Abstract
Mycobacterium tuberculosis contains mannosylated cell wall components which are important in macrophage recognition and response. The building block for the mannosyl constituents of these components is GDP-mannose, which is synthesized through a series of enzymes involved in the mannose donor biosynthesis pathway. Nothing is known about the expression levels of the genes encoding these enzymes during the course of infection. To generate transcriptional profiles for the mannose donor biosynthesis genes from virulent M. tuberculosis and attenuated Mycobacterium bovis BCG, bacteria were grown in broth culture and within human macrophages. Our results with broth-grown bacteria show that there are differences in expression of the selected genes between M. tuberculosis and BCG, with increased expression of manC in M. tuberculosis and manA in BCG during stationary-phase growth. Results for M. tuberculosis extracted from within macrophages show that whiB2 is highly expressed and manB and manC are moderately expressed during infection. Rv3256c, Rv3258c, and ppm1 have high expression levels early and decreased expression as the infection progresses. Results with BCG show that, as in M. tuberculosis, whiB2 is highly expressed throughout infection, whereas there is either low expression or little change in expression of the remaining genes studied. Overall, our results show that there is differential regulation of expression of several genes in the mannose donor biosynthesis pathway of M. tuberculosis and BCG grown in broth and within macrophages, raising the possibility that the level of mannose donors may vary during the course of infection and thereby impact the biosynthesis of mannose-containing cell wall molecules.
INTRODUCTION
Tuberculosis (TB) kills nearly 2 million people each year and has become the leading cause of death among HIV patients. Although treatments have been available for more than 80 years, inconsistent completion of antibiotic courses has led to resistance to all current anti-TB drugs among bacterial isolates (33). The emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains of TB, especially in HIV patients, poses a serious threat to the control of TB worldwide. Mycobacterium tuberculosis is the causative agent of TB and possesses a compositionally unique cell wall, which is the target of several TB therapeutics, like isoniazid and ethambutol. The last time a new drug was marketed for TB was 1963 (12), and coupled with the development of resistance, this lack of newer drugs emphasizes a critical need for the discovery of new drug targets in M. tuberculosis.
It has long been thought that M. tuberculosis has coevolved with its human host, which is the only known reservoir (29). The complex pathogenicity of this bacterium in the context of its preferred niche is only partially understood. Comparative genomics between virulent strains of M. tuberculosis and the attenuated vaccine strain Mycobacterium bovis BCG have revealed gene deletions in BCG, providing insight into some key determinants of virulence within the regions of difference (RDs) (8). It was thought that complementation of the RDs in BCG would restore complete virulence, but the result was only a partial restoration (4, 22). This clearly indicates the complexity of M. tuberculosis pathogenesis and opens the door for investigation of other factors, such as regulation of transcription, that contribute to its success as a pathogen.
The M. tuberculosis cell wall contains the highly mannosylated cell wall components phosphatidyl-myo-inositol mannosides (PIMs), lipomannan (LM), and mannose-capped lipoarabinomannan (ManLAM) (2, 9, 31), which are important in TB immunopathogenesis. The terminal mannose cap structures of higher-order PIMs and ManLAM bind to the host macrophage mannose receptor in a form of host molecular mimicry (27, 30). The terminal mannose caps as well as the mannan structures in the core of these molecules are synthesized through a variety of specific mannosyltransferases that use the donors GDP-mannose and polyprenyl phosphate mannose (PPM), which are products of the mannose donor biosynthesis pathway (11). The putative genes of this pathway in M. tuberculosis are orthologs with 100% sequence identity in the ORFs as well as in the upstream regions to those in the attenuated vaccine strain BCG and include manA (an isomerase) (20), manB (a phosphomannomutase) (15), manC (a GDP-mannose pyrophosphorylase) (14, 18), and ppm1 (polyprenol-phosphate mannose synthase) (10, 11). Additionally, there are several other neighboring genes, like whiB2 (Fe-S clustering molecule and transcriptional regulator) (26), Rv3256c and Rv3258c (both hypothetical proteins), and Rv3253c (a postulated membrane flippase), whose functions are unknown but which are potentially contributing members of the mannose donor biosynthesis pathway (Fig. 1). Although the functions of several of these genes have been elucidated in nonpathogenic species of mycobacteria, such as Mycobacterium smegmatis, the regulation of expression of these genes in vitro and within macrophages has not been determined in virulent M. tuberculosis or compared with that in BCG. Such knowledge will have direct relevance to the availability of mannose donors for building the mannosylated cell wall components.
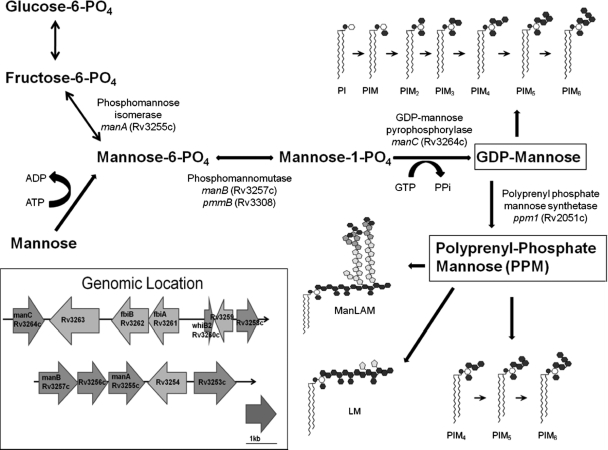
Fig. 1.
Putative mannose donor biosynthesis pathway for the building of two key mannose donor molecules, GDP-mannose and polyprenyl monophosphate mannose (PPM), which serve as substrates for mannosyltransferases. The inset shows the genomic location and arrangement of genes encoding key enzymes, manA, manB, and manC, in the pathway as well as the other genes of interest with unknown functions, Rv3253c, Rv3256c, and Rv3258c, used for the transcriptional expression study.
Expression differences between wild-type and attenuated vaccine strains of M. bovis have been reported, and it is thought that these differences may have a major impact on antigenic profiles (5). Mycobacterial gene expression work to date has been largely performed using platform technologies like microarrays, and while this approach provides an indication of large global expression changes, it lacks specificity for particular pathways postulated to contribute to virulence (25). A more in-depth look at individual pathways will provide us with greater insight into the complexities of transcriptional regulation as a major means of regulating virulence. Here we compared the regulation of expression of genes known or likely to be involved in the mannose donor biosynthesis pathway for generating mannosylated cell wall molecules between M. tuberculosis and BCG grown either in vitro or in human monocyte-derived macrophages (MDMs). We show significant differences in the expression of identical genes in the putative mannose donor biosynthesis pathway between these two mycobacterial species.
MATERIALS AND METHODS
Mycobacterial strains and growth media.
M. tuberculosis H37Rv (ATCC 27294) and M. bovis BCG-Pasteur (ATCC 35734) were grown either on Middlebrook 7H11 agar (Difco, Franklin Lakes, NJ) with 10% oleic acid, albumin, dextrose, and catalase (OADC) enrichment for 9 to 14 days (for macrophage experiments) or in Middlebrook 7H9 broth (Difco) plus 10% OADC and 0.05% Tween 80 with magnetic stirring for up to 24 days (for broth growth phase experiments) (32). Glycerol was added to both 7H11 and 7H9. For plate-grown bacteria, single-cell suspensions were obtained as previously described (28). Growth curves for broth cultures were determined by taking an OD600 (optical density at 600 nm) reading of stirred cultures grown in 7H9 plus Tween 80 every 24 h, and growth curve determination was performed in triplicate for each strain.
Isolation of MDMs.
MDMs were obtained from peripheral blood mononuclear cells (PBMCs) as previously described (27). Briefly, heparinized blood was obtained by venipuncture from purified protein derivative (PPD)-negative donors using an approved protocol by The Ohio State University Institutional Review Board. PBMCs were separated on a Ficoll cushion and were cultured for 5 days in RPMI medium containing 20% autologous serum in Teflon wells at 37°C with 5% CO2. After 5 days of growth in the Teflon wells, MDMs were made to adhere to 100- by 150-mm tissue culture dishes with medium in the presence of 10% autologous serum for 2 h; nonadherent cells were removed by washing with prewarmed RPMI, and MDMs were cultured for an additional 7 days in 20% autologous serum, allowing a total of 12 days of growth (19). At day 12, the MDMs were infected at a multiplicity of infection (MOI) of 5:1 in the presence of serum for 2 h at 37°C with 5% CO2. CFU were counted in parallel on agar plates to verify the inoculum used and thus confirm the MOI.
Bacterial lysis, RNA isolation, and real-time PCR.
Samples from broth-grown bacterial cultures were taken at predetermined growth phases and pelleted by centrifugation at 10,000 × g. Total RNA was extracted and purified by using an RNeasy mini-column (Qiagen, Valencia, CA) and 0.1-mm zirconia-silica beads (Biospec Products) coupled with DNase I (Qiagen, Valencia, CA) treatment. Isolation of bacteria from within infected macrophages was achieved by using a guanidinium thiocyanate (GTC)-based differential lysis solution as previously described (16). Bacterial RNA from within macrophages was processed by the procedure described above. RNA was reverse transcribed to cDNA using 500 U of Superscript II reverse transcriptase with 10 mM deoxynucleoside triphosphates, 10 U RNase inhibitor, 0.1 M dithiothreitol, and 3 μg of random hexamers (all from Invitrogen) for 120 min at 42°C, followed by inactivation with 1 N NaOH at 65°C for 10 min. Control reactions were performed in parallel without reverse transcriptase to verify the absence of DNA contamination. PCR was performed on the resulting cDNA using 300 mM custom-made primers with iQ SYBR green master mix (Bio-Rad) and 4% dimethyl sulfoxide. All samples were run in triplicate using the Bio-Rad CFX96 real-time system and analyzed using the 2−ΔΔCT method, and expression was determined relative to that of the housekeeping gene rpoB (13). Means and standard deviations for triplicate wells were recorded. GAPDH primers were used to identify contaminating eukaryotic RNA [5′ ACTTTGCTATCGTGGAAGGACT 3′ (forward) and 5′ GTAGAGGCAGGGATGATGTTCT 3′ (reverse)]. Bacterial-gene-specific custom primers are listed in Table 1.
Table 1.
List of specific primers used for RT-PCR
| Locus taga | Gene | Primer sequence (5′ to 3′) |
|---|---|---|
| Rv0667, BCG_0716 | rpoB | CCTGGAAGAGGTGCTCTACG (forward) |
| GGGAAGTCACCCATGAACAC (reverse) | ||
| Rv2051c, BCG_2070c | ppm1 | TGGTTGAAGTCGATCCTTCC (forward) |
| GCGAACAAGACCAGGCATATG (reverse) | ||
| Rv3253c, BCG_3282c | 53c | CCAACTACTCGCCGTTCATT (forward) |
| GCAGTTGGGTGTATGGAACC (reverse) | ||
| Rv3255c, BCG_3284c | manA | GTTCACCACCTGGATTACCG (forward) |
| AACCCTCGGTGCATAACAAG (reverse) | ||
| Rv3256c, BCG_3285c | 56c | CTGACGAGTTCGGGTTGTC (forward) |
| ACCAGATAGGCGTCATCGAG (reverse) | ||
| Rv3257c, BCG_3286c | manB | GATCACGTTGTGGATGATGG (forward) |
| GTGGATCTGCAGGCCTATGT (reverse) | ||
| Rv3258c, BCG_3287c | 58c | GTTGGTGCACGATAGCCTTT (forward) |
| ACACACAGATCCCACGAATG (reverse) | ||
| Rv3260c, BCG_3289c | whiB2 | CCATTCGAGGAACCTCTGC (forward) |
| CAGGGCGTACTCCAGACACT (reverse) | ||
| Rv3264c, BCG_3293c | manC | ACATCGCCGTTAAACACCAT (forward) |
| GTTCCTCACCCATCTGCTGT (reverse) | ||
| Rv3308, BCG_3373 | pmmB | ATACAGATCACGGCGTCACA (forward) |
| CGCTGGATATAACGGTCGAT (reverse) |
Each entry shows the locus tag designations for the same gene in M. tuberculosis and BCG. All genes listed have 100% nucleotide sequence identity, and the same primers were optimized in each organism separately.
RESULTS
Selection of transcriptional profiling candidates.
Using basic bioinformatics as well as previous work done with M. smegmatis for manA, manB, manC, and ppm1, we constructed a putative mannose donor biosynthesis pathway (Fig. 1) to depict the building of the two mannose donor molecules, GDP-mannose and PPM. Open reading frames (ORFs) for manA, manB, pmmB (another phosphomannomutase gene), manC, and ppm1 orthologues of M. tuberculosis H37Rv were obtained using the NCBI GenBank web server (http://www.ncbi.nlm.nih.gov/genome). Located near manA, manB, and manC in the M. tuberculosis genome (Fig. 1, inset) were several genes of unknown function (Rv3253c, Rv3256c, and Rv3258c) as well as whiB2, which was recently described as encoding a transcriptional regulator that is embedded in the genomic region of interest (Fig. 1, inset) (26). Further bioinformatic analysis using the basic local alignment search tool (BLAST) from NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) revealed that all of the genes of interest for our transcriptional profiling study had 100% sequence identity with those in BCG. The locus tags and names of all of these candidate genes in M. tuberculosis and BCG are listed in Table 1.
Transcriptional profiling of mannose donor biosynthesis genes of M. tuberculosis and BCG grown in broth culture.
Growth curves of M. tuberculosis and BCG were generated by growing the bacteria under identical culture conditions. Comparable time points were chosen for these two strains to analyze gene expression at the lag, log, or stationary growth phase. Custom primers were designed for each gene listed in Table 1, and because the sequences of M. tuberculosis and BCG genes were identical, the same primer sets were used for both strains under all conditions. All analyses determined expression relative to that of the housekeeping gene rpoB, which remained constant under all conditions for both strains.
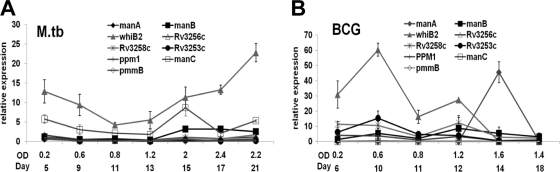
Our results for the transcriptional profile in broth show that there are differences in expression of certain genes between M. tuberculosis and BCG during the same growth phases despite their identical sequences. In M. tuberculosis (Fig. 2 A), whiB2 has relatively high expression compared to the other genes in the study. As growth continues, the expression level of whiB2 decreases during the exponential phase but then increases as growth continues further and peaks at late stationary phases. In BCG (Fig. 2B), whiB2 begins with a relatively high expression level, peaks at early log phase, and decreases as growth continues, a trend which is opposite to that seen in M. tuberculosis for the same gene. During the early log phase of growth of BCG, Rv3253c expression is higher than that of the other genes and decreases to baseline as growth continues. The most notable differences in expression between the strains were observed during the stationary phase of growth. In M. tuberculosis, manC expression spikes during this growth phase and manB has increased expression. In BCG, manA spikes during this growth phase and Rv3258c and manB show increased expression.
Fig. 2.
Transcriptional profile of putative mannose donor biosynthesis genes of M. tuberculosis (A) and BCG (B) grown in broth medium. cDNA was made from RNA extracted from both strains at their different growth phases and subjected to real-time PCR for determining the amount of transcripts of each gene as a measure of its expression. Expression of target genes is relative to that of the housekeeping gene, rpoB, and is plotted against the OD600 of the culture corresponding to the time points (in days) used for generating growth curves. Data are means ± standard deviations of values from triplicate wells. Graphs were plotted from representative experiments (n = 3).
Transcriptional profiling of mannose donor biosynthesis genes of M. tuberculosis and BCG in macrophages.
As human macrophages are the host cell niche for M. tuberculosis, we next infected these cells with M. tuberculosis and BCG and developed a reliable assay for recovering intact intracellular mycobacteria for RNA extraction. Using a specialized lysis buffer (GTC), we were able to stabilize bacterial RNA, and keep the bacteria intact while completely lysing the monolayer. Optimization required an MOI of 5:1 (bacteria to MDMs) for macrophage infection, which allowed the infected cell monolayer to remain intact for at least 5 days postinfection while still being able to generate enough bacterial RNA for cDNA synthesis, especially at early time points.
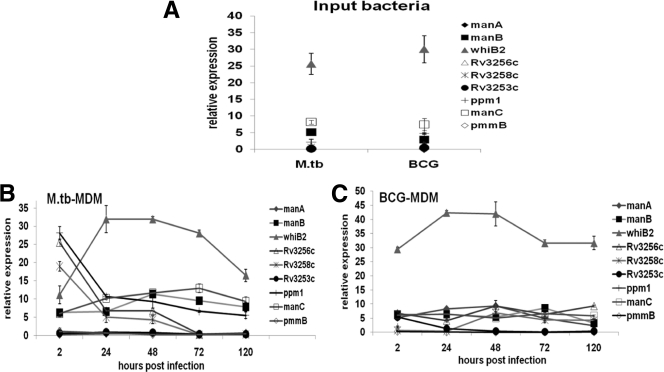
In order to carefully differentiate unique expression changes in bacteria residing within macrophages from those in the “input” bacteria for the experiment, a single-cell suspension was prepared from plate-grown bacteria and analyzed by quantitative reverse transcription-PCR. Results of input bacterial expression profiles show similar trends for both bacteria, with whiB2 being highly expressed relative to all other genes in the set (Fig. 3 A). Results from intramacrophage bacteria indicate that whiB2 is consistently highly expressed in both M. tuberculosis and BCG throughout the course of infection. In M. tuberculosis (Fig. 3B), there is an initial peak in expression for Rv3258c, Rv3256c, and ppm1 after 2 h of infection compared to input bacteria, and then expression levels for all three genes steadily decrease over time. manC and manB are moderately expressed during the course of infection. Results for BCG (Fig. 3C) show little or no change in expression of all genes in the study with the exception of whiB2, whose expression remained high throughout the course of infection.
Fig. 3.
Transcriptional profiles of putative mannose donor biosynthesis genes in M. tuberculosis and BCG from infected MDMs. Single-cell suspensions of M. tuberculosis grown on 7H11 agar plates were used to infect MDM monolayers at an MOI of 5:1. GTC lysis buffer was used to extract RNA at 24, 48, 72, and 120 h. cDNA was synthesized from RNA and subjected to real-time PCR to determine the amount of transcripts of each gene as a measure of its expression. (A) Transcriptional profile of the single-cell suspension prior to infection. (B and C) Transcriptional profiles of M. tuberculosis and BCG harvested from infected macrophages. Data are means ± standard deviations of values from triplicate wells. Graphs were plotted from representative experiments (n = 5).
DISCUSSION
In the present study, we show that there are differences in the transcriptional expression profiles of identical mannose donor biosynthetic genes between M. tuberculosis and BCG strains grown in broth culture as well as within human macrophages. One of the most noticeable differences in expression in broth between these two mycobacterial strains is that of gene encoding the Fe-S containing transcriptional regulator, whiB2 (Fig. 2). The WhiB-like molecules in the context of Mycobacterium species have been reported to be involved in crucial cellular processes like cell division, nutrient starvation, stress, antibiotic resistance, and pathogenesis (1, 6, 26). Previous work using microarrays has shown differences in expression of other WhiB-like molecules between M. tuberculosis, M. bovis, and M. bovis BCG vaccine strains (3, 7, 24). The only difference in the trend seen for whiB2 between M. tuberculosis and BCG was during growth in broth. Since no significant difference in expression of whiB2 was observed between M. tuberculosis and BCG in macrophages during the infection period or in the input bacteria (Fig. 3), this suggests that it may not play a direct role in pathogenesis. However, it will be interesting to know whether whiB2 has any effect on mycobacterial mannosylation because it is located in the neighborhood of the mannose donor biosynthesis pathway (Fig. 1, inset) and known to be essential for mycobacterial growth (23).
M. tuberculosis in broth culture during late log to early stationary growth phase showed relatively increased expression of two genes, manB and manC (Fig. 2A), encoding a phosphomannomutase and a GDP-mannose pyrophosphorylase, respectively, which are key enzymes in the mannose donor biosynthesis pathway. During the same growth phases in BCG, the genes with increased expression are manA, which encodes a phosphomannose isomerase, and Rv3258c, a hypothetical gene of unknown function (Fig. 2B). This differential regulation of expression of mannose donor-related genes between these two mycobacteria could contribute to the known structural differences in mannosylated molecules such as ManLAM (17) The structure of BCG ManLAM differs from that in M. tuberculosis in that it has a shorter mannan backbone with highly branched 3,5-linked d-Araf residues and the mannose-capping motifs in BCG are dominated by monocaps, as opposed to those in M. tuberculosis, which are dominated by di- or tricaps (21). The goal of future research will be to link changes in gene expression for the mannose donor biosynthetic enzymes with the level of biosynthesis of mannose-containing cell wall molecules produced during infection. As an example, the mannosyltransferase PimB has been shown to be expressed at higher levels in M. tuberculosis compared to M. bovis (7), and this may impact the reported differences in BCG and M. tuberculosis ManLAM (21).
Gene expression differences between M. tuberculosis and BCG in broth may relate to the fact that BCG has multiple deletions and has accumulated many mutations in the genome that have led to adaptation to growth on glycerol-containing media (5). Our current work shows that BCG has reduced expression of several genes both in broth (Fig. 2B) and within macrophages (Fig. 3C).
The expression profile of M. tuberculosis and BCG mannose donor biosynthesis genes in macrophages is of particular importance not only because macrophages are the natural host cell niche for M. tuberculosis but also because the expression profiles are highly reproducible among different donors. Because humans are a heterogeneous population, it is often speculated that donor-to-donor variation might alter bacterial gene expression, but this was not found to be the case with our study. The transcriptional expression profiles of M. tuberculosis and BCG genes from bacteria within MDMs are representative of five independent experiments with five different donors, all generating similar transcriptional profiles. These reproducible data argue that the mycobacterial transcriptional events are required for bacterial acclimation, survival, and proliferation in the host cell. In human macrophages, the distinct differences in the pattern of expression of the majority of the genes under study between M. tuberculosis (Fig. 3B) and BCG (Fig. 3C) suggest a possible link to the virulence of M. tuberculosis. It is of particular interest that expression levels of Rv3256c, Rv3258c, and ppm1 were high in M. tuberculosis 2 h after infection of macrophages and then gradually decreased. This suggests that the expression of these genes during the early period of infection is important for the entry of the bacterium into its preferred host cell environment and/or acclimation to that environment. Furthermore, the elevated expression of manB and manC at 48 and 72 h postinfection (Fig. 3B) suggests the possibility of an increased need for mannose donor molecules as the bacteria begin to replicate intracellularly after the initial adaptation period. In BCG, most of the identical genes under study were expressed at low levels (Fig. 3C) in macrophages, suggesting that BCG has less need for these genes for the purpose of infection and adaptation. Thus, the results obtained from human macrophages in our study give us new insight into the regulatory nature of transcription potentially related to virulence and provide us with an increased understanding of rational selection of bona fide drug targets for TB.
ACKNOWLEDGMENTS
We thank Shilpa Soni for technical assistance with real-time PCR development.
Funding for this work was provided by the NIH grants AI052458 and AI068846 to L.S.S.
Footnotes
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Alam M. S., Garg S. K., Agrawal P. 2009. Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. FEBS J. 276:76–93 [DOI] [PubMed] [Google Scholar]
- 2. Briken V., Porcelli S. A., Besra G. S., Kremer L. 2004. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol. Microbiol. 53:391–403 [DOI] [PubMed] [Google Scholar]
- 3. Brosch R., et al. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. U. S. A. 104:5596–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ganguly N., Siddiqui I., Sharma P. 2008. Role of M. tuberculosis RD-1 region encoded secretory proteins in protective response and virulence. Tuberculosis (Edinb.) 88:510–517 [DOI] [PubMed] [Google Scholar]
- 5. Garcia Pelayo M. C., et al. 2009. Gene expression profiling and antigen mining of the tuberculin production strain Mycobacterium bovis AN5. Vet. Microbiol. 133:272–277 [DOI] [PubMed] [Google Scholar]
- 6. Geiman D. E., Raghunand T. R., Agarwal N., Bishai W. R. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50:2836–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Golby P., et al. 2007. Comparative transcriptomics reveals key gene expression differences between the human and bovine pathogens of the Mycobacterium tuberculosis complex. Microbiology 153:3323–3336 [DOI] [PubMed] [Google Scholar]
- 8. Gordon S. V., Bottai D., Simeone R., Stinear T. P., Brosch R. 2009. Pathogenicity in the tubercle bacillus: molecular and evolutionary determinants. BioEssays 31:378–388 [DOI] [PubMed] [Google Scholar]
- 9. Guerin M. E., Kordulakova J., Alzari P. M., Brennan P. J., Jackson M. 2010. Molecular basis of phosphatidyl-myo-inositol mannoside biosynthesis and regulation in mycobacteria. J. Biol. Chem. 285:33577–33583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gurcha S. S., et al. 2002. Ppm1, a novel polyprenol monophosphomannose synthase from Mycobacterium tuberculosis. Biochem. J. 365:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guy M. R., et al. 2004. Novel prenyl-linked benzophenone substrate analogues of mycobacterial mannosyltransferases. Biochem. J. 382:905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koul A., Arnoult E., Lounis N., Guillemont J., Andries K. 2011. The challenge of new drug discovery for tuberculosis. Nature 469:483–490 [DOI] [PubMed] [Google Scholar]
- 13. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 14. Ma Y., et al. 2001. Drug targeting Mycobacterium tuberculosis cell wall synthesis: genetics of dTDP-rhamnose synthetic enzymes and development of a microtiter plate-based screen for inhibitors of conversion of dTDP-glucose to dTDP-rhamnose. Antimicrob. Agents Chemother. 45:1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCarthy T. R., et al. 2005. Overexpression of Mycobacterium tuberculosis manB, a phosphomannomutase that increases phosphatidylinositol mannoside biosynthesis in Mycobacterium smegmatis and mycobacterial association with human macrophages. Mol. Microbiol. 58:774–790 [DOI] [PubMed] [Google Scholar]
- 16. Monahan I. M., Mangan J. A., Butcher P. D. 2001. Extraction of RNA from Intracellular Mycobacterium tuberculosis: methods, considerations and applications, p. 31–42 In Parish T., Stoker N. G. (ed.), Mycobacterium tuberculosis protocols. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 17. Nigou J., Gilleron M., Puzo G. 2003. Lipoarabinomannans: from structure to biosynthesis. Biochimie 85:153–166 [DOI] [PubMed] [Google Scholar]
- 18. Ning B., Elbein A. D. 1999. Purification and properties of mycobacterial GDP-mannose pyrophosphorylase. Arch. Biochem. Biophys. 362:339–345 [DOI] [PubMed] [Google Scholar]
- 19. Olakanmi O., Britigan B. E., Schlesinger L. S. 2000. Gallium disrupts iron metabolism of mycobacteria residing within human macrophages. Infect. Immun. 68:5619–5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patterson J. H., Waller R. F., Jeevarajah D., Billman-Jacobe H., McConville M. J. 2003. Mannose metabolism is required for mycobacterial growth. Biochem. J. 372:77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prinzis S., Chatterjee D., Brennan P. J. 1993. Structure and antigenicity of lipoarabinomannan from Mycobacterium bovis BCG. J. Gen. Microbiol. 139:2649–2658 [DOI] [PubMed] [Google Scholar]
- 22. Pym A. S., Brodin P., Brosch R., Huerre M., Cole S. T. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709–717 [DOI] [PubMed] [Google Scholar]
- 23. Raghunand T. R., Bishai W. R. 2006. Mapping essential domains of Mycobacterium smegmatis WhmD: insights into WhiB structure and function. J. Bacteriol. 188:6966–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rehren G., Walters S., Fontan P., Smith I., Zarraga A. M. 2007. Differential gene expression between Mycobacterium bovis and Mycobacterium tuberculosis. Tuberculosis. (Edinb.) 87:347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rohde K. H., Abramovitch R. B., Russell D. G. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host. Microbe 2:352–364 [DOI] [PubMed] [Google Scholar]
- 26. Rybniker J., Nowag A., van G. E., Nissen N., Robinson N., Plum G., Hartmann P. 2010. Insights into the function of the WhiB-like protein of mycobacteriophage TM4-a transcriptional inhibitor of WhiB2. Mol. Microbiol. 77:642–657 [DOI] [PubMed] [Google Scholar]
- 27. Schlesinger L. S. 1993. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J. Immunol. 150:2920–2930 [PubMed] [Google Scholar]
- 28. Schlesinger L. S., Bellinger-Kawahara C. G., Payne N. R., Horwitz M. A. 1990. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J. Immunol. 144:2771–2780 [PubMed] [Google Scholar]
- 29. Smith I. 2003. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16:463–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torrelles J. B., Azad A. K., Schlesinger L. S. 2006. Fine discrimination in the recognition of individual species of phosphatidyl-myo-inositol mannosides from Mycobacterium tuberculosis by C-type lectin pattern recognition receptors. J. Immunol. 177:1805–1816 [DOI] [PubMed] [Google Scholar]
- 31. Torrelles J. B., et al. 2009. Inactivation of Mycobacterium tuberculosis mannosyltransferase pimB reduces the cell wall lipoarabinomannan and lipomannan content and increases the rate of bacterial-induced human macrophage cell death. Glycobiology 19:743–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wayne L. G. 1994. Cultivation of mycobacteria for research purposes, p. 73–83 In Bloom Barry R. (ed.), Tuberculosis: pathogenesis, protection and control. ASM Press, Washington, DC [Google Scholar]
- 33.World Health Organization. 2009 update: tuberculosis facts. World Health Organization; 2009. http://www.who.int/tb/publications/2009/tbfactsheet_2009update_one_page.pdf. [Google Scholar]