Abstract
The bacterial obligate intracellular pathogen Chlamydia trachomatis replicates within a membrane-bound vacuole termed the inclusion. From within this protective environment, chlamydiae usurp numerous functions of the host cell to promote chlamydial survival and replication. Here we utilized a small interfering RNA (siRNA)-based screening protocol designed to identify host proteins involved in the trafficking of sphingomyelin to the chlamydial inclusion. Twenty-six host proteins whose deficiency significantly decreased sphingomyelin trafficking to the inclusion and 16 proteins whose deficiency significantly increased sphingomyelin trafficking to the inclusion were identified. The reduced sphingomyelin trafficking caused by downregulation of the Src family tyrosine kinase Fyn was confirmed in more-detailed analyses. Fyn silencing did not alter sphingomyelin synthesis or trafficking in the absence of chlamydial infection but reduced the amount of sphingomyelin trafficked to the inclusion in infected cells, as determined by two independent quantitative assays. Additionally, inhibition of Src family kinases resulted in increased cellular retention of sphingomyelin and significantly decreased incorporation into elementary bodies of both C. trachomatis and Chlamydophila caviae.
INTRODUCTION
Chlamydiae are Gram-negative obligate intracellular bacteria that cause diseases with significant medical and economic impact. Distinct serologically defined variants of Chlamydia trachomatis are the leading cause of infectious blindness worldwide as well as of sexually transmitted disease in the Western world (52). Chlamydia species share a unique biphasic life cycle, alternating between infectious elementary bodies (EBs) and replicative reticulate bodies (RBs) (39). During its entire intracellular developmental cycle, C. trachomatis resides within a parasitophorous vacuole, termed an inclusion, which is not fusogenic with endocytic compartments but which acquires host-derived lipids, including sphingomyelin, cholesterol, neutral lipids, and phospholipids, via both vesicular and nonvesicular trafficking pathways (17).
Chlamydiae accept lipid traffic from a number of cellular sources, including the Golgi apparatus (12, 21, 22), multivesicular bodies (MVBs) (48), lipid droplets (13, 29), and others (23, 58). Acquisition of sphingomyelin and cholesterol from the Golgi apparatus is initiated very early in the chlamydial developmental cycle, with incorporation of fluorescent sphingolipid analogs into the bacteria by 2 h postinfection (21). Chlamydial protein synthesis is required for the initiation of this interaction, presumably by modification of the inclusion membrane to become fusogenic with vesicular traffic from the Golgi apparatus (21, 24, 53). Specificity of this pathway is further demonstrated by the preferential acquisition of sphingomyelin from a basolaterally directed pathway that segregates the immediate products of ceramide metabolism, glucosylceramide and sphingomyelin, toward apical and basolateral surfaces, respectively (37). The molecular mechanisms controlling these interactions are unknown. In an effort to define cellular components functioning in this process, we used a small interfering RNA (siRNA)-based screen for host proteins known to be involved in vesicle trafficking pathways to identify potential factors supporting sphingomyelin trafficking to the inclusion. A number of host proteins whose depletion either inhibited or enhanced sphingomyelin delivery to C. trachomatis EBs were identified. One of these, the Src family tyrosine kinase Fyn, was confirmed in additional studies.
MATERIALS AND METHODS
Organisms and cell culture.
Chlamydia trachomatis serovar L2 (LGV 434) and Chlamydophila caviae GPIC were propagated in HeLa 229 cells and purified by Renografin density gradient centrifugation as previously described (10). HeLa 229 cells were grown in RPMI 1640 medium with 10% fetal bovine serum (FBS; ATCC) and gentamicin (Gibco, Carlsbad, CA).
siRNA screen.
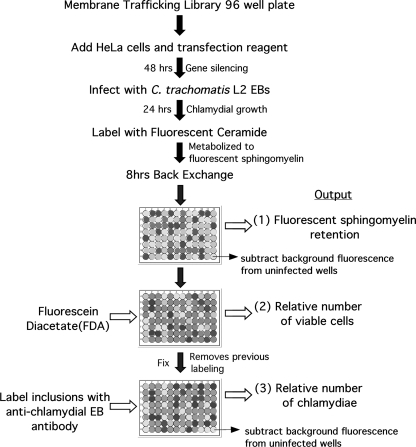
A Dharmacon (Lafayette, CO) ON-TARGETplus siRNA membrane trafficking library was rehydrated in DharmaFECT cell culture reagent and was complexed with DharmaFECT 1 transfection reagent (0.5 μl/well) for 30 min at room temperature. HeLa cells (5 × 103) in antibiotic-free RPMI 1640 plus 10% FBS were added to each well for a final siRNA concentration of 50 nM. After 48 h of gene silencing, the cultures were infected with C. trachomatis L2 or mock infected. At 24 h postinfection, plates were chilled to 4°C for 30 min and rinsed twice with cold Hanks balanced salt solution (HBSS) prior to incubation with prechilled 5 μM C6-NBD-ceramide [6-((N-(7-nitrobenz-2-oxa-1,3-diazol-4yl)amino)hexanoyl)sphingosine] (Invitrogen, Carlsbad, CA) in Dulbecco's modified Eagle medium (DMEM) supplemented with 0.034% defatted bovine serum albumin (BSA) (Sigma, St. Louis, MO) at 4°C for 30 min. Cells were then rinsed twice with HBSS and incubated for 8 h with DMEM supplemented with 0.7% defatted BSA for back-exchange (32, 33). The fluorescence associated with each well was then determined with a Safire2 plate reader (Tecan, San Jose, CA) using wavelengths of 466 nm and 536 nm for excitation and emission, respectively. Cells were then incubated with fluorescein diacetate (FDA; 1 μg/ml in HBSS) for 10 min at room temperature before fluorescence quantitation at wavelengths of 483 nm and 525 nm for excitation and emission, respectively. NBD fluorescence was apparently quenched and undetectable after FDA staining. The cultures were then fixed in methanol, eradicating any residual NBD or FDA fluorescence, and labeled with rabbit anti-L2 EB antibody that recognizes both EB and RB developmental forms, followed by an anti-rabbit Alexa Fluor 488 (Invitrogen)-conjugated secondary antibody in phosphate-buffered saline (PBS) containing 1% BSA (Sigma). Fluorescence was again determined with the plate reader. The background fluorescence from uninfected cells and non-FDA-treated cells was subtracted from the fluorescence outputs. The values from each fluorescence reading were adjusted based upon FDA fluorescence to account for siRNA-specific cell loss. A final ratio of the resulting NBD-sphingomyelin fluorescence to EB fluorescence normalized to nontargeting siRNA was calculated and used as a measure of the effect that each siRNA had on sphingomyelin trafficking to the chlamydial inclusion.
Validation of Fyn siRNA knockdown.
HeLa cells at ∼50% confluence were incubated with ON-TARGETplus Smartpool siRNA (Dharmacon) complexed with DharmaFECT 1 in antibiotic-free RPMI media supplemented with 10% FBS. Forty-eight hours posttransfection cells were mock infected or infected with C. trachomatis L2 at a multiplicity of infection (MOI) of ∼1 for 24 h. Knockdown efficiency based upon transcript level was determined by Quantigene analysis in accordance with the manufacturer's instructions (Panomics, Fremont, CA). Lysates were probed with Fyn- or GAPDH (glyceraldehyde-3-phosphate dehydrogenase)-specific RNA, and the luminescence for each sample and probe was measured using a Safire2 plate reader. The Fyn transcript was normalized to the GAPDH signal as a loading control. Fyn protein levels were determined by SDS-PAGE and immunoblotting. Proteins were transferred to nitrocellulose and probed with anti-Fyn (BD Biosciences, San Jose, CA) and anti-GAPDH antibody (Sigma), followed by horseradish peroxidase (HRP)-conjugated secondary antibody prior to chemiluminescence detection (Pierce, Rockford, IL).
Infectious progeny quantitation.
Following siRNA knockdown and C. trachomatis L2 infection, cells were lysed in water, diluted in HBSS, and replated onto fresh HeLa cell monolayers. At 24 h postinfection, the cultures were fixed and stained with rabbit anti-EB antibody followed by an anti-rabbit IgG Alexa Fluor 488-conjugated secondary antibody (Invitrogen). Inclusions were counted in 20 fields per sample using a Nikon Microphot-FXA fluorescence microscope, and the number of infectious progeny/ml was calculated.
Quantitative lipid analysis.
Following siRNA knockdown in 6-well plates (lipid extraction) or on coverslips in 24-well plates (photometer readings), cells were infected with C. trachomatis L2 or mock infected. After 24 h cells were labeled with C6-NBD-ceramide as described above. Microscope-based photometric quantitation of the fluorescent lipid content of inclusions was performed as described previously (22). A minimum of 20 inclusions per sample were analyzed, and experiments were conducted in triplicate. For quantitative lipid analysis, the medium was collected and the cells were scraped into HBSS. The lipids in the media and cells were isolated using chloroform-methanol extraction (6, 37) and dried under nitrogen. The lipid pellet was resuspended in a 2:1 chloroform-methanol solution and loaded onto 60A silica gel-coated thin-layer chromatography (TLC) plates (Whatman, Piscataway, NJ). The lipids were resolved using chloroform-methanol-water (16.25:6:1) as a solvent. Each fluorescent lipid species was visualized using a Typhoon phosphorimager, and quantitative densitometry was performed using ImageQuant software (version 5.0; GE Healthcare). Data were analyzed using GraphPad Prism 5, and significance was determined using a one-tailed Student t test.
Src family kinase inhibitor.
For analysis of sphingolipid synthesis and trafficking in uninfected cells, fluorescent lipid retention via lipid extraction was performed as described above but with a 1-h treatment with 40 μM SU6656 (Sigma) or a volume equivalent of dimethyl sulfoxide (DMSO) prior to labeling with C6-NBD-ceramide. The drug was included in the media for each subsequent step in the assay. For analysis of sphingomyelin retention by EBs, HeLa cell monolayers were infected with C. trachomatis L2 or C. caviae GPIC. Eighteen hours postinfection cells were treated with 40 μM SU6656 or DMSO for 1 h. Cells were then labeled with C6-NBD-ceramide as described above. After 3 h of back-exchange, the EBs were purified by Renografin density gradient centrifugation (10). Quantitative lipid analysis was performed as described above.
RESULTS
siRNA screen.
The investigation of sphingomyelin trafficking to the inclusion is facilitated by labeling host cells with the fluorescent sphingolipid C6-NBD-ceramide, which is the immediate precursor of glucosylceramide and sphingomyelin (33). In uninfected cells, these fluorescent lipids are trafficked from the Golgi apparatus to the plasma membrane, where they are exposed on the outer leaflet of the plasma membrane (32). There they may be extracted or “back-exchanged” by lipid acceptors such as BSA in the media. This process results in host cells with minimal fluorescent lipid content within about 4 h (21, 22). However, in chlamydia-infected cells fluorescent sphingolipids, primarily sphingomyelin, are redirected en route from the Golgi apparatus to the plasma membrane and are intercepted by the chlamydial inclusion. The fluorescent lipid not intercepted by the inclusion is trafficked to the plasma membrane. In chlamydia-infected cells, a significant portion of the fluorescent sphingomyelin is trafficked to and retained by the chlamydiae within the inclusion (21, 22). These properties were exploited and incorporated into a novel screen specifically designed to identify host proteins that are involved in sphingomyelin trafficking to the inclusion.
A commercial siRNA library (Dharmacon) targeting proteins known to be involved in membrane trafficking was screened to identify host proteins involved in sphingomyelin trafficking to the chlamydial inclusion. Protein depletion was accomplished by reverse transfection of HeLa cells with pools of siRNA targeting the transcripts of 122 host genes implicated in a wide range of cellular membrane trafficking pathways. After 48 h of gene silencing, cells were infected with C. trachomatis L2 for 24 h and then labeled with C6-NBD-ceramide. The cultures were incubated an additional 8 h to allow metabolism and export to the plasma membrane or the chlamydial inclusion. To determine the specific effect of each siRNA knockdown on sphingomyelin trafficking to the inclusion, three parameters were measured: (i) fluorescent sphingomyelin retained by infected host cells (after 8 h of back-exchange, most metabolites of C6-NBD-ceramide are exported from uninfected cells but a proportion of C6-NBD-sphingomyelin is retained by chlamydiae within infected cells), (ii) fluorescence from viable cells labeled with fluorescein diacetate to correct for any loss in cell number, and (iii) fluorescence from labeling of chlamydial developmental forms with chlamydia-specific antibodies that recognize both EBs and RBs as an indicator of chlamydial replication (Fig. 1). For outputs i and iii, backgrounds were corrected by subtracting the basal level of fluorescence from uninfected cells. All outputs were normalized to nontargeting siRNA controls. Seven gene targets were eliminated from the analysis due to apparent toxicity to HeLa cells. After correction for viable cell number, the ratio of sphingomyelin retention to chlamydial immunofluorescence was used to determine the effect of each knockdown on the ability of chlamydiae to acquire and retain sphingomyelin. Thus, the effects of siRNA knockdown on chlamydial development that are not related to sphingomyelin trafficking are minimized. Based upon the relative sphingomyelin/chlamydia ratio compared to controls the library target genes were divided into three categories (Table 1): “decreased” (<80%), “unchanged” (between 80 and 120%), and “increased” (>120%).
Fig. 1.
Flowchart of the siRNA screen for sphingomyelin trafficking to C. trachomatis in HeLa 229 cells. Details are given in Materials and Methods.
Table 1.
Identification of cellular proteins that alter sphingomyelin trafficking to the chlamydial inclusion
| Gene IDa | SM/EB (%)b |
|---|---|
| Unchanged | |
| ACTR2 | 95.8 |
| ACTR3 | 94.2 |
| ADAM10 | 118.1 |
| AMPH | 93.7 |
| AP1B1 | 110.1 |
| AP1M1 | 89.0 |
| AP1M2 | 83.9 |
| AP2A1 | 83.2 |
| AP2A2 | 109.6 |
| ARF6 | 111.4 |
| ARPC1B | 103.0 |
| ARPC2 | 103.8 |
| ARPC4 | 104.2 |
| ARRB1 | 80.1 |
| ARRB2 | 84.2 |
| ATP6V0A1 | 80.5 |
| CAV1 | 88.3 |
| CAV2 | 93.1 |
| CAV3 | 92.3 |
| CBLC | 91.7 |
| CDC42 | 99.1 |
| CIB3 | 95.2 |
| CLTA | 100.3 |
| CLTCL1 | 83.7 |
| DIAPH1 | 100.6 |
| DNM1 | 95.8 |
| DNM3 | 80.7 |
| EFS | 117.9 |
| ELKS | 100.4 |
| EPN1 | 90.7 |
| EPN2 | 94.9 |
| GIT1 | 87.8 |
| GORASP1 | 85.8 |
| GRB2 | 93.7 |
| HIP1 | 84.4 |
| HIP1R | 83.4 |
| IHPK3 | 108.7 |
| ITSN1 | 90.8 |
| LIMK1 | 80.1 |
| MAPK8IP1 | 107.9 |
| MAPK8IP3 | 88.5 |
| NEDD4 | 101.3 |
| NEDD4L | 84.9 |
| NSF | 107.0 |
| PACSIN1 | 110.3 |
| PACSIN3 | 92.7 |
| PIK3CG | 105.9 |
| PIP5K1A | 117.0 |
| PSCD3 | 110.5 |
| RAB11B | 117.2 |
| RAB3A | 119.4 |
| RAB3C | 92.4 |
| RAB4A | 92.1 |
| RAB4B | 100.1 |
| RAB5A | 112.0 |
| RAB5B | 92.8 |
| RAB5C | 111.2 |
| RAB6A | 84.1 |
| RAB6B | 110.3 |
| RAB8B | 97.2 |
| RAC1 | 110.9 |
| ROCK1 | 83.8 |
| ROCK2 | 97.2 |
| SH3GLB1 | 94.7 |
| SH3GLB2 | 94.5 |
| SNAP91 | 109.3 |
| STAU | 87.7 |
| SYNJ1 | 107.2 |
| VAMP1 | 106.0 |
| VAV2 | 81.7 |
| VIL2 | 92.8 |
| WASF1 | 101.3 |
| WASF3 | 110.3 |
| Decreased | |
| AP2B1 | 74.8 |
| ARF1 | 68.4 |
| ARFIP2 | 79.3 |
| ARPC3 | 71.9 |
| ATM | 71.6 |
| BIN1 | 68.1 |
| CBL | 71.7 |
| CBLB | 66.5 |
| CIB1 | 74.1 |
| CLTC | 78.6 |
| DDEF2 | 64.2 |
| DNM2 | 52.2 |
| EEA1 | 67.0 |
| EPN3 | 79.1 |
| EPS15 | 62.8 |
| EPS15L1 | 70.2 |
| FYN | 62.7 |
| MAP4K2 | 78.5 |
| MGC9726 | 65.0 |
| PICALM | 69.3 |
| PIK3C2G | 48.7 |
| SYNJ2 | 68.0 |
| VAMP2 | 70.8 |
| VAPA | 63.9 |
| VAPB | 66.7 |
| WAS | 69.6 |
| Increased | |
| ARPC5 | 142.3 |
| CFL1 | 200.0 |
| CIB2 | 318.2 |
| DAB2 | 128.3 |
| ITSN2 | 169.2 |
| MAPK8IP2 | 130.5 |
| PAK1 | 141.2 |
| RAB3B | 130.7 |
| RAB3D | 122.7 |
| RAB7L1 | 126.4 |
| RAB8A | 141.8 |
| RHOA | 125.4 |
| SYT1 | 129.8 |
| SYT2 | 127.0 |
| TNIK | 124.1 |
| WASF2 | 153.6 |
ID, identification. Unchanged, decreased, and increased are as defined in the text.
Values represent the means of two independent screens. SM, sphingomyelin.
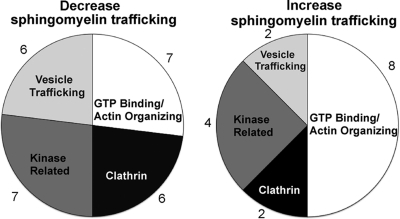
A majority (60%) of the gene targets had no significant effect on the amount of sphingomyelin trafficked to or retained by chlamydiae. Twenty-six (21%) decreased sphingomyelin trafficking to the inclusion by >20%. These can be separated into four relatively equal classes (Fig. 2): those that directly function in vesicular trafficking (e.g., VAMP2, VapA, and VapB), those that are kinase related (e.g., Atm and Fyn), those involved in clathrin-coated vesicles (e.g., CltC and Eps15), and GTP binding proteins and proteins involved in actin organization, which are grouped due to the overlapping functions of many of these proteins (e.g., Dnm2 and Was). PIK3C2G, Dnm2, and Fyn knockdowns produced the greatest decreases in sphingomyelin trafficking, with 52, 48, and 38% decreases in sphingomyelin retention, respectively. Sixteen gene targets (13%) increased the sphingomyelin retention by infected cells. These targets were also grouped into 4 subclasses as above: vesicular trafficking (e.g., Syt1 and Syt2), kinase related (e.g., Cib2 and Pak1), clathrin-coated vesicles (e.g., Dab2 and ItsN2), and GTP binding proteins/actin organization (e.g., Rab3A/Rab3D and RhoA). Cib2, Cfl1, and ItsN2 knockdowns produced the largest increases in sphingomyelin retention by infected cells, with increases of 218, 100, and 69%, respectively.
Fig. 2.
Functional categories of the candidate proteins identified in the membrane trafficking siRNA screen. A majority of the siRNAs had no significant effect on sphingomyelin trafficking to the chlamydial inclusion. Twenty-six gene targets decreased sphingomyelin trafficking to the inclusion by >20%; they can be categorized, based upon known gene function, into 4 classes. There were 16 gene targets that increased sphingomyelin retention in infected cells by >20%; they can be grouped in the same four categories. Components of the 4 categories that decreased sphingomyelin trafficking were as follows: GTP/actin, Arf1, Arfip2, Arpc3, Ddef2, Dnm2, Mgc9726, and Was; clathrin, AP2b1, Cltc, Epn3, Eps15, Eps15l1, and Picalm; kinase, Atm, Cbl, Cblb, Cib1, Fyn, Map4K2, and Pik3c2g; vesicular trafficking proteins, Bin1, Eea1, Synj2, Vamp2, VapA, and VapB. Components of the 4 categories that increased sphingomyelin retention were as follows: GTP/actin, Arpc5, CFL1, Rab3A, Rab3B, Rab3D, Rab7L1, RhoA, and Wasf2; clathrin, Dab2 and Itsn2; kinase, Cib2, Mapk8Ip2, Pak1, and TNIK; vesicular trafficking proteins, Syt1 and Syt2.
Analysis of Fyn knockdown.
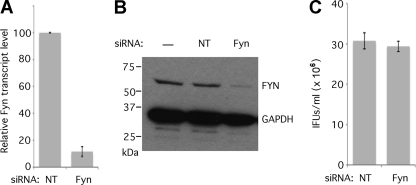
One of the targets that produced a significant decrease in sphingolipid retention, Fyn, is a member of the Src family of nonreceptor membrane-associated tyrosine kinases. The results of the initial screening were validated by confirming that knockdown of Fyn caused a specific defect in sphingomyelin trafficking to the inclusion rather than a decrease in sphingomyelin synthesis or trafficking unrelated to chlamydial infection. To validate a role for Fyn in sphingomyelin trafficking to the chlamydial inclusion, we examined first whether Fyn knockdown induces a general effect on sphingomyelin synthesis and trafficking, independent of chlamydial infection. Cells were treated with nontargeting or Fyn siRNA for 48 h, and lysates were collected for confirmation of efficient knockdown by examining Fyn transcript (Fig. 3 A) and protein levels (Fig. 3B). No significant difference in inclusion-forming unit (IFU) production was observed in Fyn knockdown cells (Fig. 3C), confirming that the decrease in sphingomyelin retention is due to inhibition of a Fyn-dependent trafficking pathway rather than a reduction in numbers of chlamydiae. Depletion of Fyn does not block the acquisition of sphingomyelin by chlamydiae sufficiently to inhibit replication.
Fig. 3.
Characterization of Fyn knockdown cells. (A) Transcript levels of Fyn mRNA (normalized to the amount of GAPDH mRNA) were quantified at 48 h posttransfection using the Quantigene2.0 system. The means of three replicate experiments are shown (error bars indicate standard errors of the means [SEM]). The difference was significant (P < 0.0001). (B) Total cellular protein was extracted and separated by SDS-PAGE, transferred by Western blotting, and probed with anti-Fyn and anti-GAPDH (loading control) antibodies. There is a significant reduction is Fyn protein levels after siRNA knockdown. NT, nontargeting siRNA. (C) Cultures infected at 48 h after knockdown by nontargeting or Fyn siRNA were lysed, and infectious progeny were replated for inclusion forming unit (IFU) determination. IFUs per field were counted for at least 20 fields, and IFUs/ml were calculated and plotted (n = 3; error bars indicate SEM). The difference was not significant.
Fyn knockdown does not affect sphingomyelin synthesis or trafficking in uninfected cells.
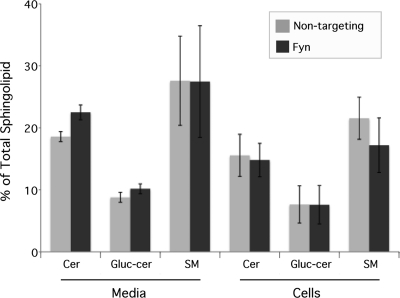
Cells were treated with nontargeting or Fyn siRNA for 48 h as described above and incubated with C6-NBD-ceramide. Fluorescent sphingolipids in the cells and media were collected, extracted, and separated by thin-layer chromatography. Densitometry of the resulting lipid bands revealed that siRNA knockdown of Fyn does not affect the synthesis of glucosylceramide and sphingomyelin from ceramide (Fig. 4). Additionally, the trafficking of sphingolipids is unaltered, as the relative percentages of each sphingolipid in the media and cellular fraction are unchanged. Thus, in the absence of chlamydial infection, sphingomyelin synthesis and trafficking were unaffected by a deficiency in Fyn, indicating that the decrease in sphingomyelin retention by chlamydiae is due to a specific defect in trafficking sphingomyelin to the inclusion.
Fig. 4.
Sphingolipid synthesis and trafficking are unaffected by Fyn knockdown in uninfected cells. HeLa cells were treated with Fyn or nontargeting siRNA. At 48 h posttransfection cells were labeled with C6-NBD-ceramide. After back-exchange, the lipids from the media and cells were extracted and separated by TLC. Relative amounts of sphingolipid species were plotted as percentages of the total fluorescent lipid content from each sample (n = 3; error bars indicate SEM). Differences were not significant. Cer, ceramide; Gluc-cer, glucosylceramide; SM, sphingomyelin.
Fyn knockdown decreases sphingomyelin retention by chlamydial inclusions.
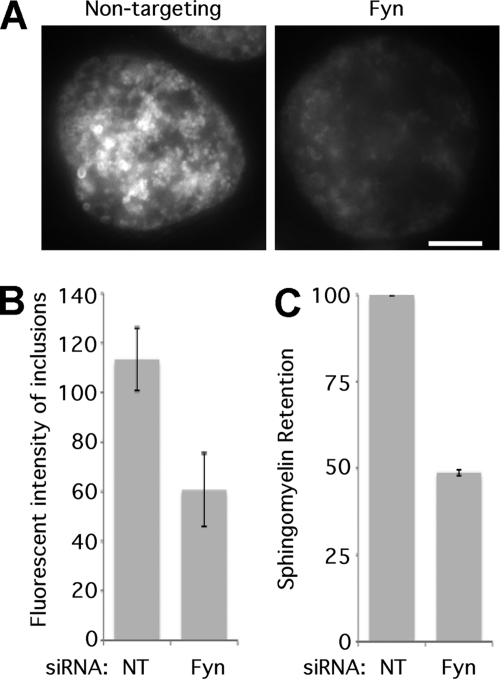
The decrease in sphingomyelin retention by chlamydial inclusions was quantified by two independent means. C6-NBD-sphingolipid retention by chlamydial inclusions was determined by using a photometric assay to measure the fluorescence associated with individual chlamydial inclusions. Inclusions in Fyn knockdown cells contained 47% less fluorescence than those in nontargeting-siRNA-treated cells (Fig. 5 A). Lipid extraction and thin-layer chromatography showed that EBs from infected Fyn knockdown cells retained 51% less fluorescent sphingomyelin than those from infected control knockdown cells (Fig. 5B and C). Analysis of total fluorescent sphingolipid content of infected knockdown and control cells indicated no significant difference in NBD-ceramide uptake by infected knockdown and control cells (data not shown). These results are consistent with the decrease originally observed in the siRNA screen.
Fig. 5.
Fyn knockdown decreases sphingomyelin trafficking to the chlamydial inclusion. (A) HeLa cells were transfected with nontargeting or Fyn siRNA and then infected with C. trachomatis L2. Cells were labeled with C6-NBD-ceramide and, after 8 h of back-exchange, were examined by fluorescence microscopy. Bar = 5 μm. The fluorescence associated with at least 20 individual inclusions was measured by microphotometry. (B) The average relative fluorescence is plotted in lux (n = 3; error bars indicate SEM). The difference was significant (P < 0.01). (C) HeLa cells were transfected with nontargeting or Fyn siRNA and then infected with C. trachomatis L2. Cells were labeled with C6-NBD-ceramide, and, after 8 h of back-exchange, lipids were extracted and the amount of sphingomyelin was determined by TLC and densitometry. Sphingomyelin in nontargeting siRNA treated cells was defined as 100%. (n = 3; error bars indicate SEM). The difference was significant (P < 0.0001).
Src family kinase inhibitors decrease sphingomyelin retention by chlamydial inclusions.
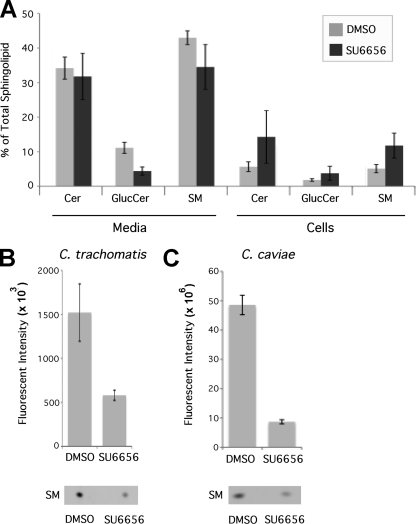
Because Fyn deficiency specifically reduced sphingomyelin trafficking to the chlamydial inclusion without disturbing normal cellular sphingolipid synthesis or trafficking, we investigated the effects of a broader inhibition of Src family kinases on sphingomyelin acquisition by chlamydiae. Chemical inhibition of Src family kinases has been shown to decrease trafficking of sphingomyelin to the plasma membrane, resulting in an increase in cellular retention of sphingomyelin (2). We first analyzed the effect of the Src family kinase inhibitor SU6656 on sphingolipid synthesis and trafficking in the absence of chlamydial infection. Monolayers were treated with SU6656 and labeled with C6-NBD-ceramide, the cultures subjected to back-exchange for 3 h, and sphingolipids in the cellular and media fractions were analyzed by lipid extraction, TLC separation, and densitometry. Inhibitor treatment increased cellular retention of each sphingolipid species, with a corresponding decrease in the sphingolipids exported to the plasma membrane and extracted into the media (Fig. 6 A).
Fig. 6.
Src family kinase inhibitors decrease sphingomyelin trafficking to the chlamydial inclusion. Uninfected (A) or infected (B and C) monolayers were treated with the Src family kinase inhibitor SU6656 or DMSO and labeled with C6-NBD-ceramide. (A) Lipids from the cellular and medium fractions of uninfected HeLa 229 monolayers were extracted and separated by TLC, and lipid fractions were analyzed by quantitative densitometry. The distribution of each species of sphingolipid was determined and plotted as the percentage of total sphingolipid (n = 3; error bars indicate SEM). Only glucosylceramide in the media produced significant differences (P < 0.05). (B and C) Cultures infected with C. trachomatis (B) or C. caviae (C) were treated with SU6656 or DMSO and labeled with C6-NBD-ceramide. After a 3-h back-exchange, EBs were purified and lipids were extracted and separated by TLC for analysis as above. The amount of sphingomyelin incorporated into EBs was normalized based upon quantitative immunoblotting for EB protein (MOMP) and plotted (n = 3; error bars indicate SEM). Differences were significant (P < 0.05).
Chlamydia spp. display unique requirements for Src family kinases throughout their developmental cycles (16, 27, 34, 35). C. trachomatis replication is severely restricted by the absence or inhibition of Src family kinases, whereas C. caviae displays no growth restriction; in fact, its growth appears to be enhanced by depletion of Src family kinases (35). To determine whether the distinct requirements of chlamydial species might be linked to the role of Src family kinases in sphingomyelin trafficking, C. trachomatis- and C. caviae-infected monolayers were treated at 18 h postinfection with SU6566 and labeled with C6-NBD-ceramide. After 3 h of back-exchange, the EBs were purified, lipids were extracted and separated by TLC, and the amount of fluorescent sphingomyelin in each sample was normalized by Western blotting for chlamydial major outer membrane protein (MOMP). Inhibitor treatment reduced the amount of sphingomyelin incorporated by EBs by about 60% for C. trachomatis L2 and about 80% for C. caviae (Fig. 6B and C), suggesting that this kinase-dependent pathway for sphingomyelin acquisition is conserved between C. trachomatis and C. caviae. Thus, although C. trachomatis and C. caviae exhibit different growth effects in the absence of Src family kinases (35), sphingomyelin trafficking appears to display similar requirements for Src family kinases in the two species. Because sphingomyelin trafficking to the inclusion is initiated early in infection, we confirmed a requirement for Fyn early in development by treatment with SU6566 at 1 h postinfection and labeling with C6-NBD-ceramide. Src family kinase inhibition blocked sphingolipid acquisition by chlamydiae even at early time points (data not shown).
DISCUSSION
Chlamydiae intercept sphingomyelin- and cholesterol-containing vesicles en route from the Golgi apparatus to the plasma membrane (12, 21, 22). Many of the interactions of the mature chlamydial inclusion are dependent upon chlamydial modification of the inclusion membrane (17). These interactions are thought to be controlled by the insertion of a family of inclusion membrane proteins (Incs) that are exposed on the cytosolic face of the inclusion membrane (4, 17, 50). Chlamydiae thus modify the inclusion membrane to intersect an exocytic pathway from which they acquire sphingomyelin that is incorporated into the bacterial cell wall (53). Sphingomyelin appears essential for chlamydial development, as inhibition of sphingolipid synthesis has a detrimental impact on chlamydial development (48, 62). Chlamydiae may have evolved various, possibly redundant, means to acquire sphingomyelin; however, the chlamydial and host proteins involved remain poorly understood. Here we have employed an siRNA library to deplete genes involved in membrane trafficking in a screen of human cells for defects in sphingomyelin trafficking to the C. trachomatis inclusion. One of these, the Src family kinase Fyn, was analyzed in greater detail by a variety of quantitative techniques to validate the efficacy of the screen and identify a requirement for Src family kinases in sphingomyelin trafficking to the chlamydial inclusion.
We have recently described a role for Fyn in dynein-dependent trafficking of the chlamydial inclusion to the microtubule organizing center (MTOC) (35, 36). Activated Fyn and other Src family kinases are recruited to localized structures, or microdomains, on the C. trachomatis inclusion membrane. These microdomains also are enriched in cholesterol and four inclusion membrane proteins, IncB, Inc101, Inc222, and Inc850. Among other possible functions, these microdomains appear to have a role in the linkage to dynein to promote the microtubule-dependent trafficking to and positioning of the C. trachomatis inclusion at the MTOC. The unique requirements of different chlamydial species for Src family kinases suggest that there are multiple roles for Src family kinases in chlamydial development. In C. trachomatis, the requirement for Src family kinases in dynein-dependent transport to the MTOC is readily distinguished from a severe growth defect in the absence of Src family kinase activity (35). Interruption of dynein-dependent trafficking by disruption of microtubules with nocodazole leads to inappropriate positioning of the C. trachomatis inclusion but does not inhibit replication. Conversely, inhibition of Src family kinase activity, even after the nascent inclusions have been allowed to traffic to the MTOC, inhibits C. trachomatis growth. These requirements are not observed in C. caviae, which does not localize to the MTOC and actually replicates to a higher titer in the absence of Src family kinases. Because transport of sphingomyelin and cholesterol to the chlamydial inclusion is dependent upon microtubules, it is possible that the effects of Fyn depletion on sphingomyelin trafficking observed here may be related to unique requirements for plus-end-directed microtubule motors (kinesin) and minus-end-directed motors (dynein) (35).
The mechanisms underlying the differential requirement(s) of chlamydial species for Src family kinases are unclear. All species of chlamydiae acquire sphingomyelin from the host cell (22, 49, 65) and are believed to have a growth requirement for sphingomyelin (48, 62). A role for Fyn in the acquisition of sphingomyelin by C. trachomatis as well as by C. caviae was established and suggests that similar mechanisms of sphingomyelin acquisition are used by both species. The different requirements for growth thus appear to be unrelated to the ability to acquire sphingomyelin. Although Fyn depletion reduced sphingomyelin trafficking to the inclusion, disruption of Fyn alone was insufficient to cause a reduction in infectious progeny formation as previously shown (35).
Typically, disruption of sphingomyelin trafficking to the inclusion does not completely abrogate sphingomyelin acquisition by chlamydiae, suggesting that alternative routes, such as multivesicular bodies or nonvesicular transport, may provide functional redundancy for an essential lipid. Inhibition of chlamydial multiplication is more dramatic in cells where sphingomyelin synthesis has been inhibited through pharmacological means (48, 62) or in temperature-sensitive cell lines conditionally unable to synthesize sphingomyelin (62), although deleterious effects on the host cell have not been completely ruled out. Serine/threonine kinases have also been implicated in sphingomyelin transport to the chlamydial inclusion based upon inhibition by rottlerin (54), an inhibitor of protein kinase C δ (PKC δ). The precise mechanisms of rottlerin inhibition remain unclear, as rottlerin has been reported to act upon multiple cellular targets (55).
In addition to Fyn, several of the targets identified in this screen have been noted in previous RNA interference screens in Drosophila melanogaster cells for host proteins involved in chlamydial replication or otherwise implicated in chlamydial pathogenesis. These include RhoA (30, 60), VapA and VapB (15), Arf1 (38), phophatidylinositol-3-phosphokinase (PI3K) (63), Bin1 (amphiphysin II) (20), and dynamin (8, 26). Recently, the resident endoplasmic reticulum (ER) proteins VapA and VapB have been identified in proximity to the inclusion membrane, suggesting recruitment of ER. Furthermore, depletion of VapA and VapB was shown to depress infectious-progeny formation (15). It has also been shown that depletion of VapA and VapB reduces levels of phophatidylinositol-4-phosphate and sphingomyelin in Golgi membranes and substantially inhibits Golgi membrane-mediated transport (42); this could explain the decrease in sphingomyelin acquisition by the inclusion in Vap-depleted cells observed here. The dynamin family of GTPase proteins is involved in the late stages of membrane vesicle fission, including clathrin-coated vesicles, and is known to function at both plasma and Golgi membranes. Knockdown of dynamin 2 produced an ∼50% decrease in sphingomyelin acquisition. Previous studies have shown that expression of a dominant negative dynamin mutant disrupts chlamydial development (8). Interestingly, Src family kinase phosphorylation of dynamin 2 leads to Golgi fragmentation (64), and inhibition of Golgi fragmentation has been shown to inhibit sphingomyelin trafficking to the inclusion and chlamydial development (25). Given the role of dynamin in vesicular trafficking, it may not be surprising that it has a prominent effect on sphingomyelin trafficking to the inclusion. Bin1 is thought to play a role in vesicular trafficking by affecting membrane curvature (59), and it has been proposed that Bin1 binds other families of proteins identified in this screen, including synaptojanin, dynamin, PI3K, and clathrin (14, 19, 46, 47). Bin1 is recruited to the Chlamydia pneumoniae inclusion and is essential for survival in macrophages (20). Knockdown of different clathrin-related proteins caused either increases or decreases in sphingomyelin acquisition by the inclusion. Clathrin-coated vesicles are known to be involved in sorting cargo in the trans-Golgi network and have been shown to be involved in sphingomyelin trafficking from the Golgi apparatus (44). Inhibition of clathrin-coated vesicles by dominant negative mutants of dynamin 1 has been shown to inhibit chlamydial development and is thought to be due to disruption of vesicular traffic (8). However, disruption of endosomal clathrin by dominant negative mutants of Eps15 had no effect on chlamydial growth (8). Eps15 is also involved in invagination and cargo sorting during MVB biogenesis (3, 45) and therefore may exert its effect by altering sphingomyelin trafficking from MVBs. Other host factors identified in this screen had not been previously shown to be involved in chlamydial pathogenesis, but their known functions lead to viable hypotheses regarding sphingomyelin acquisition by the inclusion. DDEF2 (Pap) is thought to regulate Arf1-dependent vesicle trafficking from Golgi membranes (1), and a deficiency in either protein decreases sphingomyelin trafficking to the inclusion.
The Rab family of guanine nucleotide binding proteins are important regulators of vesicle fusion (56). The finding that multiple Rab and VAMP proteins were identified in this screen is interesting given their roles in regulation of vesicle trafficking and fusion. A subset of Rabs are recruited to inclusions of all chlamydial species (Rab1, Rab4, and Rab11), while others are recruited in a species-specific manner (Rab6 and Rab10) (9, 51). Requirements for Rab1, Rab4, Rab6, Rab11, and Rab14 in chlamydial development have been identified (11, 16, 31, 41). Rab6, Rab11, and Rab14 have been implicated in sphingomyelin delivery to the inclusion (11, 31) although the recruitment of Rab14 (11) and the fragmentation of the Golgi apparatus following Rab6 and Rab11 knockdown occur much later than the observed initiation of sphingomyelin trafficking to the inclusion (25, 31). Knockdown of two of the Rab3 family members (Rab3B and Rab3D) which are involved in exocytosis of secretory vesicles (28, 56) and have not been previously implicated in chlamydial pathogenesis increased sphingomyelin retention by >20%. Knockdown of a third Rab3 family member, Rab3A, increased retention by 19%.
Depletion of some host factors caused an apparent increase in sphingomyelin retention by chlamydiae. A number of targets that increased sphingomyelin trafficking to the inclusion (e.g., RhoA, Cfl1, and Dab2) are involved in actin dynamics. The chlamydial inclusion is surrounded by an actin scaffold, which is compromised in RhoA knockdown cells, affecting inclusion membrane stability (30). One possibility might be that perturbation of the actin scaffolding surrounding the inclusion and the resulting decrease in inclusion membrane stability may facilitate acquisition of sphingomyelin-containing vesicles. Depletion of either synaptotagmin 1 or 2 resulted in increased sphingomyelin trafficking to the inclusion. Synaptotagmins are calcium binding proteins that are best known for their role in vesicular trafficking at synapses but are also expressed in nonsynaptic cells (40, 43). Synaptotagmins have extensive roles in cellular signaling pathways, including regulating exocytic vesicles from the Golgi apparatus (57) and binding the MVB marker CD63 (18); both pathways are implicated in sphingomyelin trafficking to the inclusion (5, 22, 48). Pak1 phosphorylates a light chain subunit of dynein to regulate cargo binding (61), and Pak1 activity is regulated by sphingolipid binding (7). Although sphingomyelin trafficking to the inclusion involves microtubules, it is not yet known which motor proteins are used. Directional movement along microtubules may involve opposing motors (66); thus, it is plausible that downregulating dynein motor activity could favor lipid trafficking to the inclusion.
Sphingomyelin trafficking to the chlamydial inclusion is one of the defining properties of the genus, yet the molecular mechanisms controlling this interaction are largely unknown. A number of regulators of host vesicle trafficking that disrupted chlamydial sphingomyelin acquisition were identified. Discerning how these multiple factors function together to promote lipid trafficking to the chlamydial inclusion will remain a significant challenge.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the NIAID/NIH.
We thank Janet Sager for excellent technical assistance and E. Moore, L. Bauler, E. Lutter, and A. Omsland for helpful discussion and critical review of the manuscript.
Footnotes
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Andreev J., et al. 1999. Identification of a new Pyk2 target protein with Arf-GAP activity. Mol. Cell. Biol. 19:2338–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baba A., et al. 2009. Fyn tyrosine kinase regulates the surface expression of glycosylphosphatidylinositol-linked ephrin via the modulation of sphingomyelin metabolism. J. Biol. Chem. 284:9206–9214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bache K. G., Raiborg C., Mehlum A., Stenmark H. 2003. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278:12513–12521 [DOI] [PubMed] [Google Scholar]
- 4. Bannantine J. P., Griffiths R. S., Viratyosin W., Brown W. J., Rockey D. D. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 2:35–47 [DOI] [PubMed] [Google Scholar]
- 5. Beatty W. L. 2006. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J. Cell Sci. 119:350–359 [DOI] [PubMed] [Google Scholar]
- 6. Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 7. Bokoch G. M., et al. 1998. A GTPase-independent mechanism of p21-activated kinase activation. Regulation by sphingosine and other biologically active lipids. J. Biol. Chem. 273:8137–8144 [DOI] [PubMed] [Google Scholar]
- 8. Boleti H., Benmerah A., Ojcius D. M., Cerf-Bensussan N., Dautry-Varsat A. 1999. Chlamydia infection of epithelial cells expressing dynamin and Eps15 mutants: clathrin-independent entry into cells and dynamin-dependent productive growth. J. Cell Sci. 112:1487–1496 [DOI] [PubMed] [Google Scholar]
- 9. Brumell J. H., Scidmore M. A. 2007. Manipulation of rab GTPase function by intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 71:636–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caldwell H. D., Kromhout J., Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Capmany A., Damiani M. T. 2010. Chlamydia trachomatis intercepts Golgi-derived sphingolipids through a Rab14-mediated transport required for bacterial development and replication. PLoS One 5:e14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carabeo R. A., Mead D. J., Hackstadt T. 2003. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. U. S. A. 100:6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cocchiaro J., Kumar Y., Fischer E. R., Hackstadt T., Valdivia R. H. 2008. Cytoplasmic lipid droplets are translocatedd into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U. S. A. 105:9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Heuvel E., et al. 1997. Identification of the major synaptojanin-binding proteins in brain. J. Biol. Chem. 272:8710–8716 [DOI] [PubMed] [Google Scholar]
- 15. Derre I., Swiss R., Agaisse H. 2011. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 7:e1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elwell C. A., Ceesay A., Kim J. H., Kalman D., Engel J. N. 2008. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 4:e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fields K. A., Hackstadt T. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18:221–245 [DOI] [PubMed] [Google Scholar]
- 18. Flannery A. R., Czibener C., Andrews N. W. Palmitoylation-dependent association with CD63 targets the Ca2+ sensor synaptotagmin VII to lysosomes. J. Cell Biol. 191:599–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gold E. S., et al. 2000. Amphiphysin IIm, a novel amphiphysin II isoform, is required for macrophage phagocytosis. Immunity 12:285–292 [DOI] [PubMed] [Google Scholar]
- 20. Gold E. S., et al. 2004. Amphiphysin IIm is required for survival of Chlamydia pneumoniae in macrophages. J. Exp. Med. 200:581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hackstadt T., Rockey D. D., Heinzen R. A., Scidmore M. A. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964–977 [PMC free article] [PubMed] [Google Scholar]
- 22. Hackstadt T., Scidmore M. A., Rockey D. D. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U. S. A. 92:4877–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hatch G. M., McClarty G. 1998. Cardiolipin remodeling in eukaryotic cells infected with Chlamydia trachomatis is linked to elevated mitochondrial metabolism. Biochem. Biophys. Res. Commun. 243:356–360 [DOI] [PubMed] [Google Scholar]
- 24. Heinzen R. A., Scidmore M. A., Rockey D. D., Hackstadt T. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 64:796–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heuer D., et al. 2009. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature 457:731–735 [DOI] [PubMed] [Google Scholar]
- 26. Hybiske K., Stephens R. S. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104:11430–11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jewett T. J., Dooley C. A., Mead D. J., Hackstadt T. 2008. Chlamydia trachomatis tarp is phosphorylated by src family tyrosine kinases. Biochem. Biophys. Res. Commun. 371:339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johannes L., et al. 1996. Evidence for a functional link between Rab3 and the SNARE complex. J. Cell Sci. 109:2875–2884 [DOI] [PubMed] [Google Scholar]
- 29. Kumar Y., Cocchiaro J., Valdivia R. H. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16:1646–1651 [DOI] [PubMed] [Google Scholar]
- 30. Kumar Y., Valdivia R. H. 2008. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe 4:159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lipinski A. R., et al. 2009. Rab6 and Rab11 regulate Chlamydia trachomatis development and Golgin-84-dependent Golgi fragmentation. PLoS Pathog. 5:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lipsky N. G., Pagano R. E. 1985. Intracellular translocation of fluorescent sphingolipids in cultured fibroblasts: endogenously synthesized sphingomyelin and glucocerebroside analogues pass through the Golgi apparatus en route to the plasma membrane. J. Cell Biol. 100:27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lipsky N. G., Pagano R. E. 1985. A vital stain for the Golgi apparatus. Science 228:745–747 [DOI] [PubMed] [Google Scholar]
- 34. Mehlitz A., Banhart S., Hess S., Selbach M., Meyer T. F. 2008. Complex kinase requirements for Chlamydia trachomatis Tarp phosphorylation. FEMS Microbiol. Lett. 289:233–240 [DOI] [PubMed] [Google Scholar]
- 35. Mital J., Hackstadt T. 2011. Diverse requirements for Src-family tyrosine kinases distinguish chlamydial species. mBio 2:e00031–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mital J., Miller N. J., Fischer E. R., Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell. Microbiol. 12:1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moore E. R., Fischer E. R., Mead D. J., Hackstadt T. 2008. The chlamydial inclusion preferentially intercepts basolaterally directed sphingomyelin-containing exocytic vacuoles. Traffic 9:2130–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moorhead A. M., Jung J. Y., Smirnov A., Kaufer S., Scidmore M. A. 2010. Multiple host proteins that function in phosphatidylinositol-4-phosphate metabolism are recruited to the chlamydial inclusion. Infect. Immun. 78:1990–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moulder J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Musch M. W., et al. 2007. Synaptotagmin I binds intestinal epithelial NHE3 and mediates cAMP- and Ca2+-induced endocytosis by recruitment of AP2 and clathrin. Am. J. Physiol. Gastrointest. Liver Physiol. 292:G1549–G1558 [DOI] [PubMed] [Google Scholar]
- 41. Ouellette S. P., Carabeo R. A. 2010. A functional slow recycling pathway of transferrin is required for growth of chlamydia. Front. Microbiol. 1:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. 2008. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell 19:3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perin M. S., Fried V. A., Stone D. K., Xie X. S., Sudhof T. C. 1991. Structure of the 116-kDa polypeptide of the clathrin-coated vesicle/synaptic vesicle proton pump. J. Biol. Chem. 266:3877–3881 [PubMed] [Google Scholar]
- 44. Puri V., et al. 2001. Clathrin-dependent and -independent internalization of plasma membrane sphingolipids initiates two Golgi targeting pathways. J. Cell Biol. 154:535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raiborg C., et al. 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4:394–398 [DOI] [PubMed] [Google Scholar]
- 46. Ramjaun A. R., Micheva K. D., Bouchelet I., McPherson P. S. 1997. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J. Biol. Chem. 272:16700–16706 [DOI] [PubMed] [Google Scholar]
- 47. Ren G., Vajjhala P., Lee J. S., Winsor B., Munn A. L. 2006. The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiol. Mol. Biol. Rev. 70:37–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Robertson D. K., Gu L., Rowe R. K., Beatty W. L. 2009. Inclusion biogenesis and reactivation of persistant Chlamydia trachomatis requires host cell sphingolipid biosynthesis. PLoS Pathog. 5:e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rockey D. D., Fischer E. R., Hackstadt T. 1996. Temporal analysis of the developing Chlamydia psittaci inclusion by use of fluorescence and electron microscopy. Infect. Immun. 64:4269–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rockey D. D., Scidmore M. A., Bannantine J. P., Brown W. J. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333–340 [DOI] [PubMed] [Google Scholar]
- 51. Rzomp K. A., Scholtes L. D., Briggs B. J., Whittaker G. R., Scidmore M. A. 2003. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 71:5855–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schachter J. 1999. Infection and disease epidemiology, p. 139–169In Stephens R. S.(ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC [Google Scholar]
- 53. Scidmore M. A., Rockey D. D., Fischer E. R., Heinzen R. A., Hackstadt T. 1996. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect. Immun. 64:5366–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shivshankar P., Lei L., Wang J., Zhong G. 2008. Rottlerin inhibits chlamydial intracellular growth and blocks chlamydial acquisition of sphingolipids from host cells. Appl. Environ. Microbiol. 74:1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Soltoff S. P. 2007. Rottlerin: an inappropriate and ineffective inhibitor of PKCdelta. Trends Pharmacol. Sci. 28:453–458 [DOI] [PubMed] [Google Scholar]
- 56. Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 [DOI] [PubMed] [Google Scholar]
- 57. Stojilkovic S. S. 2005. Ca2+-regulated exocytosis and SNARE function. Trends Endocrinol. Metab. 16:81–83 [DOI] [PubMed] [Google Scholar]
- 58. Su H., et al. 2004. Activation of RAf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J. Biol. Chem. 279:9409–9416 [DOI] [PubMed] [Google Scholar]
- 59. Takei K., Slepnev V. I., Haucke V., De Camilli P. 1999. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1:33–39 [DOI] [PubMed] [Google Scholar]
- 60. Thalmann J., et al. 2010. Actin re-organization induced by Chlamydia trachomatis serovar D—evidence for a critical role of the effector protein CT166 targeting Rac. PLoS One 5:e9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vadlamudi R. K., et al. 2004. Dynein light chain 1, a p21-activated kinase 1-interacting substrate, promotes cancerous phenotypes. Cancer Cell 5:575–585 [DOI] [PubMed] [Google Scholar]
- 62. van Ooij C., et al. 2000. Host cell-derived sphingolipids are required for the intracellular growth of Chlamydia trachomatis. Cell. Microbiol. 2:627–637 [DOI] [PubMed] [Google Scholar]
- 63. Verbeke P., et al. 2006. Recruitment of BAD by the Chlamydia trachomatis vacuole correlates with host-cell survival. PLoS Pathog. 2:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weller S. G., et al. 2010. Src kinase regulates the integrity and function of the Golgi apparatus via activation of dynamin 2. Proc. Natl. Acad. Sci. U. S. A. 107:5863–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wolf K., Hackstadt T. 2001. Sphingomyelin trafficking in Chlamydia pneumoniae-infected cells. Cell. Microbiol. 3:145–152 [DOI] [PubMed] [Google Scholar]
- 66. Wubbolts R., et al. 1999. Opposing motor activities of dynein and kinesin determine retention and transport of MHC class II-containing compartments. J. Cell Sci. 112(Pt. 6):785–795 [DOI] [PubMed] [Google Scholar]