Abstract
Macrophage infectivity potentiators (Mips) are a group of virulence factors encoded by pathogenic bacteria such as Legionella, Chlamydia, and Neisseria species. Mips are part of the FK506-binding protein (FKBP) family, whose members typically exhibit peptidylprolyl cis-trans isomerase (PPIase) activity which is inhibitable by the immunosuppressants FK506 and rapamycin. Here we describe the identification and characterization of BPSS1823, a Mip-like protein in the intracellular pathogen Burkholderia pseudomallei. Recombinant BPSS1823 protein has rapamycin-inhibitable PPIase activity, indicating that it is a functional FKBP. A mutant strain generated by deletion of BPSS1823 in B. pseudomallei exhibited a reduced ability to survive within cells and significant attenuation in vivo, suggesting that BPSS1823 is important for B. pseudomallei virulence. In addition, pleiotropic effects were observed with a reduction in virulence mechanisms, including resistance to host killing mechanisms, swarming motility, and protease production.
INTRODUCTION
Burkholderia pseudomallei is a motile, Gram-negative bacillus and the causative agent of the disease melioidosis. Melioidosis is endemic in Southeast Asia and Northern Australia. Infection typically occurs by inoculation of the organism through skin lesions, but infection by inhalation or ingestion of the organism has also been reported (10). Clinical presentation of melioidosis in humans varies from disseminated acute septicemia to localized chronic infection (4). Pneumonic infection occurs in 60% of acute cases, resulting in significantly higher mortality rates (34). B. pseudomallei is listed as a category B agent by the U.S. Centers for Disease Control and Prevention (37). There is currently no vaccine available for prophylaxis, and intrinsic antibiotic resistance makes treatment regimens complex.
Although the virulence mechanisms employed by B. pseudomallei have been extensively studied in recent years (1), many remain poorly defined. As an intracellular organism, B. pseudomallei is able to invade, replicate, and spread directly from cell to cell (23, 24). In addition, bacteria can evade phagosome-lysosome fusion and destroy the phagosome membrane (17). However, the mechanisms used by B. pseudomallei to avoid clearance are largely unknown.
FK506-binding proteins (FkBPs) are ubiquitous in eukaryotes and prokaryotes; they typically possess peptidylprolyl cis-trans isomerase (PPIase) activity and catalyze the folding of proline-containing proteins. PPIase activity is inhibitable upon binding to the immunosuppressants FK506 and rapamycin (38). Although PPIases are widely distributed in bacteria, the functions of these proteins are poorly understood. In some bacteria, PPIases have been shown to play a role in virulence and have been termed macrophage infectivity potentiators (Mips) (7, 20, 27, 30). The best studied Mip is a 24-kDa FKBP from Legionella pneumophila, which has been shown to play a role in the invasion of human macrophages and virulence in guinea pigs (7, 8). Although Mips have been shown to be required for virulence in several pathogens, the cellular target(s) of Mip is yet to be elucidated. Furthermore, because of the potentially diverse functions of Mips, it is unclear whether the Legionella Mip provides a paradigm for extrapolating the functions of Mips in other bacteria.
This study reports the identification of a Mip-like protein encoded by B. pseudomallei which possesses PPIase activity and is inhibitable by rapamycin. The Mip-like protein is required for intracellular survival and for virulence in a BALB/c mouse model of infection. In addition, inactivation of the Mip-like gene has pleiotropic effects on several known virulence mechanisms, providing new information on the role of bacterial Mips in disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. All strains were grown in LB broth at 37°C overnight with agitation, unless otherwise stated. Antibiotics were used at the following final concentrations: kanamycin, 50 μg/ml; ampicillin, 50 μg/ml; chloramphenicol, 30 μg/ml; and gentamicin, 10 μg/ml to 30 μg/ml.
Table 1.
Bacterial strains used in this study
| Species and strain | Description | Reference or source |
|---|---|---|
| E. coli | ||
| BL21(DE3) | BL21 with a λ DE3 lysogen | Invitrogen |
| S17-1 λpir | S17-1 with a λ prophage carrying the pir gene | 35 |
| HB101(pRK2013) | HB101 containing pRK2013 Kmr | 13 |
| B. pseudomallei | ||
| AI | K96243 derivative; unmarked deletion ΔamrA; Gms | S. Harding, Dstl |
| AI ΔBPSS1823 | K96243 derivative; unmarked deletion ΔamrA ΔBPSS1823; Gms | This study |
| ΔBPSS1823 (PBBR-1823) | K96243 derivative; unmarked deletion ΔamrA ΔBPSS1823::pBBR1Mip; Gms Kmr | This study |
Construction of an expression plasmid for production of recombinant BPSS1823.
The open reading frame BPSS1823 was amplified by PCR using B. pseudomallei strain K96243 genomic DNA as a template and the primers pET.F (CATATGACAGTCGTCACCACC) and pET.R (GGATCCTCAGACGTCGAGCAGTTC). The PCR product was inserted into the NdeI/BamHI site of the pET15b expression plasmid (Novagen). The construct was transformed into Escherichia coli strain BL21(DE3) to allow expression of His6-tagged BPSS1823 recombinant protein.
Purification of recombinant BPSS1823 protein.
A single colony of E. coli BL12(DE3) harboring the expression construct was used to inoculate 2 liters of LB broth. This was incubated at 37°C with agitation until the absorbance reached 0.4 to 0.6 at 600 nm. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM and growth continued at 20°C with agitation for 4 h. Cells were harvested by centrifugation at 8,000 × g for 15 min at 4°C and then disrupted by sonication. Cell debris was pelleted by centrifugation at 8,000 × g for 30 min at 4°C. The supernatant was loaded onto a 1-ml Histrap FF column (GE Healthcare) and the recombinant protein eluted in 100 mM imidazole. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and purity examined by staining with Coomassie brilliant blue (Pierce Biotechnology). The protein concentration was determined using a bicinchoninic acid assay (Pierce Biotechnology). Imidazole was removed from the purified protein by dialysis against 10 mM phosphate-buffered saline (PBS) and samples frozen at −80°C until use.
Peptidylprolyl isomerase assay.
The peptidylprolyl cis-trans isomerase activity of recombinant BPSS1823 protein was determined by a protease-coupled assay as described previously (14). Briefly, 10 nM BPSS1823 protein was incubated for 6 min at 10°C in 1.2 ml 35 mM HEPES buffer (pH 7.8) with succinyl-Ala-Phe-Pro-Phe-p-nitroanilide (10 mg/ml; Bachem). Chymotrypsin (Sigma) was added to the cuvette at a final concentration of 0.8 mg/ml and mixed. Hydrolysis of the substrate was measured at 390 nm using a Shimadzu 1800 UV/visible spectrophotometer at 1-s intervals until there was no further change in absorbance. For inhibition measurements, recombinant BPSS1823 protein was preincubated with various concentrations of rapamycin from 30 nM to 1 nM for 6 min prior to the addition of substrate. At least three independent readings were taken at each data point. All data fitting and statistical analyses were performed using SPSS v16.0 (IBM).
The pseudo-first-order rate constant was calculated using the equation ln (A∞ − At) = −kobst + ln (A∞ − A0); data from 10 to 50 s (which were always after the lag phase and before substrate became limiting) were taken, and kobs was calculated by linear regression. The enzymatic rate was determined by comparing the observed rate to the uncatalyzed rate using the equation kenz = kobs − kuncat. The specificity constant kcat/Km for the enzyme was calculated using the equation (kcat/Km) = (kenz/[PPIase]) (18); data were taken using 1 nM, 5 nM, and 10 nM BPSS1823 and were fit using linear regression. Data for inhibitor assays were fit to the equation (43) using least-squares nonlinear fitting. v0 and KIapp were fit using initial estimates based on the raw data, and [E] was kept constant.
Mutant strain construction.
B. pseudomallei deletion mutants were constructed as previously described (27). A 453-bp upstream flanking region including the start codon and a 311-bp downstream region including the stop codon were amplified from B. pseudomallei K96243 genomic DNA using primer pairs LFF/LFR (TCTAGAGCCGCCGACCTTTACATT/AGATCTGCTCGAATCGAACTTCTG) and RFF/RFR (AGATCTCTCGTGTTCGAAGTCGAA/TCTAGACCAGTTGGCTGTTGTCGG). Restriction sites were engineered into the primers to allow ligation of the flanks and insertion into the XbaI site of pDM4. The pDM4 construct was transformed into E. coli S17 λpir and conjugated into B. pseudomallei strain AI. Merodiploid integrants were identified using antibiotic selection and plated onto LB agar lacking sodium chloride but containing 10% sucrose. SacB counterselection was used to select for excision of vector DNA, resulting in an unmarked deletion. Colonies were screened for chloramphenicol sensitivity and analyzed by PCR. Southern hybridization, using a digoxigenin (DIG)-labeled upstream flanking region to probe, was used to confirm a 171-bp deletion of BPSS1823, and the strain was termed B. pseudomallei AI ΔBPSS1823.
Complementation studies.
The open reading frame BPSS1823 was amplified by PCR using B. pseudomallei strain K96243 genomic DNA as a template and the primers PBBR.F (GAATTCATGACAGTCGTCACCACC) and PBBR.R (TCTAGATCAGACGTCGAGCAGTTC). The PCR product was inserted into the EcoRI/XbaI restriction sites of pBBR1-MCS2. The complementation construct was transformed into E. coli S17 λpir and conjugated into B. pseudomallei AI ΔBPSS1823 with the helper strain E. coli HB101(pRK2013). Conjugates were selected for resistance on LB agar containing 700 μg/ml kanamycin and 50 μg/ml ampicillin and confirmed by colony PCR. For future experiments, the complemented mutant strain was grown in LB broth containing 200 μg/ml kanamycin and 1 mM IPTG to induce expression of BPSS1823.
Infection of cell lines.
J774A.1 murine macrophages or A549 human epithelial cells were seeded onto a 24-well tissue culture plate at a concentration of 4 × 105 cells/ml in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% l-glutamine and 10% fetal calf serum and incubated at 37°C with 5% CO2 for approximately 16 h. B. pseudomallei strains were grown at 37°C overnight then adjusted in Leibovitz L-15 medium with 10% fetal calf serum to an absorbance of 0.35 to 0.4 at 590 nm. Bacteria were serially diluted in L-15 medium, and 1 ml was added to the cells at a multiplicity of infection (MOI) of 1 or 10 and incubated at 37°C for 30 min or 1 h. Further dilutions were plated onto LB agar at the time of infection to allow for determination of the starting inoculum. Bacteria were removed, and infected cells incubated with L-15 containing 30 μg/ml gentamicin for 30 min at 37°C. Antibiotic medium was removed, serially diluted in PBS, and plated onto LB agar to confirm extracellular killing. Cells were then incubated with 10 μg/ml gentamicin for 24 h. At 0, 2, 4, and 24 h postinfection, cells were lysed with 1 ml distilled water (dH2O), serially diluted in PBS, and plated onto LB agar to determine intracellular numbers.
Adhesion to A549 epithelial cells.
A549 cells and bacteria were prepared as described above. Cytochalasin D (Sigma) was added to approximately 1 × 106 cells at a final concentration of 1 μg/ml and incubated at 37°C with 5% CO2 for 30 min. Cytochalasin D was added to approximately 1 × 107 CFU/ml bacteria at a final concentration of 1 μg/ml. One milliliter of treated bacteria was added to the pretreated cells at an MOI of 1:10 and incubated at 37°C for 1 h. The cells were then washed 3 times with warm PBS to remove nonadherent bacteria. Cells were lysed with 1 ml dH2O, serially diluted in PBS, plated onto LB agar, and incubated at 37°C overnight. Cytochalasin D was present throughout the assay.
Exposure to low pH.
B. pseudomallei strains were grown at 37°C overnight, adjusted to an absorbance of 0.01 at 590 nm, and grown for 2 h at 37°C with agitation. One hundred microliters of adjusted bacterial culture was inoculated into 10 ml LB broth at pH 4 or pH 7 (adjusted with HCl) and incubated at 37°C overnight with agitation. At 0, 3, and 24 h postinoculation, 100 μl of bacterial culture was removed, serially diluted, plated onto LB agar, and incubated at 37°C overnight.
Motility assay.
B. pseudomallei strains were grown at 37°C overnight. One microliter of overnight culture was stabbed into 0.3% motility agar using a sterile inoculating loop and the plates incubated at 37°C overnight. Bacterial spread was measured using a Scienceware vernier caliper (Sigma).
Electron microscopy.
B. pseudomallei strains were grown at 37°C overnight. Two milliliters of culture was pelleted at 15,000 × g for 5 min. Samples were fixed in 4% formalin for 24 h. Samples were stained with 2% (wt/vol) uranyl acetate and examined in an FEI CM12 transmission electron microscope operating at 80 kV, and images were captured using a 1MP Keenview digital camera.
Protease assay.
An overnight culture of B. pseudomallei was diluted 1:50 and grown at 37°C. Following 24 h of growth, 1 ml bacterial culture was removed and pelleted at 15,000 × g for 5 min. One hundred microliters of supernatant was added to 100 μl azocasein (5 mg/ml; Sigma) and incubated at 37°C for 1 h. The reaction was stopped with 10% trichloroacetic acid (Sigma) and nonhydrolyzed azocasesin pelleted at 10,000 × g for 15 min. The supernatant was added to 500 mM NaOH and read using a WPA Colorwave colorimeter (model C07500) at 440 nm.
Animals.
Groups of six female BALB/c age-matched mice were housed together with free access to food and water and subjected to a 12-h light/dark cycle. All studies involving animals were carried out according to the requirements of the Animal (Scientific Procedures) Act (1986) and the Codes of Practice for the Housing and Care of Animals used in Scientific Procedures (1989). For challenge with B. pseudomallei, animals were handled under biosafety level III containment.
B. pseudomallei challenge.
Groups of six mice were challenged with 6.3 × 106 CFU of B. pseudomallei AI or 2.5 × 106 CFU of B. pseudomallei AI ΔBPSS1823 intraperitoneally, and infection was monitored for 5 weeks. Humane endpoints were strictly observed so that animals presenting predetermined clinical signs indicative of a lethal infection were culled.
Isolation of bacteria from murine spleens.
Following challenge with B. pseudomallei, remaining survivors were humanely culled. The spleens were aseptically removed and homogenized in 1 ml sterile PBS. Dilutions of the homogenates were plated onto LB agar to determine bacterial load.
Modeling of BPSS1823.
The structure of BPSS1823 protein was modeled using MODELLER version 9.8 (12). Three structures (1FD9, 1FKB, and 1ROT) were selected as templates. A structure-based sequence alignment for these structures was produced using MAMMOTH-mult (31) and edited by hand. Structure-based alignment of the sequence of BPSS1823 was performed using JOY (32) and FUGUE (40). Ten models were prepared using the high-quality VTFM optimization and MD/SA optimization options. Models were scored according to MODELLER energy score, and Ramachandran plot quality was judged by RAMPAGE (28).
Statistical analysis.
For intracellular infection and pH exposure assays, a two-way analysis of variance (ANOVA) and Bonferroni's posttest were used to determine statistical significance between groups. For motility assays, a one-way ANOVA and Bonferroni's multiple-comparison test were used. For protease assays, an unpaired Student t test was used. Survival curves were compared using a log rank (Mantel-Cox) test. Significances are indicated as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Statistical analyses were performed using either GraphPad Prism version 4.0 or Microsoft Office Excel 2003.
RESULTS
Burkholderia pseudomallei encodes a Mip-like protein.
The open reading frame BPSS1823 from B. pseudomallei K96243 encodes a polypeptide of 113 amino acids annotated as a peptidylprolyl cis-trans isomerase (http://www.sanger.ac.uk/Projects/B_pseudomallei/). BLAST searches against a nonredundant NCBI protein database revealed sequence similarity between BPSS1823 and Mip proteins from a variety of bacterial intracellular pathogens.
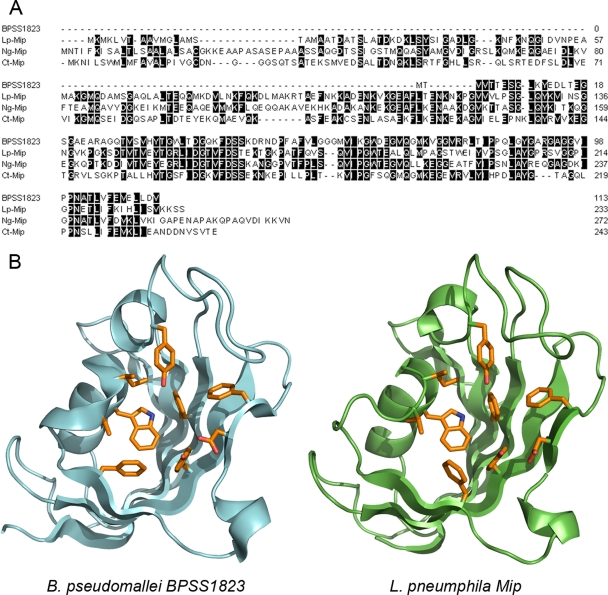
BPSS1823 has 40%, 45%, and 42% amino acid identity to L. pneumophila Mip, Neisseria gonorrhoeae Mip, and Chlamydia trachomatis Mip, respectively (Fig. 1A). BPSS1823 does not contain a putative N-terminal dimerization domain but has high homology to the C-terminal PPIase domain possessed by other Mips, suggesting that it could have PPIase activity. In addition, BPSS1823 possesses most residues required for PPIase activity in human FKBP12 (3, 21, 29).
Fig. 1.
BPSS1823 encodes a Mip-like protein. (A) Sequence alignment of BPSS1823 and L. pneumophila (Lp), T. cruzi (Tc), and N. gonorrhoeae (Ng) Mips. Identical amino acids are shaded in gray. (B) Overview of the modeled structure of BPSS1823 (green) in comparison with L. pneumophila Mip (structure 1FD9, cyan). The nine most conserved amino acids in the active site are shown in orange.
To verify that BPSS1823 is likely to encode a Mip homologue, we modeled the structure of the protein (Fig. 1B). This model predicts that, like L. pneumophila Mip, BPSS1823 adopts a classical FKBP fold and has an active site that strikingly resembles the L. pneumophila Mip active site. In addition, all of the residues that are highly conserved in the active site of FKBPs appear to be conserved in sequence and space. This model gave us further confidence that BPSS1823 is a bona fide Mip homologue and that it might have a role similar to that of the Mips in other organisms.
The B. pseudomallei Mip-like protein exhibits rapamycin-inhibitable PPIase activity.
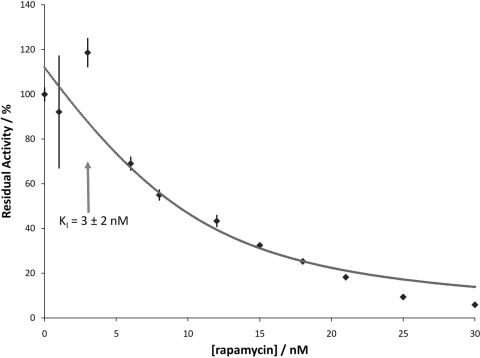
Purified recombinant His-tagged BPSS1823 protein had a molecular mass, determined by mass spectrometry, of 14,436 Da. Size exclusion chromatography (data not shown) demonstrated that this protein is monomeric, consistent with the observation of the lack of an N-terminal dimerization domain in the sequence. Mip proteins from other bacteria have been shown to have PPIase activity which can be inhibited upon binding to FK506 and rapamycin (7, 20, 26, 30, 33). Recombinant BPSS1823 protein was tested for PPIase activity in an enzyme-coupled assay by measuring cis-trans isomerization of the tetrapeptide Suc-Ala-Phe-Pro-Phe-p-nitroanilide (14). Using this substrate, the maximal activity of a highly purified enzyme fraction had a calculated specificity constant kcat/Km of 6.7 × 106 ± 0.4 × 106 M−1 s−1. To examine the effect of rapamycin on the PPIase activity of BPSS1823, recombinant protein was incubated with increasing concentrations of rapamycin. The PPIase activity of BPSS1823 protein is inhibited by nanomolar concentrations of rapamycin, with a KI of 3 ± 2 nM (Fig. 2).
Fig. 2.
Inhibition of PPIase activity of recombinant BPSS1823 by rapamycin. Increasing concentrations of rapamycin lead to dose-dependent inhibition of PPIase activity. The predicted inhibition curve for the fit data is shown. A KI of 3 nM was calculated.
BPSS1823 is required for intracellular survival or replication within, but not adhesion to, eukaryotic cells.
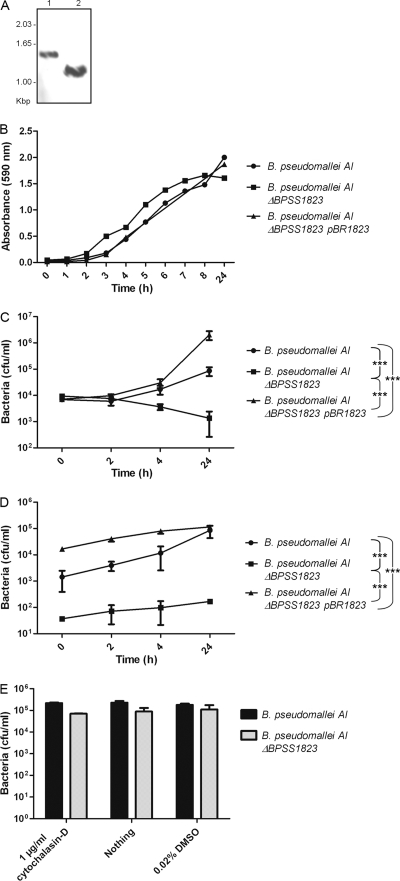
To evaluate the role of BPSS1823 in B. pseudomallei, an in-frame deletion mutant was made in B. pseudomallei strain AI and the deletion confirmed by Southern hybridization (Fig. 3A). The parent strain or the ΔBPSS1823 mutant strain was used to infect phagocytic (J774A.1) or nonphagocytic (A549) cells. In J774A.1 macrophages, the numbers of parent bacteria or ΔBPSS1823 mutant bacteria recovered 1 h after infection were similar. However, significantly fewer ΔBPSS1823 mutant bacteria were recovered from cells at 24 h postinfection (Fig. 3C) (P < 0.001). In A549 epithelial cells, the number of ΔBPSS1823 mutant bacteria recovered 1 h after infection was significantly lower than the number of parent bacteria (Fig. 3D) (P < 0.01). In addition, while the intracellular numbers of the parent strain increased 60-fold over 24 h, almost no replication of the mutant strain was observed (Fig. 3D) (P < 0.001). Reintroduction of the wild-type gene in trans fully restored the ability of the ΔBPSS1823 mutant to survive and grow within both cell lines (Fig. 3C and D) (P < 0.001), confirming that the defect was specific to BPSS1823 in vitro and not due to polar effects. Furthermore, the ΔBPSS1823 mutant did not exhibit reduced growth in LB broth at neutral pH (Fig. 3B) or increased sensitivity to gentamicin (data not shown).
Fig. 3.
Intracellular survival kinetics of B. pseudomallei AI, B. pseudomallei AI ΔBPSS1823, and B. pseudomallei AI ΔBPSS1823 (PBBR-1823). (A) Southern hybridization of B. pseudomallei genomic DNA using a BPSS1823-specific DNA probe. Lane 1, wild-type genomic DNA digested with BamHI and ClaI (1.55 kbp); lane 2, mutant genomic DNA digested with BamHI and ClaI (1.38 kbp). (B) Growth of bacteria in neutral LB broth. Values are from a single experiment. (C) Intracellular survival in J774 macrophage-like cells (MOI of 1). (D) Intracellular survival in A549 epithelial cells (MOI of 10). (E) Adhesion to A549 epithelial cells. Values are the means from triplicate experiments ± standard errors. P values are shown for the comparison of intracellular bacteria at 24 h postinfection.
We investigated whether BPSS1823 also played a role in adherence to A549 cells. Phagocytosis was inhibited preinfection using cytochalasin D, and nonadherent bacteria were removed by washing with PBS. The number of adherent bacteria was determined at 1 h after incubation of bacteria with cells, and no significant difference between the parent and ΔBPSS1823 mutant strains was observed (Fig. 3, E).
BPSS1823 is involved in resistance of B. pseudomallei to low pH.
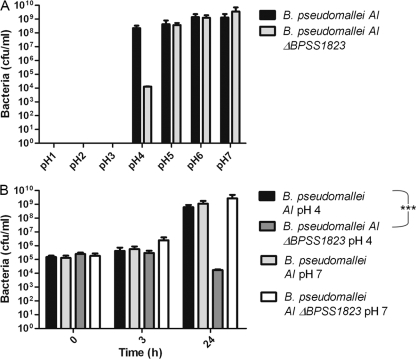
To further characterize the role of BPSS1823 in intracellular survival, the ΔBPSS1823 mutant strain was exposed to a range of environmental stresses, including osmotic stress (NaCl), peroxide stress (H2O2), and a range of pH conditions. There was no difference in the survival of parent or ΔBPSS1823 mutant bacteria under osmotic or peroxide stress (data not shown). While the parent strain grew to a concentration of 107 to 109 CFU/ml in LB medium adjusted to pH 4, 5, 6, or 7, the growth of the ΔBPSS1823 mutant was significantly reduced by 24 h of growth at pH 4 (Fig. 4) (P < 0.001). Neither the parent nor the ΔBPSS1823 mutant was able to grow in medium at pH 3 or below (Fig. 4A).
Fig. 4.
Growth of B. pseudomallei AI and B. pseudomallei AI ΔBPSS1823 at different pHs. (A) Bacteria grown in media adjusted to pH 1 to 7 for 24 h. Values are the means from duplicate experiments ± standard errors. (B) Bacteria grown at pH 4 or 7 at 0, 3, and 24 h postinoculation. Values are the means from triplicate experiments ± standard errors. P values are shown for the comparison of intracellular bacteria at 24 h postinfection.
Deletion of BPSS1823 renders B. pseudomallei immotile and reduces protease production.
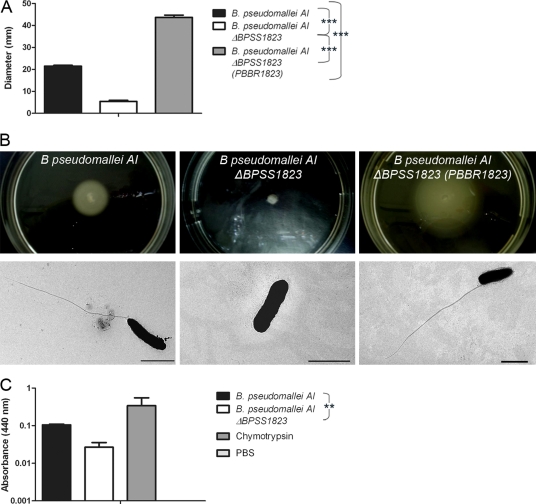
PPIases have been shown to assist in the folding and chaperoning of outer membrane proteins (41). Therefore, membrane-associated virulence mechanisms such as swarming motility and protease secretion were examined in the ΔBPSS1823 mutant. While inoculation of B. pseudomallei AI into 0.3% agar resulted in a mean bacterial spread of 21.4 mm, inoculation with the ΔBPSS1823 mutant resulted in localized growth of 5.4 mm at the site of inoculation and significantly less bacterial spread (Fig. 5A and B) (P < 0.001). In addition, unlike B. pseudomallei AI, the ΔBPSS1823 mutant did not produce flagella (Fig. 5B). Complementation of the ΔBPSS1823 mutant strain fully restored bacterial motility and flagellum formation, resulting in significantly increased bacterial spread compared to that of both B. pseudomallei AI ΔBPSS1823 and B. pseudomallei AI (Fig. 5A and B) (P < 0.001).
Fig. 5.
Swarming motility and protease production of B. pseudomallei AI and B. pseudomallei AI ΔBPSS1823. (A) Diameters of bacterial spread through 0.3% agar. (B) Photographs of bacterial spread through 0.3% agar and representative electron micrographs showing flagella. Scale bar, 2 μm. (C) Protease activity of bacteria using azocasein as a substrate. Values are the means from triplicate experiments ± standard errors. P values are shown for the comparison of strains.
Secreted protease activity was determined by using azocasein as a substrate (2). While both strains exhibited protease activity, hydrolysis of azocasein was 4-fold less in the mutant strain (Fig. 5C) (P < 0.01). This indicates that BPSS1823 is required for production of putative virulence mechanisms in B. pseudomallei, such as swarming motility and protease production.
BPSS1823 is required for full virulence of B. pseudomallei in a murine model of infection.
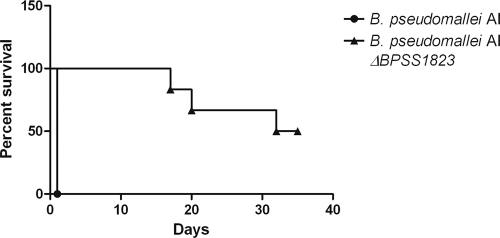
The role of BPSS1823 in B. pseudomallei virulence in vivo was investigated by challenging BALB/c mice via the intraperitoneal route with 6.2 × 106 CFU B. pseudomallei AI or 2.5 × 106 CFU B. pseudomallei AI ΔBPSS1823. All mice challenged with B. pseudomallei AI had succumbed to infection by 1 day postchallenge. In contrast, animals challenged with B. pseudomallei AI ΔBPSS1823 had significantly increased survival, with a mean time to death of >35 days (Fig. 6) (P < 0.001). The mice were monitored for 5 weeks postchallenge, survivors culled, and spleens aseptically removed. Colonies showing morphology typical of B. pseudomallei were recovered from a spleen from one out of three surviving mice, with a bacterial burden of <3 × 102 CFU/ml. Therefore, deletion of BPSS1823 significantly attenuated B. pseudomallei in mice, but low levels of viable bacteria were isolated from one mouse.
Fig. 6.
B. pseudomallei AI ΔBPSS1823 is significantly attenuated in a BALB/c mouse model of infection. Intraperitoneal infection of BALB/c mice (n = 6) with 6.2 × 106 CFU B. pseudomallei AI or 2.5 × 106 CFU B. pseudomallei AI ΔBPSS1823 is shown.
DISCUSSION
Previous studies have shown that Mips are important virulence determinants in several intracellular pathogens (7, 20, 26, 30, 33). Despite the importance of Mip for bacterial pathogenesis, little is known about its specific role or intracellular target. In this study, we describe the identification of a Mip-like protein from B. pseudomallei which is important for virulence. In addition, we show for the first time that a functional Mip is important for enabling a more diverse range of virulence-associated functions than previously reported for other Mips, including bacterial motility, protease production, and acid tolerance.
L. pneumophila Mip is a dimeric outer membrane lipoprotein containing an N-terminal dimerization and chaperone domain and a C-terminal PPIase domain (36). BPSS1823 shows significant sequence identity (>40%) to the L. pneumophila Mip PPIase domain. Three-dimensional modeling of BPSS1823 indicated that the structure is highly conserved (Fig. 1B) and that all of the amino acids that are believed to contribute most significantly to enzyme activity are present (3, 7, 21, 29) These observations were confirmed by nuclear magnetic resonance (NMR) and X-ray determination of the structure of BPSS1823 (34a). These observations strongly suggest that BPSS1823 is a functional orthologue of Mip.
The kcat/Km of L. pneumophila Mip is reported to be 1.2 × 106, (25). We have shown that recombinant BPSS1823 exhibits PPIase activity which is >5-fold higher than that of L. pneumophila Mip (kcat/Km = 6.7 × 106 ± 0.4 × 106 M−1 s−1). As the same substrate (Suc-Ala-Phe-Pro-Phe-p-nitroanilide) was used to analyze PPIase activity in both cases, this observation is unlikely to be due to a difference in substrate specificity; instead, these data may indicate at the importance of PPIase activity for the function of BPSS1823. Furthermore, this enzyme activity is inhibitable by rapamycin, confirming that BPSS1823 belongs to the FKBP family of PPIases. Previous studies have questioned the importance of PPIase activity of L. pneumophila Mip because its variants showing a strongly reduced PPIase activity could complement L. pneumophila strains for intracellular survival in U937 cells and Acanthamoeba castellani (44). However, subsequent work on a parvulin-like PPIase indicated that vanishingly low levels of enzyme activity might suffice to ensure protection against loss of PPIase function (16). Consequently, targeting the PPIase domain of L. pneumophila Mip with activity-neutralizing monoclonal antibodies inhibited Legionella infection of cells, and FK506 or rapamycin inhibited transmigration of L. pneumophila across NCl-H292 lung epithelial cells (19, 42). In addition, removal of the PPIase domain of L. pneumophila Mip attenuated virulence in guinea pig model of infection (25). The importance of PPIase activity for Mip-associated virulence and the availability of licensed PPIase inhibitors suggest that Mips represent novel antimicrobial targets for therapeutics (3). Further work to establish the role of PPIase activity in BPSS1823's role in virulence is required.
Inactivation of L. pneumophila Mip resulted in reduced replication within macrophages and protozoa and attenuated virulence in a guinea pig model of infection (7, 8, 9). We report that the deletion of BPSS1823 in B. pseudomallei results in reduced intracellular survival or replication within eukaryotic cells and significant attenuation in a BALB/c mouse model of infection. Although intracellular growth of the mutant strain was fully restored in vitro by complementation (Fig. 3C and D), care must be taken when extrapolating these data to in vivo studies. The defects in intracellular growth may be partially explained by the observation that the ΔBPSS1823 mutant was more sensitive to low-pH conditions. Following bacterial infection of host cells, the phagosome acidifies to between pH 4 and 5 (11). Therefore, BPSS1823 may act on a protein that protects against acid stress, providing resistance to intracellular host killing mechanisms. In addition, deletion of BPSS1823 resulted in reduced swarming motility and protease production. Previous studies have reported that flagella from B. pseudomallei are involved in invasion of cell lines and virulence in a BALB/c mouse model (5, 6, 22). Secreted proteases have also been shown to be important for B. pseudomallei pathogenesis in a rat model of lung infection but not in a Swiss mouse model (15, 39). It may be hypothesized that BPSS1823 acts to fold or export proteins required for formation of the flagellar complex or production of extracellular proteases. While deletion of BPSS1823 did not render B. pseudomallei avirulent, this can be explained by our in vitro data which indicate that the mutant exhibits defective rather than a lack of infection of cells and virulence mechanisms. Therefore, it could be suggested that BPSS1823 is required for acute infection in BALB/c mice. Additional studies are required to validate the association between BPSS1823 and B. pseudomallei virulence, in particular the contribution of PPIase activity to the protein function.
We have shown that BPSS1823 encodes a Mip-like protein in B. pseudomallei which modulates a broader range of virulence-associated phenotypes than previously reported with other bacterial Mips. While the exact mechanism by which BPSS1823 functions remains unclear, the pleiotropic effects on virulence provide novel insights into the role of Mips in general. In addition, the identification of a Mip-like protein in B. pseudomallei indicates its potential as a target for development of novel antimicrobials to treat melioidosis.
ACKNOWLEDGMENTS
I.H.N. and M.S.-T. were supported by funding from the United Kingdom Ministry of Defense. K.E.K. was supported by a BBSRC CASE award with Dstl.
We are grateful to Tom Laws for assistance with statistical analysis, to Kerry Anderson for carrying out mass spectrometry, and to Simon Smith for carrying out electron microscopy.
Footnotes
Published ahead of print on 22 August 2011.
REFERENCES
- 1. Adler N. R. L., et al. 2009. The molecular and cellular basis of pathogenesis in melioidosis: how does Burkholderia pseudomallei cause disease? FEMS Microbiol. Rev. 33:1079–1099 [DOI] [PubMed] [Google Scholar]
- 2. Brock F. M., Forsberg C. W., Buchanansmith J. G. 1982. Proteolytic activity of rumen microorganisms and effects of proteinase-inhibitors. Appl. Environ. Microbiol. 44:561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ceymann A., et al. 2008. Solution structure of the Legionella pneumophila Mip-rapamycin complex. BMC Struct. Biol. 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng A. C., Currie B. J. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chua K. L., Chan Y. Y., Gan Y. H. 2003. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 71:1622–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chuaygud T., Tungpradabkul S., Sirisinha S., Chua K. L., Utaisincharoen P. 2008. A role of Burkholderia pseudomallei flagella as a virulent factor. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl. 1):S140–S144 [DOI] [PubMed] [Google Scholar]
- 7. Cianciotto N. P., Eisenstein B. I., Mody C. H., Toews G. B., Engleberg N. C. 1989. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect. Immun. 57:1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cianciotto N. P., Eisenstein B. I., Mody C. H., Engleberg N. C. 1990. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J. Infect. Dis. 162:121–126 [DOI] [PubMed] [Google Scholar]
- 9. Cianciotto N. P., Fields B. S. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. U. S. A. 89:5188–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Currie B. J., Jacups S. P. 2003. Intensity of rainfall and severity of melioidosis, Australia. Emerg. Infect. Dis. 9:1538–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Downey G. P., et al. 1999. Phagosomal maturation, acidification, and inhibition of bacterial growth in nonphagocytic cells transfected with Fc gamma RIIA receptors. J. Biol. Chem. 274:28436–28444 [DOI] [PubMed] [Google Scholar]
- 12. Eswar N., et al. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 5:5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Figurski D. H., Helinski D. R. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fischer G., Bang H., Mech C. 1984. Detection of enzyme catalysis for cis-trans isomerization of peptide-bonds using proline-containing peptides as substrates. Biomed. Biochim. Acta 43:1101–1111 [PubMed] [Google Scholar]
- 15. Gauthier Y. P., Thibault F. M., Paucod J. C., Vidal D. R. 2000. Protease production by Burkholderia pseudomallei and virulence in mice. Acta Trop. 74:215–220 [DOI] [PubMed] [Google Scholar]
- 16. Gemmill T. R., Wu X. Y., Hanes S. D. 2005. Vanishingly low levels of Ess1 prolyl-isomerase activity are sufficient for growth in Saccharomyces cerevisiae. J. Biol. Chem. 280:15510–15517 [DOI] [PubMed] [Google Scholar]
- 17. Harley V. S., Dance D. A. B., Tovey G., McCrossan M. V., Drasar B. S. 1998. An ultrastructural study of the phagocytosis of Burkholderia pseudomallei. Microbios 94:35–45 [PubMed] [Google Scholar]
- 18. Harrison R. K., Stein R. L. 1990. Substrate specificities of the peptidyl prolyl cis-trans isomerase activities of cyclophilin and FK-506 binding-protein—evidence for the existence of a family of distinct enzymes. Biochemistry 29:3813–3816 [DOI] [PubMed] [Google Scholar]
- 19. Helbig J. H., et al. 2003. The PPIase active site of Legionella pneumophila Mip protein is involved in the infection of eukaryotic host cells. Biol. Chem. 384:125–137 [DOI] [PubMed] [Google Scholar]
- 20. Horne S. M., Kottom T. J., Nolan L. K., Young K. D. 1997. Decreased intracellular survival of an fkpA mutant of Salmonella typhimurium Copenhagen. Infect. Immun. 65:806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ikura T., Ito N. 2007. Requirements for peptidyl-prolyl isomerization activity: a comprehensive mutational analysis of the substrate-binding cavity of FK506-binding protein 12. Protein Sci. 16:2618–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inglis T. J. J., Robertson T., Woods D. E., Dutton N., Chang B. J. 2003. Flagellum-mediated adhesion by Burkholderia pseudomallei precedes invasion of Acanthamoeba astronyxis. Infect. Immun. 71:2280–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones A. L., Beveridge T. J., Woods D. E. 1996. Intracellular survival of Burkholderia pseudomallei. Infect. Immun. 64:782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kespichayawattana W., Rattanachetkul S., Wanun T., Utaisincharoen P., Sirisinha S. 2000. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect. Immun. 68:5377–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kohler R., et al. 2003. Biochemical and functional analyses of the Mip protein: influence of the N-terminal half and of peptidylprolyl isomerase activity on the virulence of Legionella pneumophila. Infect. Immun. 71:4389–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leuzzi R., et al. 2005. Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol. Microbiol. 58:669–681 [DOI] [PubMed] [Google Scholar]
- 27. Logue C. A., Peak I. R. A., Beacham I. R. 2009. Facile construction of unmarked deletion mutants in Burkholderia pseudomallei using sacB counter-selection in sucrose-resistant and sucrose-sensitive isolates. J. Microbiol. Methods 76:320–323 [DOI] [PubMed] [Google Scholar]
- 28. Lovell S. C., et al. 2003. Structure validation by C alpha geometry: phi, psi and C beta deviation. Proteins 50:437–450 [DOI] [PubMed] [Google Scholar]
- 29. Löw C., et al. 2010. Crystal structure determination and functional characterization of the metallochaperone SlyD from Thermus thermophilus. J. Mol. Biol. 398:375–390 [DOI] [PubMed] [Google Scholar]
- 30. Lundemose A. G., Kay J. E., Pearce J. H. 1993. Chlamydia trachomatis Mip-like protein has peptidyl-prolyl cis-trans isomerase activity that is inhibited by FK506 and rapamycin and is implicated in initiation of chlamydial infection. Mol. Microbiol. 7:777–783 [DOI] [PubMed] [Google Scholar]
- 31. Lupyan D., Leo-Macias A., Ortiz A. R. 2005. A new progressive-iterative algorithm for multiple structure alignment. Bioinformatics 21:3255–3263 [DOI] [PubMed] [Google Scholar]
- 32. Mizuguchi K., Deane C. M., Blundell T. L., Johnson M. S., Overingon J. P. 1998. JOY: protein sequence-structure representation and analysis. Bioinformatics 14:617–623 [DOI] [PubMed] [Google Scholar]
- 33. Moro A., Ruizcabello F., Fernandezcano A., Stock R. P., Gonzalez A. 1995. Secretion by Trypanosoma cruzi of a peptidyl-prolyl cis-trans isomerase involved in cell infection. EMBO J. 14:2483–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mukhopadhyay A., Lee K. H., Tambyah P. A. 2004. Bacteraemic melioidosis pneumonia: impact on outcome, clinical and radiological features. J. Infect. 48:334–338 [DOI] [PubMed] [Google Scholar]
- 34a. Norville I. H., et al. 2011. The structure of a Burkholderia pseudomallei immunophilin-inhibitor complex reveals new approaches to antimicrobial development. Biochem. J. 437:413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Penfold R. J., Pemberton J. M. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145–146 [DOI] [PubMed] [Google Scholar]
- 36. Riboldi-Tunnicliffe A., et al. 2001. Crystal structure of Mip, a prolylisomerase from Legionella pneumophila. Nat. Struct. Biol. 8:779–783 [DOI] [PubMed] [Google Scholar]
- 37. Rotz L. D., Khan A. S., Lillibridge S. R., Ostroff S. M., Hughes J. M. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schreiber S. L. 1991. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251:283–287 [DOI] [PubMed] [Google Scholar]
- 39. Sexton M. M., Jones A. L., Chaowagul W., Woods D. E. 1994. Purification and characterization of a protease from Pseudomonas pseudomallei. Can. J. Microbiol. 40:903–910 [DOI] [PubMed] [Google Scholar]
- 40. Shi J. Y., Blundell T. L., Mizuguchi K. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310:243–257 [DOI] [PubMed] [Google Scholar]
- 41. Vertommen D., Ruiz N., Leverrier P., Silhavy T. J., Collet J. F. 2009. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9:2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wagner C., et al. 2007. Collagen binding protein Mip enables Legionella pneumophila to transmigrate through a barrier of NCI-H292 lung epithelial cells and extracellular matrix. Cell. Microbiol. 9:450–462 [DOI] [PubMed] [Google Scholar]
- 43. Williams J. W., Morrison J. F. 1979. The kinetics of reversible tight-binding inhibition. Methods Enzymol. 63:437–467 [DOI] [PubMed] [Google Scholar]
- 44. Wintermeyer E., et al. 1995. Influence of site-specifically altered Mip proteins on intracellular survival of Legionella pneumophila in eukaryotic cells. Infect. Immun. 63:4576–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]