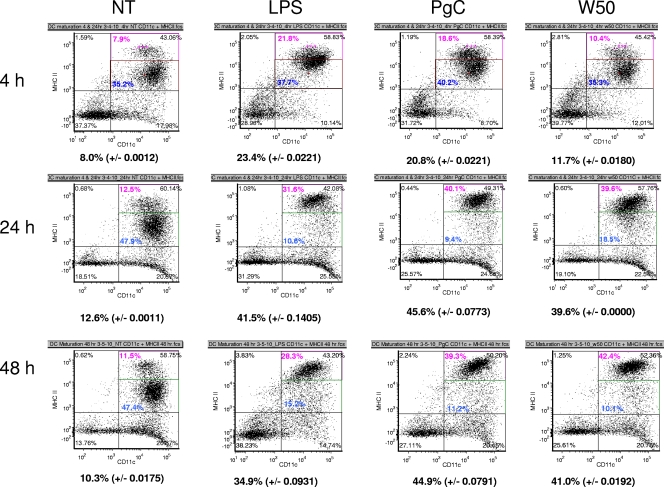
Immature DCs reside in peripheral tissues of the host, and the encounter of DCs with antigens results in their maturation and the subsequent increased expression of surface markers, such as CD11c, CD40, and CD86, in conjunction with a higher expression of MHC-II. Therefore, the expression of markers in immature DCs following infection with encapsulated and nonencapsulated P. gingivalis strains W50 and PgC was examined. As a positive control, the strongly immunogenic LPS was used. As shown in Fig. 2 (also see Fig. S3A and B in the supplemental material), encapsulated P. gingivalis induces expression of the markers in a time-dependent manner. However, the ability of the nonencapsulated PgC strain to induce maturation of DCs is increased 2-fold for all the maturation markers tested following a 4-h challenge. The maturation ability of W50 was comparable to that of the control (no treatment at 4 h), while the maturation ability of the nonencapsulated strain was similar to that of the positive control, E. coli LPS (Fig. 2). Following incubation for 24 h, DCs infected with the nonencapsulated PgC strain expressed more CD40 than DCs matured in the presence of the encapsulated W50 bacteria (37% versus 27%); however, the expression of other markers was similar in DCs infected with the two bacterial strains. This trend continued, and at 48 h, the expression of the maturation markers was similar for the two strains and comparable to the results for the positive control, LPS (Fig. 2; also see Fig. S3A, B, and C in the supplemental material).
Fig. 2.
Maturation of DCs. Immature DCs from day 8 were infected with bacteria at an MOI of 100:1. LPS was used as a positive control, and no treatment (NT) served as a negative control. Plates were incubated with PgC (nonencapsulated strain) and W50 (wild-type encapsulated bacteria) for various times (4, 24, and 48 h). Following incubation, flow cytometry analysis was done using samples incubated with isotype control or double-stained CD11c-PE plus MHC-II–APC antibodies. The subset of cells with the highest expression of double-stained maturation markers was considered to be the activated/mature DCs. Flow cytometry data for CD11c–MHC-II are shown here, and data for CD40 and CD86 are shown in Fig. S3A and B in the supplemental material. The percentages shown below each cytogram indicate the average value obtained for the most highly double-positive cells for each sample.
Capsule enhances P. gingivalis virulence.
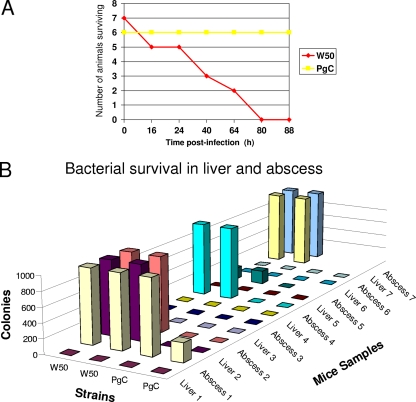
The final, most important aim was to determine the role of the capsule in the in vivo virulence of P. gingivalis. Infection of mice with 3 × 1010 W50 bacteria/animal resulted in the death of all animals in that group before 88 h postinfection (Fig. 3 A). Mortality was rapid. Two mice did not survive for 16 h postinfection, and another two did not survive for 40 h postinfection. By 80 h postinfection, all mice were dead (Fig. 3A). Importantly, all animals challenged with the capsule-deficient mutant, PgC, survived (Fig. 3A). Examination of other bacterial doses (1 × 1010 and 3 × 109 W50 bacteria/animal, respectively) resulted in survival of all challenged animals (results not shown). In addition to a higher mortality rate, animals challenged with 3 × 1010 W50 bacteria/animal exhibited a higher rate of secondary lesion formation; four out of seven challenged mice had large lesions on the abdominal site, while none of the animals challenged with the same dose of PgC had large abdominal lesions. Based on the above-described results, which suggested a reduced ability of the nonencapsulated bacteria to survive in the presence of host cells, the ability of the bacteria to survive when present in infected animals was also examined. Although the animals were challenged through inoculation at a dorsal site, previous work has shown that the bacteria can disseminate to various parts of the animal's body (19, 37). Microbiological examination of animal organs (liver), as well as blood and abdominal sites (abscesses), revealed that the encapsulated W50 strain had a greater ability to survive in animals than the nonencapsulated PgC strain (Fig. 3B; also see Tables S2 and S3 in the supplemental material). Virulence potential was also examined using strain W83 and its isogenic, capsule-deficient mutant EpsC (9). Within 88 h postinfection with 3 × 1010 bacteria/animal, all animals were euthanized due to their critical health status (results not shown), and examination of the numbers of live bacteria showed that significantly more bacterial cells survived in animals challenged with the parental strain W83 than in those challenged with the capsule-deficient mutant strain (see Table S4 in the supplemental material).
Fig. 3.
Role of capsule in P. gingivalis virulence. Animals were infected with 3 ×1010 cells of the P. gingivalis parental (W50) strain and its capsule-deficient mutant (PgC). (A) Survival of animals during 88 h postinfection. (B) Survival of bacteria in animals infected with encapsulated W50 and nonencapsulated PgC. Survival was determined by the number of colonies obtained following plating of tissues derived from infected animals on blood agar plates. The axis labeled “Colonies” shows the number of colonies observed on blood plates; the “Strains” axis indicates samples from animals infected with P. gingivalis W50 and P. gingivalis PgC; and the “Mice samples” axis indicates the mouse from which the tissue sample was derived.
Thus, overall, the results demonstrate that the capsule increases the virulence potential of P. gingivalis in the mouse abscess model.
DISCUSSION
Our results demonstrate that the capsule alters the transcriptional response of host immune cells to infection with P. gingivalis. At both time points, 1 h and 8 h, a significant number of genes had lower expression in cells challenged with the encapsulated bacteria than in cells challenged with the nonencapsulated one. These data are consistent with results reported by Brunner et al. (9), who also showed reduced expression of several cytokines in response to infection with encapsulated P. gingivalis W83 compared to their expression in the nonencapsulated isogenic mutant strain, using human gingival fibroblasts. Interestingly, the cells challenged with the nonencapsulated strain also triggered higher levels of the cytokine signaling suppressors SOCS1 and SOCS3, indicating that although higher levels of cytokines may be triggered by the nonencapsulated strain, the signaling role of the cytokines may be attenuated by the suppressors, thus ultimately reducing the proinflammatory capacity of the bacteria. Also, although it is clear that the nonencapsulated strain leads to a more robust macrophage response, this may lead to increased inflammation, as well as elevated recruitment of phagocytic cells, as evidenced by the drastic upregulation of chemokines, which in turn may clear the bacteria. Additional comprehensive studies are needed to examine the differential host response to encapsulated and nonencapsulated bacteria.
The microbial capsule is well known for its antiphagocytic abilities. Indeed, both bacteria and fungi expressing capsules have lower rates of phagocytosis (38, 51, 52). Our phagocytosis results are in agreement with other reports in that P. gingivalis expressing a capsule is internalized at a lower rate by macrophages and DCs than the nonencapsulated strain (45).
Interestingly, the nonencapsulated strain, although capable of being internalized by both macrophages and DCs very rapidly and in high numbers, was also rapidly killed. The cells were exposed to the strains for 30 min, and that was sufficient to allow killing of the nonencapsulated bacteria. Our experiments were done under anaerobic conditions, thus eliminating the effect of the capsule on oxidative stress. It has been shown that the Cryptococcus neoformans capsule contributes to resistance to oxidative stress (51, 52), and possibly, this is also the major mechanism accounting for the increased survival of encapsulated P. gingivalis in the presence of host immune cells. This mechanism may have further implications with regard to survival of internalized bacteria. Oxidative stress is the major immune effector of nonimmune cells as well, including epithelial, fibroblast, and endothelial cells, and thus, the presence and expression of a capsule under such conditions is a subject for future investigation. The P. gingivalis capsule may also contribute to increased survival by reducing the bactericidal effect of defensins, small antimicrobial peptides produced by a variety of host cells. Indeed, a recent study demonstrated capsule-dependent increased resistance of P. gingivalis to defensins (26).
The adaptive response has been shown to be downregulated in response to encapsulated P. gingivalis (49). The IgA response in particular was significantly elevated in animals challenged with the nonencapsulated strain 381, while no significant response to the encapsulated ATCC 53977 was detected (6). These results indicate that encapsulated bacteria evade adaptive immunity and, specifically, the humoral response. To further investigate the role of the capsule in this process, its effect on the maturation of DCs was examined. DCs are the most peripherally located cells of the immune system, and their role in the oral mucosa has been demonstrated (11). DCs residing in peripheral tissues ingest antigens which in turn stimulate their maturation, as exemplified by the high expression of various membrane molecules (CD11c, CD40, CD80, CD86, CD83, and HLA-DR) and the secretion of cytokines. Importantly, the mature subsets of DCs prime T cells, polarizing them toward Th1 (T-helper cell), Th2, or Th17 phenotypes (33). T-cell activity (Th2) is required for B-cell development and antibody production, and thus, any alteration of their function will lead to reduced humoral response and ultimately have an effect on the outcome of periodontal disease (11, 12).
As the capsule shields the outermost antigenic structures involved in the interaction of the bacterium with DCs, its presence will likely have an effect on the maturation of DCs. Indeed, our results showed that the nonencapsulated P. gingivalis PgC strain was able to induce the maturation of DCs more rapidly than the encapsulated W50 strain. These results suggest that the P. gingivalis capsule inhibits the host immune response, thus allowing the bacteria to escape host immune defenses and ultimately promoting bacterial survival and growth. Our results are in contrast to the conclusion of Vernal et al. (47), who were not able to discriminate between the rate of maturation induced by the encapsulated and nonencapsulated bacterial strains. We did discriminate due to the shorter time periods used in our study. A significant difference is the fact that different strains were used for the previous study; while W83 served as the K1 encapsulated strain, P. gingivalis 33277 was used as the nonencapsulated bacterium. It is probable that there are other differences between strains W83 and 33277 that account for the reduced activation of DCs. Indeed, the genome of P. gingivalis 33277 has been determined and multiple differences at the genetic level have been identified (35). Therefore, the use of an isogenic mutant to isolate the role of the capsule in the maturation of DCs is the best way to show definitive results focusing only on the role of the capsule.
The results of several previous experiments have suggested that the capsule plays a role in the virulence potential of P. gingivalis in the mouse abscess model (15, 29). However, significant differences in virulence were obtained among the encapsulated strains, thus leaving the role of the capsule in virulence inconclusive (29). Our study, done using a defined isogenic mutant, shows that the nonencapsulated PgC strain is less virulent than the encapsulated W50 strain, thus demonstrating that indeed the capsule plays a role in the virulence of P. gingivalis in the mouse abscess model. These results are consistent not only with previous reports using the rodent abscess model but also with a recent publication demonstrating reduced virulence of the nonencapsulated PgC strain compared to that of encapsulated W50 bacteria in the fruit fly Drosophila melanogaster model of infection (26). A model of infection in which the outcome is bacterium initiated but host driven was used in the study, as is observed in the case of periodontal disease. The signs of disease that were measured, such as lesion formation and, ultimately, animal death, rely on a host inflammatory response and thus are consistent with our above-described studies involving the analysis of host response to the bacteria.
The outcome of the virulence study was also consistent with the results of our survival studies. A drastic reduction of the numbers of live nonencapsulated bacteria recovered from animals compared to the numbers of encapsulated ones was observed in various organs, as well as in the blood of the infected animals. The enhanced activation of the immune system would suggest a greater inflammatory response, and activation of the host would also lead to recruitment of immune cells capable of eliminating the invading bacteria. As the nonencapsulated strain is more susceptible to killing by the cells of the immune system, lower numbers of nonencapsulated bacteria would be expected in the infected animals. Therefore, our transcriptional response study was also consistent with the outcome of our virulence study.
Our studies were also consistent with studies that have used other animal models, including the alveolar bone loss model in which encapsulated bacteria were shown to have increased virulence potential (6, 17, 49). Several other studies, however, suggest higher virulence of nonencapsulated bacteria based on the fact that they have higher rates of attachment to various surfaces, as well as a higher hemagglutination capability (1, 13, 14, 28). However, to increase the chances of attachment, capsule expression can be reduced to facilitate colonization, and work focusing on the regulatory mechanism is under way in other laboratories (2). Examination of capsule expression in in vivo conditions, as well as verification of its role using the alveolar bone loss model and isogenic mutants, is needed to further define its role in virulence.
In summary, our findings indicate that the capsule plays multiple roles in the virulence of P. gingivalis. These include reduction of the host immune response to the bacterium, reduced phagocytosis, and increased bacterial survival. Our study opens avenues for more detailed analysis of the various ways capsules can promote virulence, as well as pointing out the importance of work that will examine capsule expression at various stages of infection. Having been demonstrated as an important virulence factor, it also sets the stage for further examination of the role of the P. gingivalis capsule using more periodontitis-like conditions, as well as humanlike models. As an important virulence factor, it may also serve as a target for the development of therapeutic strategies.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the contribution of Jiayan He to this project.
This work was supported by NIH-NIDCR grants R01DE016124 and R01DE018039 from the National Institute of Dental and Craniofacial Research awarded to Janina Lewis. Also, this investigation was supported by the Medical Research Council (United Kingdom), grant no. G0501478. Support for the flow cytometry resources used in this study, provided from NIH grant P30CA16059, is gratefully acknowledged.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 12 September 2011.
REFERENCES
- 1. Aduse-Opoku J., et al. 2006. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect. Immun. 74:449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alberti-Segui C., Arndt A., Cugini C., Priyadarshini R., Davey M. E. 2010. HU protein affects transcription of surface polysaccharide synthesis genes in Porphyromonas gingivalis. J. Bacteriol. 192:6217–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amano A., et al. 2000. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 79:1664–1668 [DOI] [PubMed] [Google Scholar]
- 4. Amano A., Nakagawa I., Kataoka K., Morisaki I., Hamada S. 1999. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J. Clin. Microbiol. 37:1426–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anaya-Bergman C., et al. 2010. Porphyromonas gingivalis ferrous iron transporter FeoB1 influences sensitivity to oxidative stress. Infect. Immun. 78:688–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baker P. J., Dixon M., Evans R. T., Roopenian D. C. 2000. Heterogeneity of Porphyromonas gingivalis strains in the induction of alveolar bone loss in mice. Oral Microbiol. Immunol. 15:27–32 [DOI] [PubMed] [Google Scholar]
- 7. Birkedal-Hansen H. 1993. Role of cytokines and inflammatory mediators in tissue destruction. J. Periodontal Res. 28:500–510 [DOI] [PubMed] [Google Scholar]
- 8. Birkedal-Hansen H., et al. 1993. Matrix metalloproteinases: a review. Crit. Rev. Oral Biol. Med. 4:197–250 [DOI] [PubMed] [Google Scholar]
- 9. Brunner J., et al. 2010. The capsule of Porphyromonas gingivalis reduces the immune response of human gingival fibroblasts. BMC Microbiol. 10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coyne M. J., Chatzidaki-Livanis M., Paoletti L. C., Comstock L. E. 2008. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. U. S. A. 105:13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cutler C. W., Jotwani R. 2006. Dendritic cells at the oral mucosal interface. J. Dent. Res. 85:678–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cutler C. W., Teng Y. T. 2007. Oral mucosal dendritic cells and periodontitis: many sides of the same coin with new twists. Periodontol. 2000 45:35–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davey M. E., Duncan M. J. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J. Bacteriol. 188:5510–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dierickx K., et al. 2003. Adhesion of Porphyromonas gingivalis serotypes to pocket epithelium. J. Periodontol. 74:844–848 [DOI] [PubMed] [Google Scholar]
- 15. Ebersole J. L., Kesavalu L., Schneider S. L., Machen R. L., Holt S. C. 1995. Comparative virulence of periodontopathogens in a mouse abscess model. Oral Dis. 1:115–128 [DOI] [PubMed] [Google Scholar]
- 16. Eick S., Rodel J., Einax J. W., Pfister W. 2002. Interaction of Porphyromonas gingivalis with KB cells: comparison of different clinical isolates. Oral Microbiol. Immunol. 17:201–208 [DOI] [PubMed] [Google Scholar]
- 17. Evans R. T., et al. 1992. Periodontopathic potential of two strains of Porphyromonas gingivalis in gnotobiotic rats. Arch. Oral Biol. 37:813–819 [DOI] [PubMed] [Google Scholar]
- 18. Ezzo P. J., Cutler C. W. 2003. Microorganisms as risk indicators for periodontal disease. Periodontol. 2000 32:24–35 [DOI] [PubMed] [Google Scholar]
- 19. Fletcher H. M., et al. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geissmann F., et al. 2010. Development of monocytes, macrophages, and dendritic cells. Science 327:656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glaros T., Larsen M., Li L. 2009. Macrophages and fibroblasts during inflammation, tissue damage and organ injury. Front. Biosci. 14:3988–3993 [DOI] [PubMed] [Google Scholar]
- 22. Graves D. 2008. Cytokines that promote periodontal tissue destruction. J. Periodontol. 79:1585–1591 [DOI] [PubMed] [Google Scholar]
- 23. Graves D. T., Cochran D. 2003. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J. Periodontol. 74:391–401 [DOI] [PubMed] [Google Scholar]
- 24. Grenier D., Mayrand D. 1987. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J. Clin. Microbiol. 25:738–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holt S. C., Ebersole J. L. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72–122 [DOI] [PubMed] [Google Scholar]
- 26. Igboin C. O., Tordoff K. P., Moeschberger M. L., Griffen A. L., Leys E. J. 2011. Porphyromonas gingivalis-host interactions in a Drosophila melanogaster model. Infect. Immun. 79:449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katz J., Ward D. C., Michalek S. M. 1996. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol. Immunol. 11:309–318 [DOI] [PubMed] [Google Scholar]
- 28. Kremer B. H., van Steenbergen T. J. 2000. Peptostreptococcus micros coaggregates with Fusobacterium nucleatum and non-encapsulated Porphyromonas gingivalis. FEMS Microbiol. Lett. 182:57–62 [DOI] [PubMed] [Google Scholar]
- 29. Laine M. L., van Winkelhoff A. J. 1998. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol. Immunol. 13:322–325 [DOI] [PubMed] [Google Scholar]
- 30. Lefever S., Vandesompele J., Speleman F., Pattyn F. 2009. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res. 37:D942–D945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Y. C., Lerner U. H., Teng Y. T. 2010. Cytokine responses against periodontal infection: protective and destructive roles. Periodontol. 2000 52:163–206 [DOI] [PubMed] [Google Scholar]
- 32. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 33. Mazzoni A., Segal D. M. 2004. Controlling the Toll road to dendritic cell polarization. J. Leukoc. Biol. 75:721–730 [DOI] [PubMed] [Google Scholar]
- 34. McCaffrey R. L., et al. 2004. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc. Natl. Acad. Sci. U. S. A. 101:11386–11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naito M., et al. 2008. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 15:215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakagawa I., et al. 2002. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J. Periodontal Res. 37:425–432 [DOI] [PubMed] [Google Scholar]
- 37. Neiders M. E., et al. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J. Periodontal Res. 24:192–198 [DOI] [PubMed] [Google Scholar]
- 38. O'Riordan K., Lee J. C. 2004. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17:218–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ozmeric N., Preus N. R., Olsen I. 2000. Genetic diversity of Porphyromonas gingivalis and its possible importance to pathogenicity. Acta Odontol. Scand. 58:183–187 [DOI] [PubMed] [Google Scholar]
- 40. Pattyn F., Robbrecht P., De P. A., Speleman F., Vandesompele J. 2006. RTPrimerDB: the real-time PCR primer and probe database, major update 2006. Nucleic Acids Res. 34:D684–D688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quin L. R., Moore Q. C., III, McDaniel L. S. 2007. Pneumolysin, PspA, and PspC contribute to pneumococcal evasion of early innate immune responses during bacteremia in mice. Infect. Immun. 75:2067–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raffatellu M., et al. 2006. Capsule-mediated immune evasion: a new hypothesis explaining aspects of typhoid fever pathogenesis. Infect. Immun. 74:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Socransky S. S., Haffajee A. D., Cugini M. A., Smith C., Kent R. L., Jr 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134–144 [DOI] [PubMed] [Google Scholar]
- 44. Spandidos A., Wang X., Wang H., Seed B. 2010. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38:D792–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sundqvist G., Figdor D., Hanstrom L., Sorlin S., Sandstrom G. 1991. Phagocytosis and virulence of different strains of Porphyromonas gingivalis. Scand. J. Dent. Res. 99:117–129 [DOI] [PubMed] [Google Scholar]
- 46. Tripp C. H., Ebner S., Ratzinger G., Romani N., Stoitzner P. 2010. Conditioning of the injection site with CpG enhances the migration of adoptively transferred dendritic cells and endogenous CD8+ T-cell responses. J. Immunother. 33:115–125 [DOI] [PubMed] [Google Scholar]
- 47. Vernal R., et al. 2009. Differential cytokine expression by human dendritic cells in response to different Porphyromonas gingivalis capsular serotypes. J. Clin. Periodontol. 36:823–829 [DOI] [PubMed] [Google Scholar]
- 48. Walter C., et al. 2004. Porphyromonas gingivalis strain-dependent activation of human endothelial cells. Infect. Immun. 72:5910–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilensky A., et al. 2009. Strain-dependent activation of the mouse immune response is correlated with Porphyromonas gingivalis-induced experimental periodontitis. J. Clin. Periodontol. 36:915–921 [DOI] [PubMed] [Google Scholar]
- 50. Ximenez-Fyvie L. A., Haffajee A. D., Socransky S. S. 2000. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27:648–657 [DOI] [PubMed] [Google Scholar]
- 51. Zaragoza O., et al. 2008. Capsule enlargement in Cryptococcus neoformans confers resistance to oxidative stress suggesting a mechanism for intracellular survival. Cell Microbiol. 10:2043–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zaragoza O., et al. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 68:133–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.