Abstract
Funded by the National Institute of Allergy and Infectious Diseases, the Pathosystems Resource Integration Center (PATRIC) is a genomics-centric relational database and bioinformatics resource designed to assist scientists in infectious-disease research. Specifically, PATRIC provides scientists with (i) a comprehensive bacterial genomics database, (ii) a plethora of associated data relevant to genomic analysis, and (iii) an extensive suite of computational tools and platforms for bioinformatics analysis. While the primary aim of PATRIC is to advance the knowledge underlying the biology of human pathogens, all publicly available genome-scale data for bacteria are compiled and continually updated, thereby enabling comparative analyses to reveal the basis for differences between infectious free-living and commensal species. Herein we summarize the major features available at PATRIC, dividing the resources into two major categories: (i) organisms, genomes, and comparative genomics and (ii) recurrent integration of community-derived associated data. Additionally, we present two experimental designs typical of bacterial genomics research and report on the execution of both projects using only PATRIC data and tools. These applications encompass a broad range of the data and analysis tools available, illustrating practical uses of PATRIC for the biologist. Finally, a summary of PATRIC's outreach activities, collaborative endeavors, and future research directions is provided.
A RESOURCE FOR INFECTIOUS-DISEASE RESEARCH
The National Institute of Allergy and Infectious Diseases (NIAID) established the Bioinformatics Resource Centers (BRCs) to provide scientists with genomics-centric resources for NIAID category A, B, and C priority microbial pathogens (a complete list of these priority pathogens is provided at the NIAID Biodefense and Related Programs website: http://www.niaid.nih.gov/topics/biodefenserelated/biodefense/research/pages/cata.aspx) (22). Originally, NIAID funded eight BRCs to provide annotated genomic and related data on microbes causing emerging and re-emerging infectious diseases, including bacterial, viral, and eukaryotic pathogens, as well as invertebrate vectors of infectious-disease agents. The Pathosystems Resource Integration Center (PATRIC), one of the original eight BRCs, stored and integrated data on six different bacterial and viral pathogens (40). In 2009, NIAID reorganized the BRC program through a competitive renewal for four BRCs, each one with a discrete yet all-encompassing organismal focus: bacteria, viruses, eukaryotic pathogens, and invertebrate vectors (with one exception: the Influenza Resource Database [IRD] specifically focuses on the influenza virus). PATRIC was awarded the bacterial BRC (http://www.patricbrc.org).
All bacteria with a focus on the NIAID priority watch list.
PATRIC integrates and annotates all genomic and associated data available from most of the major bacterial lineages, allowing comparative analysis of the NIAID priority infectious agents with closely related free-living, symbiotic, and commensal species (see “Annotation FAQs” at http://enews.patricbrc.org/faqs/, which links to all FAQs subjects). With an emphasis on consistency in comparative genomic analysis, PATRIC has standardized annotation of all available bacterial genomes using the RAST (rapid annotation using subsystems technology) system (5), a product of the Fellowship of Interpretation of Genomes (FIG) SEED team, which is a component of the PATRIC team. RAST, which predicts genes, assigns gene functions, and reconstructs metabolic pathways, is powered by a robust assembly of subsystems that have been curated based on evaluation of hundreds of prokaryotic genomes and the clustering of common protein families encoded within these genomes (FIGfams). As of 1 July 2010, PATRIC had annotated 2,865 bacterial genomes using RAST (Note: the “All Bacteria” homepage at http://wwww.patricbrc.org/portal/portal/patric/Taxon?cType=taxon&cId=2 lists the current annotation statistics, including eight genome and protein sequence statistics and 43 genomic features). As it is anticipated that the growing number of sequenced prokaryotic genomes will continue to improve the quality of SEED subsystems, PATRIC will continue to update RAST-based gene, protein, and protein family annotations, as well as providing historical information to track future amendments.
In addition to the RAST-based annotations, PATRIC preserves and provides the historical annotations present at GenBank (RefSeq), as well as the annotations created by the specialists at the previous BRCs, referred to as “Legacy BRC” on the PATRIC site. Importantly, the different annotation methods allow comparison of many genomes using all three approaches. However, given the breadth of coverage of bacterial genomes using RAST, the Legacy BRC annotations are generally the least complete source at PATRIC, because annotation efforts by the previous BRCs ended in 2009. As such, PATRIC houses 355 genomes with annotations from the former BRCs. From GenBank, annotations from 3,230 genomes are currently included, allowing comparison of different annotation schemes across most bacterial genomes. Finally, our cyberinfrastructure technology enables PATRIC to support additional annotations that specific communities implement for their focal organisms, such as curated MetaCyc data (12). Thus, it is anticipated that comparative approaches to genome annotation will continue in the near future.
Organisms, genomes, and comparative genomics.
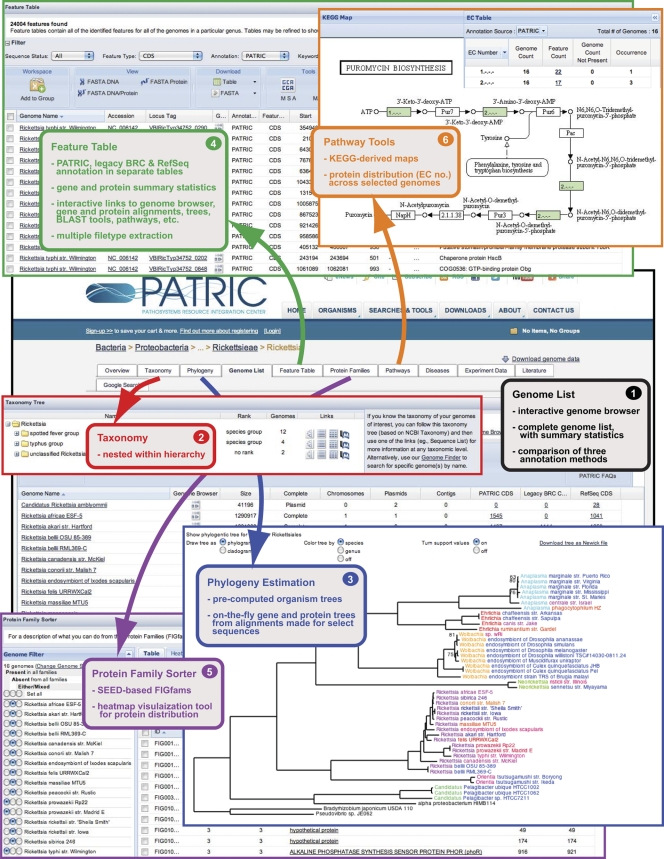
The PATRIC website is primarily organism-centric, with various levels of genomic data and associated information related to each included organism. While the PATRIC homepage lists the 22 watch list genera for easy access to data associated with many pathogenic species, compilation and organization of all relevant data for “All Bacteria” are standardized according to bacterial (NCBI) taxonomy, with options for viewing sets of genomes within the hierarchical bacterial tree. Thus, specific “Overview” pages can be accessed for selected taxa within the bacterial tree (e.g., genus, family, order, class, etc.). The “Overview” page contains genome (and associated data) information for all available genomic sequences (closed and incomplete) within a selected taxon and also lists the most recent PubMed articles pertinent to the study of the focal taxon. Each “Overview” page also contains six search tools (Genome Finder, Feature Finder, Comparative Pathway Tool, Protein Family Sorter (PFS), Gene Ontology (GO) Search, and Enzyme Commission (EC) Search) that allow quick directed searches without navigating further into the more detailed pages that house specific data for each organism. The “Genome List” page (Fig. 1, box 1) provides the compiled genomes (closed and incomplete, chromosomal and plasmid) for a given taxon, with statistics for all three different annotation methods and direct links to an interactive genome browser based on JBrowse (37, 38). The “Taxonomy” page (Fig. 1, box 2) provides classification schemes that are listed at NCBI, with assigned NCBI taxonomic identifiers used to relate associate data for each organism across the website. The “Phylogeny” page (Fig. 1, box 3) illustrates precomputed trees generated for higher-level groups (typically at the order level), which are based on concatenated alignments of multiple conserved protein families (50, 51). The methods used to estimate organism phylogenies are more detailed than the trees generated from individual gene and protein alignments within other pages of the website (see “Phylogeny FAQs”).
Fig. 1.
Schema depicting major genomic and comparative genomic tools available from an organism “Overview” homepage. This example illustrates the Rickettsia genomes compiled at PATRIC. The “Genome List” (box 1) provides statistics across three different annotation methods (RAST, legacy BRC, and RefSeq), with each genome linked to an interactive genome browser tool. The “Taxonomy” page (box 2) provides classification schemes from the NCBI taxonomy database, taxonomic identifiers specific to each organism used to associate related data across the website. The “Phylogeny” page (box 3) demonstrates the precomputed trees estimated for higher-level groups (typically at the order level), which are based on concatenated alignments of conserved protein families. Each “Locus Tag” leads to unique pages for each CDS that provide links to NCBI (corresponding RefSeq locus tags), FASTA-formatted protein and nucleotide files, Uniprot mapping data for proteins, and direct interaction with the genome browser tool. The “Protein Families” page (box 5) lists the SEED-derived FIGfams (31) generated for any selection of genomes using the genome filter tool. An interactive heat map visualization tool gives a bird's-eye view of both protein distribution across multiple genomes and relative conservation of synteny (see Fig.3A). Finally, the “Pathways” page (box 6) provides the metabolic pathways that are encoded within a selected taxon, integrating information from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (33). All pathways can be visualized for each of the three different annotation methods, and pathway conservation across multiple genomes can be evaluated.
Several pages encompass the majority of genomic data and present convenient platforms for comparative genomic analysis. The “Feature Table” (Fig. 1, box 4) provides the tabulation of information for each protein-encoding sequence (CDS), as well as noncoding RNAs, within a selected genome and can be visualized for each of the three different annotation methods. All columns contain user-defined sorting options, and selection of “Locus Tag” leads to specific pages for each CDS that list additional information, including links to NCBI (corresponding RefSeq locus tags), FASTA-formatted protein and nucleotide files, Uniprot mapping data for proteins, and direct interaction with the genome browser tool. Recent implementation of a “Compare Region Viewer” allows synteny analysis across all genomes encoding a selected CDS (see Fig. S1 in the supplemental material). A video tutorial for navigating a typical “Feature Table” illustrates its functionality (see “Feature Table FAQs”). The “Protein Families” page (Fig. 1, box 5) lists the orthologous groups of proteins generated across a selected number of input genomes, with SEED-derived FIGfams used for clustering conserved families (31). A genome filter tool allows user-defined inclusion/exclusion of genomes, and the annotated FIGfams are provided with the number of included genomes (and sequences) and length range for sequences within the protein clusters. An interactive two-dimensional (2-D) heat map visualization tool is also provided to give a bird's-eye (pan-proteome) view of both protein distribution across multiple genomes and relative conservation of synteny. A demonstration of the full range of the PFS, as applied to a typical genomics-driven experimental design, is illustrated in the following section. Finally, the “Pathways” page (Fig. 1, box 6) lists the cellular function and metabolic pathways that are encoded within a selected taxon, integrating information from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (33). Pathways are classified according to major biological roles (e.g., carbohydrate metabolism, translation, biosynthesis of secondary metabolites, etc.) and are assigned identifications from a list of 137 unique cellular pathways. All pathways can be visualized for each of the three different annotation methods, and all annotation schemes can be simultaneously superimposed over pathway maps. For evaluation of pathway conservation across multiple genomes, components within KEGG maps (depicted by EC numbers) are color coded according to a spectrum depicting gene presence/absence across analyzed genomes.
Application to comparative genomics: erythritol utilization in Brucella.
In conjunction with the tools mentioned above, PATRIC's compilation of all public bacterial genomes provides a powerful platform for comparative genomic analysis. Such in silico experiments often shed light on factors implicated in pathogenicity, including their evolutionary trajectories and functions across diverse bacterial lineages. We selected a previously identified virulence factor associated with brucellosis to illustrate this experimental design. Originally isolated from infected bovine fetal tissues (39), the four-carbon sugar erythritol is the preferred carbon and energy source of Brucella spp. Subsequent experiments showed that erythritol stimulated in vitro growth of B. abortus and enhanced infections caused by a second species, B. melitensis (27). It is thought that erythritol uptake is linked to spontaneous abortion, a complication of Brucella infection in some hosts. Animals with low placental concentrations of erythritol do not have the overwhelming infection that is seen in species with high concentrations (39). Seminal studies on the biochemical pathway for erythritol catabolism in B. abortus (42, 43) led to a genetic characterization of the genes involved in this metabolism (36).
Four genes in the Brucella ery operon (eryABCD) encode enzymes that have been characterized in erythritol catabolism: erythritol kinase (EryA), erythritol phosphate dehydrogenase (EryB), d-erythrulose 4-phosphate dehydrogenase (EryC), and erythritol transcriptional regulator (EryD) (36). The ery operon has also been found in closely related bacteria (including some nonpathogenic species), suggesting a broader biological utilization for this sugar source. For example, genes involved in erythritol transport were recently identified in the legume symbiont Rhizobium leguminosarum (55), in which the ery genes play a role in root nodule formation. Discovery of the transporter operon (eryEFG), found adjacent to the catabolic operon in R. leguminosarum, led to the identification and reannotation of genes adjacent to the ery operon in Brucella. A third adjacent operon (deoR-tpiA2-rpiB) was also identified by Yost et al. (55) as possibly being important in erythritol catabolism. As the experiments demonstrating importance of this operon in erythritol catabolism have not yet been published, this operon was excluded from the present analysis.
Regarding Brucella spp., Brucella ovis and a vaccine strain, Brucella abortus S19, are known for their inability to oxidize erythritol. Tsolis et al. (46) identified four genes in B. ovis (eryA, eryD, eryF, and eryG) with mutations rendering them pseudogenes. Additionally, Crasta et al. (13) identified a 703-bp deletion that interrupts the coding regions of eryC and eryD in B. abortus S19. With 41 Brucella genomes now sequenced, we wanted to examine the genes considered important in erythritol catabolism and identify similar and perhaps additional problems that might exist in the newly available genomes. Given the presence of these genes in other bacteria, we extended our analysis to include all members of the order Rhizobiales, which, aside from Brucella and Rhizobium, contains an interesting assortment of pathogens, symbionts, and free-living members (51).
In the examination of the erythritol catabolism among Brucella spp., eight proteins were analyzed in detail, including a protein whose annotation recently changed from “hypothetical protein” to “hypothetical lipoprotein component of the erythritol ABC transporter.” Using the PFS suite of tools available at PATRIC, as well as the multiple-sequence alignment viewer tool BLAST (1) and the Genome Browser tool (Fig. 2), we were able to identify mutations in seven of these eight proteins (see Fig. S2A in the supplemental material). Although all mutations found are listed, we stress that some mutations found in single genomes (e.g., those of B. ovis and B. abortus S19) do not have supporting experimental evidence and could be sequencing or assembly errors. However, more weight should be given to mutations shared by phylogenetically related genomes, because sequencing and assembly errors are less likely to be conserved across various genomes. With this in mind, we were able to identify some mutations that are phylogenetically shared. Brucella ceti strains M13/05/01 and M644/93/1, which are monophyletic within the B. ceti clade, share two single-base-pair deletions, resulting in premature stop codons that affect eryA and the hypothetical lipoprotein component of the erythritol ABC transporter. An additional shared single-base-pair deletion that affects all nine members of the B. ceti clade is found in eryF. Brucella ovis and Brucella sp. strain NVSL 07-2006 are members of the same clade, yet they share only one of the mutations known to occur in B. ovis, a single-base-pair deletion that results in an altered start site for eryG. As B. ovis has mutations that alter four proteins, it is difficult to say if this single shared deletion renders strain NVSL 07-2006 incapable of catabolizing erythritol.
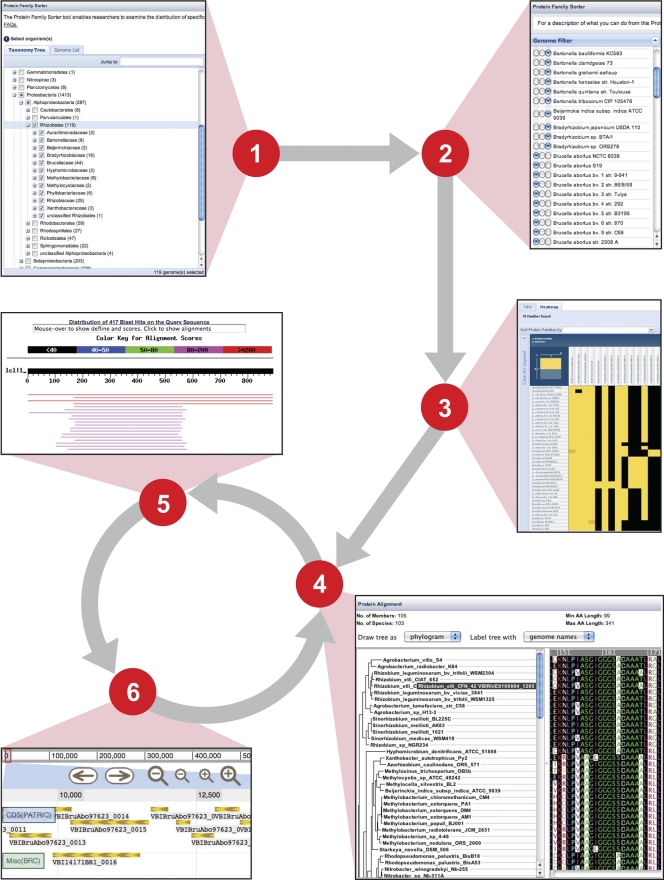
Fig. 2.
Experimental design for evaluating the conservation and distribution of erythritol catabolic and transport genes across 107 Rhizobiales genomes. Steps 1 to 4 illustrate the functionality of the PATRIC Protein Family Sorter (PFS) tool. (Step 1) From either the “Taxonomy Tree” or the “Genome List,” any number of genomes can be selected for analysis. (Step 2) The “Genome Filter” tool allows the evaluation of FIGfam membership (e.g., present or absent in all selected genomes, patchy distribution across genomes), and an “Advanced Filter” tool enables the retrieval of more refined FIGfam lists based on specific terms (e.g., “Product Descriptions,” “Perfect Families,” and/or the number of proteins or genomes per protein family). (Step 3) The interactive “Protein Family Heat-map” provides an overview of the distribution of proteins across a selected set of genomes. A reference genome can be selected to anchor the display of the protein families, and each individual column or row within the heat map can be moved to adjust the display. All protein sequences for each FIGfam can be extracted from the heat map in a variety a ways (see “Protein Family Heatmap FAQs”). (Step 4) Once a FIGfam is captured, proteins can be selected and evaluated using the “Integrated Protein Tree and Alignment” option. This displays the sequences in the “Multiple Sequence Alignment Viewer” tool, which combines an estimated phylogeny (left) with the full sequence alignment (right). (Step 5) Using BLAST tools within PATRIC, full-length sequences from the alignment can be used as queries in searches against all genomes for sequences not included within the FIGfam, such as highly divergent proteins, split ORFs, and truncations (BLASTP) and pseudogenes not annotated as CDSs in the genomes (TBLASTN). (S6) For sequences detected outside the FIGfam, the “Genome Browser” tool can be used to evaluate potential pseudogenes (i.e., validation of point and frameshift mutations) as well as areas of low sequence coverage or poor quality. Steps 4 to 6 can be iterative in evaluating the relative conservation of a protein family across a set of diverse genomes.
One interesting finding involves B. abortus strains S19 and NCTC 8038, for which phylogeny estimation suggests monophyly within the B. abortus clade (see Fig. S2B in the supplemental material). While these may be the same strain, these genome sequences were generated by different teams: S19 by the Virginia Bioinformatics Institute (13) and NCTC 8038 by the Broad Institute (http://www.broadinstitute.org/annotation/genome/brucella_group/MultiHome.html). Curiously, the 703-bp deletion affecting both eryC and eryD (see above) is not present in the NCTC 8038 genome, which has complete open reading frames for these genes. It is currently unknown if these sequences represent two different isolations within the B. abortus S19 strain. If so, then there appears to be some variability in the presence of this deletion among isolates of this important vaccine strain. The only mutation that S19 and NCTC 8038 share is a single-base-pair deletion that results in a truncated eryG.
Looking more broadly across the order Rhizobiales, all proteins putatively involved in erythritol catabolism and transport were identified and compiled using several PATRIC tools. With the PFS, a visual representation of the presence or absence of these proteins in a heat map view was created, with the bacterial families within the order and the operons of interest annotated (Fig.3A). Analysis of these proteins showed that the ery catabolism operon is present across all members of the families Brucellaceae, Phyllobacteriaceae, and Aurantimonidaceae, but it is only sporadically found in Rhizobiaceae and Bradyrhizobiaceae genomes. This operon, and any associated transport genes, is completely missing from the families Bartonellaceae, Xanthobacteriaceae, Methylobacteriaceae, and Beijerinckiaceae. Using the 2-D heat map view, it is evident that some genomes within the Rhizobiaceae have all proteins within this operon annotated, while some are missing components. This genomic distribution has been described previously, as it has been suggested that the erythritol operon is used for root nodule formation by the non-Brucella organisms (55). Our bioinformatics analysis presents a platform for testing the hypothesis that a complete ery operon and associated transporter genes are essential for root nodule formation.
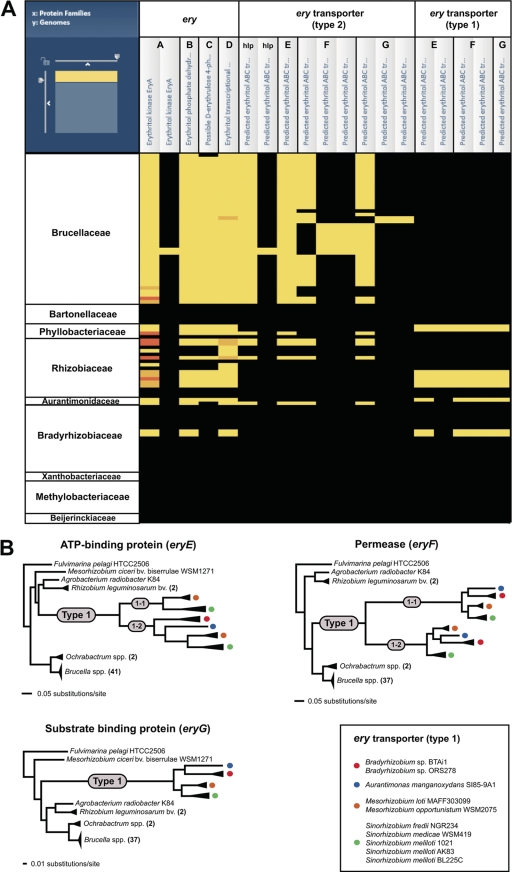
Fig. 3.
Phylogenomic analysis of erythritol catabolic and transport genes across 107 Rhizobiales genomes. These results summarize the comparative genomics experimental design, which primarily utilizes the PATRIC Protein Family Sorter (PFS) tool (Fig.2). (A) Heat map depiction of the distribution of erythritol catabolic (eryA-D) and transport (hypothetical lipoprotein [hlp] and eryE-G) proteins. The x axis of the map lists the annotated Ery protein families (simplified at top), with individual components (including duplications and split CDSs) enclosed within black boxes. The y axis shows the genomes, with taxon names simplified and arranged at the family level. Black regions indicate no representative proteins assigned to the protein family; bright yellow regions indicate one representative protein assigned to the protein family. Other colors depict multiple representatives per protein family, with increasing membership ranging from dark yellow to dark orange. (B) Phylogenetic analysis of the type 2 and type 1 ery transport proteins. Alignments, performed using MUSCLE v3.6 (17, 18), and generated trees, estimated using FastTree v.2 (34), were visualized simultaneously using the PATRIC Multiple Sequence Alignment Viewer (see Fig. S2C in the supplemental material). The phylograms for EryE, EryF, and EryG are simplifications of the larger trees and depict the evolution of type 1 transporter components from the type 2 family. Smaller gray circles illustrate the duplication of the type 1 components into type 1-1 and type 1-2 (EryE and EryF only). All taxa encoding type 1 components are represented with colored circles, which are explained in the inset at bottom right.
An unexpected result of our analysis was the identification of a second set of genes putatively involved in erythritol transport. While the Brucellaceae and some of the genomes in other families have a type 2 erythritol ABC transporter, a genetically distinct system is encoded within the genomes of other families (Fig. 3A). In order to examine the evolutionary origin of the genes encoding these two divergent transporters, protein sequences from similarly named components (e.g., the permease component of either transporter 1 or 2) were assembled using the above-mentioned tools (see Fig. S2C in the supplemental material). Trees for all three components of the similarly named transporter proteins were generated (Fig. 3B). From this analysis, it is clear that in all three cases the Brucella proteins appear to be part of a broadly conserved ancestral family (type 2) and that a less conserved erythritol transport system (type 1) evolved from within this group. Because the transporter gene trees do not corroborate the Rhizobiales species tree (see Fig. S2B in the supplemental material), it is likely that horizontal transfer events have facilitated the dissemination of the type 1 erythritol transport system genes throughout Rhizobiales evolution. The biological relevance of diverse transport systems for erythritol and their possible correlations with pathogenicity (type 2) and symbiosis (type 1) remain to be elucidated.
Recurrent integration of community-derived associated data.
In addition to the acquisition, annotation, integration, and bioinformatics processing of genome-scale data sets, PATRIC provides “awareness” of community-derived research and information associated with each bacterial organism. Principally, these genome-associated data are organized into three categories: disease, experimental data, and literature (Fig. 4). All of this information is made available to the researcher in a recurring and contextualized manner, such that it is continually updated (contingent on PATRIC and corresponding website updates) and provided at useful locations throughout the website. Thus, this feature provides the infectious-disease research community with an invaluable integration of research data and metadata from a multitude of sources, enabling sophisticated and comprehensive analyses across any bacterial taxon of interest at a single website with consistent tools and interfaces.
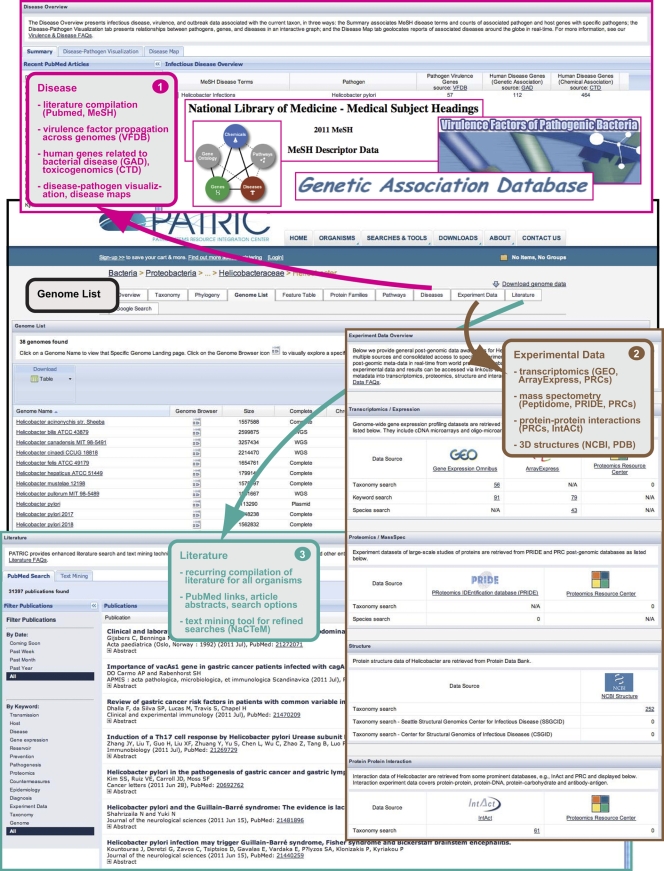
Fig. 4.
Schema depicting the integrated community-derived associated data available from an organism “Overview” homepage. Navigation from the Helicobacter “Genome List” (outlined in black) is illustrated. Disease information (box 1) can be summarized into four main categories: Literature (PubMed article compilation and MeSH terms for database searching [32]), virulence factors (data from the Virulence Factor Database [VFDB] [54] is used to identify all putative homologs present within other bacterial genomes), human genes associated with disease (Genetic Association Database [8, 57] and Comparative Toxicogenomics Database [14]), and disease-pathogen data (interactive graphics for relationships between pathogens, diseases, virulence genes, and disease-associated host genes, as well as interactive global health maps [11] illustrating recent reports and outbreaks of bacterial diseases). “Experimental Data” (box 2) encompasses transcriptomic data (GEO [6, 7], ArrayExpress [26], and Proteomics Resource Centers [PRCs] [56]), proteomics data from mass spectrometry (Peptidome [25], PRIDE [48] and the PRCs), protein-protein interaction data from the PRCs and IntACt (4), and protein 3-D structure data from NCBI and Protein Data Bank (PDB) (10). “Literature” (box 3) is primarily comprised of a recurrent compilation of literature and web text resources pertaining to each organism (PubMed abstracts and links to articles), with a search tool that allows filtering by keywords, dates, etc. An integrated text-mining tool (UK National Text Mining Centre [NaCTeM]) allows efficient recall of relevant documents through the identification of key entities from the search text (i.e., genes, proteins, metabolites, drugs, diseases, symptoms, etc.).
For disease-related information (Fig.4, box 1), a catalog of PubMed literature relevant to associated diseases is provided. Additionally, medical subject headings (MeSH) disease terms are listed, allowing direct access to the National Library of Medicine MeSH Descriptor Database (32). Candidate virulence factors can be evaluated based on a strategy that integrates data from the Virulence Factor Database (VFDB) (54). Briefly, virulence factors listed at the VFDB are compiled at PATRIC and used to identify all putative homologs present within other bacterial genomes. Information is also provided on human genes associated with each disease, including genetic and chemical evidence. Integrating data from the Genetic Association Database (8, 57), the “Genetic Association Source” table lists human genes that have been shown to have some genetic association with a bacterial disease. Similarly, data from the Comparative Toxicogenomics Database (14) is integrated in the “Comparative Toxicogenomics Source,” which lists human genes associated with a bacterial disease that have been characterized via chemical treatment or exposure. Both the “Genetic Association Source” and the “Comparative Toxicogenomics Source” provide additional information about the human genes from NCBI as well as GeneCards, a comprehensive and authoritative compendium of annotative information pertaining to human genes (35). Finally, two additional tools round out the integrated information pertinent to bacterial diseases. The “Disease-Pathogen Visualization” page provides an interactive, graphical image of the relationships between pathogens, diseases, virulence genes, and disease-associated host genes. The “Disease Map” page provides a real-time global view of recent reports and outbreaks of bacterial diseases, with geolocation superimposed on an interactive global health map (11). An example of a PATRIC disease map shows the high activity index in Europe of reported Escherichia coli infections during the recent outbreak of the German enterohemorrhagic/verocytotoxin-producing E. coli (EHEC/VTEC) strain (see Fig. S3 in the supplemental material).
A major undertaking for PATRIC is to provide a summary of the wide range of experimental data found in a variety of databases for all bacteria (Fig.4, box 2). This information, collectively referred to as postgenomic data, encompasses transcriptomic data primarily from microarrays (in addition to serial analysis of gene expression [SAGE] and RNA-Seq), proteomics data from mass spectrometry, protein-protein interaction data, and protein 3-D structure data (X ray and nuclear magnetic resonance [NMR]). At the species and strain levels, these data are sometimes difficult to find at the associated databases. PATRIC recurrently searches select external databases using several keywords (i.e., organism name, NCBI taxonomic identifier, etc.) specific to each source and provides links to data that are continually updated at these repositories. Thus, PATRIC provides a summary of the number and types of data available at NCBI's GEO (Gene Expression Omnibus) (6, 7), EBI's ArrayExpress, (26), and the legacy NIAID-funded Proteomics Resource Centers (PRCs) (56). Mass spectrometry data are accessed from Peptidome, (25), PRIDE (48), and the PRCs. Current knowledge on protein-protein interactions is also retrieved from the PRCs, as well as IntACt (4). Finally, PATRIC links to protein 3-D structure data from the NCBI and the Protein Data Bank (PDB) (10).
A continual challenge for PATRIC is to provide the user with a robust and real-time list of literature and web text resources pertaining to each organism (Fig.4, box 3). Relevant articles (and abstracts when available) from PubMed are listed chronologically, with direct links to PubMed provided. Literature compilations may be filtered by date and keyword for winnowing down large lists. A more direct way to reduce irrelevant results while increasing the recall of relevant documents is to use the text-mining tool, which implements technology developed in conjunction with the UK National Text Mining Centre (NaCTeM), another component of the PATRIC team. This process displays search results based on indexes of UK Medline abstracts, identifying key entities from the search text (i.e., genes, proteins, metabolites, drugs, diseases, symptoms, etc.). Results are summarized by entity type and allow progressive filtering. Abstracts are provided with key entities highlighted in different colors and contain direct links to PubMed.
Application to annotation driven by data integration: drug and vaccine targeting.
The computed proteomes of all PATRIC genomes provide rich data sets for large-scale computa-tional analyses. One of PATRIC's major focal areas of research is the design and execution of experiments that integrate multiple levels of information from community databases for improving bacterial genome annotation (i.e., adding information beyond standard automated annotation). Importantly, while the data integrated from the community may pertain to selected high-profile pathogens, PATRIC's analysis pipelines work to propagate this information across all bacterial genomes when gene and protein homology supports such an approach. In theory, this strategy of refining functional gene and protein annotations will expand our knowledge of the factors directly involved at the interface between host and pathogen, e.g., virulence factor identification, antibiotic resistance and synthesis gene characterization, drug and vaccine targeting, etc. The following example illustrates this approach for the development of a drug targeting classification for all bacterial genomes.
With the list of drug and vaccine targets in the infectious-disease research community rapidly growing (53), we hypothesize that this information, combined with the comprehensive proteome of PATRIC genomes, may be utilized to propose novel antibacterial drug targets. The logic in our approach presumes that previously determined drug targets in some bacterial species might provide reasonable candidate targets for other species if structural and functional data are similar across bona fide and candidate targets. Aside from sequence-based criteria, we elected to incorporate information from protein 3-D structure into our experimental design, as there is a tendency for approved and pending bacterial drug targets to have associated structural data (NMR, cryo-electron microscopy, X-ray crystallography, etc.). We also considered the human genome in our analysis, distinguishing between drug targets with high similarity to human proteins and those with no significant human-encoded counterparts. The latter distinction is important, as selection of drug targets with some degree of similarity to human proteins would require more careful design to avoid effective targeting of both host and pathogen proteins.
To illustrate the PATRIC's potential for large-scale drug target annotation, the workflow is divided into two processes. First, a data set was created containing significant similarity between a set of position-specific scoring matrices (PSSMs) (23) from NCBI's Protein Clusters (28) and (i) protein sequences encoded within the human genome (47), (ii) proteins previously annotated as drug targets (29, 52), and (iii) proteins with associated 3-D structure information (47) (see Fig. S4A in the supplemental material). A high PSSM score within a region of a sequence (query) is a good indication of a comparable biological role of this region to the domain, family, or motif characterized by the PSSM (9). Sequence similarity across query proteins and the PSSMs was evaluated using reverse-position-specific BLAST (RPSBLAST) (30) with an E-value cutoff of 0.001. This resulted in a diverse set of annotated proteins and, importantly, substantially limited the number of possible matches for transferring annotations to bacterial genes. In the second step (Fig. S4B), the set of protein sequences (total = 2,771,151) encoded within 800 bacterial genomes (794 species) was used in RPSBLAST searches against the data set constructed in the first step, with the identical search strategy and significance threshold. This resulted in the identification of bacterial genes encoding proteins with regions of significant similarity to at least human proteins, previously described drug targets, or proteins with associated structural data (n = 454,842, or 16.4% of query proteins). Many of these bacterial proteins scored a match for two or all three of these specific groups identified using the PSSMs (see Fig. S4C).
The result of propagating information from host, prior drug targets, and structure to novel bacterial proteins is shown for 22 NIAID category A, B, and C priority microbial pathogens (Table 1). A modest number of proteins (n = 40,180) encoded within these 22 genomes scored significant matches to the PSSMs described above, with slightly more having significant similarity to domains within human proteins (55.2%). This attests to the nature of protein conservation, particularly domain architecture, even across diverse organisms such as bacteria and vertebrates. However, of the 18,013 proteins lacking significant similarity to human proteins, only 19.7% lacked PSSMs matching previously defined drug targets and/or proteins with associated structural data. Thus, our analysis winnowed down a robust list to strictly prokaryotic protein domains with existing drug target analogs (n = 12), relevant structural information (n = 7,290), or both (n = 7,155), all of which provide candidate drug targets that can be utilized with minimal regard for host proteins. Regarding the bacterial proteins having significant similarity to human protein domains, the majority (97.8%) also contained matches to PSSMs with existing drug target analogs (n = 352), relevant structural information (n = 3,791), or both (n = 17,546). Of the latter class, the majority of proteins (67.4%) have matches to approved (versus under development) drug targets, suggesting that many of the existing drug targets may be applicable to pathogens with similarly functioning proteins encoded in their genomes.
Table 1.
Drug-targeting attributes characterized within the genomes of 22 NIAID category A, B, and C priority microbial pathogensa
| Organismb | No. of proteins withc: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No human homologs |
Human homologs |
||||||||||
| N | NS | NAS | ND | NDS | H | HS | HA | HAS | HD | HDS | |
| BAC | 215 | 481 | 83 | 453 | 39 | 216 | 23 | 900 | 1 | 461 | |
| BAR | 63 | 158 | 19 | 98 | 13 | 139 | 8 | 301 | 1 | 156 | |
| BOR | 34 | 96 | 13 | 70 | 6 | 94 | 1 | 162 | 115 | ||
| BRU | 172 | 337 | 43 | 324 | 21 | 176 | 14 | 701 | 1 | 278 | |
| BUR | 291 | 471 | 63 | 555 | 36 | 238 | 29 | 1043 | 1 | 367 | |
| CAM | 70 | 197 | 27 | 145 | 13 | 122 | 8 | 333 | 1 | 169 | |
| CHL | 32 | 79 | 14 | 54 | 6 | 98 | 203 | 2 | 116 | ||
| CLO | 107 | 412 | 46 | 461 | 30 | 177 | 11 | 671 | 1 | 340 | |
| COX | 53 | 142 | 22 | 127 | 14 | 165 | 18 | 388 | 1 | 182 | |
| EHR | 31 | 75 | 7 | 43 | 9 | 114 | 9 | 200 | 1 | 106 | |
| ESC | 469 | 812 | 69 | 5 | 590 | 45 | 277 | 36 | 797 | 1 | 402 |
| FRA | 67 | 179 | 27 | 141 | 18 | 170 | 28 | 423 | 1 | 226 | |
| HEL | 56 | 154 | 31 | 102 | 9 | 108 | 5 | 266 | 1 | 151 | |
| LIS | 118 | 353 | 40 | 314 | 22 | 166 | 9 | 550 | 1 | 313 | |
| MYC | 66 | 256 | 43 | 256 | 18 | 144 | 12 | 951 | 2 | 314 | |
| RIC | 36 | 80 | 9 | 53 | 13 | 123 | 9 | 181 | 1 | 102 | |
| SAL | 419 | 722 | 64 | 3 | 551 | 47 | 264 | 32 | 730 | 1 | 391 |
| SHI | 520 | 743 | 69 | 1 | 559 | 41 | 261 | 36 | 773 | 1 | 394 |
| STA | 104 | 286 | 41 | 211 | 15 | 148 | 9 | 544 | 1 | 240 | |
| STR | 72 | 221 | 27 | 188 | 6 | 120 | 3 | 412 | 1 | 228 | |
| VIB | 229 | 440 | 55 | 492 | 25 | 222 | 13 | 613 | 1 | 321 | |
| YER | 332 | 596 | 58 | 3 | 498 | 32 | 249 | 16 | 692 | 1 | 340 |
A selected species was used for each genus. Results for all species within the 22 genera are provided in Fig. S4C in the supplemental material.
BAC, Bacillus anthracis Sterne; BAR, Bartonella henselae Houston-1; BOR, Borrelia burgdorferi B31; BRU, Brucella abortus bv 1 9-941; BUR, Burkholderia mallei ATCC 23344; CAM, Campylobacter jejuni 1336; CHL, Chlamydophila pneumoniae AR39; CLO, Clostridium difficile 630; COX, Coxiella burnetii CbuG Q212; EHR, Ehrlichia canis Jake; ESC, Escherichia coli O157-H7 EC4115; FRA, Francisella tularensis subsp. holarctica 257; HEL, Helicobacter pylori 2017; LIS, Listeria monocytogenes 08-5578; MYC, Mycobacterium tuberculosis H37Rv; RIC, Rickettsia typhi Wilmington; SAL, Salmonella enterica subsp. enterica serovar Typhi Ty2; SHI, Shigella dysenteriae 1012; STA, Staphylococcus aureus subsp aureus MRSA252; STR, Streptococcus pneumoniae 70585; VIB, Vibrio cholerae 121291; YER, Yersinia pestis Angola.
S, significant similarity to a protein with associated 3-D structure in the Protein Data Bank; A, significant similarity to an approved drug target; D, significant similarity to a drug target under experimental testing; N, no significant similarity to a human protein; H, significant similarity to a human protein.
While currently under development, the novel set of bacterial genes annotated with drug-targeting attributes will become available to all PATRIC researchers in a future release. Similar “reverse annotation” strategies are also being employed for the curation of antibiotic synthesis and resistance genes, as well as a vast set of virulence factors defined by a novel controlled vocabulary. All of these data will be propagated across all genomes at PATRIC in a manner consistent with the provision of other associated data across the website. Improvements to genomic annotation generated from the strategy outlined above will drive the design and development of new resources at PATRIC, which will facilitate comprehensive comparative analyses for infectious-disease research.
A USER-CENTERED APPROACH FOR PATRIC
The community-derived information that is integrated into PATRIC is provided through a practical, rich interface that delivers access to all the relevant data from these key public external sources. Advancing the user's experience and research capability at PATRIC is a driving force; therefore, we formally apply the structured, user-centered process known as usability engineering (24) to improve users' experience with the site. Specifically, we actively involve representative researchers and other stakeholders in formulating user-centered requirements, design, and evaluation and continue their involvement through the PATRIC operational releases, thereby ensuring a highly usable site derived from real user experience (44). To create functional areas of the website, we iteratively cocreate conceptual design sketches with researchers that organize insights from domain analysis activities and user-centered requirements. We thoroughly analyze results from these early evaluations and use them to create detailed designs that use modern technologies to provide a user-centered experience.
Throughout the development and refinement of PATRIC, we have identified three keystone design principles from the field of human-computer interaction that are well suited to serve the infectious-disease researcher community. We employed each of these principles throughout the PATRIC website. The first pertains to information integration. This approach stresses seamless accession of all organisms, tasks/tools, and data throughout the website without forcing users to go repeatedly to different pages or website areas. Second, the progressive filtering method is implemented, supporting numerous levels of filtering and drill-down, e.g., over all PATRIC data, on a single organism, on a single genome, etc. Finally, a context sensitivity approach offers options (controls, filters, tools, etc.) that are appropriate to the user's current scope (e.g., as instantiated in filters, task areas, and tabs on PATRIC's data browser page). In sum, to meet the challenge of clearly and efficiently delivering a comprehensive collection of integrated data for infectious-disease research, PATRIC's user-centered design approach has produced a usable, friendly web interface.
OUTREACH AND FORTHCOMING DEVELOPMENTS
Recently, PATRIC has utilized the above-mentioned tools, analysis platforms, and other resources in bioinformatics investigations pertaining to various aspects of infectious-disease research, including virulence factors (2, 19, 20), comparative genomics (21, 41, 49), large-scale phylogenetics (50, 51), human–bacterial-pathogen protein interaction networks (16), text mining (3; S. Pyysalo et al., presented at the 2010 Workshop on Biomedical Natural Language Processing, ACL 2010, Uppsala, Sweden, 15 July 2010), and data integration (44). Our efforts have also been utilized in various collaborations generating experimental research (15, 45). As such, with the recurrent expansion of the scope of information integration, PATRIC's infrastructure will continue to grow through developments driven by various collaborations with the infectious-disease research community, education and outreach activities, community engagement and feedback, and continuing PATRIC-driven research. Three aspects of PATRIC's future are described below.
Driving Biological Projects program.
PATRIC conducts several activities to engage the infectious-disease research community and to drive development of further infrastructure. One important example is the Driving Biological Projects (DBPs) program. Via DBPs, we collaborate with groups within the infectious-disease research community to produce large-scale data in order to define, cocreate, develop, and deploy the infrastructure needed to support further novel data types (such as RNA-Seq) and respective integrated analyses by the community. These are competitively awarded projects that are reviewed by PATRIC's scientific working group and awarded as PATRIC subcontracts. Through this process, PATRIC further evolves into a resource that can provide researchers with analysis capabilities and integrative access to new and evolving types of data.
In 2010, PATRIC awarded two subcontracts in the inaugural round of the DBPs program (see http://enews.patricbrc.org/feature/call-for-dbp-proposals/). The first project will focus on comparative transcriptome, proteome, and phenotype microarray analysis of five divergent Clostridium difficile strains to facilitate the understanding of mechanisms of C. difficile pathogenesis. The result of large-scale data analysis and comparisons will help verify and update C. difficile genome annotations and aid in obtaining a comprehensive overview of C. difficile core, divergent, and strain-specific genes and pathways involved in pathogenesis. In addition to its value for the C. difficile research community, this work will help expand the PATRIC data model (e.g., integration of Biolog data) by joint development, testing, and deployment of novel tools, such as RNA-Seq analysis pipeline and visualization. These tools will be directly applicable to other bacterial projects.
The second project will aim to provide PATRIC with essential information for displaying genes characteristic of nontyphoidal Salmonella enterica serovar Typhimurium, particularly those that contribute to survival in a variety of environments, including various host species. This will be accomplished primarily through a combination of high-throughput screening and sequencing approaches and unique resources developed to annotate the S. Typhimurium genome with fitness data. The generation of S. Typhimurium transcriptomes from bacteria growing in defined environments (including rich and minimal media, at stationary phase, and under conditions that induce virulence pathways) will yield basal reference profiles to help standardize, as well as streamline, the massive amount of high-throughput transcriptomics data from impending studies. Novel tools and infrastructure developed in concert with the DBPs will be incorporated into PATRIC in future releases. Future calls for DBPs will be posted at the PATRIC homepage.
PATRIC workshops.
We conduct additional outreach through delivery of workshops designed to educate researchers in how to maximally benefit from PATRIC's broad resources. Workshops include lectures on in silico experimental designs and bioinformatics tools and methods, as well as demonstrations of various analyses that can be performed using the PATRIC website. The scope of the workshops includes pathogens, as well as other bacterial species, and especially makes use of the comparative tools described in the examples outlined above and in recent publications (for example, see references 21, 41, 49, and 50). Workshops are conducted on a recurrent basis and will undergo changes in content as new developments are instituted at PATRIC. Our team also participates in various scientific meetings and conferences, and numerous presentations have been given. Web pages listing information on past and future presentations (see http://enews.patricbrc.org/category/presentations) as well as general PATRIC news feeds (see http://www.patricbrc.org) are updated on a regular basis.
Future additions to PATRIC.
Many new capabilities are already planned for PATRIC to improve the user experience and to provide the most comprehensive resource for computational analyses directed toward understanding bacterial pathogenesis and for development of antibacterial drugs, diagnostics, and vaccines. In the future, PATRIC researchers will be able to analyze and compare their own data against available data for all bacterial genomes. A complete list of future developments is beyond the scope of this introductory article but includes a more versatile multiple-sequence viewer, access to metagenomics data and annotation tools, and improved and more integrated text-mining capabilities. This growing suite of tools will enable complex analyses through workflows. Forthcoming developments at PATRIC will ensure that it meets the varied needs of the infectious-disease research community, especially teams working to develop antibacterial drugs and vaccines.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the constructive criticism provided by the two anonymous reviewers.
This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272200900040C awarded to B.W.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 6 September 2011.
The authors have paid a fee to allow immediate free access to this paper.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ammerman N. C., Gillespie J. J., Neuwald A. F., Sobral B. W., Azad A. F. 2009. A typhus group-specific protease defies reductive evolution in rickettsiae. J. Bacteriol. 191:7609–7613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ananiadou S., et al. 2011. Named entity recognition for bacterial Type IV secretion systems. PLoS One 6:e14780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aranda B., et al. 2010. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 38:D525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aziz R. K., et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrett T., et al. 2011. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 39:D1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrett T., et al. 2009. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37:D885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Becker K. G., Barnes K. C., Bright T. J., Wang S. A. 2004. The genetic association database. Nat. Genet. 36:431–432 [DOI] [PubMed] [Google Scholar]
- 9. Beckstette M., Homann R., Giegerich R., Kurtz S. 2006. Fast index based algorithms and software for matching position specific scoring matrices. BMC Bioinformatics 7:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berman H. M., et al. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brownstein J. S., Freifeld C. C., Reis B. Y., Mandl K. D. 2008. Surveillance Sans Frontieres: Internet-based emerging infectious disease intelligence and the HealthMap project. PLoS Med. 5:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caspi R., et al. 2010. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 38:D473–D479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crasta O. R., et al. 2008. Genome sequence of Brucella abortus vaccine strain S19 compared to virulent strains yields candidate virulence genes. PLoS One 3:e2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis A. P., et al. 2011. The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. 39:D1067–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dreher-Lesnick S. M., et al. 2010. Analysis of Rickettsia typhi-infected and uninfected cat flea (Ctenocephalides felis) midgut cDNA libraries: deciphering molecular pathways involved in host response to R. typhi infection. Insect Mol. Biol. 19:229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dyer M. D., et al. 2010. The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS One 5:e12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edgar R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gillespie J. J., et al. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4:e4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gillespie J. J., et al. 2010. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect. Immun. 78:1809–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gillespie J. J., et al. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One 3:e2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Greene J. M., et al. 2007. National Institute of Allergy and Infectious Diseases bioinformatics resource centers: new assets for pathogen informatics. Infect. Immun. 75:3212–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gribskov M., McLachlan A. D., Eisenberg D. 1987. Profile analysis: detection of distantly related proteins. Proc. Natl. Acad. Sci. U. S. A. 84:4355–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hix D., Hartson H. R. 1993. Developing user interfaces: ensuring usability through product and process. John Wiley & Sons, Inc [Google Scholar]
- 25. Ji L., et al. 2010. NCBI Peptidome: a new repository for mass spectrometry proteomics data. Nucleic Acids Res. 38:D731–D735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kapushesky M., et al. 2010. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res. 38:D690–D698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keppie J., Williams A. E., Witt K., Smith H. 1965. The role of erythritol in the tissue localization of the brucellae. Br. J. Exp. Pathol. 46:104–108 [PMC free article] [PubMed] [Google Scholar]
- 28. Klimke W., et al. 2009. The National Center for Biotechnology Information's Protein Clusters Database. Nucleic Acids Res. 37:D216–D223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knox C., et al. 2011. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 39:D1035–D1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marchler-Bauer A., et al. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30:281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer F., Overbeek R., Rodriguez A. 2009. FIGfams: yet another set of protein families. Nucleic Acids Res. 37:6643–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neveol A., Shooshan S. E., Mork J. G., Aronson A. R. 2007. Fine-grained indexing of the biomedical literature: MeSH subheading attachment for a MEDLINE indexing tool. AMIA Annu. Symp. Proc. 553–557 [PMC free article] [PubMed] [Google Scholar]
- 33. Ogata H., et al. 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 27:29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Price M. N., Dehal P. S., Arkin A. P. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Safran M., et al. 2010. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010:baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sangari F. J., Aguero J., Garcia-Lobo J. M. 2000. The genes for erythritol catabolism are organized as an inducible operon in Brucella abortus. Microbiology 146(Pt. 2):487–495 [DOI] [PubMed] [Google Scholar]
- 37. Skinner M. E., Holmes I. H. 2010. Setting up the JBrowse genome browser. Curr. Protoc. Bioinformatics 32:9.13.1–913.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Skinner M. E., Uzilov A. V., Stein L. D., Mungall C. J., Holmes I. H. 2009. JBrowse: a next-generation genome browser. Genome Res. 19:1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith H., et al. 1962. Foetal erythritol: a cause of the localization of Brucella abortus in bovine contagious abortion. Nature 193:47–49 [DOI] [PubMed] [Google Scholar]
- 40. Snyder E. E., et al. 2007. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 35:D401–D406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sobral B. W., Wattam A. R. 2011. Comparative genomics and phylogenomics of the Brucella, p. 13–36 In Lopez-Goni I., O'Callaghan D. (ed.), Brucella: molecular microbiology and genetics. Horizon Scientific Press, Norwich, United Kingdom [Google Scholar]
- 42. Sperry J. F., Robertson D. C. 1975. Erythritol catabolism by Brucella abortus. J. Bacteriol. 121:619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sperry J. F., Robertson D. C. 1975. Inhibition of growth by erythritol catabolism in Brucella abortus. J. Bacteriol. 124:391–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sullivan D. E., Gabbard J. L., Jr., Shukla M., Sobral B. 2010. Data integration for dynamic and sustainable systems biology resources: challenges and lessons learned. Chem. Biodivers. 7:1124–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sutten E. L., et al. 2010. Anaplasma marginale type IV secretion system proteins VirB2, VirB7, VirB11, and VirD4 are immunogenic components of a protective bacterial membrane vaccine. Infect. Immun. 78:1314–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsolis R. M., et al. 2009. Genome degradation in Brucella ovis corresponds with narrowing of its host range and tissue tropism. PLoS One 4:e5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Venter J. C., et al. 2001. The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- 48. Vizcaino J. A., et al. 2010. The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 38:D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wattam A. R., et al. 2009. Analysis of ten Brucella genomes reveals evidence for horizontal gene transfer despite a preferred intracellular lifestyle. J. Bacteriol. 191:3569–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Williams K. P., et al. 2010. Phylogeny of gammaproteobacteria. J. Bacteriol. 192:2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Williams K. P., Sobral B. W., Dickerman A. W. 2007. A robust species tree for the alphaproteobacteria. J. Bacteriol. 189:4578–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wishart D. S., et al. 2008. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36:D901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wishart D. S., et al. 2006. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34:D668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang J., Chen L., Sun L., Yu J., Jin Q. 2008. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 36:D539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yost C. K., Rath A. M., Noel T. C., Hynes M. F. 2006. Characterization of genes involved in erythritol catabolism in Rhizobium leguminosarum bv. viciae. Microbiology 152:2061–2074 [DOI] [PubMed] [Google Scholar]
- 56. Zhang C., et al. 2008. An emerging cyberinfrastructure for biodefense pathogen and pathogen-host data. Nucleic Acids Res. 36:D884–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Y., et al. 2010. Systematic analysis, comparison, and integration of disease based human genetic association data and mouse genetic phenotypic information. BMC Med. Genomics 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.