Abstract
Cholera toxin (CT) is endocytosed and transported by vesicle carriers to the endoplasmic reticulum (ER). The catalytic CTA1 subunit then crosses the ER membrane and enters the cytosol, where it interacts with its Gsα target. The CTA1 membrane transversal involves the ER chaperone BiP, but few other host proteins involved with CTA1 translocation are known. BiP function is regulated by ERdj3, an ER-localized Hsp40 chaperone also known as HEDJ. ERdj3 can also influence protein folding and translocation by direct substrate binding. In this work, structural and functional assays were used to examine the putative interaction between ERdj3 and CTA1. Cell-based assays demonstrated that expression of a dominant negative ERdj3 blocks CTA1 translocation into the cytosol and CT intoxication. Binding assays with surface plasmon resonance demonstrated that monomeric ERdj3 interacts directly with CTA1. This interaction involved the A12 subdomain of CTA1 and was further dependent upon the overall structure of CTA1: ERdj3 bound to unfolded but not folded conformations of the isolated CTA1 subunit. This was consistent with the chaperone function of ERdj3, as was the ability of ERdj3 to mask the solvent-exposed hydrophobic residues of CTA1. Our data identify ERdj3 as a host protein involved with the CT intoxication process and provide new molecular details regarding CTA1-chaperone interactions.
INTRODUCTION
Many toxins share a structural organization that consists of a catalytic A subunit and a cell-binding B subunit (32). These extracellular toxins attack targets within the eukaryotic cytosol and must therefore cross a membrane barrier in order to function. Some AB toxins, such as diphtheria toxin (DT), access the cytosol from acidified endosomes. Other AB toxins move from the plasma membrane to the endoplasmic reticulum (ER) before passage into the cytosol by a process involving the quality control system of ER-associated degradation (ERAD) (14, 21). For both endosome and ER translocation sites, holotoxin disassembly precedes or occurs concurrently with A chain entry into the cytosol.
The process of ERAD-mediated toxin translocation is not completely defined, but some details have been elucidated for ERAD interactions with the catalytic A1 subunit of cholera toxin (CTA1). The single disulfide bond linking CTA1 to the rest of the cholera holotoxin (CT) is reduced at the resident redox state of the ER (22). Reduced CTA1 dissociates from the holotoxin with the aid of protein disulfide isomerase (PDI) (39, 45), and unfolding of the isolated CTA1 polypeptide then facilitates its passage into the cytosol through the Sec61 and/or Derlin-1 protein-conducting channels (5, 11, 33, 34). Most ERAD substrates are efficiently degraded in the cytosol by the ubiquitin-proteasome system (47). CTA1 avoids this fate because it only has two lysine residues to serve as potential ubiquitin attachment sites (14, 30, 31). The translocated pool of CTA1 thus persists in the cytosol long enough to modify its Gsα target. A hydrophobic region in the C-terminal A13 subdomain of CTA1 was originally thought to activate the ERAD system (14, 20), but more recent work has shown that the A13 subdomain is not required for CTA1 entry into the cytosol (44). We and others have proposed an alternative model in which partial unfolding of CTA1 at 37°C serves as the trigger for ERAD (1, 25, 30, 44). Unfolding occurs spontaneously after CTA1 dissociation from the holotoxin (30). The role of CTA1 thermal instability in toxin-ERAD interactions is a current focus of study, but this work is restricted in part by the limited number of ERAD factors known to interact with CTA1.
ERdj3/HEDJ is an ER-localized Hsp40 chaperone and a component of ERAD (7, 29, 35, 37, 50). It can be found in a multiprotein complex with BiP (15, 27, 35, 49), an Hsp70 chaperone involved with CTA1 translocation (48). ERdj3 can also be found in a complex with the catalytic A1 subunit of Shiga toxin, another AB type toxin that enters the cytosol from the ER (49). Furthermore, overexpression or disruption of ERdj3 function generates cellular resistance to Shiga toxin (29, 50). We therefore hypothesized that CTA1 interacts with ERdj3 during the translocation process. Cell-based assays and structural studies were performed in order to test this prediction. The cell-based assays provided functional evidence for the role of ERdj3 in CTA1 translocation and CT intoxication. The structural studies demonstrated a direct, conformation-specific interaction between ERdj3 and CTA1: ERdj3 bound to the disordered conformation of CTA1 at 37°C but did not bind to the folded conformations of CTA1 present at 10°C or in the CT holotoxin. Consistent with its role as a chaperone, ERdj3 binding to CTA1 masked the solvent-exposed hydrophobic residues of CTA1. This work identifies ERdj3 as an ERAD factor involved with the CT intoxication process and further indicates that CTA1 thermal instability plays an important role in toxin-ERAD interactions.
MATERIALS AND METHODS
Materials.
The pcDNA3.1 vector and cell culture reagents were purchased from Invitrogen (Carlsbad, CA). His-tagged CTA1, CTA11–168, and CTA11–133 constructs were purified as previously described (25). CT and DT were purchased from List Biologicals (Campbell, CA). [35S]methionine was purchased from Perkin Elmer (Waltham, MA), and digitonin was purchased from Calbiochem (La Jolla, CA). bis-ANS [4,4′-bis(1-anilinonaphthalene 8-sulfonate)] and other chemicals were purchased from Sigma-Aldrich (St. Louis, MO). The anti-CTA antibody was purchased from Sigma-Aldrich, the anti-PDI and anti-Hsp90 antibodies were purchased from Enzo Life Sciences (Plymouth Meeting, PA), and the anti-ERdj3 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Expression plasmids.
Generation of the pHED-3 and pcDNA3.1/ssCTA1 plasmids has been previously described (43, 50). pHED-2 encodes full-length ERdj3 with the native stop codon. The gene was amplified from a cDNA library with upstream primer 5′-GGCCTCACAGGGCCGGGTGGGCTGG-3′ and downstream primer 5′-ATATCCTTGCAGTCCATTGTATACCTTC-3′. The resulting product was ligated into plasmid pCR3.1-Topo/V5/His (Invitrogen). Plasmid pGEX-HED2 encodes the mature ERdj3 protein with the native stop codon fused to the carboxyl terminus of glutathione S-transferase (GST). The ERdj3 gene was amplified with primers 5′-GGCCTGGAATTCGGGGCGGTGATTGCCGGACGAG-3 and 5′-GGTCGACTCGAGCTCTCAATATCCTTGCAGTCCATTG-3′. The PCR product was ligated into pCR3.1-Topo/V5/His and sequenced to ensure fidelity. This plasmid was digested with EcoRI and XhoI to release the ERdj3 insert, which was then ligated into plasmid pGEX-6.1 that had been digested with EcoRI and XhoI.
pHED-2 encodes an untagged ERdj3 protein which will be expressed as a soluble protein in the ER lumen (6, 35). In contrast, pHED-3 encodes an ERdj3 protein with a C-terminal V5-His tag which will be expressed as a membrane-anchored protein in the ER (50). As shown here, the membrane-anchored form of ERdj3 acted as a dominant negative construct, possibly because the propensity of ERdj3 to form dimers (16) allowed the membrane-embedded ERdj3 to titrate free ERdj3 away from the soluble space of the ER lumen.
Cell culture and transfection.
CHO cells were grown in Ham's F-12 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA). Growth conditions were 37°C and 5% CO2 in a humidified incubator. For transfection, cells were seeded to a 6-well plate and grown overnight to 80% confluence. The cells were then exposed to 1 μg of DNA for each plasmid and 5 μl of Lipofectamine (Invitrogen) for 3 h according to the manufacturer's instructions. Following an overnight incubation, the transfected cells were lifted from the 6-well plate, diluted, transferred to 24-well plates, and grown overnight to 80% confluence. Toxicity assays were then performed at 48 h posttransfection. For translocation assays, the transfected cells were kept in 6-well plates and used at 16 to 24 h posttransfection.
Toxicity assays.
CHO cells transfected with plasmid expression vectors encoding nothing (empty pcDNA3.1 vector), wild-type ERdj3 (wtERdj3; pHED-2), or dominant negative ERdj3 (dnERdj3; pHED-3) were challenged with various concentrations of CT for 2 h. The cells were washed with phosphate-buffered saline (PBS) and incubated with 0.25 ml of ice-cold HCl-ethanol (EtOH) (1:100) for 15 min at 4°C. Supernatants were placed in microcentrifuge tubes and allowed to air dry. Cyclic AMP (cAMP) levels were then determined using an 125I-cAMP competition assay as per the manufacturer's instructions (GE Healthcare, Piscataway, NJ). Background cAMP levels from unintoxicated cells transfected with the empty pcDNA3.1 vector were subtracted from the experimental values before comparative analysis. The maximum cAMP response was arbitrarily set to 100%, and all other values were expressed as a fraction of that value.
To monitor the activity of DT, transfected cells were challenged with various concentrations of toxin for 4 h. The cells were then placed in methionine-free medium for 30 min, followed by a 15-min incubation in methionine-free medium supplemented with 10 μCi [35S]methionine/ml. Radiolabeled cells were washed with 10% trichloroacetic acid in PBS for 30 min at 4°C, followed by a second wash for 15 min. The cells were then solubilized in 0.2 N NaOH, and counts per minute from radiolabeled proteins in the cell extracts were quantified with a Beckman Coulter (Brea, CA) LS 6500 multipurpose scintillation counter. For each condition, data were expressed as percentages of the maximal value obtained from unintoxicated control cells that had been transfected with the corresponding vector.
Translocation assay.
Transfected cells were incubated for 1 h in 0.5 ml of methionine-free medium, washed with PBS, and incubated for another hour in 0.5 ml of methionine-free medium supplemented with 150 μCi [35S]methionine/ml. The radiolabeled cells were then washed with PBS, lifted from the wells with 0.5 mM EDTA in PBS, and transferred to microcentrifuge tubes for a 5-min spin at 5,000 × g. The supernatant was discarded, and the cell pellet was resuspended in 100 μl of 0.04% digitonin in HCN buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 2 mM CaCl2, 10 mM N-ethylmaleimide, and a 1:20 dilution of protease inhibitor cocktail from Sigma). After 10 min on ice, the samples were spun at 16,000 × g for 10 min. The supernatant (i.e., cytosolic fraction) was collected and transferred to a fresh microcentrifuge tube. One milliliter of lysis buffer was added to the pellet (i.e., membrane fraction), and 900 μl of lysis buffer was added to the supernatant fraction. Lysis buffer consisted of 25 mM Tris-Cl (pH 7.4), 20 mM NaCl, 1% deoxycholic acid, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, and 1 μg/ml leupeptin. After 20 min at 4°C, an anti-CTA antibody was added to each 1-ml fraction for an overnight incubation. Protein A-conjugated Sepharose beads were subsequently used to immunoprecipitate the antigen-antibody complexes. The immunoisolated samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 15% polyacrylamide gels. PhosphorImager analysis was used to visualize and quantify the immunoisolated pools of CTA1. The background was subtracted from all samples prior to data calculations. The percentage of cytosolic CTA1 was defined as % cytosol = (supernatant/[pellet + supernatant]) × 100.
For Western blot analysis, a parallel set of unlabeled cells transfected with the empty pcDNA3.1 vector was separated into pellet and supernatant fractions as described above. Equivalent volumes of each fraction were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes which were probed with anti-Hsp90 and anti-PDI antibodies at 1:20,000 and 1:5,000 dilutions, respectively.
Purification of ERdj3.
Escherichia coli harboring the pGEX-HED2 expression plasmid was grown in 1 liter of Luria-Bertani broth at 37°C to an A600 of 0.6, at which point the culture was shifted to 18°C and induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 18 h. The cells were then pelleted and resuspended in extraction buffer comprised of PBS, 1% Triton X-100, 0.1 mg/ml lysozyme, 10 μl/ml protease inhibitor cocktail, and 0.1 μl/ml DNase I. Cells were lysed with 3 freeze-thaw cycles at −80°C and 37°C for 15 min each. The lysate was centrifuged to remove cellular debris, and the resulting supernatant was incubated for 20 min at 37°C with 50 mM Tris (pH 7.2), 2 mM ATP, and 10 mM MgSO4 before syringe filtration with a 0.2-μm filter. The filtered material was applied to a GSTrap column attached to an ÄKTA protein purifier (GE Healthcare). GST-tagged ERdj3 was eluted off the column with PBS (pH 7.3) containing 20 mM reduced glutathione. Protein concentrations in the collected 1-ml fractions were determined by Bradford assay. In addition to GST-tagged ERdj3, GST and GST-cleaved ERdj3 were also collected in the eluted fractions. The isolation of native ERdj3 was possible because (i) bacterial proteases can cause premature cleavage of the GST tag as described in the manufacturer's instructions (GE Healthcare) and (ii) ERdj3 can form dimers (16), allowing some of the cleaved ERdj3 to be retained with GST-tagged ERdj3 on the GSTrap column. The identities of both native and GST-tagged ERdj3 were confirmed by Western blot analysis with anti-ERdj3 and anti-GST antibodies. Material eluted from the GSTrap column was applied to a glutathione spin column (Pierce Biotechnology, Rockford, IL) which retained GST and GST-tagged ERdj3, thereby allowing a pool of purified, native ERdj3 to be collected in the flowthrough.
Surface plasmon resonance.
Experiments were performed with a Reichert (Depew, NY) SR7000 surface plasmon resonance (SPR) refractometer. For SPR experiments with His-tagged CTA1 sensor slides, the toxin was captured on a nickel-nitrilotriacetic acid (NTA) slide that was rinsed with 2 M NaCl and 10 mM NaOH. The slide was then rinsed with water, activated with 0.08 mg/ml of 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (Thermo Scientific, Waltham, MA), and 0.02 mg/ml of N-hydroxysuccinimide (NHS) (Pierce Biotechnology) for 10 min, rinsed with water again, and covered with 1 mg/ml of Nα,Nα-bis(carboxymethyl)-l-lysine hydrate in 20 mM sodium acetate, pH 5.2. After overnight incubation, the slide was rinsed with water and covered with 1 M ethanolamine, pH 8.5, for 10 min. The slide was then rinsed with water and allowed to dry. After mounting the activated slide in the SPR instrument, 0.01 M PBS with 0.05% Tween 20 (PBST), pH 7.4, was perfused over the plate, followed by perfusion of 40 mM nickel sulfate for 5 min. His-tagged CTA1 (diluted in 20 mM sodium acetate, pH 5.5) was then perfused over the plate for 5 min followed by PBST, pH 7.4, for 5 to 10 min. After establishment of a new baseline measurement corresponding to the mass of the sensor-bound CTA1, ERdj3 diluted in PBST to appropriate concentrations (1,600, 800, 400, 200, and 100 ng/ml) was flowed over the CTA1 sensor slide for 5 min, followed by a 5-min PBST (pH 7.4) wash. For qualitative experiments, ERdj3 was used at a single concentration of 1,600 ng/ml.
For SPR experiments with ERdj3-coated sensor slides, PBST (pH 7.4) was perfused over a Reichert SPR sensor slide with a mixed self-assembled monolayer that was mounted in the SPR instrument. The slide was activated by flowing over 0.08 mg/ml of EDC and 0.02 mg/ml of NHS for 5 min followed by a 5-min perfusion of ERdj3 antibody diluted in 20 mM sodium acetate, pH 5.5, at approximately 200 ng/ml. Ethanolamine (1 M; pH 8.5) was then injected for 5 min followed by PBST (pH 7.4) for 5 to 10 min to establish a stable baseline signal corresponding to the mass of the sensor-bound antibody. Purified GST-ERdj3 diluted in PBST, pH 7.4, was then flowed over the bound antibody for 5 min. The temperature was dropped to 10°C after establishment of a stable baseline, and 40 μl/ml PreScission protease (GE Healthcare) was perfused over the slide for approximately 3 min until a corresponding signal indicating binding of the protease was established. At this point, the flow was stopped overnight for on-slide cleavage of the GST tag. PBST was then perfused over the plate at 37°C to remove the protease and cleaved GST. A new baseline corresponding to the mass of bound ERdj3 was established for the plate. CTA1 or CTA11–168 diluted in PBST, pH 7.4, was then perfused over the plate at a concentration of 1,600, 800, 400, 200, or 100 ng/ml. For qualitative experiments, CT was used at 1,600 ng/ml, CTA1 was used at 1,600 ng/ml or 400 ng/ml, and CTA11–133 was used at 400 ng/ml.
The flow rate for all steps was 41 μl/min. Reichert LabView software was used for data collection. The BioLogic (Campbell, Australia) Scrubber 2 software and WaveMetrics (Lake Oswego, OR) Igor Pro software were used to analyze the data and generate figures. Binding affinities between CTA1 and ERdj3 were calculated in the Scrubber 2 software using a 1:1 Langmuir fit.
bis-ANS assay.
A reduced CTA1/CTA2 heterodimer (8 μM) was incubated with 50 μM bis-ANS in the absence or presence of 8 μM ERdj3. Fluorescence spectra were recorded at 18°C or after a stepwise increase in temperature to 25°C, 30°C, 37°C, or 42°C. Experiments were performed using a Jasco (Easton, MD) J-810 spectrofluoropolarimeter with an excitation wavelength of 380 nm. Excitation and emission slits were 2 nm and 5 nm, respectively.
RESULTS
A functional role for ERdj3 in CT intoxication.
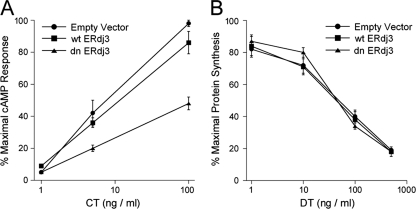
To examine the role of ERdj3 in CT intoxication, CHO cells transiently transfected with expression vectors encoding nothing (empty vector), wild-type ERdj3 (wtERdj3), or dominant negative ERdj3 (dnERdj3) were exposed to various concentrations of CT for 2 h. Cell extracts were then generated and screened for cAMP content with an 125I-cAMP competition assay. As shown in Fig. 1A, the expression of dnERdj3 generated substantial resistance to CT. A half-maximal effective concentration (EC50) of 8 ng CT/ml was calculated for cells transfected with the empty vector, while an EC50 of about 100 ng/ml was calculated for cells transfected with dnERdj3. Thus, in relation to the EC50, cells expressing dnERdj3 were approximately 12-fold more resistant to CT than the control cells. Overexpression of wtERdj3, which was confirmed by Western blotting (not shown), did not generate substantial resistance to CT. Neither dnERdj3 nor wtERdj3 expression affected cellular susceptibility to DT, which enters the cytosol from acidified endosomes (Fig. 1B). As cells with compromised ERdj3 function were resistant to CT, ERdj3 appeared to play a functional role in the CT intoxication process.
Fig. 1.
Role of ERdj3 in CT intoxication. (A) CHO cells transfected with plasmid expression vectors encoding nothing (Empty Vector), wild-type ERdj3 (wtERdj3), or dominant negative ERdj3 (dnERdj3) were exposed to the stated concentrations of CT for 2 h. The extent of intoxication was determined from the elevated levels of intracellular cAMP. Results are presented as percentages of the maximal cAMP response for all tested conditions. (B) CHO cells transfected with plasmid expression vectors encoding nothing, wtERdj3, or dnERdj3 were exposed to the stated concentrations of DT for 4 h. The extent of intoxication was determined from the incorporation of radiolabeled methionine into newly synthesized proteins. Results are presented as percentages of the maximal signal obtained from unintoxicated cells. Data in both panels represent the means ± standard errors of the means from 4 independent experiments with triplicate samples for each condition.
ERdj3 is involved with CTA1 translocation from the ER to the cytosol.
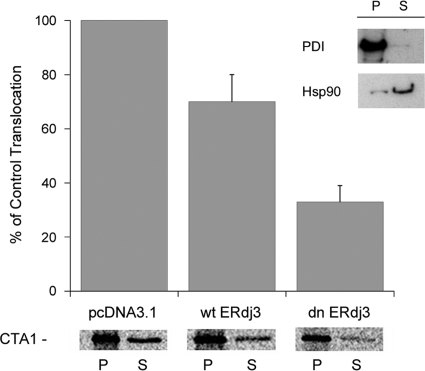
CT intoxication can be blocked by an inhibition of CTA1 translocation from the ER to the cytosol (5, 25, 40, 42). We accordingly examined the putative role of ERdj3 in CTA1 translocation. CTA1 was expressed directly in the ER of CHO cells with a plasmid-based transfection system in which the coding sequence of CTA1 was appended with an N-terminal signal sequence for cotranslational delivery to the ER lumen. Previous work has shown that this signal sequence targets the entire detectable pool of CTA1 to the ER (41), and similar experimental systems have been used by multiple investigators to study toxin translocation from the ER (9, 19, 34, 36, 46, 49). The signal sequence is proteolytically cleaved from ER-localized CTA1, and the mature CTA1 subunit is then exported from the ER to the cytosol (41). Cells were cotransfected with the CTA1-encoding plasmid and an additional expression vector encoding nothing (empty vector), wtERdj3, or dnERdj3. Passage of CTA1 from the ER to the cytosol was monitored through selective permeabilization of the plasma membrane with digitonin. This process can be used to separate the cell extract into two fractions containing either intact organelles or the cytosol. Western blot analysis demonstrated the fidelity of our fractionation protocol: ER resident protein PDI was detected in the organelle/pellet fraction, while cytosolic protein Hsp90 was found in the cytosolic/supernatant fraction (Fig. 2, inset). The presence of a minor, membrane-associated pool of Hsp90 was consistent with previous reports (12, 17, 40).
Fig. 2.
Role of ERdj3 in CTA1 translocation. CHO cells were cotransfected with plasmids encoding an ER-localized CTA1 construct and either nothing (pcDNA3.1), wtERdj3, or dnERdj3. After metabolic labeling, cell extracts were separated into organelle (pellet [P]) and cytosolic (supernatant [S]) fractions by selective permeabilization of the plasma membrane with digitonin. The distribution of CTA1 immunoprecipitated from each fraction was visualized and quantified by SDS-PAGE with PhosphorImager analysis. The averages ± standard deviations from 3 independent experiments are presented in the graph. The inset shows a Western blot control documenting the distributions of soluble ER protein PDI and cytosolic protein Hsp90 in the pellet and supernatant fractions.
Coexpression of ER-localized CTA1 with wtERdj3 or dnERdj3 allowed us to examine the role of ERdj3 in CTA1 translocation (Fig. 2). Cotransfected cells were radiolabeled with [35S]methionine for 1 h at 37°C before digitonin was added at 4°C to permeabilize the plasma membrane. Membrane and cytosolic fractions collected after centrifugation of the permeabilized cells were subjected to anti-CTA antibody immunoprecipitation. The immunoisolated material was then resolved by SDS-PAGE and visualized/quantified by PhosphorImager analysis. With this method, we found that coexpression of CTA1 with dnERdj3 substantially inhibited the movement of CTA1 from the ER to the cytosol. Cells expressing dnERdj3 exported 67% less CTA1 to the cytosol than control cells cotransfected with the empty vector. Cells cotransfected with wtERdj3 also exhibited a defect in CTA1 translocation, as these cells exported 30% less CTA1 to the cytosol than control cells. Overexpression of a wild-type protein, as well as a dominant negative protein, can disrupt homeostasis and cellular processes. We believe this occurred in our transfection experiment and further note that overexpression of wild-type ERdj3 has been reported to disrupt intoxication with Shiga toxin (29). Collectively, our results indicated that ERdj3 plays a functional role in CTA1 translocation and that ERdj3 dysfunction blocks CT intoxication by inhibiting CTA1 export to the cytosol.
CTA1 thermal instability dictates the extent of CTA1-ERdj3 interaction.
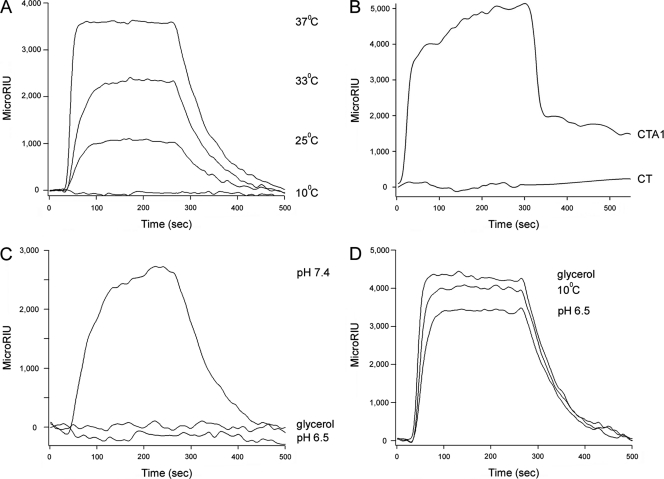
ERdj3 operates in ER quality control by regulating BiP-substrate interactions and by direct binding to ERAD substrates (2, 7, 8, 15, 16, 24, 35). SPR was accordingly used to detect a physical interaction between ERdj3 and CTA1 (Fig. 3). For this experiment, a His-tagged CTA1 construct (44) was appended to an SPR sensor slide as described in Materials and Methods. When ERdj3 was perfused over the CTA1-His6 sensor slide at 37°C, a strong interaction between CTA1-His6 and ERdj3 was detected. Substantially weaker interactions between CTA1-His6 and ERdj3 were detected at 33°C and 25°C, while no interaction between CTA1 and ERdj3 occurred at 10°C (Fig. 3A). The isolated CTA1 subunit is in a folded conformation at 10°C but loses a substantial amount of its native structure with increasing temperature, so at 37°C the toxin contains a disordered tertiary structure and a partially perturbed secondary structure (30). Thus, consistent with its role as a chaperone, ERdj3 preferentially interacted with the unfolded conformations of CTA1.
Fig. 3.
Recognition of disordered CTA1 conformations by ERdj3. (A) ERdj3 was perfused over a CTA1-coated SPR sensor slide at pH 7.4 and the indicated temperatures. (B) CT and CTA1 were perfused over an ERdj3-coated SPR sensor slide at pH 7.4 and 37°C. (C) ERdj3 was perfused over a CTA1-coated SPR sensor slide at 37°C in pH 7.4 buffer, in pH 7.4 buffer containing 10% glycerol, or in pH 6.5 buffer. (D) ERdj3 was perfused over an SPR sensor slide appended with denatured CTA1 at 10°C in pH 7.4 buffer, at 37°C in pH 7.4 buffer containing 10% glycerol, and at 37°C in pH 6.5 buffer. For all experiments, ligand (1,600 ng/ml) was removed from the perfusion buffer approximately 300 s into the experiment. One of at least two representative experiments is presented in each panel.
CTA1 thermal instability is apparent only after its dissociation from the CT holotoxin (30). Thus, as ERdj3 recognizes only the unfolded conformation of CTA1, there should be no interaction between ERdj3 and the CT holotoxin. We perfused CT over an ERdj3-coated SPR sensor slide to test this prediction (Fig. 3B). ERdj3 did not bind to the CT holotoxin at 37°C, which indicated that CTA1-ERdj3 interactions are initiated only after CTA1 dissociation from the holotoxin.
Acidic pH and 10% glycerol both inhibit CTA1 thermal denaturation and CTA1 translocation to the cytosol (3, 25). As assessed by SPR, both of these conditions also inhibited the interaction between CTA1 and ERdj3 at 37°C (Fig. 3C). The loss of CTA1-ERdj3 interaction at an acidic pH and in the presence of 10% glycerol again suggested that ERdj3 recognizes a region of disordered structure in the dissociated CTA1 subunit. These observations provided further evidence for the functional role of CTA1 thermal instability in toxin-ERAD interactions.
To ensure that ERdj3 could function under all of the tested conditions, we irreversibly denatured CTA1-His6 by heating the toxin at 60°C for 10 min (30). ERdj3 could interact with this denatured CTA1 construct at 10°C, at 37°C in pH 6.5 buffer, or at 37°C with 10% glycerol in the buffer (Fig. 3D). Our SPR experiments thus demonstrated a direct, conformation-sensitive interaction between ERdj3 and CTA1.
ERdj3 recognizes the A12 subdomain of CTA1.
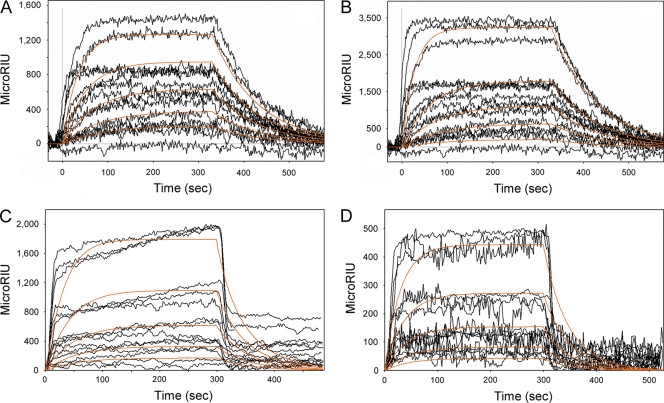
Toxin-ERAD interactions were originally thought to involve the hydrophobic, C-terminal A13 subdomain of CTA1 (14, 20). However, we found that the A13 subdomain is not required for CTA1 translocation to the cytosol (44). This suggested that CTA1-ERAD interactions can occur in the absence of the CTA13 subdomain. We accordingly used SPR to examine the interaction between ERdj3 and CTA11–168, a His-tagged CTA1 construct lacking most of the A13 subdomain. Previous work has shown that CTA11–168 is a folded protein with the same thermal unfolding profile as full-length CTA1 (1, 3, 44). CTA11–168 also exhibited the same general affinity for ERdj3 as full-length CTA1 (Fig. 4, Table 1). Thus, the CTA13 subdomain was not necessary for an interaction with ERdj3. Further analysis indicted that ERdj3 binds to CTA1 and CTA11-168 in a monomeric state: a 1:1 Langmuir fit was used for the SPR data, and CTA1-ERdj3 interactions were roughly equivalent regardless of whether ERdj3 was in the perfusion buffer or was bound as a monomer to the SPR sensor slide.
Fig. 4.
ERdj3 affinity for CTA1 and CTA11–168. (A and B) ERdj3 was perfused at concentrations of 1,600, 800, 400, 200, and 100 ng/ml over SPR sensor slides coated with CTA1 (A) or CTA11–168 (B). ERdj3 was removed from the perfusion buffer 300 s into the experiment. (C and D) CTA1 (C) or CTA11–168 (D) was perfused at concentrations of 1,600, 800, 400, 200, and 100 ng/ml over an ERdj3-coated SPR sensor slide. Toxin was removed from the perfusion buffer 300 s into the experiment. For all experiments, perfusion was at pH 7.4 and 37°C. Measurements collected from three independent experiments per condition are shown. The orange lines represent best-fit curves derived from the raw data using 1:1 binding models.
Table 1.
Binding affinities between ERdj3 and either CTA1 or CTA11–168a
| Sensor | Ligand | Ka (1/ms) | Kd (1/s) | KD (nM) |
|---|---|---|---|---|
| ERdj3 | CTA1 | 140,000 | 0.015 | 110 |
| CTA1 | ERdj3 | 90,000 | 0.010 | 114 |
| CTA11–168 | ERdj3 | 62,232 | 0.010 | 163 |
| ERdj3 | CTA11–168 | 87,000 | 0.034 | 390 |
Ka, association rate constant; Kd, dissociation rate constant; KD, equilibrium dissociation constant.
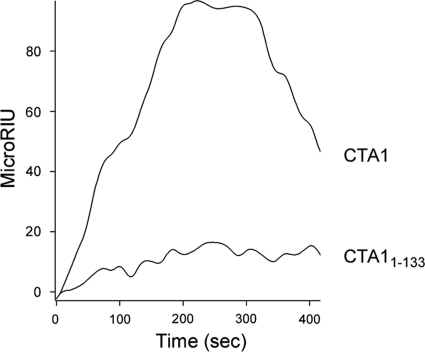
We also used SPR to examine the binding affinity between ERdj3 and CTA11-133, a CTA1 construct lacking both the A12 and A13 subdomains (3). The A12 subdomain is an extended linker which wraps around one side of CTA1 and connects the A13 subdomain to the catalytic core of the toxin, the A11 subdomain (51). CTA11-133 is less stable than either CTA11-168 or full-length CTA1 (3), yet ERdj3 did not exhibit any substantial interaction with CTA11-133 at 37°C (Fig. 5). Indeed, the minimal binding of CTA11-133 to ERdj3 was insufficient to calculate a reproducible KD (equilibrium dissociation constant) value for the toxin-chaperone interaction (n = 5). Our collective SPR data thus suggested that ERdj3 recognizes an unfolded region in the A12 subdomain of CTA1 (amino acid residues 133 to 161).
Fig. 5.
ERdj3 affinity for CTA11–133. CTA1 and CTA11–133 were perfused at a concentration of 400 ng/ml over an ERdj3-coated SPR sensor slide at pH 7.4 and 37°C. Ligand was removed from the perfusion buffer 300 s into the experiment. One of two representative experiments is shown.
ERdj3 masks the hydrophobic residues of thermally unfolded CTA1.
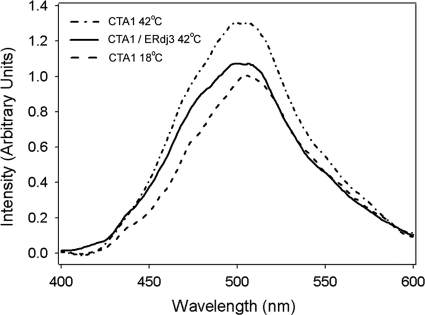
BiP maintains CTA1 in a translocation-competent state by preventing toxin aggregation (48). ERdj3 is likewise thought to prevent the aggregation of misfolded proteins by binding and masking the solvent-exposed hydrophobic residues of its client proteins (15, 35). To determine if ERdj3 performed this function when bound to CTA1, we used bis-ANS to probe the hydrophobic properties of CTA1 in the absence or presence of ERdj3. bis-ANS is nonfluorescent in water but becomes fluorescent in nonpolar environments, such as the solvent-exposed hydrophobic regions of an unfolded protein (13, 18). As shown in Fig. 6, bis-ANS produced a greater fluorescent intensity when incubated with CTA1 at 42°C than when incubated with CTA1 at 18°C. A CTA1-ERdj3 sample at a 1:1 molar ratio incubated with bis-ANS at 42°C generated a significantly lower ANS fluorescence intensity than CTA1 alone at the same concentration. The interaction of ERdj3 with CTA1 thus strongly inhibited bis-ANS binding to the exposed hydrophobic residues present in the 42°C structure of the isolated CTA1 subunit. Although CTA1 is destabilized at temperatures below 42°C (3, 25, 30), we could not detect an ANS signal above the 18°C baseline value until the temperature was raised to 42°C. We attribute this observation to the established propensity of ANS to increase the thermostability of its bound substrate (10, 26). Thus, ERdj3 binding to CTA1 appears to involve a significant hydrophobic component. This interaction likely maintains unfolded CTA1 in a translocation-competent state.
Fig. 6.
ERdj3 masks the hydrophobic residues of thermally unfolded CTA1. CTA1 (8 μM) treated with 50 μM bis-ANS was either placed at 18°C, heated to 42°C, or heated to 42°C in the presence of 8 μM ERdj3. Fluorescence spectra were then recorded with an excitation wavelength of 380 nm.
DISCUSSION
ERAD plays an important role in toxin passage from the ER to the cytosol, but only a few host proteins involved with toxin-ERAD interactions have been identified. As a result, many of the molecular details regarding toxin translocation are uncharacterized. The paucity of ERAD proteins known to interact with CTA1 has also hindered the evaluation of competing models for the translocation event. A prevailing model of ERAD-mediated toxin translocation treats CTA1 as a stable protein that must be actively unfolded by PDI (45). This model views CTA1 as a unique ERAD substrate. In contrast, our structural studies have formed the basis for an alternative model in which toxin-ERAD interactions are dictated by the unstable nature of the isolated CTA1 subunit (3, 25, 30). Here, we documented the functional and physical interactions between CTA1 and ERdj3. The interplay between these two proteins suggests that CTA1 is processed as a typical ERAD substrate.
Expression of dnERdj3 blocked CTA1 movement from the ER to the cytosol and inhibited CT intoxication. Cells transfected with dnERdj3 required a 12-fold-higher concentration of CT to reach the EC50 than cells transfected with an empty vector. Toxin resistance most likely resulted from the 67% drop in CTA1 translocation in cells expressing dnERdj3. A partial inhibition of toxin translocation was also detected in cells expressing wtERdj3, although in this case we did not record a corresponding block of CT intoxication. This discrepancy likely resulted from the different modes of toxin delivery in the translocation assay and in the intoxication assay. With the intoxication assay, all cells were exposed to CT, but only a portion of the cells were transfected with wtERdj3. In contrast, the cotransfection protocol of our translocation assay ensured that most cells expressing CTA1 would also express wtERdj3. We were thus able to detect an effect of wtERdj3 on CTA1 translocation but not on CT intoxication. The limited inhibitory effect of wtERdj3 overexpression on CTA1 translocation was apparently insufficient to impact the intoxication results. In contrast, a strong block of CTA1 translocation in the subpopulation of dnERdj3-expressing cells was sufficient to inhibit overall cAMP production in the entire population of intoxicated cells. These results establish a functional role for ERdj3 in the CT intoxication process.
Disruption of ERdj3 function is likely to trigger the unfolded protein response (UPR), a stress mechanism that enhances ERAD activity (4). However, engagement of the UPR cannot account for toxin resistance in cells expressing dnERdj3: we have found that CT sensitization, rather than resistance, occurs in cells with an active UPR (A. Grabon, N. VanBennekom, and K. Teter, unpublished data). Dixit et al. have likewise shown that treatment with tunicamycin, a UPR-inducing agent, results in cellular sensitization to CT (11). The disruption of CTA1-ERdj3 interactions thus appears to be directly responsible for toxin resistance.
Direct binding of ERdj3 to CTA1 was detected by SPR. This interaction did not require the presence of ATP, which was consistent with the known binding properties of ERdj3 (24). A strong interaction between CTA1 and ERdj3, with a KD value of ≈0.1 μM, was detected when CTA1 was in a partially unfolded state at 37°C, but the interaction was progressively suppressed as CTA1 gained more of its native structure with the lowering of the temperature from 33°C to 25°C to 10°C. Likewise, the interaction between ERdj3 and CTA1 was eliminated when glycerol or acidic pH was used to stabilize the structure of CTA1 at 37°C. Control experiments with denatured CTA1 demonstrated that ERdj3 could function at low temperatures, at acidic pH values, and in the presence of 10% glycerol. Finally, ERdj3 did not interact with the folded CTA1 polypeptide present in CT at 37°C. These observations, in conjunction with the results of our CTA1 structural studies (3, 25, 30), strongly suggest that the interaction between ERdj3 and CTA1 occurs after CTA1 dissociates from the holotoxin and assumes a partially unfolded conformation.
ERdj3 bound to both CTA1 and CTA11–168 with similar affinities (Table 1), so CTA1-ERdj3 interactions do not require the CTA13 subdomain. This was consistent with the translocation-competent state of CTA11–168 (44). Instead, ERdj3 appeared to recognize a region in the A12 subdomain of CTA1: the chaperone did not bind to CTA11–133, a construct lacking both the A12 and A13 subdomains. The loss of affinity cannot be attributed to the stability of CTA11–133, as ERdj3 binds to disordered toxin conformations and CTA11–133 is less stable than either CTA1 or CTA11–168 (3). Thus, ERdj3 appears to recognize a disordered region in the A12 subdomain of CTA1 or, alternatively, a disordered region in the A11 subdomain that is structurally linked to the A12 subdomain.
With our previous use of SPR, we were able to demonstrate that Hsp90 binds to CTA1 as a dimer (40). Here, we found that ERdj3 binds to CTA1 and CTA11–168 in a monomeric state. A previous study reported that the association between ERdj3 and denatured luciferase involves a dimer of ERdj3 (16). It thus appears that ERdj3-substrate interactions can involve either monomeric or dimeric forms of ERdj3, depending on the particular client protein.
Since unfolded conformations of CTA1 are recognized by ERdj3, the interaction between CTA1 and ERdj3 represents a normal substrate-chaperone interaction. This, however, is a departure from a current model of CTA1 translocation in which PDI is required to unfold the supposedly stable CTA1 polypeptide (45). Moreover, PDI has been proposed to bypass the ERAD system and deliver unfolded CTA1 directly to the Derlin-1 pore for export to the cytosol (5, 28). Our recent structural studies have challenged this model by demonstrating that PDI does not act as a CTA1 “unfoldase” (39) and that CTA1 will spontaneously unfold at physiological temperatures after its dissociation from the rest of the toxin (30). Thus, CTA1 does not masquerade as a misfolded protein to facilitate ERAD-mediated toxin translocation; it actually is an unfolded protein. The isolated, disordered CTA1 subunit is accordingly recognized by ERdj3 and likely other components of the ERAD system for export to the cytosol in a process that mimics standard ERAD-substrate interactions. In accordance with this model, the stabilization of the CTA1 structure has been shown to inhibit CTA1-ERdj3 interactions (Fig. 3), CTA1 translocation (3, 25, 38), and CT intoxication (25, 38).
For many ERAD substrates, ERdj3 and BiP work in a sequential fashion (2, 7, 15, 23, 35). ERdj3 initially binds to the substrate, BiP-ATP is recruited to the ERdj3-substrate complex, and ERdj3 stimulates the hydrolysis of BiP-bound ATP to ADP and then dissociates from the BiP-substrate complex. We were unable to reconstitute the putative transfer of CTA1 from ERdj3 to BiP with our SPR system. Additional host factors missing from our in vitro SPR system may be required to facilitate the interaction between BiP and the ERdj3-CTA1 complex. In support of this possibility, Winkeler et al. (48) noted that the inhibition of CTA1 aggregation by purified BiP was enhanced in the presence of an ER extract. Our work suggests that ERdj3 was one, but possibly not the only, ER factor responsible for enhancing BiP activity. ERdj3 likewise represents one component of ERAD complexes that interact with the catalytic A1 subunit of Shiga toxin (49) and incompletely folded immunoglobulin heavy chains (27).
This work establishes ERdj3 as an ERAD component involved with the CT intoxication process and indicates that thermal instability in the isolated CTA1 subunit plays an important role in toxin-ERAD interactions. ERdj3 is also active in translocation of the Shiga toxin A1 subunit, although the molecular basis of this interaction has not been elucidated (49, 50). ERdj3 is one of only a few ERAD components with demonstrated function in the cytosolic entry of multiple ER-translocating toxins and thus represents a possible target for the development of broad-spectrum antitoxin therapeutics.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant R01 AI073783 to K. Teter from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Ampapathi R. S., et al. 2008. Order-disorder-order transitions mediate the activation of cholera toxin. J. Mol. Biol. 377:748–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Awad W., Estrada I., Shen Y., Hendershot L. M. 2008. BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc. Natl. Acad. Sci. U. S. A. 105:1164–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banerjee T., et al. 2010. Contribution of subdomain structure to the thermal stability of the cholera toxin A1 subunit. Biochemistry 49:8839–8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernales S., Papa F. R., Walter P. 2006. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22:487–508 [DOI] [PubMed] [Google Scholar]
- 5. Bernardi K. M., Forster M. L., Lencer W. I., Tsai B. 2008. Derlin-1 facilitates the retro-translocation of cholera toxin. Mol. Biol. Cell 19:877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bies C., et al. 1999. A Scj1p homolog and folding catalysts present in dog pancreas microsomes. Biol. Chem. 380:1175–1182 [DOI] [PubMed] [Google Scholar]
- 7. Brodsky J. L. 2007. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation). Biochem. J. 404:353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buck T. M., Kolb A. R., Boyd C. R., Kleyman T. R., Brodsky J. L. 2010. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol. Biol. Cell 21:1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castro M. G., McNamara U., Carbonetti N. H. 2001. Expression, activity and cytotoxicity of pertussis toxin S1 subunit in transfected mammalian cells. Cell. Microbiol. 3:45–54 [DOI] [PubMed] [Google Scholar]
- 10. Celej M. S., Dassie S. A., Freire E., Bianconi M. L., Fidelio G. D. 2005. Ligand-induced thermostability in proteins: thermodynamic analysis of ANS-albumin interaction. Biochim. Biophys. Acta 1750:122–133 [DOI] [PubMed] [Google Scholar]
- 11. Dixit G., Mikoryak C., Hayslett T., Bhat A., Draper R. K. 2008. Cholera toxin up-regulates endoplasmic reticulum proteins that correlate with sensitivity to the toxin. Exp. Biol. Med. (Maywood) 233:163–175 [DOI] [PubMed] [Google Scholar]
- 12. Forster M. L., et al. 2006. Protein disulfide isomerase-like proteins play opposing roles during retrotranslocation. J. Cell Biol. 173:853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hawe A., Sutter M., Jiskoot W. 2008. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 25:1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hazes B., Read R. J. 1997. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry 36:11051–11054 [DOI] [PubMed] [Google Scholar]
- 15. Jin Y., Awad W., Petrova K., Hendershot L. M. 2008. Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 27:2873–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin Y., Zhuang M., Hendershot L. M. 2009. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry 48:41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaiser E., Pust S., Kroll C., Barth H. 2009. Cyclophilin A facilitates translocation of the Clostridium botulinum C2 toxin across membranes of acidified endosomes into the cytosol of mammalian cells. Cell. Microbiol. 11:780–795 [DOI] [PubMed] [Google Scholar]
- 18. Lakowicz J. R. 1999. Principles of fluorescence spectroscopy, 2nd ed. Kluwer Academic/Plenum Publishers, New York, NY [Google Scholar]
- 19. LaPointe P., Wei X., Gariepy J. 2005. A role for the protease-sensitive loop region of Shiga-like toxin 1 in the retrotranslocation of its A1 domain from the endoplasmic reticulum lumen. J. Biol. Chem. 280:23310–23318 [DOI] [PubMed] [Google Scholar]
- 20. Lencer W. I., Tsai B. 2003. The intracellular voyage of cholera toxin: going retro. Trends Biochem. Sci. 28:639–645 [DOI] [PubMed] [Google Scholar]
- 21. Lord J. M., Roberts L. M., Lencer W. I. 2005. Entry of protein toxins into mammalian cells by crossing the endoplasmic reticulum membrane: co-opting basic mechanisms of endoplasmic reticulum-associated degradation. Curr. Top. Microbiol. Immunol. 300:149–168 [DOI] [PubMed] [Google Scholar]
- 22. Majoul I., Ferrari D., Soling H. D. 1997. Reduction of protein disulfide bonds in an oxidizing environment. The disulfide bridge of cholera toxin A-subunit is reduced in the endoplasmic reticulum. FEBS Lett. 401:104–108 [DOI] [PubMed] [Google Scholar]
- 23. Marcinowski M., et al. 2011. Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18:150–158 [DOI] [PubMed] [Google Scholar]
- 24. Marcus N. Y., Marcus R. A., Schmidt B. Z., Haslam D. B. 2007. Contribution of the HEDJ/ERdj3 cysteine-rich domain to substrate interactions. Arch. Biochem. Biophys. 468:147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Massey S., et al. 2009. Stabilization of the tertiary structure of the cholera toxin A1 subunit inhibits toxin dislocation and cellular intoxication. J. Mol. Biol. 393:1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matulis D., Baumann C. G., Bloomfield V. A., Lovrien R. E. 1999. 1-Anilino-8-naphthalene sulfonate as a protein conformational tightening agent. Biopolymers 49:451–458 [DOI] [PubMed] [Google Scholar]
- 27. Meunier L., Usherwood Y. K., Chung K. T., Hendershot L. M. 2002. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13:4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moore P., Bernardi K. M., Tsai B. 2010. The Ero1alpha-PDI redox cycle regulates retro-translocation of cholera toxin. Mol. Biol. Cell 21:1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakanishi K., et al. 2004. Localization and function in endoplasmic reticulum stress tolerance of ERdj3, a new member of Hsp40 family protein. Cell Stress Chaperones 9:253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pande A. H., et al. 2007. Conformational instability of the cholera toxin A1 polypeptide. J. Mol. Biol. 374:1114–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodighiero C., Tsai B., Rapoport T. A., Lencer W. I. 2002. Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep. 3:1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandvig K., van Deurs B. 2002. Membrane traffic exploited by protein toxins. Annu. Rev. Cell Dev. Biol. 18:1–24 [DOI] [PubMed] [Google Scholar]
- 33. Saslowsky D. E., et al. 2010. Intoxication of zebrafish and mammalian cells by cholera toxin depends on the flotillin/reggie proteins but not Derlin-1 or -2. J. Clin. Invest. 120:4399–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmitz A., Herrgen H., Winkeler A., Herzog V. 2000. Cholera toxin is exported from microsomes by the Sec61p complex. J. Cell Biol. 148:1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen Y., Hendershot L. M. 2005. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol. Biol. Cell 16:40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simpson J. C., et al. 1999. Ricin A chain utilises the endoplasmic reticulum-associated protein degradation pathway to enter the cytosol of yeast. FEBS Lett. 459:80–84 [DOI] [PubMed] [Google Scholar]
- 37. Sitia R., Braakman I. 2003. Quality control in the endoplasmic reticulum protein factory. Nature 426:891–894 [DOI] [PubMed] [Google Scholar]
- 38. Taylor M., et al. 2011. A therapeutic chemical chaperone inhibits cholera intoxication and unfolding/translocation of the cholera toxin A1 subunit. PLoS One 6:e18825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taylor M., Banerjee T., Ray S., Tatulian S. A., Teter K. 2011. Protein disulfide isomerase displaces the cholera toxin A1 subunit from the holotoxin without unfolding the A1 subunit. J. Biol. Chem. 286:22090–22100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taylor M., et al. 2010. Hsp90 is required for transfer of the cholera toxin A1 subunit from the endoplasmic reticulum to the cytosol. J. Biol. Chem. 285:31261–31267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Teter K., Allyn R. L., Jobling M. G., Holmes R. K. 2002. Transfer of the cholera toxin A1 polypeptide from the endoplasmic reticulum to the cytosol is a rapid process facilitated by the endoplasmic reticulum-associated degradation pathway. Infect. Immun. 70:6166–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teter K., Holmes R. K. 2002. Inhibition of endoplasmic reticulum-associated degradation in CHO cells resistant to cholera toxin, Pseudomonas aeruginosa exotoxin A, and ricin. Infect. Immun. 70:6172–6179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teter K., Jobling M. G., Holmes R. K. 2004. Vesicular transport is not required for the cytoplasmic pool of cholera toxin to interact with the stimulatory alpha subunit of the heterotrimeric G protein. Infect. Immun. 72:6826–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teter K., Jobling M. G., Sentz D., Holmes R. K. 2006. The cholera toxin A13 subdomain is essential for interaction with ADP-ribosylation factor 6 and full toxic activity but is not required for translocation from the endoplasmic reticulum to the cytosol. Infect. Immun. 74:2259–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsai B., Rodighiero C., Lencer W. I., Rapoport T. A. 2001. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104:937–948 [DOI] [PubMed] [Google Scholar]
- 46. Veithen A., Raze D., Locht C. 2000. Intracellular trafficking and membrane translocation of pertussis toxin into host cells. Int. J. Med. Microbiol. 290:409–413 [DOI] [PubMed] [Google Scholar]
- 47. Vembar S. S., Brodsky J. L. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Winkeler A., Godderz D., Herzog V., Schmitz A. 2003. BiP-dependent export of cholera toxin from endoplasmic reticulum-derived microsomes. FEBS Lett. 554:439–442 [DOI] [PubMed] [Google Scholar]
- 49. Yu M., Haslam D. B. 2005. Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect. Immun. 73:2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu M., Haslam R. H., Haslam D. B. 2000. HEDJ, an Hsp40 cochaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 275:24984–24992 [DOI] [PubMed] [Google Scholar]
- 51. Zhang R. G., et al. 1995. The three-dimensional crystal structure of cholera toxin. J. Mol. Biol. 251:563–573 [DOI] [PubMed] [Google Scholar]