Abstract
Recognition of microbial patterns by host receptors is the first step in a multistep sequence leading to neutrophil-dependent host resistance. Although the role of membrane-bound sensors in bacterial recognition has been examined in detail, the importance of cytosolic sensors in the lungs is largely unexplored. In this context, there is a major lack of understanding related to the downstream signaling mediators, such as cells and/or molecules, during acute extracellular Gram-negative bacterial pneumonia. In order to determine the role of NOD-like receptors (NLRs), we used an experimental Escherichia coli infection model using mice deficient in the gene coding for the NLR adaptor, receptor-interacting protein 2 (RIP2). RIP2−/− mice with E. coli infection displayed higher bacterial burden and reduced neutrophil recruitment and tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), macrophage inflammatory protein 2 (MIP-2), and CXCL5/LIX expression, along with attenuated histopathological changes in the lungs. Decreased IL-17A levels were observed, along with lower numbers of IL-17A-producing T cells, in RIP2−/− mice after infection. RIP2−/− mice also show reduced IL-6 and IL-23 levels in the lungs, along with decreased activation of STAT3 after infection. Furthermore, activation of NF-κB and mitogen-activated protein kinases (MAPKs) and expression of intercellular adhesion molecule 1 (ICAM-1) and vascular cell adhesion molecule 1 (VCAM-1) in the lungs of infected RIP2−/− mice were attenuated following infection. Although neutrophil mobilization to the blood was impaired in RIP2−/− mice following infection, the expression of CD62P, CD11a/18, CD11b, and CXCR2 on blood and lung neutrophils was not altered between infected wild-type (WT) and RIP2−/− mice. Thus, RIP2 contributes to neutrophil-dependent host defense against an extracellular Gram-negative pathogen via (i) IL-17A regulation and (ii) neutrophil mobilization to the blood.
INTRODUCTION
Lower respiratory tract infections cause more deaths than HIV, ischemic heart disease, and diarrheal diseases (7, 40). Gram-negative bacterial pathogens are a common cause of nosocomial infections (48). Host defense against microbes is composed of two evolutionarily distinct arms, the innate and adaptive immune systems. Pattern recognition receptors (PRRs) are critical to induce effective innate and/or adaptive immune responses (2). PRRs, such as Toll-like receptors (TLRs) and NOD-like receptors (NLRs), can recognize pathogen-associated molecular patterns (PAMPs) in microbes. This recognition triggers a cascade of events leading to the activation of transcription factors, production of chemokines/cytokines, upregulation of cell adhesion molecules, phagocytic cell infiltration, and subsequent clearance of the microbes. Neutrophils are the primary innate immune cells to migrate toward the site of bacterial infection to augment host defense via complex signaling cascades (1, 14). However, excessive neutrophil recruitment and activation can lead to eventual mortality associated with acute lung injury (ALI) or acute respiratory distress syndrome (ARDS). In this regard, investigation of molecular and cellular mechanisms involved in neutrophil trafficking to the lungs is essential to designing improved therapies.
The adaptor molecule receptor-interacting protein 2 (RIP2) is a caspase recruitment domain (CARD) containing serine/threonine kinase (4, 28). A recent report shows that RIP2 also has tyrosine kinase activity (56). Human RIP2 is a 61-kDa protein with 531 amino acids and is located on chromosome 8 (4). The murine homolog has 86% identity at the molecular level and 84% identity at the amino acid level and is located on chromosome 4 (4). RIP2 induces downstream signaling via the CARD (28). However, the role of RIP2 in TLR and NLR signaling is still debatable. For example, Park et al., have demonstrated that RIP2 contributes to innate immune mechanisms via both NOD1 and NOD2 signaling, but not through the TLR signaling (45). Other studies have revealed that RIP2 functions downstream of TLRs to mediate NF-κB and mitogen-activated protein kinase (MAPK) activation (4, 18). In this context, RIP2 has been shown to be recruited to TLR2, TLR3, and TLR4 but not TLR9 in order to activate NF-κB, p38 MAPK, Jun N-terminal protein kinase (JNK), and extracellular signal-regulated kinase (ERK) (28). Additional studies have shown that lipopolysaccharide (LPS)-mediated TLR4 signaling involves RIP2, although its role in the signaling is not dependent on its kinase activity (32). Studies with pneumococcus have revealed that NOD2 recognition of Streptococcus pneumoniae involves RIP2 signaling (43).
RIP2 plays an essential role in mucosal immunity to intracellular pulmonary pathogens, such as Listeria monocytogenes (12), Legionella pneumophila (3), Chlamydophila pneumoniae (53), and Mycobacterium tuberculosis (44). Another study, using intraperitoneal infection with Pseudomonas aeruginosa and Escherichia coli after LPS prestimulation, showed higher inflammation in the peritoneum through NOD1/NOD2 signaling cascades and induced lethality during Pseudomonas infection via bacterial dissemination (46). Recent studies show that peptidoglycan (PGN) obtained from Gram-negative bacteria can induce NOD1 signaling in lung epithelium, which protects mice after S. pneumoniae infection (33, 51). Despite these elegant studies, the role of RIP2 in acute extracellular Gram-negative bacterial pneumonia has not been explored. Although recent reports suggest that interleukin-17A (IL-17A) plays an essential role against Gram-negative pathogens (60), the importance of RIP2 in IL-17A production during acute bacterial pneumonia has not been explored. Therefore, our hypothesis is that RIP2 is critical for the host defense during Gram-negative pneumonia by regulating IL-17A production. Additional experimental data demonstrate that RIP2 controls neutrophil mobilization to the blood. Our findings provide new mechanistic insights into the importance of RIP2 in mediating lung inflammation and host defense against a Gram-negative bacterial pathogen, E. coli.
MATERIALS AND METHODS
Mice.
RIP2−/− mice were generated as described earlier (28). All gene-deficient mice used in our experiments are on the C57BL6 background after backcrossing with C57BL/6 mice at least 10 generations. Because of this, C57BL6 mice were used as the wild-type (WT) controls. Eight- to 10-wk-old female mice were used, and animal studies were approved by the Louisiana State University Animal Care and Use Committee.
Intratracheal (i.t.) instillation of bacteria.
Bacteria were prepared for mouse inoculation as described in our previous reports (25). E. coli (ATCC 25922) was grown in Trypticase soy broth (TSB) at 37°C overnight under constant agitation. Bacteria were harvested by centrifugation, washed two times in isotonic saline, and resuspended in saline at a concentration of 20 × 106 CFU/ml. Mice were anesthetized by intraperitoneal (i.p.) injection of a mixture containing 120 mg/kg ketamine and 16 mg/kg xylazine. The trachea was surgically exposed, and a total volume of 50 μl of saline containing 106 E. coli CFU/mouse was administered. In control experiments, 50 μl of saline was instilled into the trachea. The initial mouse inoculums were confirmed by plating serial 10-fold dilutions on MacConkey and tryptic soy agar (TSA) plates. For enumeration of bacterial CFU in the lungs, whole lungs were homogenized in 2 ml sterile saline for 30 s, and 20 μl of the resulting homogenates was plated by serial 10-fold dilutions on MacConkey and TSA plates. Bacterial colonies were counted after incubation at 37°C for 18 to 24 h.
Cell counts.
Bronchoalveolar lavage fluid (BALF) was collected to obtain airway cells for enumeration. The trachea was exposed, intubated, and instilled with phosphate-buffered saline (PBS) containing heparin and dextrose in 0.8-ml aliquots 4 times. Approximately 3 ml of BALF was retrieved per mouse. Cytospin samples were subsequently prepared from BALF cells and stained with Diff-Quick (Fisher). Total leukocytes in BALF were determined using a hemocytometer, whereas leukocyte subsets were examined by direct counting of stained slides based on cell nuclear morphology. Neutrophil recruitment to the lung parenchyma was assessed by myeloperoxidase (MPO) activity as described in our earlier reports (27).
Lung pathology.
The murine lungs were perfused from the right ventricle of the heart with 10 ml isotonic saline at 24 h postinfection. Lungs were removed and fixed in 4% phosphate-buffered formalin. Fixed tissue samples were processed in paraffin blocks, and fine sections (5 μm thick) were cut and stained with hematoxylin and eosin (H&E). Assessments of histopathology were performed in a blinded fashion by a veterinary pathologist according to the following scoring scale: 0, no inflammatory cells (macrophages or neutrophils) present in section; 1, <5% of the section is infiltrated by inflammatory cells; 2, 5 to 10% of the section is infiltrated by inflammatory cells; and 3, >10% of the section is infiltrated by inflammatory cells.
Cytokines and chemokines.
Cell-free BALF was used for the determination of tumor necrosis factor α (TNF-α) (eBioscience), IL-6 (eBioscience), macrophage inflammatory protein 2 (MIP-2) (R&D Systems), and LIX (R&D Systems) proteins by enzyme-linked immunosorbent assay (ELISA). For the determination of IL-17A and IL-23 (eBioscience), lung homogenates were used. The minimum detection limit is 2 pg/ml of cytokine or chemokine protein.
Western blotting.
Tissue proteins were extracted by homogenizing the lungs in a lysis buffer cocktail containing 0.1% Triton X-100 in PBS, complete protease and phosphatase inhibitor cocktail (Thermo Scientific), and 1 mM dithiothreitol (DTT), followed by centrifugation at maximum speed using a microcentrifuge at 4°C. The resulting supernatants were used for Western blotting. Whole-cell proteins were fractioned by 10 to 12% SDS-PAGE and transferred onto Immobilon-P (Millipore) polyvinylidene difluoride membranes. Membranes were blocked and probed with RIP2, phospho-RIP2 (Ser176), phospho-IKKα/β (Ser176/180), NF-κB, phospho-NF-κB (Ser536), Iκβα, ICAM-1, VCAM-1, phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), phospho-p38 MAPK (Thr180/Tyr182), phospho-MAPK/JNK (Thr183/Tyr185), STAT3, phospho-STAT3 (Ser727; Thr705), and acetyl-STAT3 (Lys685) (1:1,000 dilution). Horseradish peroxidase (HRP)-conjugated appropriate secondary antibodies were used, and the ECL enhanced chemiluminescence system was used to detect signals (Amersham). To demonstrate equal protein loading on gels, the blots were stripped and reprobed with antibody (Ab) specific for total p38 MAPK or GAPDH (glyceraldehyde-3-phosphate dehydrogenase). The intensity of immunoreactive bands was determined using a gel digitizing software (UN-SCAN-IT gel) from Silk Scientific, Inc.
NF-κB DNA binding.
Nuclear proteins were extracted from the lung tissue at 6 and 24 h post-E. coli administration, as described in our previous publications (9, 10). A total of 7.5 μg nuclear extract was mixed with binding buffer, added to the precoated plate, and incubated for 1 h at room temperature according to the manufacturer's protocol (TransAM ELISA kit). Excess Ab was removed with wash buffer, and plates were incubated with NF-κB/p65 antibody for 1 h. Plates were washed to remove excess Ab, and HRP-conjugated anti-rabbit IgG was added. Plates were incubated for 1 h and subsequently read at 450 nm after adding the color developing reagent.
In vitro experiments with BMDMs and BMDCs.
Marrow cells were isolated from bone marrow of RIP2−/−, and WT (control) mice and differentiated into macrophages for 7 days by adding macrophage colony-stimulating factor (M-CSF) and into dendritic cells (DCs) by adding IL-4 and granulocyte-macrophage colony-stimulating factor. A total of 106 cells/well was used for each group at each time point for infection with E. coli at a multiplicity of infection (MOI) of 0.1 and incubated at 37°C with slow agitation. At 10, 20, 30, 60, and 360 min, cell pellets and supernatant were collected and cell pellets of BMDMs were processed for Western blot analysis to determine the expression of RIP2 and activation of NF-κB and MAPKs. Supernatants from both bone marrow-derived macrophages (BMDMs) and bone marrow-derived DCs (BMDCs) were used for the determination of cytokines/chemokines.
Flow cytometry.
A total of 50 μl of whole blood or perfused lung digest of WT and RIP2 −/− mice challenged with E. coli or saline (control) was aliquoted into tubes and Fc blocked. A total of 4 μl of mouse conjugated anti-mouse Gr-1/Ly6G, P-selectin/CD62P, LFA-1 (CD11a/CD18), CD11b, and CXCR-2 (R&D Systems) antibodies was added to tubes. Samples were vortexed and incubated for 30 min at room temperature in the dark. Cells were washed by adding 2 ml of 1× PBS and centrifuged at 200 × g for 8 min. Red blood cells (RBCs) were lysed by adding 2 ml of NH4Cl lysing buffer to each sample tube, mixed well, and incubated at room temperature for 10 min. Then the samples were centrifuged immediately at 200 × g for 8 min, and the supernatant was removed. Cells were washed twice with PBS, fixed by adding 200 μl of cold 1% formaldehyde–PBS, and stored at 1 to 6°C for fluorescence-activated cell sorting (FACS) analysis.
Determination of IL-17A-producing cells using flow cytometry.
The procedure used to analyze IL-17A-producing cells was reported earlier (47, 50). Lung samples were obtained from both wild-type and RIP2−/− mice 6 h after E. coli infection (106 CFU/animal). They were minced, digested with collagenase, made into a single-cell suspension, and stimulated with phorbol myristate acetate (PMA; 50 ng/ml), ionomycin (750 ng), and GolgiStop (7 μl/10 ml) for 5 h. After stimulation, cells were surface stained with markers for IL-17A-producing T cells (γδ, NK, and CD4 cells). Following incubation, cells were washed, fixed, and permeabilized for intracellular staining. Permeabilized cells were subsequently incubated with IL-17A Ab for intracellular staining. Finally, cells were washed and resuspended for flow cytometry analysis.
Exogenous IL-17A (rIL-17) administration.
RIP2−/− mice were treated intratracheally (i.t.) with recombinant IL-17A (rIL-17A) (1 μg/animal) 1 h after E. coli infection (106 CFU/50 μl/animal), and the control mice were treated with an equal volume of PBS. At 24 h postinfection, BALF was collected and processed for cellular enumeration and the determination of CFU.
Statistical analysis.
Data are expressed as means ± standard error (SE). Data were analyzed by one-way or two-way analysis of variance (ANOVA) followed by Bonferroni's post hoc correction for multiple comparisons. Statistical calculations were performed using InStat software and GraphPad Prism 4.0 (GraphPad Software, Inc., San Diego, CA). Differences were considered statistically significant when P was <0.05 for comparisons between WT and knockout (KO) mice.
RESULTS
RIP2 expression and activation in the lungs are induced upon E. coli infection.
To investigate whether RIP2 expression is altered during E. coli pneumonia, WT (C57BL/6) mice were infected i.t. with E. coli at 106 CFU/animal, and lungs were removed at 6 and 24 h postinfection and processed for Western blotting (Fig. 1A). Notably, we observed increases in expression and activation of RIP2 upon infection compared to saline-challenged controls (Fig. 1A). Furthermore, we found enhanced RIP2 expression and activation in bone marrow-derived macrophages (BMDMs) following E. coli challenge (see Fig. 4A).
Fig. 1.
Importance of RIP2 in host defense during Gram-negative pneumonia. (A) RIP2 expression in C57BL/6 (WT) mice at 6 and 24 h post-E. coli infection. WT mice were infected i.t. with E. coli, and lung samples were processed for Western blotting using Abs against the activated or regular form of RIP2. This is a representative blot from 2 separate experiments. n = 3 mice in each group in each experiment; *, P < 0.05 between infected and saline-challenged mice. The letter “P” in a circle represents phospho-RIP2. (B) Impaired bacterial clearance in the lungs in RIP2−/− mice. Lung homogenates at 6 and 24 h after infection were used to enumerate the bacterial CFU (n = 5 or 6 mice/group/time point). Asterisks indicate significant differences between WT (C57BL/6) and RIP2−/− (KO) mice (P < 0.05). (C and D) Attenuated neutrophil recruitment to the lungs of RIP2−/− mice following E. coli infection. Mice were inoculated with E. coli (106 CFU/mouse), BALF was obtained at 6 and 24 h postinfection, and cell (total and differential) enumeration was performed to determine cellular recruitment to the lung (n = 5 or 6 mice/group/time point; P < 0.05). (E) Reduced MPO in the lungs of RIP2−/− mice after E. coli infection. Lung MPO was determined in lung homogenates (n = 4 or 5 mice/group/time point; P < 0.05). (F) Attenuated lung histology in RIP2−/− mice following E. coli inoculation. Mice were inoculated with E. coli (106 CFU/mouse), lungs were obtained at 24 h postinfection and stained with H&E, and inflammation in histological sections was scored by a veterinary pathologist as described in Materials and Methods. These are representative sections of 6 mice under each condition with identical results (magnification, ×200). (G to J) Impaired cytokine and chemokine responses in the airspaces following E. coli infection. BALF was collected from the lungs after i.t. instillation of E. coli (106 CFU/mouse) at designated time points. TNF-α (G), IL-6 (H), MIP-2 (I), and LIX (J) levels in BALF were quantified by sandwich ELISA. Asterisks indicate significant differences between WT and KO mice (P < 0.05; n = 5 to 7 mice/group/time point).
Fig. 4.
RIP2 expression and phosphorylation, NF-κB and MAPK activation, ICAM-1 expression, and IL-6 and IL-23 production in macrophages and dendritic cells. (A and B) Decreased RIP2 expression and phosphorylation in bone marrow-derived macrophages (BMDMs) upon E. coli infection. (C) Reduced activation of NF-κB and MAPKs in BMDMs obtained from RIP2−/− mice following infection with E. coli. Representative Western blots from 3 separate experiments/blots are shown. (D) Densitometric analysis of NF-κB and MAPK activation in BMDMs of WT and RIP2 −/− mice. Relative densities normalized against GAPDH are representative of 3 independent experiments/blots. (E and F) Attenuated IL-6 and IL-23 levels in culture supernatants obtained from BMDMs (E) and BMDCs (F) of and RIP2−/− mice following E. coli infection. Experiments were performed in triplicate wells. For the experiments shown in panels A to F, 5 or 6 mice/group were used; *, P < 0.05 compared with RIP2−/− (KO) mice.
RIP2 limits bacterial burden in the lungs.
To determine whether RIP2 is important for bacterial clearance in the lungs, RIP2−/− mice and WT controls were instilled i.t. with E. coli (106 CFU/mouse) and sacrificed at 6 and 24 h postinfection for bacterial culture. At 24 h after infection, the bacterial burden in the lungs was higher in RIP2−/− mice than that in WT controls (Fig. 1B). However, there was no bacterial dissemination observed in either RIP2−/− or WT mice with this bacterial dose (data not shown).
RIP2 contributes to neutrophil trafficking to the lungs.
Since there was attenuated bacterial clearance in RIP2−/− mice following E. coli infection, we determined whether RIP2 deficiency affects neutrophil transmigration to the lungs following infection. Mice were infected with E. coli, and BALF was collected at designated time points. At 6 and 24 h postinfection, there was a reduction in total leukocyte and neutrophil counts in the airspaces of RIP2−/− mice (Fig. 1C and D). Consistent with this, MPO activity in lung homogenates of RIP2−/− mice was reduced at 6 and 24 h postinfection (Fig. 1E). We next examined whether RIP2 deficiency affects pathological consequences in the lungs during Gram-negative bacterial pneumonia. RIP2−/− and WT mice were infected with E. coli i.t. at a dose of 106/mouse, and the lungs were processed for histopathological studies 24 h after infection. Unlike WT mice, RIP2−/− mice showed reduced inflammation, as evidenced by attenuated inflammatory cell accumulation and alveolar edema compared to WT controls (Fig. 1F).
RIP2−/− mice show decreased expression of proinflammatory cytokines and chemokines in the lungs.
Neutrophil recruitment to the infected site is mediated by cytokines and chemokines. Therefore, the next step was to determine whether RIP2 has a dominant role in the regulation of cytokine and chemokine expression. Sandwich ELISA was used to determine the levels of cytokines (TNF-α and IL-6) and chemokines (MIP-2 and LIX) in BALF. Figure 1 shows reduced levels of TNF-α and IL-6 in RIP2−/− mice compared to their controls postinfection (panels G to J). The level of the potent neutrophil chemokine MIP-2 was reduced in BALF of RIP2−/− mice at 24 h postinfection, although the LIX levels were reduced only at 6 h (Fig. 1G to J). These findings show that RIP2 regulates the expression of proinflammatory cytokines and chemokines.
IL-17A is a cytokine mainly produced by Th17 cells and has been implicated to be critical in the production of cytokines and neutrophil chemokines, such as IL-1β, granulocyte colony-stimulating factor (G-CSF), and MIP-2 (60). Since cytokine/chemokine production can be mediated through IL-17A, we determined IL-17A levels in lung homogenates of RIP2−/− mice upon E. coli infection. IL-17A levels were substantially reduced in the lungs of RIP2−/− mice (Fig. 2A). Since IL-17A production can be induced by IL-23-dependent or independent signaling cascades (22, 24), IL-23 levels were determined in the lungs after bacterial infection. RIP2−/− mice show attenuated levels of IL-23 in the lungs compared to WT mice following infection (Fig. 2B). These results indicate that RIP2 regulates the IL-23/IL-17A axis.
Fig. 2.
Production of IL-17A and IL-23 and activation of STAT3 in the lungs following E. coli infection. (A and B) Reduced IL-17A and IL-23 levels in lung parenchyma of RIP2−/− mice following infection. Whole-lung homogenates of WT and RIP2−/− mice at 6 and 24 h post-E. coli infection were used to determine the IL-17A and IL-23 levels by ELISA (n = 5 to 7 mice/group/time point). (C) Decreased STAT3 activation in the lungs of RIP2−/− mice following E. coli infection. STAT3 activation was determined by Western blotting of lung homogenates obtained from the lungs of RIP2−/− and WT mice after E. coli infection. (n = 4 to 6/group/time point). (D) Densitometric analysis of expression of phospho-STAT3 (Ser and Thr) and acetyl-STAT3 (Ac-STAT3) using blots from 3 separate experiments. The data obtained were normalized against total STAT3 and expressed as means ± SE. Asterisks denote the differences between WT and RIP2−/− (KO) mice (P < 0.05; for panels D and E, n = 5 to 7/group).
RIP2 deficiency reduces STAT3 activation in the lungs.
STAT3 activation and IL-6 production are two critical events for the production of IL-17A (59). Recent studies have shown that IL-6 regulates the activation of STAT3 in the lungs after E. coli infection (49). In this context, we found reduced IL-6 expression and phosphorylated STAT3 levels in the lungs of RIP2−/− mice following E. coli infection (Fig. 2C and D). Our results correlate with earlier studies which demonstrate that serine and tyrosine phosphorylation, as well as acetylation of STAT3, is critical for the development of Th17 cells (19, 34). Our results also indicate that RIP2 is important for the activation of STAT3 by tyrosine phosphorylation and acetylation but not serine phosphorylation of STAT3 (Fig. 2C and D).
RIP2 deficiency impairs activation of NF-κB and MAPKs and expression of cellular adhesion molecules in the lungs.
Expression of proinflammatory mediators is regulated by a number of transcription factors in which NF-κB is well characterized in the context of infection and inflammation. RIP2 activates numerous transcription factors, including NF-κB, that contribute to immune and inflammatory responses (36). Therefore, we examined the activation of NF-κB in lung homogenates of RIP2−/− and WT mice by NF-κB DNA binding assay and Western blotting. We found that NF-κB activation was reduced in the lungs of RIP2−/− mice at 6 and 24 h following bacterial infection (Fig. 3A to C).
Fig. 3.
Activation of NF-κB and MAPKs and upregulation of ICAM-1 and VCAM-1 in the lungs during E. coli infection. (A and B) Attenuated NF-κB activation in the lungs of RIP2−/− mice following infection. Whole-lung homogenates of RIP2−/− and WT mice at 6 and 24 h after E coli infection were used for the NF-κB binding assay, the proteins were run on an SDS-PAGE gel, and membranes were blotted with the appropriate Abs to determine activation of NF-κB and MAPKs, as well as expression of ICAM-1 and VCAM-1 (B). This blot is representative of 3 independent experiments/blots. OD, optical density. (C) Densitometric analysis of NF-κB and MAPK activation, as well as ICAM-1 and VCAM-1 expression. Densitometry was performed from 3 separate blots/experiments. The results obtained were normalized against GAPDH and expressed as means ± SE. n = 6 to 9/group; *, P < 0.05 compared to RIP2−/− (KO) mice.
Activation of MAPKs is involved in the expression of cytokines/chemokines. Activation of p38, ERK, and JNK in RIP2−/− and WT mice upon E. coli challenge was determined by Western blot analysis. There was reduced activation of p38 and JNK observed at 6 h in the E. coli-infected lungs of RIP2−/− mice compared to WT mice (Fig. 3A to C). These observations exhibit an important role for RIP2 in the activation of p38 and JNK but not ERK.
Cytokines and chemokines produced in response to infection promote upregulation of cellular adhesion molecules on resident cells, such as endothelium and myeloid cells, such as leukocytes (14). To investigate whether RIP2 deficiency decreases the expression of the cellular adhesion molecules, E. coli-infected lungs were processed for Western blotting. ICAM-1 and VCAM-1 expression was attenuated in the lungs of RIP2−/− mice compared to WT mice after infection (Fig. 3A to C).
RIP2-deficient BMDMs show reduced activation of NF-κB and MAPKs and IL-23 and IL-6 levels following E. coli infection.
In order to further validate our in vivo findings, we utilized bone marrow-derived macrophages (BMDMs) to determine the activation of transcription factors, including NF-κB and MAPKs, and the expression of cytokines (IL-6 and IL-23). BMDMs obtained from RIP2−/− mice demonstrate decreased activation of NF-κB and MAPKs (Fig. 4C), production of IL-6 and IL-23 (Fig. 4D), and expression of ICAM-1 when BMDMs are infected with E. coli at an MOI of 0.1.
RIP2−/− mice show reduced IL-6 and IL-23 production by dendritic cells post-E. coli infection.
Since IL-23 has been shown to be primarily produced by dendritic cells as well (30, 58), we determined whether RIP2 has any role in IL-23 production during E. coli infection in dendritic cells. Using BMDCs, we observed increased IL-6 and IL-23 production by BMDCs following infection (Fig. 4E). These cytokine levels were significantly reduced in RIP2−/− DCs, demonstrating a role for RIP2 in IL-6 and IL-23 production during E. coli infection (Fig. 4E).
RIP2 deficiency decreases IL-17A-producing cells in the lungs in response to E. coli infection.
Although we observed reduced IL-17A levels in the lungs of RIP2−/− mice following E. coli infection, the T cell subsets which produce IL-17A in the lungs during infection were not determined. In order to determine the cell types that are involved in RIP2-mediated IL-17A production, we performed flow cytometry using intracellular and surface staining. Interestingly, we found that natural killer (NK) cells and γδ cells were the predominant sources of IL-17A production in the lungs at 6 h following E. coli infection (Fig. 5A and B). Furthermore, the numbers of IL-17A-producing cells were significantly lower in RIP2−/− mice following infection (Fig. 5A and B).
Fig. 5.
RIP2 regulates neutrophil accumulation and bacterial clearance in the lungs during E. coli infection. (A and B) Reduced IL-17A-producing T cells (NK T cells and γδ cells) in the lungs of RIP2−/− mice upon E. coli infection. Flow cytometric analysis was performed with cells obtained from the digested lungs as described in Materials and Methods. (C and D) Enhanced neutrophil recruitment in airspaces (C) and bacterial clearance (D) following exogenous administration of rIL-17A in E. coli-infected RIP2−/− mice. Mice were infected with 106 CFU/mouse i.t. and administered rIL-17A 1 h later, and BALF was collected at 24 h postinfection. For the experiments shown in panels A to D, a total of 7 to 9 mice/group was used. *, P < 0.05 compared to RIP2−/− (KO) mice.
Exogenous IL-17A administration restores neutrophil numbers and improves bacterial clearance in RIP2 −/− mice following E. coli infection.
Since we found reduced IL-17A levels associated with lower neutrophil numbers and impaired bacterial clearance in lungs of RIP2 −/− mice following E. coli infection, we determined whether administration of exogenous recombinant IL-17A rescues the neutrophil numbers and bacterial clearance in RIP2−/− mice. We treated RIP2−/− mice i.t. with rIL-17A 1 h post-E. coli infection and determined neutrophil numbers and lung CFU. We observed increased neutrophil numbers in the BALF of RIP2−/− mice following rIL-17A treatment, compared to RIP2−/− mice upon E. coli infection (Fig. 5C). Moreover, rIL-17A treatment restored bacterial clearance by RIP2−/− mice following infection (Fig. 5D).
RIP2 is essential for neutrophil mobilization during E. coli infection.
A large pool of mature neutrophils is stored in the bone marrow. These neutrophils can rapidly be mobilized to the blood during infection and/or inflammation, resulting in substantial increase in circulating neutrophil numbers (60). Since we observed reduced neutrophil recruitment in the lungs of RIP2−/− mice during E. coli infection, this could also be due to attenuated release of neutrophils to the blood from the marrow. To investigate this, we examined the numbers of neutrophils in the blood using flow cytometry. Although the neutrophil numbers in the blood remained the same in RIP2−/− and WT mice following saline challenge, the mobilization/release of neutrophils to the blood was impaired in RIP2−/− mice after E. coli infection (Fig. 6A and B).
Fig. 6.
RIP2−/− mice show reduced neutrophil counts in the blood following E. coli infection. (A and B) Flow cytometric analysis was performed with blood obtained from WT and RIP2−/− mice at 24 h after i.t. E. coli infection (106 CFU/mouse). Blood cells were stained with antibodies against Gr-1/Ly6G. A total of 3 mice/group/time point was used in each of 3 independent experiments (n = 6 to 9 mice/group; P < 0.05). Asterisks denote significant differences between WT and KO mice. (C to F) Expression of P-selectin/CD62L, LFA-1, CD11b and CXCR2 by flow cytometric analysis of blood and lung neutrophils obtained from WT and KO mice at 24 h after i.t. E. coli (106 CFU/mouse) infection. The neutrophil population was gated and analyzed for the expression of adhesion molecules. For panels C to F, n = 6 to 8 mice/group/time point; P < 0.05 between WT and RIP2−/− (KO) mice.
RIP2 deficiency does not affect the expression of P-selectin/CD62P, LFA-1, CD11b, and CXCR2 on lung and blood neutrophils during E. coli pneumonia.
Neutrophil migration is a multistep process which is associated with the expression of cellular adhesion molecules expressed on resident and myeloid cells, such as neutrophils. To determine whether attenuated neutrophil accumulation in the lungs of RIP2−/− mice was due to decreased expression of cell adhesion molecules on neutrophils, we determined expression of CD62P, LFA-1 (CD11a/CD18), CD11b, and CXCR2 on neutrophils obtained from blood and lungs following E. coli infection. We did not observe substantial neutrophils in the lungs after digestion (data not shown). We did not see differences in the expression of these cellular adhesion molecules on neutrophils between RIP2−/− and WT mice (Fig. 6C to F). These observations exclude the possibility that expression of CD62P, CD11b, LFA-1, and CXCR2 on neutrophils was a mechanism for attenuated neutrophil recruitment to the lungs during E. coli infection (Fig. 6C to F).
DISCUSSION
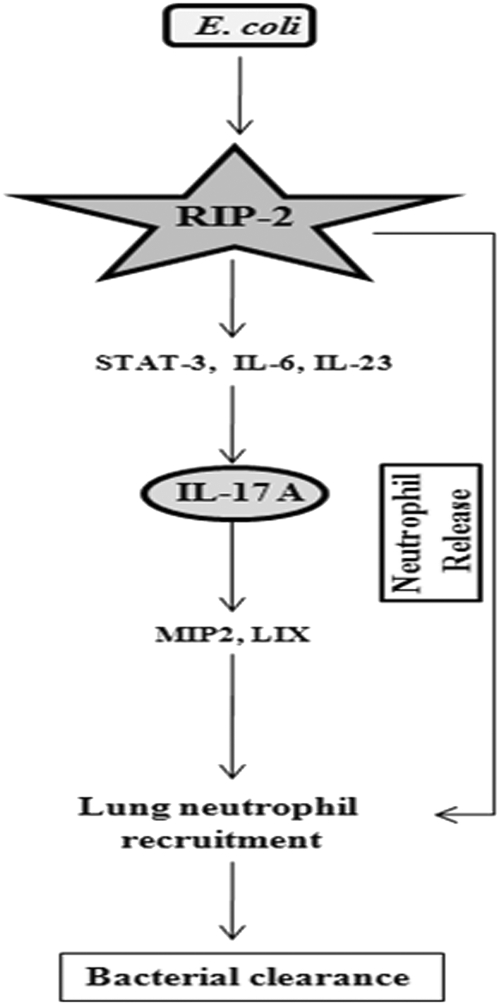
The immune system has complex mechanisms by which it is capable of sensing and responding to microbes. Pattern recognition receptors, such as TLRs and NLRs, contribute to multifaceted innate immune response (2, 16). However, the signaling cascades induced by interaction of microbes with these receptors are complex. Although numerous studies from our laboratory (10, 26, 27) and others (8, 41) have explored the TLR-mediated signaling events in detail, limited studies have delineated the role of NLRs in the context of pulmonary bacterial infections. In particular, the role of NLRs in Gram-negative extracellular pathogens has not been investigated. Although IL-17A is critical for the host defense against Gram-negative bacteria (60), whether RIP2 can regulate IL-17A in the lungs during acute bacterial pneumonia is not clear. In this regard, we determined the importance of RIP2 in an experimental model of acute bacterial pneumonia using E. coli. Our observations identify roles of RIP2 signaling in neutrophil accumulation in the lungs initiating innate immunity against Gram-negative pneumonia. First, in the absence of RIP2 signaling, lung infection with E. coli showed decreased neutrophil accumulation in the lungs. Second, RIP2 also caused innate host defense against this pathogen via IL-23 and IL-17A production (Fig. 7).
Fig. 7.
Schematic depicting the role of RIP2 in neutrophil recruitment and release during E. coli infection. E. coli infection induces IL-17A production through RIP2-mediated STAT3 activation and IL-6 and IL-23 production. IL-17A in turn regulates the production of CXC chemokines (MIP-2 and LIX) and neutrophil release, resulting in neutrophil recruitment and subsequent bacterial clearance in the lungs. Furthermore, RIP2 plays an essential role in neutrophil release from the marrow during pulmonary infection with E. coli.
The RIP family members have been identified as a group of kinases that can induce NF-κB activation (36). RIP2's tissue distribution is broad, and it is highly expressed at the mRNA level in numerous tissues, such as placenta, kidney, pancreas, and brain (36). RIP2 mRNA was upregulated upon activation of murine macrophages by LPS (12, 28). Our data show that E. coli infection regulates the expression and activation of RIP2 in the lungs and in macrophages. These data are in agreement with previous reports showing that stimulation with TLR ligands, such as LPS, lipoteichoic acid (LTA), and peptidoglycan (PGN), in mice increases the expression of RIP2 (12, 28), although RIP2 activation was not determined by earlier studies. Subsequent studies (28) have demonstrated that RIP2 deficiency impairs both TLR- and NLR-mediated signaling, although Park et al. have shown that RIP2 is involved in NOD1 and NOD2 signaling but not TLR signaling (45).
It has been well documented that bone marrow and resident cells in the lungs produce neutrophil chemoattractants (15). In this regard, hematopoietic cells produce neutrophil chemoattractants, including keratinocyte cell-derived chemokine (KC) and MIP-2, whereas the resident cells produce the neutrophil chemoattractants, LIX and lungkine. We reported that KC produced by both myeloid and resident cells is important for neutrophil-dependent inflammation during Klebsiella pneumonia (9). However, the expression of pattern recognition receptors and their adaptors on these cell types is not well documented. We demonstrated in our earlier studies that MD-2 in both cell types is important for neutrophil-mediated inflammation in the lungs. Additional investigations have shown that MyD88 derived from hematopoietic cells is more important for LPS-induced bronchial constriction and expression of TNF-α and IL-12p40 (42), whereas both hematopoietic and resident cell-derived forms of MyD88 are important for LPS-induced neutrophil influx (49, 54). Shimada et al. (53) have reported that hematopoietic cell-derived RIP2 is important for the clearance of C. pneumophila in the lungs, although the role of these cells in neutrophil sequestration to the lungs was not examined (53). Recent studies have shown that lung epithelium also plays a central role in protecting the host against bacterial challenge (31). In particular, it has been shown that Gram-negative bacterial PGN stimulates NOD1 signaling in lung epithelium and causes resistance to S. pneumoniae infection (33, 51). The role of the cell-type-derived RIP2 in neutrophil recruitment to the lungs during acute bacterial pneumonia caused by extracellular Gram-negative pathogens needs to be determined by future investigations.
The initial crucial mechanism that supports mucosal host defense in the lung against pathogens is successful recruitment of neutrophils from the bloodstream (57). Neutrophil accumulation within capillaries and migration into lung parenchyma and subsequently to the alveolar spaces during lung infection is a multistep process that involves shape/size changes of neutrophils, retention in capillaries, adhesion to endothelium, and transmigration into the alveolus (6, 57). In this regard, using neutrophil depletion, we previously reported that neutrophils are critical to control bacterial clearance in the lungs following E. coli infection (5). In our study, however, we found reduced neutrophil recruitment, cytokine/chemokine production, and VCAM-1 and ICAM-1 expression (14). Neutrophils bind to ICAM-1, E-selectin, and VCAM-1 expressed on endothelium. In particular, ICAM-1 and VCAM-1 are not constitutively expressed on endothelium, but these cell adhesion molecules can be upregulated by LPS and other inflammatory mediators, such as TNF-α (14, 57). The reduction of TNF-α in the lungs at both early (6-h) and late (24-h) time points in response to E. coli infection suggests that TNF-α may have caused this upregulation in addition to LPS. Although neutrophil recruitment to the site of infection depends on the expression of cellular adhesion molecules, such as P-selectin, LFA-1, CD11b, and CXCR2 on neutrophils, we did not see changes of these molecules on neutrophils between RIP2−/− and WT mice after infection. These findings indicate that cellular adhesion molecule expression on resident or other myeloid cells plays an important role in neutrophil recruitment to the lungs during E. coli infection.
IL-17A has been shown to be critical for the neutrophil-mediated host defense during Gram-negative bacterial infection (22, 60). The primary T cells that produce IL-17A during bacterial infection have been recognized as γδ, NK, and CD4+ cells (17, 47). IL-17A cell differentiation and recruitment to tissues are critical events to regulate the inflammation via Th17 cells (60). Although there are reports on the role of RIP2 in Th1/Th2 differentiation, the role of RIP2 in Th17 differentiation is not well understood (12, 21, 28). Although IL-17A has a plethora of proinflammatory functions, the key function of IL-17A is to stimulate the production of proinflammatory cytokines and chemokines, such as IL-1β, G-CSF, and MIP-2 (60). It is also clear that G-CSF produced by IL-17A is important for neutrophil release from the marrow (60), and E. coli infection induces IL-17A-mediated neutrophil recruitment (52). Our findings clearly show reduced IL-17A production, along with lower numbers of IL-17A-producing cells (NK and γδ), in the lungs of RIP2 −/− mice following E. coli infection. Additional experiments showed that exogenous administration of RIP2 −/− mice with rIL-17A partially rescued neutrophil-mediated host defense. Several possibilities could be attributed to these findings: (i) more IL-17A may be needed for complete rescue, and (ii) RIP2 may have an IL-17A-independent role in neutrophil-mediated host defense. Further studies are warranted to examine these possibilities. Our findings also demonstrate that RIP2 indeed regulates IL-17A production and neutrophil influx in the lungs via MIP-2 and/or LIX production.
Th17 cell development involves the activation of transcription factor STAT3, which can be regulated through IL-6 (13, 35) and IL-23 (29, 39). STAT3 is a latent cytoplasmic transcription factor which migrates to the nucleus and exerts its transcriptional activity upon stimulation (29, 39). STAT3 can be activated via phosphorylation of serine 727 and tyrosine 705 residues, as well as acetylation of the Lys 685 residue (11, 55). Our data support STAT tyrosine phosphorylation and acetylation in the lungs following E. coli infection. IL-17A can be produced in an IL-23-dependent manner and independent manner (22, 24). Consistent with this, our results show that RIP2−/− mice exhibit reduced IL-6 along with attenuated levels of IL-23, IL-17A, and STAT3 activation in the lungs following E. coli infection. We found that BMDMs and BMDCs produce substantial IL-23 in response to E. coli infection, although BMDCs produce more IL-23 than macrophages. These results are in agreement with the report demonstrating that both BMDMs and BMDCs can produce IL-23 in response to Streptococcus pneumoniae infection (58). The results validate previous observations that IL-17A production is regulated through the IL-6/IL-23 axis in other pathological settings (49). Since we found reduced neutrophil numbers in the blood of RIP2−/− mice and IL-17A levels in the lungs following E. coli infection, these observations suggest that RIP2-dependent IL-17A signaling contributes to neutrophil release to the blood.
Furthermore, one key question arising from our studies is how NLR-mediated signaling is activated upon E. coli infection. The majority of NOD proteins have specific domains composed of leucine-rich repeats (LRRs) for ligand binding (6). Several diverse bacterial NOD1 and NOD2 ligands can activate the RIP2 signaling cascades, including peptidoglycan, a component of Gram-negative bacterial cell wall and muramyldipeptide (MDP), a structure found in all bacteria (6, 20, 23, 38). Although the mechanisms involved in the recognition of extracellular pathogen by an intracellular receptor (NOD2) are not clear, at least three different mechanisms could be attributed to the recognition of pathogens and/or their components: (i) bacterial MDP can be released to the cytosol and stimulate the intracellular receptors; (ii) E. coli cells and/or their products can be leaked into the cytosol from the phagosomes; and (iii) cross talk between TLRs and NLRs could occur after initial bacterial recognition by TLRs. Future studies are required to identify the exact mechanisms of host recognition of extracellular pathogens by NLRs.
In conclusion, our data reveal the complex nature of the innate immune system to recognize extracellular pathogens. Based on our findings, we propose that E. coli activation of RIP2 leads to IL-17A production via IL-6, IL-23, and STAT3 activation. In turn, IL-17A stimulates the production of CXC chemokines (MIP-2 and LIX), as well as neutrophil release, to cause neutrophil-mediated host defense against this bacterial pathogen (Fig. 7). Although we did not investigate the role of RIP2 in the induction of adaptive immune responses, recent studies have shown such a role for RIP2. For example, RIP2 has a central role in T cell-mediated acquired responses against bacterial pathogens (4, 12, 28). Furthermore, RIP2-deficient T cells have severely reduced NF-κB activation, IL-2 production, proliferation, and differentiation (12, 28).
Observations from the current study have both clinical and therapeutic potential in terms of novel therapeutic targets to modulate host defense during bacterial infections. In this regard, targeting a single adaptor molecule, such as RIP2, to augment the host defense could be a viable strategy. Since RIP2 is implicated in several other diseases, including graft rejection, Crohn's disease, collagen-induced arthritis, and type I diabetes (4, 37), RIP2 can be used as a potential therapeutic target to augment host defense during bacterial infections of the lungs.
ACKNOWLEDGMENTS
We thank Jay Kolls and Kong Chen for help in Th17 cytokine staining. We thank Pete Mottram at LSU for critical reading of the manuscript. We also thank lung biology lab members Himanshu Raje, Jin Liliang, and K. Jeyagowri for helpful discussions and critical reading of the manuscript.
This work was supported by a Scientist Award from the FAMRI (YCSA-062466); and grants from the NIH (R01 HL-091958 and R01 HL-091958S1 via ARRA) to S.J.
Footnotes
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Abraham E. 2003. Neutrophils and acute lung injury. Crit. Care Med. 31:S195–S199 [DOI] [PubMed] [Google Scholar]
- 2. Akira S., Uematsu S., Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801 [DOI] [PubMed] [Google Scholar]
- 3. Archer K. A., Ader F., Kobayashi K. S., Flavell R. A., Roy C. R. 2010. Cooperation between multiple microbial pattern recognition systems is important for host protection against the intracellular pathogen Legionella pneumophila. Infect. Immun. 78:2477–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnold I. C., Paul W. D., Genhong C. 2005. Rip2: a key molecule that regulates both innate and acquired immunity. Curr. Med. Chem. 4:35–42 [Google Scholar]
- 5. Balamayooran G., Batra S., Balamayooran T., Cai S., Jeyaseelan S. 2011. Monocyte chemoattractant protein 1 regulates pulmonary host defense via neutrophil recruitment during Escherichia coli infection. Infect. Immun. 79:2567–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balamayooran T., Balamayooran G., Jeyaseelan S. 2010. Review: Toll-like receptors and NOD-like receptors in pulmonary antibacterial immunity. Innate Immun. 16:201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beutz M. A., Abraham E. 2005. Community-acquired pneumonia and sepsis. Clin. Chest Med. 26:19–28 [DOI] [PubMed] [Google Scholar]
- 8. Bhan U., et al. 2010. Cooperative interactions between TLR4 and TLR9 regulate interleukin 23 and 17 production in a murine model of gram negative bacterial pneumonia. PLoS One 5:e9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai S., Batra S., Lira S. A., Kolls J. K., Jeyaseelan S. 2010. CXCL1 regulates pulmonary host defense to Klebsiella infection via CXCL2, CXCL5, NF-kB, and MAPKs. J. Immunol. 185:6214–6225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai S., Batra S., Shen L., Wakamatsu N., Jeyaseelan S. 2009. Both TRIF- and MyD88-dependent signaling contribute to host defense against pulmonary Klebsiella infection. J. Immunol. 183:6629–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Catlett-Falcone R., Dalton W. S., Jove R. 1999. STAT proteins as novel targets for cancer therapy. Curr. Opin. Oncol. 11:490. [DOI] [PubMed] [Google Scholar]
- 12. Chin A. I., et al. 2002. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature 416:190–194 [DOI] [PubMed] [Google Scholar]
- 13. Cho M.-L., et al. 2006. STAT3 and NF-kB signal pathway is required for IL-23-mediated IL-17 production in spontaneous arthritis animal model IL-1 receptor antagonist-deficient mice. J. Immunol. 176:5652–5661 [DOI] [PubMed] [Google Scholar]
- 14. Cowburn A. S., Condliffe A. M., Farahi N., Summers C., Chilvers E. R. 2008. Advances in neutrophil biology. Chest 134:606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Craig A., Mai J., Cai S., Jeyaseelan S. 2009. Neutrophil recruitment to the lungs during bacterial pneumonia. Infect. Immun. 77:568–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Creagh E. M., O'Neill L. A. J. 2006. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 27:352–357 [DOI] [PubMed] [Google Scholar]
- 17. Cua D. J., Tato C. M. 2010. Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 10:479–489 [DOI] [PubMed] [Google Scholar]
- 18. Dorsch M., et al. 2006. Identification of a regulatory autophosphorylation site in the serine-threonine kinase RIP2. Cell. Signal. 18:2223–2229 [DOI] [PubMed] [Google Scholar]
- 19. Egwuagu C. E. 2009. STAT3 in CD4+ T helper cell differentiation and inflammatory diseases. Cytokine 47:149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Girardin S. E., et al. 2003. Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 300:1584–1587 [DOI] [PubMed] [Google Scholar]
- 21. Hall H. T. L., et al. 2008. RIP2 contributes to Nod signaling but is not essential for T cell proliferation, T helper differentiation or TLR responses. Eur. J. Immunol. 38:64–72 [DOI] [PubMed] [Google Scholar]
- 22. Happel K. I., et al. 2003. Cutting edge: roles of Toll-like receptor 4 and IL-23 in IL-17 expression in response to Klebsiella pneumoniae infection. J. Immunol. 170:4432–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Inohara N., et al. 2003. Host recognition of bacterial muramyl dipeptide mediated through NOD2. J. Biol. Chem. 278:5509–5512 [DOI] [PubMed] [Google Scholar]
- 24. Ivanov S., et al. 2007. Functional relevance of the IL-23-IL-17 axis in lungs in vivo. Am. J. Respir. Cell Mol. Biol. 36:442–451 [DOI] [PubMed] [Google Scholar]
- 25. Jeyaseelan S., et al. 2005. Toll-IL-1 receptor domain-containing adaptor protein is critical for early lung immune responses against Escherichia coli lipopolysaccharide and viable Escherichia coli. J. Immunol. 175:7484–7495 [DOI] [PubMed] [Google Scholar]
- 26. Jeyaseelan S., et al. 2007. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF)-mediated signaling contributes to innate immune responses in the lung during Escherichia coli pneumonia. J. Immunol. 178:3153–3160 [DOI] [PubMed] [Google Scholar]
- 27. Jeyaseelan S., et al. 2006. Toll/IL-1R domain-containing adaptor protein (TIRAP) is a critical mediator of antibacterial defense in the lung against Klebsiella pneumoniae but not Pseudomonas aeruginosa. J. Immunol. 177:538–547 [DOI] [PubMed] [Google Scholar]
- 28. Kobayashi K., et al. 2002. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416:194–199 [DOI] [PubMed] [Google Scholar]
- 29. Korn T., Bettelli E., Oukka M., Kuchroo V. K. 2009. IL-17 and Th17 cells. Annu. Rev. Immunol. 27:485–517 [DOI] [PubMed] [Google Scholar]
- 30. Langrish C. L., et al. 2004. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol. Rev. 202:96–105 [DOI] [PubMed] [Google Scholar]
- 31. LeibundGut-Landmann S., Weidner K., Hilbi H., Oxenius A. 2011. Nonhematopoietic cells are key players in innate control of bacterial airway infection. J. Immunol. 186:3130–3137 [DOI] [PubMed] [Google Scholar]
- 32. Lu C., et al. 2005. Participation of Rip2 in lipopolysaccharide signaling is independent of its kinase activity. J. Biol. Chem. 280:16278–16283 [DOI] [PubMed] [Google Scholar]
- 33. Lysenko E. S., et al. 2007. Nod1 signaling overcomes resistance of S. pneumoniae to opsonophagocytic killing. PLoS Pathog. 3:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma C. S., et al. 2008. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J. Exp. Med. 205:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mathur A. N., et al. 2007. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J. Immunol. 178:4901–4907 [DOI] [PubMed] [Google Scholar]
- 36. McCarthy J. V., Ni J., Dixit V. M. 1998. RIP2 is a novel NF-kB-activating and cell death-inducing kinase. J. Biol. Chem. 273:16968–16975 [DOI] [PubMed] [Google Scholar]
- 37. McCully M. L., Fairhead T., Blake P. G., Madrenas J. 2008. The future of RIP2/RICK/CARDIAK as a biomarker of the inflammatory response to infection. Expert Rev. Mol. Diagn. 8:257–261 [DOI] [PubMed] [Google Scholar]
- 38. McDonald C., Inohara N., Núñez G. 2005. Peptidoglycan signaling in innate immunity and inflammatory disease. J. Biol. Chem. 280:20177–20180 [DOI] [PubMed] [Google Scholar]
- 39. McKenzie B. S., Kastelein R. A., Cua D. J. 2006. Understanding the IL-23 IL-17 immune pathway. Trends Immunol. 27:17–23 [DOI] [PubMed] [Google Scholar]
- 40. Mizgerd J. P. 2006. Lung infection—a public health priority. PLoS Med. 3:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morris A. E., Liggitt H. D., Hawn T. R., Skerrett S. J. 2009. Role of Toll-like receptor 5 in the innate immune response to acute P. aeruginosa pneumonia. Am. J. Physiol. Lung Cell. Mol. Physiol. 297:L1112–L1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Noulin N., et al. 2005. Both hemopoietic and resident cells are required for MyD88-dependent pulmonary inflammatory response to inhaled endotoxin. J. Immunol. 175:6861–6869 [DOI] [PubMed] [Google Scholar]
- 43. Opitz B., et al. 2004. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J. Biol. Chem. 279:36426–36432 [DOI] [PubMed] [Google Scholar]
- 44. Pandey A. K., et al. 2009. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 5:e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park J.-H., et al. 2007. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 178:2380–2386 [DOI] [PubMed] [Google Scholar]
- 46. Park J.-H., Kim Y.-G., Nunez G. 2009. RICK promotes inflammation and lethality after Gram-negative bacterial infection in mice stimulated with lipopolysaccharide. Infect. Immun. 77:1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Passos S. T., et al. 2010. IL-6 promotes NK cell production of IL-17 during toxoplasmosis. J. Immunol. 184:1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peleg A. Y., Hooper D. C. 2010. Hospital-acquired infections due to Gram-negative bacteria. N. Engl. J. Med. 362:1804–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinton L. J., et al. 2008. Alveolar epithelial STAT3, IL-6 family cytokines, and host defense during Escherichia coli pneumonia. Am. J. Respir. Cell Mol. Biol. 38:699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rangel-Moreno J., et al. 2011. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat. Immunol. 12:639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ratner A. J., Aguilar J. L., Shchepetov M., Lysenko E. S., Weiser J. N. 2007. Nod1 mediates cytoplasmic sensing of combinations of extracellular bacteria. Cell. Microbiol. 9:1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shibata K., Yamada H., Hara H., Kishihara K., Yoshikai Y. 2007. Resident Vδ1+γδ T cells control early infiltration of neutrophils after Escherichia coli infection via IL-17 production. J. Immunol. 178:4466–4472 [DOI] [PubMed] [Google Scholar]
- 53. Shimada K., et al. 2009. The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS Pathog. 5:e1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Skerrett S. J., et al. 2004. Respiratory epithelial cells regulate lung inflammation in response to inhaled endotoxin. Am. J. Physiol. Lung Cell. Mol. Physiol. 287:L143–L152 [DOI] [PubMed] [Google Scholar]
- 55. Song L., Turkson J., Karras J. G., Jove R., Haura E. B. 2003. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene 22:4150–4165 [DOI] [PubMed] [Google Scholar]
- 56. Tigno-Aranjuez J. T., Asara J. M., Abbott D. W. 2010. Inhibition of RIP2's tyrosine kinase activity limits NOD2-driven cytokine responses. Genes Dev. 24:2666–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wagner J. G., Roth R. A. 2000. Neutrophil migration mechanisms, with an emphasis on the pulmonary vasculature. Pharmacol. Rev. 52:349–374 [PubMed] [Google Scholar]
- 58. Wang J., Ma J., Charboneau R., Barke R., Roy S. 2011. Morphine inhibits murine dendritic cell IL-23 production by modulating Toll-like receptor 2 and Nod2 signaling. J. Biol. Chem. 286:10225–10232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang X. O., et al. 2007. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282:9358–9363 [DOI] [PubMed] [Google Scholar]
- 60. Ye P., et al. 2001. Requirement of interleukin 17 receptor signaling for lung Cxc chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 194:519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]