Abstract
A Yersinia pestis-human protein interaction network is reported here to improve our understanding of its pathogenesis. Up to 204 interactions between 66 Y. pestis bait proteins and 109 human proteins were identified by yeast two-hybrid assay and then combined with 23 previously published interactions to construct a protein-protein interaction network. Topological analysis of the interaction network revealed that human proteins targeted by Y. pestis were significantly enriched in the proteins that are central in the human protein-protein interaction network. Analysis of this network showed that signaling pathways important for host immune responses were preferentially targeted by Y. pestis, including the pathways involved in focal adhesion, regulation of cytoskeleton, leukocyte transendoepithelial migration, and Toll-like receptor (TLR) and mitogen-activated protein kinase (MAPK) signaling. Cellular pathways targeted by Y. pestis are highly relevant to its pathogenesis. Interactions with host proteins involved in focal adhesion and cytoskeketon regulation pathways could account for resistance of Y. pestis to phagocytosis. Interference with TLR and MAPK signaling pathways by Y. pestis reflects common characteristics of pathogen-host interaction that bacterial pathogens have evolved to evade host innate immune response by interacting with proteins in those signaling pathways. Interestingly, a large portion of human proteins interacting with Y. pestis (16/109) also interacted with viral proteins (Epstein-Barr virus [EBV] and hepatitis C virus [HCV]), suggesting that viral and bacterial pathogens attack common cellular functions to facilitate infections. In addition, we identified vasodilator-stimulated phosphoprotein (VASP) as a novel interaction partner of YpkA and showed that YpkA could inhibit in vitro actin assembly mediated by VASP.
INTRODUCTION
Plague has caused three worldwide pandemics that claimed millions of lives. It remains a threat to humans due to bioterrorism concerns and outbreaks in Asia, Eurasia, Africa, and America every year (17, 61). The etiologic agent, Yersinia pestis, is a facultative intracellular pathogen that can be readily engulfed and killed by polymorphonuclear leukocytes (PMNs) but survives and replicates in macrophages by inhibiting the acidification of phagosomes (64, 65). Three major forms of clinical plague infection are bubonic, septicemic, and pneumonic plague. After onset, plague progresses with extraordinary speed, which is largely facilitated by the suppression of host innate immune responses at the early stage of infection (15, 42). Yersinia bacilli taken up by the tissue macrophages proliferate rapidly and synthesize F1 capsules, the type III secretion system (T3SS), pH 6 antigen, and various other virulence-associated factors required for pathogenesis in mammalian hosts (82). Upon escaping from macrophages, Y. pestis acquires the ability to inhibit phagocytosis by PMNs, a key virulence strategy to suppress the innate immune responses against the pathogen. The virulence of Y. pestis in mammalian hosts includes abilities to inhibit innate immune responses, such as inhibition of phagocytosis by PMNs and macrophages (3, 24), induction of apoptosis of the infected cells (43, 55), and inhibition of the secretion of proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and gamma interferon (IFN-γ) (14, 69). Three human-pathogenic Yersinia species share a 70-kb plasmid that encodes the T3SS, by which the bacteria inject a set of virulent effectors called Yersinia outer membrane proteins (Yops) into the mammalian host cells (20). The molecular mechanisms of how these Yop effectors function during infection remain a focus of intensive research (33, 51, 81).
Large-scale protein-protein interaction (PPI) networks, both interspecies and intraspecies, have been reported in recent years. The published PPI networks of humans and some model organisms provide valuable references for investigating protein interaction networks between pathogens and their hosts (41, 46, 59, 66, 71). Several virus-host infection networks have been reported, such as those of hepatitis C virus (HCV), Epstein-Barr virus (EBV), and Kaposi sarcoma-associated herpesvirus (KSHV) (18, 22, 73). Dayer et al. have integrated publicly available human-pathogen PPIs for 190 strains of various pathogens, including viral, bacterial, and protozoan pathogens, to provide a global view of pathogenesis strategies used by different pathogens (26).
These reports reveal that pathogens tend to interact with hubs (proteins interacting with a large number of partners) and bottlenecks (proteins connecting with many functional modules) in the human PPI network, and distinct pathogens preferentially interfere with a different spectrum of cellular pathways to facilitate infection and dissemination. More recently, the interaction networks between human proteins and those of bacterial pathogens Y. pestis, Bacillus anthracis, and Francisella tularensis have been published (27). Using a random yeast two-hybrid (Y2H) approach, 3,073 human-B. anthracis, 1,383 human-F. tularensis, and 4,059 human-Y. pestis PPIs were identified, and the conserved modules among the three pathogen-human protein interaction networks were computed, yielding 39 to 64 conserved protein interaction modules between different pairs of pathogens.
To gain a comprehensive overview of the Y. pestis-human protein interactions that might occur during plague infection, a different strategy was used to identify PPIs between Y. pestis and human proteins. A total of 153 potential virulence-associated Y. pestis proteins were chosen as bait proteins to perform direct Y2H screening, and the Y2H-generated PPI data set was combined with the published interactions to construct a Y. pestis-human PPI (YHPI) network. We found that Y. pestis was highly prone to interact with hub and bottleneck proteins essential for normal cellular functions in the human PPI network. The Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) (4) pathways enriched in human proteins targeted by Y. pestis proteins (HYTs) highlight a number of important pathways in immune response, including the Toll-like receptor (TLR) and mitogen-activated protein kinase (MAPK) signaling pathways and pathways involved in leukocytes' transendoepithelial migrations, focal adhesion, and cytoskeletal regulation. Our results may point to important areas of research to guide future functional characterization to unravel the molecular mechanism of Y. pestis pathogenesis in humans.
MATERIALS AND METHODS
Y2H screening.
Gateway cloning techniques were used to construct the plasmids expressing bait proteins (77). PCR primers incorporating attB recombination sites (forward, 5′-GGGGACAACTTTGTACAAAAAAGTTGGCATG; and reverse, 5′-GGGGACAACTTTGTACAAGAAAGTTGG) followed by open reading frame (ORF)-specific sequences were designed to amplify each target gene using genomic DNA of Y. pestis strain 91001 (biovar microtus) or strain 82009 (biovar orientalis) as a template (see Table S1 in the supplemental material). The amplified products were cloned into pDONER 91001 using BP Clonase enzyme mix (Invitrogen), and the presence of the correct inserts was confirmed by DNA sequencing. Target genes in entry clones were then inserted into pDEST32 destination vectors using the LR Clonase enzyme mix (Invitrogen) to generate plasmids expressing bait proteins fused with the DNA-binding domain (DBD) of GAL4.
For Y2H library screening, a GAL4-based ProQuest Y2H system was used (Invitrogen). DBD-ORF plasmids were first transformed into the yeast (Saccharomyces cerevisiae) strain AH109. The bait proteins that activate HIS3, ADE2, and lacZ reporter genes after cotransforming AH109 with empty prey plasmid pDEST22 were excluded from the Y2H cDNA library screening. Yeast AH109 cells were sequentially transformed with the bait plasmids and a human spleen cDNA library according to the ProQuest two-hybrid system user manual. At least 1 × 106 transformants were screened for each bait protein. Yeast transformants were plated onto high-stringency dropout medium (synthetic complete [SC] −Trp −Leu −His −Ade) and incubated for 5 to 10 days at 30°C. The positive colonies grown on SC −Trp −Leu −His −Ade plates were restreaked onto new plates to grow for another 3 days. Filter-lift assays were used to evaluate lacZ activity following the manufacturer's instructions. Positive clones that activated at least two of the three reporter genes (HIS3, ADE2, or lacZ) were chosen and sequenced. Interactions were further validated by retransformation assays using purified plasmids.
Bioinformatics analysis.
Cytoscape version 2.6.1 was used to compute the topological parameters of human proteins integrated in the reference human PPI network with the aid of the NetworkAnalyzer plugin (6). The Mann-Whitney U test was chosen to test the significance of the observed difference between the parameters k (degree), b (betweenness), and l (shortest path) of HYTs and human proteins (designated H). A permutation test was applied to test the significance of the differences in the number of hubs and bottlenecks between a data set generated from random sampling of 109 proteins (HR [described in more detail below]) and the H data set (78).
Functional annotation and analysis.
Cellular pathway data were retrieved from KEGG and were used to annotate human gene functions (4). The pathway enrichment analysis was performed using web-based DAVID (35) bioinformation tools (http://david.abcc.ncifcrf.gov), and the significance of the enrichment was tested by Fisher's exact test followed by Benjamini and Hochberg multiple test correction. Enrichment of the GO term was analyzed using the BiNGO plugin (50) of Cytoscape, tested using a hypergeometric test followed by Benjamini and Hochberg multiple test correction.
Cell culture, transfection, and plasmids.
HEK-293T cells were grown in Dulbecco's modified Eagle's medium (HyClone) containing 10% fetal bovine serum (FBS) and 2 mM l-glutamine at 37°C in a 5% CO2 incubator. Cells were transfected using MegaTran 1.0 (OriGene). Coding sequences of Y. pestis proteins used in the glutathione S-transferase (GST) pulldown assay were cloned into pHG-GWA (16) using the Gateway recombination cloning technology and expressed as N-terminal GST-tagged fusions in Escherichia coli BL21λ(DE3). Human genes were cloned into pFLAG-CMV2 (Clontech). ypcd1.05, ypcd1.45, ypcd1.61, ypo2940, and ypo3877 ORFs were cloned into pCMV-Myc (Clontech) for expression of the corresponding proteins in NF-κB luciferase reporter assays. HEK-293T cells were transfected with pNF-κB-Luc, pRL-TK, and the plasmids expressing Y. pestis proteins. pGFP-C2-VASP, which expresses human protein VASP (vasodilator-stimulated phosphoprotein), was a generous gift from Klemens Rottner, and the VASP gene was reintroduced into pFlag-CMV-2 (Sigma) and pGEX4T-1 (GE). The ypkA gene of Y. pestis was PCR amplified using the genomic DNA of Y. pestis 91001 biovar microtus as a template and cloned into pCMV-Myc (Clontech) and pGEX4T-1 (GE).
GST pulldown assays.
Expression of GST-tagged Y. pestis proteins was induced with 0.01 mM IPTG (isopropyl-1-thio-β-d-galactopyranoside), and bacteria were harvested and then suspended in lysis buffer (0.5% NP-40, 20 mM Tris-HCl [pH 8.0], 180 mM NaCl, 1 mM EDTA) supplemented with Roche complete protease inhibitor cocktail. Bacterial cells were then lysed by sonication, and cleared lysates were mixed with glutathione-Sepharose 4B beads (GE Healthcare) at 4°C and washed thoroughly with lysis buffer. HEK-293T cells were transfected with plasmids expressing human genes, and after 2 days, the cells were harvested, washed with ice-cold phosphate-buffered saline (PBS), and lysed in lysis buffer. The cell lysates were precleared and incubated at 4°C overnight with Sepharose 4B beads bound with either GST or GST-tagged Yersinia proteins. Beads were washed extensively, and bound proteins were boiled in Laemmli buffer, separated by SDS-PAGE, and analyzed by immunoblotting with antibodies for GST or FLAG epitope tag (Santa Cruz).
NF-κB luciferase reporter assays.
HEK-293T cells were seeded onto a 96-well plate and cotransfected with 100 ng of pNF-κB-Luc (Stratagene) plus 2 ng of Renilla luciferase expression vector pRL-TK (Promega) with pCMV-Myc empty vector or pCMV-Myc derivates expressing the indicated Y. pestis protein. After 2 days of incubation, the cells were treated with TNF-α (10 ng/ml) for 1 h, and luciferase activity was determined using the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions.
Actin polymerization assay.
Recombinant GST-VASP and GST-YpkA proteins were purified from E. coli using glutathione-Sepharose 4B beads according to the instructions of the manufacturers. Actin polymerization assays were conducted as previously described (34). Actin polymerization was measured by monitoring the fluorescence intensity of G-actin and pyrene-labeled G-actin mixture (5:1) in fresh G-buffer (5 mM Tris-HCl [pH 7.5], 0.2 mM ATP, 0.2 mM dithiothreitol [DTT], 0.5 mM CaCl2) at a 1 μM final concentration in the presence of one of the following components—GST, GST-YpkA, or GST-VASP (YpkA plus VASP)—at a 0.1 μM final concentration. The G-actin mixture and proteins were precleared at 100,000 × g for 1 h. Actin polymerization was initiated by adding 10× actin polymerization buffer (500 mM KCl, 20 mM MgCl2, 10 mM ATP). Fluorescence intensity was monitored every 1 min at an excitation wavelength of 365 nm and emission wavelength of 405 nm.
RESULTS AND DISCUSSION
Construction of Y. pestis-human interaction network.
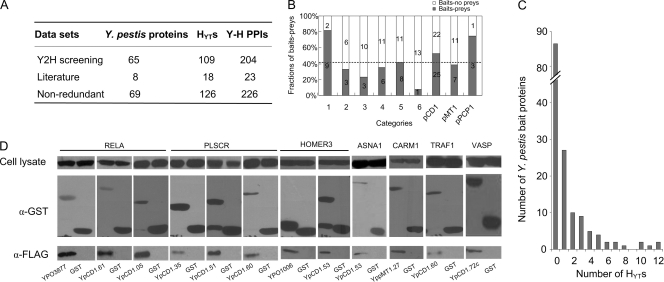
Y. pestis contains approximately 4,000 genes (58). The majority of these genes are involved in bacterial metabolism and growth, which are not likely to interact directly with host proteins during infection. A total of 153 virulence-associated proteins were selected according to the annotated genome of Y. pestis CO92 (58) and the current literature regarding Yersinia pathogenesis (see Table S1 in the supplemental material). Bait proteins were selected according to the following criteria (with the numbers of bait proteins in each category indicated in parentheses): (i) all published virulence factors, such as the type III Yop effectors, pH 6 antigen, Pla protease, Ail, and F1 antigen (n=12); (ii) genes that are intact in fully virulent CO92 but are disrupted in 91001 (n=9), a biovar microtus strain that is avirulent to humans but highly virulent to mice (58, 70); (iii) genes that are actively regulated in response to various adverse in vitro stimuli or inside infected cells (n=13) (31, 32, 47, 82); (iv) genes encoding proteins that trigger antibody responses in infected rodents or humans (n=17) (44, 45); (v) some genes that encode putative membrane protein, exported protein, or surface structures according to the annotated genome of CO92; (vi) other genes, including some genes that are annotated to be related to pathogenicity, detoxification, and mobility; and (vii) genes located in the three virulence-associated plasmids pCD1, pMT1, and pPCP1 (n=69). The full-length sequences of all of the bait proteins except YPCD1.72c (YpkA) were cloned into pDEST22 using Gateway recombination technology (77). The 5′-terminal (1 to 1,500 bp) and 3′-terminal (358 to 2,199 bp) sequences of YPCD1.72c were cloned into pDEST22, respectively. YPCD1.26c (YopM) was eliminated because it self-activated the reporter in the yeast indicator strain. Each bait protein was screened at least once against the human spleen cDNA library. A total of 319 Y. pestis-human protein-protein interactions (YHPI), involving 79 bait proteins and 159 human proteins, were identified from the first round of Y2H screening (see Table S2 in the supplemental material). To reduce the number of false-positive interactions, each pair of 319 interactions was retested by yeast cotransformation assays. Up to 204 interactions between 65 Y. pestis proteins and 109 human proteins were validated, representing a validation rate of 63.9% (Fig. 1A; see Table S3 in the supplemental material). Yop effectors were the most intensively studied virulence proteins in Yersinia, for which a lot of cellular targets in host cells had been identified (see Table S4 in the supplemental material). Interactions that were identified by Y2H screening but not validated by an independent assay were not included in the “known interactions.” Unfortunately, we could not gain those interaction pairs in our Y2H screens except for YpkA-β-actin (ACTB) (37). The relatively low overlap with the known Yersinia-host protein interactions might reflect the unsaturation of our Y2H screening, and the use of a few fragments of one Yersinia protein as bait to screen against human cDNA libraries of different tissues might improve the overlap rate. The number of bait proteins that yielded prey and no prey in each category are shown in Fig. 1B, and about 43.1% of the bait proteins yielded prey in the Y2H screening. The bait category of “known virulence factors” has the highest prey-yielding percentage (82%). Three bait proteins that are intact in CO92 but mutated in 91001 (YPO0594, YPO0962, and YPO2309) were found to interact with human proteins in our Y2H screenings. YPO0594 is a conserved hypothetical protein containing a C-terminal vWA domain that is found in cell adhesion and extracellular matrix proteins, and this protein is deserved to be investigated for its roles in Y. pestis pathogenesis. Eight of 19 membrane proteins or surface structures were found to interact with prey proteins of the human cDNA library, similar to the average prey-yielding percentage of all of the bait proteins from different categories. The category of “other genes,” which includes genes predicted to be related to pathogenicity, detoxification, or mobility (Fig. 1 to 6), gives the lowest prey-yielding percentage. To validate the protein-protein interactions in our PPI data set using a method independent of Y2H, GST-pulldown assays were performed to test 24 pairs of interactions (11.8% of the Y2H data set). Of the 24 pairs (75%), 18 were confirmed by GST pulldown assays (see Table S5 in the supplemental material), suggesting the reliability of our Y2H data set (Fig. 1D). This validation rate is comparable to that of a previously published data set of HCV-human interactomes (84.6%) (22) and higher than that of the EBV-human and KSHV-human interactomes (42% and 48%, respectively) (18, 73).
Fig. 1.
YHPI data set. (A) Summary of the YHPI data set. HYTs are the human proteins targeted by Y. pestis; H represents the human proteins, and Y represents Y. pestis proteins. (B) Number of bait proteins yielding prey (blue bars) and with no prey (white bars) in each category. Bars: 1, known virulence factor; 2, intact in CO92 but mutated in 91001; 3, actively regulated in response to unfavorable conditions; 4, triggering antibody response; 5, predicted membrane protein, surface structures, and so on; 6, other genes. pCD1, pMT1, and pPCP1 represent the genes carried in pCD1, pMT1, and pPCP1, respectively. (C) Distribution of Y. pestis proteins according to the number of HYTs interacting with a certain bait protein. (D) GST pulldown validation of Y. pestis-human interactions. α-GST, anti-GST; α-FLAG, anti-FLAG. The results for 12 of the 18 pairs of positive interactions are shown.
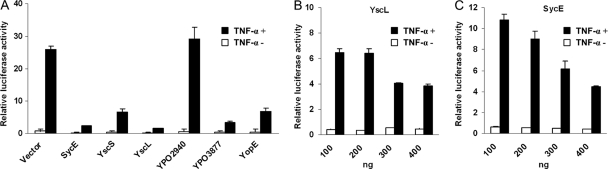
Fig. 6.
Inhibition of NF-κB activation by Y. pestis proteins. (A) HEK-239T cells were transfected with NF-κB dual-luciferase reporter plasmid together with an empty vector, pCMV-Myc, or one of the plasmids expressing the Y. pestis SycE, YscC, YscL, YopE, YPO2940, and YPO3877 proteins. After 2 days, the luciferase activities were determined for cells treated with or without 10 ng/ml TNF-α for 1 h before the determination. (B and C) Dose-dependent inhibition of NF-κB activation by YscL and SycE, respectively. The results represent the mean values of the relative luciferase activities from triplicate determinations.
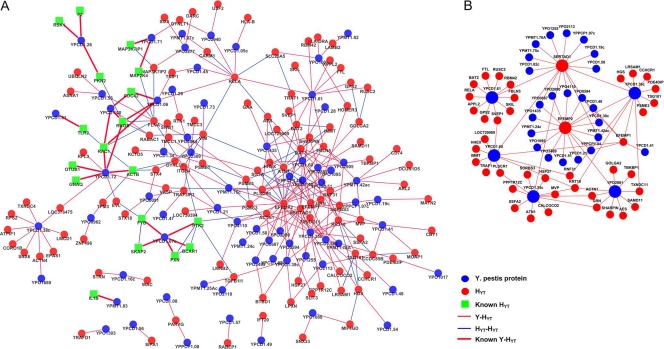
The distribution of bait proteins according to the number of yielded prey proteins in the Y2H assay is shown in Fig. 1C. Among the 153 bait proteins, 87 did not yield any prey under our experimental conditions. The majority of the rest (70%) interacted with 1 to 3 human proteins. However, some bait proteins have as many as 10 interaction partners, such as YPCD1.35c, YPCD1.36c, YPCD1.61, YPCD1.60, and YPO2881 (Fig. 2B). One-third of the HYTs (42 proteins) were connected to two or more Y. pestis bait proteins in our YHPI network. Several HYTs, such as EFEMP2 and SERTAD1, interacted with 16 out of the 69 Y. pestis proteins (Fig. 2B). Interestingly, SETD2 and HOXD8 also interacted with 5 out of 11 HCV proteins (22). In addition, HOMER3 and GRN were found to interact with 9 out of 85 EBV proteins (18). It seems common for pathogens to utilize multiple proteins to interact with the same host protein, implying that those human proteins may be critical for pathogenesis and are possibly involved in various stages of bacterial and viral infection. We noted that many interactions in our data set involve Y. pestis proteins that have not been thought to be secreted out of bacteria or surface exposed. For instance, YPCD1.05c (SycE) and YPCD1.51 (YscB) are chaperone proteins, and YPCD1.38 (TyeA) and YPCD1.37c (SycN) are proteins that participate in the formation of the YopN/TyeA/YscB/SycN complex that blocks the secretion of Yops under the noninducing condition (21); YscS is an inner membrane protein and YscL is a cytosolic protein, and these proteins play a regulatory role with YscN ATPase (21). Whether these proteins are accessible to host proteins are questionable; however, a lot of interaction partners of these proteins were identified in Y2H screens. It has been acknowledged that Y2H experiments are prone to false-positive results that have no biological significance. Thus, it is inevitable that a portion of the Y2H-identified interactions were false-positive results. However, we guess that not all of the interactions that currently seem hard to understand were false positive, some of which might be cues for the future studies. After all, many unrevealed mechanism in pathogenesis are waiting to be illuminated.
Fig. 2.
Graphical view of the YHPI network. (A) Graph of the YHPI network by combining the interactions identified in the present Y2H study with the 23 published interactions between Y. pestis and human proteins. (B) Graph of interactions involving high-degree nodes in the YHPI network, including both Y. pestis proteins and HYTs that interact with multiple HYTs and multiple Y. pestis proteins. Blue and red circles represent Y. pestis proteins and HYTs, respectively. Green squares represent known HYTs. Red lines represent interactions between Y. pestis and HYTs, and bold red lines represent the previously published interactions. Blue lines represent interactions between HYTs. The larger nodes are proteins that interact with over 10 partners.
A YHPI network was then constructed using 204 pairs of interactions identified in our Y2H assay and 23 pairs of known interactions (Fig. 2A; see Table S4 in the supplemental material). According to the definition of “known interactions” described above, we have not included interactions reported by Dyer et al. (27) in this YHPI network. The resulting network consisted of 226 interactions involving 69 Y. pestis bait proteins and 126 human proteins. A human-human protein-protein interaction (H-H PPI) network constructed by de Chassey et al. (22) was used as a reference to determine the relationships between different human proteins and to calculate the topological parameters of each human protein. This reference network consists of 44,223 PPIs between 9,522 distinct human proteins (22), and all of the interactions were physical and direct binary PPIs retrieved from BIND (7), BioGRID (13), DIP (80), GeneRIF (48), HPRD (60), IntAct (39), MINT (19), and Reactome (74). These 9,522 human proteins represent approximately 30% of the human proteome, and the rest of the human proteins that have no known cellular partners are therefore not included in this network. Interestingly, 109 HYTs (corresponding to 86.5% of the total number of 126) can be found in the H-H PPI network, suggesting that Y. pestis bait proteins preferentially interact with the human proteins that have known connections with other cellular partners (P < 0.0001, Fisher's exact test). This result coincides well with the findings with interactomes for HCV, EBV, and human-pathogen PPI networks by Dyer et al. (18, 26, 73).
Y. pestis preferentially interacts with hubs and bottlenecks of the human PPI network.
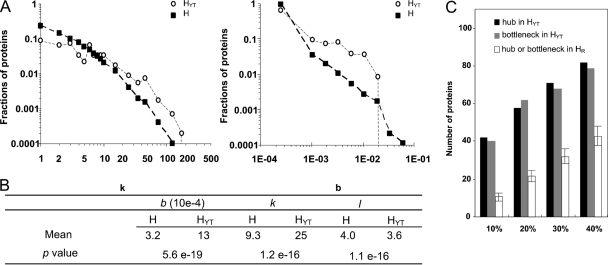
To understand better the properties of human proteins that interact with Y. pestis proteins, the topological features of the YHPI network were analyzed. Biological networks are generally characterized as small world and scale free, and they can be depicted by the parameters “degree” (k), “betweenness” (b), and “shortest path” (l) to describe the centrality of a protein in a network (28). The degree of a given protein denotes the number of proteins that directly interact with it in a network; thus, it indicates the local centrality of the protein. The shortest path is the minimum distance between two proteins in a network, and the smaller the l, the greater the topological proximity of a protein to other nodes in a network (36). Betweenness denotes the number of shortest paths that pass through a given protein; thus, b reflects the global centrality of a protein. Generally, proteins interacting with a large number of partners (with higher degrees) are designated hubs, whereas those that lie on many shortest paths are designated bottlenecks (with higher betweenness). The parameters k, b, and l for HYTs (human proteins that are targeted by Y. pestis) and human proteins (H) were calculated using the NetworkAnalyzer plugin of Cytoscape (6). The distributions of degree and betweenness for H and HYTs were plotted in log-log scale, and both the degree and the betweenness distributions of HYTs were shown to be more biased toward high-degree/betweenness proteins than those for H (Fig. 3A). The average k and b of HYTs are significantly higher than those of H integrated in the H-H PPI network (for k, P=1.2 × 10−16; for b, P=5.6 × 10−19; and for l, P=1.1 × 10−16; Mann-Whitney U test). However, the average l is significantly lower than that of H (Fig. 3B), indicating that Y. pestis proteins tend to interact with highly connected human proteins that have higher local or global centrality in the human PPI network.
Fig. 3.
Topological analysis of the YHPI network. (A) Log-log distributions of degree and betweenness for H and HYTs in the H-H PPI network. (B) Averages of degree (k), betweenness (b), and shortest path length (l) for all human proteins (H) and for HYTs integrated in the H-H PPI network. The Mann-Whitney U test was used to test the significance of the differences in k, b, and l between H and HYTs. (C) Comparison of the numbers of hubs or bottlenecks in HYTs with those in HR, which represents the data set generated by random sampling of H from the H-H interaction network. The x axis indicates the cutoffs for the classification of a protein as a hub protein or as a bottleneck protein (10%, 20%, 30%, or 40%), and the y axis indicates the number of hub or bottleneck proteins in the indicated data sets. The average number of hub or bottleneck proteins in 1,000 HR data sets is shown with the standard deviations.
To determine if this tendency of HYTs with higher centrality in the human PPI network is statistically significant, the percentages of the hubs and bottlenecks in HYTs were compared with those of data sets generated by random sampling from the human PPI network. First, all nodes in the human PPI network were sorted by k values (when the number of hubs is calculated) or b values (when the number of bottlenecks is calculated). Proteins with k values in the top 10% are classified as hubs, whereas those with b values in the top 10% are classified as bottlenecks (Fig. 3C). The number of hubs or bottlenecks in a data set is then calculated by counting the number of proteins with k or b values higher than the 10% cutoff values. “HR” represents a data set generated by randomly sampling 109 proteins from the human PPI network, and the average number of hubs or bottlenecks in 1,000 HR data sets was calculated. The number of proteins above various cutoff values (20%, 30%, or 40%) (Fig. 3C), was also analyzed. Both hubs and bottlenecks are significantly enriched in HYT compared with HR (empirical P < 0.001, permutation test). Hub proteins usually play important roles in human cellular functions and are central in certain functional modules in an organism. Bottleneck proteins are critical for many cellular pathways, and they can link distinct functional modules together and facilitate cells to exert biological process synergistically. The interactomes previously reported for EBV, HCV, and KSHV also found that proteins with higher centrality are more frequently targeted by pathogen proteins. These findings indicate that microbial pathogens have evolved to interfere preferentially with the essential nodes in the host PPI network.
Resistance of phagocytosis by Y. pestis and host cytoskeleton pathways.
To analyze further the YHPI network, the specific cellular pathways enriched in HYTs were studied using the DAVID web-based bioinformatics approach (35). Enrichment of the KEGG pathway was tested by Fisher's exact test followed by the Benjamini and Hochberg multiple test correction. Five pathways were found to be significantly enriched (P < 0.05) (Table 1), especially those important in immune surveillance and innate immune response, such as the leukocyte transendothelial migration, TLR, and MAPK signaling pathways. The rest of the enriched pathways, related to focal adhesion and regulation of actin cytoskeleton, are known to be involved in phagocytosis. The capability of Y. pestis to interfere with these pathways is intimately correlated with the plague infection. The HCV infection protein network reported that two of the pathways interfered with by Y. pestis, the MAPK signaling and focal adhesion pathways, were also enriched for HCV proteins. Besides those two pathways, 14 pathways, including those of insulin, Jak/STAT, and transforming growth factor β (TGF-β), which were associated with clinical disorders of chronically infected patients, were found to be enriched for HCV proteins. Comparison of the pathways interfered with by HCV and Y. pestis suggests that the bacterial and viral pathogens could attack a distinct spectrum of cellular pathways, which might reflect the specific lifestyle and pathogenesis of different pathogens.
Table 1.
KEGG pathway enrichment analysis of HYTs
| Pathway type and KEGG term | No. of HYTsa | P valueb | Known HYTs | Newly identified HYTs |
|---|---|---|---|---|
| Immune system | ||||
| Leukocyte transendothelial migration | 11 (7) | 3.14E−06 | RHOA, RAC1, CDC42, BCAR1, PTK2, ACTB, PXN | VASP, ACTN4, ACTN1, SIPA1 |
| Toll-like receptor signaling | 7 (5) | 1.05E−02 | MAP2K4, IL1B, RAC1, MAP3K7IP1, TLR2 | MAP3K7IP2, RELA |
| Cell communication | ||||
| Focal adhesion | 14 (7) | 1.36E−06 | RAC1, BCAR1, PXN, PTK2, CDC42, ACTB, RHOA | FLNA, ITGB4, ACTN1, ACTN4, PARVG, EVL, LAMB2 |
| Signaling | ||||
| MAPK | 9 (6) | 3.63E−02 | MAP2K4, RAC1, RSK1, MAP3K7IP1, CDC42, IL1B | FLNA, RELA, MAP3K7IP2 |
| Cell motility | ||||
| Regulation of actin cytoskeleton | 11 (8) | 5.24E−04 | RHOA, RAC1, CDC42, BCAR1, F2, PTK2, ACTB, PXN | ACTN4, ACTN1, ITGB4 |
Numbers in parentheses indicate the numbers of HYTs that have been previously reported.
Enrichment of the KEGG pathway was tested by Fisher's exact test followed by the Benjamini and Hochberg multiple test correction (P < 5E−02).
Focal adhesion plays vital roles in various cellular processes, including phagocytosis, a host defense mechanism that Y. pestis has to avoid to begin its extracellular life in the host. Our YHPI network reveals that Y. pestis proteins interfere in the regulation of focal adhesion and the actin cytoskeleton at multiple points, which is in accordance with the functional impairment of phagocytes by Y. pestis. Thus, a subnetwork that contains interactions associated with the Y. pestis ability for antiphagocytosis (Fig. 4A) was generated by combining PPIs involving the focal adhesion and cytoskeletal regulation pathways (Y. pestis-FC subnetwork).
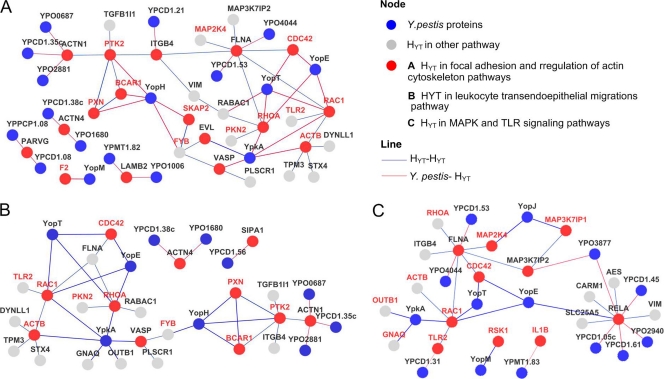
Fig. 4.
Subnetworks of the YHPI network. Human proteins in corresponding pathways, their first neighbors, and the relationship between the proteins in the YHPI network are contained in each subnetwork. (A) Y. pestis-FC subnetwork; (B) Y. pestis-MT subnetwork; (C) Y. pestis-LTM subnetwork.
In the FC subnetwork, 7 HYTs were identified to interact with Y. pestis proteins in the current study, and the other HYTs were human targets that have been previously reported for Y. pestis. Among them, BCAR1 (10), PTK2 (62), FYB (23), SKAP2, and PXN (11) are important components of the focal adhesion complex, all of which are substrates of YopH (YPCD1.67c), a potent tyrosine phosphatase delivered into the host cells by the Y. pestis T3SS. Dephosphorylation of these proteins by YopH leads to the disruption of focal adhesion complexes and impairment of phagocytosis (62). Phagocytosis is also controlled by small GTP-binding proteins (G proteins), such as CDC42, RAC1, and RHOA (12, 30). YopT (YPCD1.20), YopE (YPCD1.06), and YpkA (YPCD1.72c) have been shown to disrupt the host cytoskeleton by interacting with CDC42, RAC1, and RHOA (Fig. 4A), as well as contributing to the antiphagocytic activity of Y. pestis (2, 8, 83).
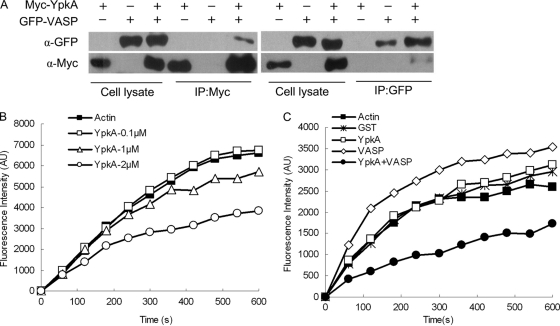
Vasodilator-stimulated phosphoprotein (VASP) and EVL are newly identified cellular targets of YpkA in the Y. pestis-FC subnetwork. They belong to the Enabled/VASP (Ena/VASP) family, which plays important roles in the regulation of cytoskeleton dynamics (40). The mammalian Ena/VASP proteins are substrates of serine/threonine kinase such as cyclic AMP (cAMP)-dependent protein kinase A (PKA) and protein kinase C (PKC) (29, 79). YpkA, a Yersinia T3SS effector, harbors an N-terminal serine/threonine kinase catalytic domain, a C-terminal guanidine nucleotide dissociation inhibitor (GDI) domain, and an actin-binding domain consisting of C-terminal 20 amino acids of YpkA (25, 37, 63). Activation of its kinase activity within the host cell depends on binding with β-actin (ACTB), and deletion of the actin-binding domain obliterates its kinase activity, as well as the interaction between YpkA and actin (37). We determined that VASP, EVL, ACTB, RPL3, and LOC376475 interact with the C-terminal fragment of YpkA, which harbors an intact kinase domain, GDI domain, and actin-binding domain. However, no prey proteins were obtained in Y2H screening when using the N-terminal fragment of YpkA as bait, which is deprived of the actin-binding domain and functional GDI domain. Binding activity between YpkA and VASP was confirmed by GST-pulldown assay (Fig. 1D). We further performed a coimmunoprecipitation assay by overexpressing Myc-YpkA and GFP-VASP in HEK-293 cells and immunoprecipitating them using anti-Myc or anti-green fluorescent protein (GFP) antibodies. It was found that YpkA could be immunoprecipitated by the antibody against GFP and vice versa, confirming the binding activities between YpkA and VASP (Fig. 5A).
Fig. 5.
YpkA interacts with VASP and inhibits VASP-mediated activation of actin assembly. (A) Overexpressed YpkA and VASP interact with each other in vivo. HEK-293T cells were transfected with Myc-YpkA- and GFP-VASP-expressing plasmids as indicated. Supernatants of the cell lysates were subjected to immunoprecipitation (IP) with anti-Myc (α-Myc) or anti-GFP (α-GFP) antibodies followed by immunoblotting with the indicated antibodies. (B) YpkA inhibits actin polymerization in a concentration-dependent manner. Actin polymerization was measured by monitoring the fluorescence intensity (in arbitrary units [AU]) of G-actin and a pyrene-labeled G-actin mixture (5:1) in fresh G-buffer in the presence of GST-YpkA at 0.1, 1, and 2 μM final concentrations. (C) YpkA inhibits VASP-mediated enhancement of actin polymerization. Actin polymerization was measured as described above in the presence of GST, GST-YpkA, GST-VASP, or YpkA plus VASP at a 0.1 μM final concentration.
Previous studies have shown that VASP could promote actin polymerization (5, 9, 72). In order to see whether YpkA could affect the physiological function of VASP, we performed an actin polymerization assay using pyrene-labeled actin in the presence of the purified YpkA and VASP recombinant proteins (34). The results showed that the presence of GST-YpkA could inhibit spontaneous actin polymerization in a dose-dependent manner (Fig. 5B), whereas GST protein alone had no effect (data not shown). Addition of GST-VASP to the G-actin buffer at a 0.1 μM final concentration resulted in great enhancement of actin polymerization compared to the spontaneous actin polymerization (Fig. 5C). However, simultaneous addition of both GST-YpkA and VASP (each at a 0.05 μM final concentration) dramatically inhibited the VASP-mediated actin polymerization (Fig. 5C), although YpkA alone at this concentration has no obvious effect on the actin polymerization. These results indicated that YpkA may interfere with the VASP-induced actin polymerization.
We also found that YPO1006 and YPMT1.82 interacted with LAMB2, an extracellular matrix protein implicated in cell motility and proliferation. YPO1006 is a leucine-rich protein, and YPMT1.82 encodes a chaperone for the F1 capsules (49). Given that LAMB2 is an extracellular matrix structural constituent and both of the Yersinia proteins are located on the bacterial cell surface, we speculated that these interactions might play roles in bacterial adhesion. In addition, both YPO4044 and YPCD1.53 interacted with FLNA. YPO4044 is a putative fimbrial protein, whereas YPCD1.53 is a T3SS structural protein. FLNA is important in binding to small GTPase (56), actin filament (57), and Fc-γ receptor I complex and plays a positive regulatory role in the IκB kinase/NF-κB cascade.
The HYTs in the FC subnet play various roles in cytoskeletal actin regulation, from the G protein-coupled receptor (GPCR) subunit GANQ (54) and small G protein (CDC42, RAC1, and RHOA) (52, 67, 76) to the structural constituent of cytoskeleton (ACTB) (38). This indicates that Y. pestis can interfere at multiple points in a single cellular process, possibly to exert functional redundancy to ensure successful modulation of the cellular process. Multiple interactions between Y. pestis and human proteins may possibly synergistically perturb the signal transductions of phagocytosis, thereby enabling Y. pestis to exploit the cytoskeletal remodeling essential for phagocytosis.
Intercepting the innate immune response via LTM, TLR, and MAPK signaling pathways.
Leukocyte migration from blood into the sites of infection is a vital immune surveillance strategy (68). Various cytokines can be stimulated and secreted during infection with plague to recruit leukocytes to the infection sites. The YHPI network revealed that 11 HYTs involved in leukocyte transendoepithelial migrations (LTM) were targeted by 10 Y. pestis proteins (Fig. 4B). This suggests that Y. pestis might affect the neutrophil/PMN migration to the infection sites by interfering with the LTM pathway. Clinical manifestations indicate that Y. pestis replicates very fast inside hosts, and high bacterial loads are often found in liver, spleen, lymph nodes, or blood of mice infected with plague, largely due to the severe suppression of the host immune response (1, 17, 42). Our findings suggest that Y. pestis may actively modulate the recruitment of leukocytes to avoid being killed and thus replicate massively. This notion is supported by previous results showing that dendritic cells pulsed by Y. pestis failed to migrate through the endothelial cells (75). In addition, robust neutrophil recruitment to the lungs could not be observed 48 h after infection, implying that leukocyte migration is delayed (15).
TLRs and MAPK cascade play pivotal roles in host defense in response to microbial infections. Activation of the TLR signaling pathway triggers downstream cascades that ultimately result in the activation of NF-κB and initiation of p38 and Jun N-terminal protein kinase (JNK) MAP kinase signaling pathways. Distinct groups of MAPKs (extracellular signal-regulated kinase 1/2 [ERK-1/2], JNK, p38 proteins, and ERK-5) can be activated by specific MAPK kinases in response to distinct extracellular stimuli. Interestingly, all HYTs (except RSK1, known to interact with YopM) involved in the MAPK signaling pathway (see Fig. S1 in the supplemental material) participate in the JNK and p38 pathways that play central roles in immune responses, suggesting that these Y. pestis-human PPIs are highly relevant to the host response to infection. Due to the overlapping of HYTs in these two pathways, we combined them to construct a subnetwork (Fig. 4C) that targets human TLR and MAPK (TM) signaling pathways (Y. pestis-TM subnetwork).
The majority of HYTs (MAP2K4, RSK1, MAP3K7IP1, CDC42, RAC1, and IL1B) in the MAPK signaling pathway are known human targets of Y. pestis. For example, YopJ, YopE, YopT, and YpkA all interact, directly or indirectly, with the MAPK pathway to modulate the host immune response (53, 69). FLNA, RELA, and MAP3K7IP2 are newly identified to interact with Y. pestis in this subnetwork.
To determine whether the identified Y. pestis-human interactions are functionally relevant, the activation of NF-κB in HEK-293T cells expressing Yersinia proteins that were identified to interact with RELA (p65 subunit of NF-κB) was analyzed using a luciferase reporter assay. Except for YPO2940, expression of YPCD1.05 (SycE), YPCD1.06 (YopE), YPCD1.45 (YscK), YPCD1.61 (YscL), and YPO3877 in cells significantly inhibited TNF-α-induced NF-κB activation (Fig. 6A), and the inhibition activities of YscL and SycE showed dose dependence (Fig. 6B and C). In addition, the interactions of RELA with YPO3877, YscL, and SycE were confirmed by GST pulldown assays, suggesting that these interactions are functionally relevant (see Table S5 in the supplemental material). Although RELA could be pulled down by bacterially expressed GST-tagged YPO2940, no influence on NF-κB activation could be detected in cells expressing YPO2940. The reasons for these conflicting results are unknown, and further experiments have to be done to get more confident results. The molecular mechanisms through which these Yersinia proteins inhibit NF-κB activation merit future investigation.
Y. pestis and viruses share common human targets.
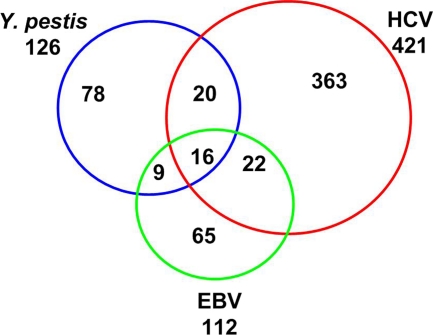
Only very limited studies have use Y2H to examine the interactome between pathogen and host proteins until now. The recently published three interaction networks between human and bacterial pathogens by Dyer et al. reported interactomes of Y. pestis, B. anthracis, and F. tularensis by using a random yeast two-hybrid (Y2H) approach (27). However, no validation for these interactions has been done. The human-Y. pestis interaction network reported in this work contains more than 4,000 interactions between 1,218 Y. pestis proteins and 2,108 human proteins, a much larger data set than ours (27). Comparison of Dyer's data set to ours found that 27 of 124 HYTs were included in 2,108 human target proteins in Dyer's work (see Table S7 in the supplemental material); however, the Y. pestis-human protein interaction pairs involved are distinct from our data set. Only 22 of 153 Yersinia bait proteins in our study were contained in Dyer's data set, and no common interaction existed for these 22 Y. pestis proteins in the two data sets, probably and partially due to the fact that different approaches were adopted in the two studies (random Y2H versus direct Y2H). Three virus-human interactomes involving HCV, EBV, and KSHV had been reported (18, 22, 73). Among them, the KSHV-human interactome was constructed based on the bioinformatics prediction combined with coimmunoprecipitation (Co-IP) and Y2H validation and contains only 20 interactions between 8 KSHV and 20 human proteins (73), which render it obviously unsuitable for the detailed comparison with our YHPI network. The PPI data set of interactomes between human and HCV or EBV proteins were all generated by direct Y2H screening against a human cDNA library (18, 22). The numbers of bait proteins (pathogen proteins), prey proteins (human targets), and PPIs were 11, 421, and 481 for the HCV-human interactome and 40, 112, and 148 for the EBV-human interactome, respectively (see Table S6 in the supplemental material). These numbers are comparable with the YHPI network, which contains 225 PPIs between 66 Y. pestis proteins and 124 human proteins; therefore, we compared these two published virus-host interactomes with our YHPI network. Bacterial and viral pathogens have different niches and life cycles in the host; therefore, one would expect that their cellular targets and strategies for causing infection should be largely different. However, our analysis indicated that many proteins targeted by Yersinia are also modulated by HCV and/or EBV (Fig. 7) (18, 22). A total of 67 human proteins interact with proteins from at least two of the three pathogens. Y. pestis tends to interfere with the proteins involved in cytoskeletal rearrangement and cell adhesion, whereas HCV and/or EBV preferentially interact with extracellular components (see Fig. S2 in the supplemental material). This distinction may reflect that adhesion and disruption of cytoskeleton are necessary for Y. pestis pathogenesis, but viruses need to anchor themselves to the host cell surface through extracellular matrix before entry into the target cells. In addition, a high fraction of proteins (16/127 HYTs) interacted with all three pathogens (Table 2). Among these 16 common targets, PLSCR1, FBLN5, and EFEMP2 are involved in blood coagulation and wound healing, whereas LAMB2, VIM, and SLIT3 are involved in cell motility. Future studies may unravel the molecular mechanisms through which these interactions aid the infections. The present PPI data sets between humans and pathogens (73, 18, 22) are obviously far from complete, and much more common cellular targets will be identified for various groups of pathogens in the future. These may aid in the design of therapeutic agents to control a wide spectrum of infections.
Fig. 7.
Common human targets of EBV, HCV, and Y. pestis. Shown is a Venn diagram of human proteins interacting with three different pathogens. The total numbers of human proteins interacting with the specific pathogens are labeled below the pathogen names. Blue circle, HYTs; red circle, human proteins that interact with HCV proteins; green circle, human proteins that interact with EBV proteins.
Table 2.
Human proteins that interact with HCV, EBV, and Y. pestis proteins
| Gene name | Product | Functional annotation |
|---|---|---|
| ACTN1 | Actin, alpha 1 | F-actin cross-linking protein thought to anchor actin to a variety of intracellular structures |
| CCHCR1 | Coiled-coil alpha-helical rod protein 1 | May be a regulator of keratinocyte proliferation or differentiation |
| EFEMP1 | EGFa-containing fibulin-like extracellular matrix protein 1 | Extracellular matrix glycoprotein |
| EFEMP2 | EGF-containing fibulin-like extracellular matrix protein 2 | Extracellular matrix glycoprotein implicated in blood coagulation, activation of complement, and determination of cell fate during development |
| FBLN5 | Fibulin 5 | Promotes adhesion of endothelial cells and could be a vascular ligand for integrin receptors |
| GRN | Granulin | Functions in normal development, wound healing, and tumorigenesis |
| HOMER3 | Homer homolog 3 | May be involved in cell growth and belongs to the homer family of dendritic proteins |
| LAMB2 | Laminin, beta 2 (laminin S) | Implicated in a wide variety of biological processes, including cell adhesion, differentiation, migration, signaling, neurite outgrowth, and metastasis |
| PLSCR1 | Phospholipid scramblase 1 | May play roles in antiviral response of interferon and contribute to cytokine-regulated cell proliferation and differentiation |
| PSME3 | Proteasome (prosome, macropain) activator subunit 3 | Subunit of the PA28-gamma proteasome regulator, inhibits apoptosis after DNA damage, and may also be involved in cell cycle regulation |
| RNF31 | Ring finger protein 31 | Unknown; contains a RING finger, a motif present in a variety of functionally distinct proteins and known to be involved in protein-DNA and protein-protein interactions |
| SERTAD1 | SERTA domain containing 1 | Acts at E2F-responsive promoters to integrate signals provided by PHD- and/or bromo domain-containing transcription factors, stimulates E2F-1/DP-1 transcriptional activity, and renders the activity of cyclin D1/CDK4 resistant to the inhibitory effects of p16(INK4a) |
| SLIT3 | Slit homolog 3 | May act as molecular guidance cue in cellular migration |
| TRAF3IP3 | TRAF3-interacting protein 3 | May function as an adapter molecule that regulates TRAF3-mediated JNK activation |
| TXNDC11 | Thioredoxin domain containing 11 | Regulator involved in DUOX protein folding |
| VIM | Vimentin | Responsible for maintaining cell shape, integrity of the cytoplasm, and stabilizing cytoskeletal interactions and is also involved in the immune response |
EGF, epidermal growth factor.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daoguo Zhou (Purdue University, West Lafayette, IN), Feng Shao (National Institute of Biological Sciences, China), and Yue Zhang (State University of New York at Stony Brook, Stony Brook, NY) for critical review of the manuscript.
This work was supported by the National Basic Research Program of China (2012CB518704), the National Natural Science Foundation of China (no. 30970121, 30930001, and 31170122), and the National Natural Science Foundation of China for Distinguished Young Scholars (no. 30525025).
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 12 September 2011.
REFERENCES
- 1. Agar S. L., et al. 2009. Characterization of the rat pneumonic plague model: infection kinetics following aerosolization of Yersinia pestis CO92. Microbes Infect. 11:205–214 [DOI] [PubMed] [Google Scholar]
- 2. Aili M., Isaksson E. L., Hallberg B., Wolf-Watz H., Rosqvist R. 2006. Functional analysis of the YopE GTPase-activating protein (GAP) activity of Yersinia pseudotuberculosis. Cell. Microbiol. 8:1020–1033 [DOI] [PubMed] [Google Scholar]
- 3. Andersson K., et al. 1996. YopH of Yersinia pseudotuberculosis interrupts early phosphotyrosine signalling associated with phagocytosis. Mol. Microbiol. 20:1057–1069 [DOI] [PubMed] [Google Scholar]
- 4. Aoki-Kinoshita K. F., Kanehisa M. 2007. Gene annotation and pathway mapping in KEGG. Methods Mol. Biol. 396:71–92 [DOI] [PubMed] [Google Scholar]
- 5. Applewhite D. A., et al. 2007. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol. Biol. Cell 18:2579–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Assenov Y., Ramirez F., Schelhorn S. E., Lengauer T., Albrecht M. 2008. Computing topological parameters of biological networks. Bioinformatics 24:282–284 [DOI] [PubMed] [Google Scholar]
- 7. Bader G. D., Betel D., Hogue C. W. 2003. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 31:248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barz C., Abahji T. N., Trulzsch K., Heesemann J. 2000. The Yersinia Ser/Thr protein kinase YpkA/YopO directly interacts with the small GTPases RhoA and Rac-1. FEBS Lett. 482:139–143 [DOI] [PubMed] [Google Scholar]
- 9. Barzik M. 2005. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 280:28653–28662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Black D. S., Bliska J. B. 1997. Identification of p130Cas as a substrate of Yersinia YopH (Yop51), a bacterial protein tyrosine phosphatase that translocates into mammalian cells and targets focal adhesions. EMBO J. 16:2730–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Black D. S., Marie-Cardine A., Schraven B., Bliska J. B. 2000. The Yersinia tyrosine phosphatase YopH targets a novel adhesion-regulated signalling complex in macrophages. Cell. Microbiol. 2:401–414 [DOI] [PubMed] [Google Scholar]
- 12. Boquet P. 2000. Small GTP binding proteins and bacterial virulence. Microbes Infect. 2:837–843 [DOI] [PubMed] [Google Scholar]
- 13. Breitkreutz B. J., et al. 2008. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 36:D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brubaker R. R. 2003. Interleukin-10 and inhibition of innate immunity to yersiniae: roles of Yops and LcrV (V antigen). Infect. Immun. 71:3673–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bubeck S. S., Cantwell A. M., Dube P. H. 2007. Delayed inflammatory response to primary pneumonic plague occurs in both outbred and inbred mice. Infect. Immun. 75:697–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Busso D., Delagoutte-Busso B., Moras D. 2005. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 343:313–321 [DOI] [PubMed] [Google Scholar]
- 17. Butler T. 2009. Plague into the 21st century. Clin. Infect. Dis. 49:736–742 [DOI] [PubMed] [Google Scholar]
- 18. Calderwood M. A., et al. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U. S. A. 104:7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chatr-Aryamontri A., et al. 2007. MINT: the Molecular INTeraction database. Nucleic Acids Res. 35:D572–D574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cornelis G. R., et al. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cornelis G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811–825 [DOI] [PubMed] [Google Scholar]
- 22. de Chassey B., et al. 2008. Hepatitis C virus infection protein network. Mol. Syst. Biol. 4:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deleuil F., Mogemark L., Francis M. S., Wolf-Watz H., Fallman M. 2003. Interaction between the Yersinia protein tyrosine phosphatase YopH and eukaryotic Cas/Fyb is an important virulence mechanism. Cell. Microbiol. 5:53–64 [DOI] [PubMed] [Google Scholar]
- 24. Du Y., Rosqvist R., Forsberg A. 2002. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 70:1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dukuzumuremyi J. M., et al. 2000. The Yersinia protein kinase A is a host factor inducible RhoA/Rac-binding virulence factor. J. Biol. Chem. 275:35281–35290 [DOI] [PubMed] [Google Scholar]
- 26. Dyer M. D., Murali T. M., Sobral B. W. 2008. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 4:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dyer M. D., et al. 2010. The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS One 5:e12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goh K. I., Oh E., Jeong H., Kahng B., Kim D. 2002. Classification of scale-free networks. Proc. Natl. Acad. Sci. U. S. A. 99:12583–12588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gomez T. M., Robles E. 2004. The great escape; phosphorylation of Ena/VASP by PKA promotes filopodial formation. Neuron 42:1–3 [DOI] [PubMed] [Google Scholar]
- 30. Hall A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509–514 [DOI] [PubMed] [Google Scholar]
- 31. Han Y., et al. 2005. DNA microarray analysis of the heat- and cold-shock stimulons in Yersinia pestis. Microbes Infect. 7:335–348 [DOI] [PubMed] [Google Scholar]
- 32. Han Y., et al. 2007. Comparative transcriptomics in Yersinia pestis: a global view of environmental modulation of gene expression. BMC Microbiol. 7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hentschke M., et al. 2010. Yersinia virulence factor YopM induces sustained RSK activation by interfering with dephosphorylation. PLoS One 5:e13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Higashide W., Dai S., Hombs V. P., Zhou D. 2002. Involvement of SipA in modulating actin dynamics during Salmonella invasion into cultured epithelial cells. Cell. Microbiol. 4:357–365 [DOI] [PubMed] [Google Scholar]
- 35. Huang D. W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 36. Jeong H., Mason S. P., Barabasi A. L., Oltvai Z. N. 2001. Lethality and centrality in protein networks. Nature 411:41–42 [DOI] [PubMed] [Google Scholar]
- 37. Juris S. J., Rudolph A. E., Huddler D., Orth K., Dixon J. E. 2000. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc. Natl. Acad. Sci. U. S. A. 97:9431–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Juris S. J., Shao F., Dixon J. E. 2002. Yersinia effectors target mammalian signalling pathways. Cell. Microbiol. 4:201–211 [DOI] [PubMed] [Google Scholar]
- 39. Kerrien S., et al. 2007. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 35:D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krause M., Dent E. W., Bear J. E., Loureiro J. J., Gertler F. B. 2003. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell Dev. Biol. 19:541–564 [DOI] [PubMed] [Google Scholar]
- 41. LaCount D. J., et al. 2005. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438:103–107 [DOI] [PubMed] [Google Scholar]
- 42. Lathem W. W., Crosby S. D., Miller V. L., Goldman W. E. 2005. Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. U. S. A. 102:17786–17791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lemaitre N., Sebbane F., Long D., Hinnebusch B. J. 2006. Yersinia pestis YopJ suppresses tumor necrosis factor alpha induction and contributes to apoptosis of immune cells in the lymph node but is not required for virulence in a rat model of bubonic plague. Infect. Immun. 74:5126–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li B., et al. 2005. Protein microarray for profiling antibody responses to Yersinia pestis live vaccine. Infect. Immun. 73:3734–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li B., et al. 2009. High-throughput identification of new protective antigens from a Yersinia pestis live vaccine by enzyme-linked immunospot assay. Infect. Immun. 77:4356–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li S., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303:540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu H., et al. 2009. Transcriptional profiling of a mice plague model: insights into interaction between Yersinia pestis and its host. J. Basic Microbiol. 49:92–99 [DOI] [PubMed] [Google Scholar]
- 48. Lu Z., Cohen K. B., Hunter L. 2007. GeneRIF quality assurance as summary revision. Pac. Symp. Biocomput. 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. MacIntyre S., et al. 2001. An extended hydrophobic interactive surface of Yersinia pestis Caf1M chaperone is essential for subunit binding and F1 capsule assembly. Mol. Microbiol. 39:12–25 [DOI] [PubMed] [Google Scholar]
- 50. Maere S., Heymans K., Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449 [DOI] [PubMed] [Google Scholar]
- 51. McCoy M. W., Marre M. L., Lesser C. F., Mecsas J. 2010. The C-terminal tail of Yersinia pseudotuberculosis YopM is critical for interacting with RSK1 and for virulence. Infect. Immun. 78:2584–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mejia E., Bliska J. B., Viboud G. I. 2008. Yersinia controls type III effector delivery into host cells by modulating Rho activity. PLoS Pathog. 4:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mukherjee S., et al. 2006. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312:1211–1214 [DOI] [PubMed] [Google Scholar]
- 54. Navarro L., et al. 2007. Identification of a molecular target for the Yersinia protein kinase A. Mol. Cell 26:465–477 [DOI] [PubMed] [Google Scholar]
- 55. Noel B. L., Lilo S., Capurso D., Hill J., Bliska J. B. 2009. Yersinia pestis can bypass protective antibodies to LcrV and activation with gamma interferon to survive and induce apoptosis in murine macrophages. Clin. Vaccine Immunol. 16:1457–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ohta Y., Suzuki N., Nakamura S., Hartwig J. H., Stossel T. P. 1999. The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. U. S. A. 96:2122–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pal Sharma C., Goldmann W. H. 2004. Phosphorylation of actin-binding protein (ABP-280; filamin) by tyrosine kinase p56lck modulates actin filament cross-linking. Cell Biol. Int. 28:935–941 [DOI] [PubMed] [Google Scholar]
- 58. Parkhill J., et al. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523–527 [DOI] [PubMed] [Google Scholar]
- 59. Parrish J. R., et al. 2007. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 8:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peri S., et al. 2004. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 32:D497–D501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Perry R. D., Fetherston J. D. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Persson C., Carballeira N., Wolf-Watz H., Fallman M. 1997. The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130Cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. EMBO J. 16:2307–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Prehna G., Ivanov M. I., Bliska J. B., Stebbins C. E. 2006. Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell 126:869–880 [DOI] [PubMed] [Google Scholar]
- 64. Pujol C., Bliska J. B. 2005. Turning Yersinia pathogenesis outside in: subversion of macrophage function by intracellular yersiniae. Clin. Immunol. 114:216–226 [DOI] [PubMed] [Google Scholar]
- 65. Pujol C., et al. 2009. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect. Immun. 77:2251–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rual J. F., et al. 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437:1173–1178 [DOI] [PubMed] [Google Scholar]
- 67. Shao F., et al. 2003. Biochemical characterization of the Yersinia YopT protease: cleavage site and recognition elements in Rho GTPases. Proc. Natl. Acad. Sci. U. S. A. 100:904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Simon S. I., Green C. E. 2005. Molecular mechanics and dynamics of leukocyte recruitment during inflammation. Annu. Rev. Biomed. Eng. 7:151–185 [DOI] [PubMed] [Google Scholar]
- 69. Sing A., et al. 2002. Yersinia V-antigen exploits toll-like receptor 2 and CD14 for interleukin 10-mediated immunosuppression. J. Exp. Med. 196:1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Song Y., et al. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11:179–197 [DOI] [PubMed] [Google Scholar]
- 71. Stelzl U., et al. 2005. A human protein-protein interaction network: a resource for annotating the proteome. Cell 122:957–968 [DOI] [PubMed] [Google Scholar]
- 72. Trasak C., et al. 2007. Yersinia protein kinase YopO is activated by a novel G-actin binding process. J. Biol. Chem. 282:2268–2277 [DOI] [PubMed] [Google Scholar]
- 73. Uetz P., et al. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239–242 [DOI] [PubMed] [Google Scholar]
- 74. Vastrik I., et al. 2007. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 8:R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Velan B., et al. 2006. Discordance in the effects of Yersinia pestis on the dendritic cell functions manifested by induction of maturation and paralysis of migration. Infect. Immun. 74:6365–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Von Pawel-Rammingen U., et al. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737–748 [DOI] [PubMed] [Google Scholar]
- 77. Walhout A. J., et al. 2000. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328:575–592 [DOI] [PubMed] [Google Scholar]
- 78. Welch W. J. 1990. Construction of permutation tests. J. Am. Stat. Assoc. 85:693–698 [Google Scholar]
- 79. Wentworth J. K., Pula G., Poole A. W. 2006. Vasodilator-stimulated phosphoprotein (VASP) is phosphorylated on Ser157 by protein kinase C-dependent and -independent mechanisms in thrombin-stimulated human platelets. Biochem. J. 393:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xenarios I., et al. 2002. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 30:303–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ye Z., et al. 2009. Gr1+ cells control growth of YopM-negative Yersinia pestis during systemic plague. Infect. Immun. 77:3791–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhou D., et al. 2006. Genome-wide transcriptional response of Yersinia pestis to stressful conditions simulating phagolysosomal environments. Microbes Infect. 8:2669–2678 [DOI] [PubMed] [Google Scholar]
- 83. Zumbihl R., et al. 1999. The cytotoxin YopT of Yersinia enterocolitica induces modification and cellular redistribution of the small GTP-binding protein RhoA. J. Biol. Chem. 274:29289–29293 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.