Abstract
Malaria remains a devastating disease despite efforts at control and prevention. Extensive studies using mostly rodent infection models reveal that successful Plasmodium parasite transmission by the African mosquito vector Anopheles gambiae depends on finely tuned vector-parasite interactions. Here we investigate the transcriptional response of A. gambiae to geographically related Plasmodium falciparum populations at various infection intensities and different infection stages. These responses are compared with those of mosquitoes infected with the rodent parasite Plasmodium berghei. We demonstrate that mosquito responses are largely dependent on the intensity of infection. A major transcriptional suppression of genes involved in the regulation of midgut homeostasis is detected in low-intensity P. falciparum infections, the most common type of infection in Africa. Importantly, genes transcriptionally induced during these infections tend to be phylogenetically unique to A. gambiae. These data suggest that coadaptation between vectors and parasites may act to minimize the impact of infection on mosquito fitness by selectively suppressing specific functional classes of genes. RNA interference (RNAi)-mediated gene silencing provides initial evidence for important roles of the mosquito G protein-coupled receptors (GPCRs) in controlling infection intensity-dependent antiparasitic responses.
INTRODUCTION
Malaria is a devastating parasitic disease that remains an enormous public health burden, especially in sub-Saharan Africa, where the deadliest of the malaria parasites, Plasmodium falciparum, is transmitted mainly by Anopheles gambiae mosquitoes. Extensive laboratory studies of mosquito-parasite interactions have exploited genomics and postgenomics technologies for gene expression profiling and functional characterization to generate new knowledge. It has been established that parasite development in the mosquito midgut is the most vulnerable stage of the entire transmission cycle. This appears to be due largely to robust mosquito responses that effectively reduce the number of parasites able to invade the midgut and establish infections in the hemolymph (8, 22, 30). Therefore, detailed understanding of the vector-parasite interactions could lead to novel strategies that can supplement current control methods.
Recent studies have identified mosquito genes that affect P. falciparum and the rodent malaria parasite, Plasmodium berghei, which is often used as a laboratory model, similarly; however, such conserved responses can only partly explain the vector-parasite interactions in natural settings (6, 8, 11, 20, 21). In fact, A. gambiae is not a natural vector of P. berghei, and the intensities of P. berghei infection are much higher than those commonly observed in nature with P. falciparum (2). Differences in P. falciparum infection levels have been associated with several genomic loci and have been shown to depend on vector-parasite genotype-by-genotype interactions (12, 16, 29).
Evidence is now accumulating that input parasite densities affect the progression of the infection as well as mosquito signaling pathways and mortality; however, the actual impact of infection intensities on mosquito responses is still unclear, and very few studies have considered infection intensities in the analysis of infection data (7, 24, 26). Here we have carried out A. gambiae infections with geographically related P. falciparum parasites sampled from naturally infected children in order to investigate the impact of different infection intensities on the mosquito midgut transcriptome. The results reveal that responses to parasites are dependent on the intensity of infection. Functional annotation and phylogenetic analyses of the genes identified have been used to draw critical conclusions about the nature of these responses. Reverse genetics analysis by RNA interference (RNAi) has been performed to identify genes with important roles during infection. Four rhodopsin-like G protein-coupled receptors (GPCRs) are shown to be involved in these responses, with functions that differ for different parasite species and infection intensities. Our results suggest that vector-parasite coevolution and coadaptation have led to fine-tuning of the mosquito responses to differing intensities of infection with human Plasmodium parasites so as to minimize the impact of these responses on mosquito fitness and permit low-intensity infection, respectively.
MATERIALS AND METHODS
Ethics statement.
Procedures that involved human subjects were approved by the Ethical Review Committee of the Organisation de Coordination pour la Lutte contre les Endémies en Afrique Centrale (OCEAC) (0608-05/SG/CAB/SAF/LRP), the Cameroon National Ethics Committee, and the WHO Research Ethics Review Committee. Written informed consent was obtained from the parents or other legal guardians of the study participants. The mouse work was carried out in strict accordance with the United Kingdom Animals (Scientific Procedures) Act of 1986. The protocols for maintenance of mosquitoes by blood feeding and for infection of mosquitoes with P. berghei by blood feeding on parasite-infected mice were approved and carried out under a United Kingdom Home Office License. The procedures are of mild to moderate severity, and the numbers of animals used are minimized by incorporation of the most economical protocols. Opportunities for reduction, refinement, and replacement of animal experiments are constantly monitored, and new protocols are implemented following approval by the Imperial College Ethical Review Committee.
Mosquito strains.
The Yaoundé strain of A. gambiae was used in all of the assays reported. This strain was established at OCEAC, Cameroon, from populations of A. gambiae sensu stricto (the M molecular form and the Forest chromosomal form) caught in the area of Yaoundé and selected for efficient feeding on Parafilm membranes (28). No additional genetic selection for malaria parasite infection was ever carried out. The strain retains a high degree of genetic diversity compared to that of other inbred laboratory colonies and remains a reasonable approximation of local field mosquitoes (20).
Parasitological survey.
Surveys for P. falciparum gametocyte carriers were conducted in collaboration with a local medical team in primary schools in Mfou, a town near Yaoundé, Cameroon, where malaria is endemic. Blood samples were collected by finger prick from 5- to 11-year-old children, and thick blood smears were stained with 10% Giemsa stain, followed by microscopic examination for the presence of P. falciparum. Densities of sexual and asexual parasites were estimated by assuming a standard number of 8,000 white blood cells (WBC)/μl of blood and counting against 1,000 WBC. Asymptomatic gametocyte carriers were enrolled as volunteers upon parental consent. Children infected with mixed Plasmodium species were excluded.
A. gambiae infections with P. falciparum and P. berghei.
Mosquito infections used for expression profiling were carried out by membrane feeding. Two groups of mosquitoes from the same rearing culture were used per replicate infection. Each mosquito in the first group was fed on blood sampled by venipuncture from a P. falciparum gametocyte carrier or by heart puncture from a BALB/c mouse infected with the P. berghei ANKA 15cy1A (2.34) strain, while each mosquito in the second (control) group was fed on the same blood following a 12-min incubation at 42 to 43°C for gametocyte inactivation. To eliminate transmission-blocking factors, infected blood serum was replaced by nonimmune serum: human AB serum for P. falciparum infections and serum from noninfected BALB/c mice for P. berghei infections. Infections were performed as described previously (20). To determine infection levels, mosquito guts were dissected 8 to 10 days after blood feeding and were stained with 2% Mercurochrome before microscopic examination. The same P. falciparum infection protocol was used for the RNAi experiments.
Microarray hybridizations and analysis.
The Anopheles MMC2 microarray platform was used for all hybridizations presented, encompassing 7,246 A. gambiae genes according to AGAMP3.4 (release date, June 2007). For each infection or mock infection (with heat-inactivated parasites), total RNA was prepared by Trizol extraction (Invitrogen) from 30 to 50 mosquito midguts dissected in phosphate-buffered saline (PBS) solution at 4°C and was then processed for microarray hybridizations, data acquisition, and filtering as described previously (30). Normalization by the locally weighted linear regression (Lowess) method was performed using GeneSpring GX, version 7.3 (Agilent Technologies). Genes exhibiting expression in at least 66% of all the biological replicates were subjected to one-way analysis of variance (ANOVA) (P ≤ 0.05) based on errors calculated by a cross-gene error model. Conditional tree analysis of gene expression profiles was performed using the pvclust package implemented in R, according to the Pearson correlation coefficient. The uncertainty of hierarchical clustering analysis was analyzed by normal bootstrap resampling (1,000 iterations). Significant differences in gene expression between test and control groups were evaluated by combining an expression ratio cutoff of 0.7 on a log2 scale and one-way ANOVA statistical tests (with P values of ≤0.025) across infection stages. Coexpression clusters were produced using Cluster, version 3.0 (10), according to the Pearson correlation score. Clusters were visualized with Java TreeView, version 1.0.13.
Functional, phylogenetic, and chromosomal colocalization analysis.
Associated Gene Ontology (GO) terms, InterPro (IPR) domains, and orthology calls between A. gambiae genes and Aedes aegypti or Drosophila melanogaster were according to the AgamP3.4 gene set (release date, June 2007). For all analyses, statistically significant differences in ortholog frequency for the various data sets were detected by applying a Bonferroni corrected hypergeometric distribution to determine the probability of the occurrence compared to the overall transcriptome of the same infection stage. These data were calculated using GeneMerge (5) for functional terms and Excel (Microsoft Corp., Redmond, WA) for the remaining analysis.
Gene silencing in adult A. gambiae mosquitoes.
RNAi-mediated gene silencing experiments were performed and analyzed as described previously (20). Briefly, target sequences were amplified with specific PCR primers flanked at their 5′ ends by T7 promoter sequences (underlined in primer sequences), which were used to produce double-stranded RNAs (dsRNAs) by using the MEGAscript kit (Ambion) according to the manufacturer's instructions. Primer sequences were as follows: GPGR1 forward primer, 5′-TAATACGACTCACTATAGGGATGTCTGCCTTCCGGTATTAT-3′; GPGR1 reverse primer, 5′-TAATACGACTCACTATAGGGTGGAATGTTGCGCATCGC-3′; GPGR2 forward primer, 5′-TAATACGACTCACTATAGGGATGCCCTTGAGCGAGTTC-3′; GPGR2 reverse primer, 5′-TAATACGACTCACTATAGGGTGCCCGGTGCCTGAGG-3′; GPRNPY3 forward primer, 5′-TAATACGACTCACTATAGGGCCGTCCTACACGTTCATGC-3′; GPRNPY3 reverse primer, 5′-TAATACGACTCACTATAGGGATCTCGATCCGCCTCTTCT-3′; GPRCCK1 forward primer, 5′-TAATACGACTCACTATAGGGGTGTCGGTGGCGGTCTG-3′; GPRCCK1 reverse primer, 5′-TAATACGACTCACTATAGGGCGCGGCACCAGTGGGA-3′.
Data and statistical analysis.
Infection intensity data from the RNAi experiments were analyzed primarily using restricted maximum likelihood (REML) variance components analysis by fitting a mixed-effect model, where the knockdown or control status was treated as the fixed effect and a random effect was introduced for the biological replicate. This method was chosen in order to take into account the high (and mostly unpredicted) variability between biological replicates in infections with P. falciparum field isolates, which include different parasite isolates in each infection (sometimes more than one isolate per infection), child-to-child blood type differences, differences in the medical histories of infected children, etc. In addition, the method takes into account the variable genetic makeup of individual mosquitoes of the Yaoundé strain. Normality of the data was achieved following logarithmic transformation and was confirmed with the Shapiro-Wilk test. In order to include noninfected midguts (which had a value of zero) in the analysis, 1 was added to the total number of oocysts in each mosquito prior to logarithmic transformation [log10(n + 1)]. The geometric mean of log-transformed data was computed; the resulting value (x) was log10 detransformed (10x); and 1 was deducted from the final value (10x − 1) to calculate the geometric mean of the raw data. Meta-analysis of the standardized mean differences with 95% confidence intervals using fixed- and random-effect models was also carried out using comprehensive meta-analysis software (BioStat, version 2) to compare means and variations between biological replicates. The infection data were also analyzed with the nonparametric Mann-Whitney U test (GraphPad Prism, version 5). Differences in infection prevalence were analyzed using the chi-square goodness-of-fit test (GenStat software).
RESULTS
Infections of mosquitoes with African P. falciparum isolates and P. berghei.
We assessed the impact of rodent and human Plasmodium infections on A. gambiae responses by using analogous experimental designs (see Fig. S1A in the supplemental material). Mosquitoes from a laboratory colony originating from populations of the Yaoundé area in Cameroon (28) were fed either on human blood donated by naturally infected asymptomatic P. falciparum gametocyte carriers from the Yaoundé area or on the blood of mice infected with P. berghei. Mosquito midguts were dissected at three time points, corresponding to distinct infection stages: 1 to 3 h postinfection (hpi), representing the postfertilization and zygote development stages (T1); 22 to 25 hpi, corresponding to the peak of ookinete invasion of the mosquito midgut (T2); and 48 to 52 hpi, corresponding to the onset of oocyst development following midgut invasion (T3).
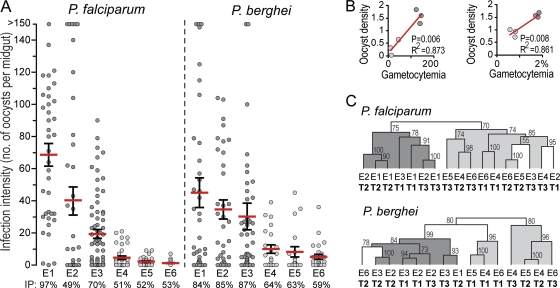
Six independent replicate infections were performed for each parasite, resulting in infection levels that differed substantially (Fig. 1A; see also Table S1 in the supplemental material). These infections were ranked in order of mean infection intensity (highest to lowest) and were identified as P. falciparum E1 to E6 and P. berghei E1 to E6. A strong positive correlation was detected between starting gametocyte densities and mosquito infection intensities for both parasites (Fig. 1B).
Fig. 1.
Infection of A. gambiae with Plasmodium. (A) Intensities recorded for six replicate P. falciparum and P. berghei infections (E1 to E6), ranked in order of mean infection intensity. Red horizontal lines indicate the arithmetic means of oocyst numbers, and error bars indicate standard errors. IP, infection prevalence (percentage of mosquito midguts showing at least one oocyst). (B) Correlation between gametocytemia and oocyst density (number of oocysts per midgut). The level of gametocytemia is presented as the number of gametocytes per microliter of host blood for P. falciparum infections and as a percentage of the red blood cells counted for P. berghei infections and is plotted against the corresponding log10 of the mean oocyst density. (C) Cladograms show conditional analysis of gene expression profiles according to the Pearson correlation coefficient. Bootstrap values from normal resampling (1,000 iterations) are shown above the nodes on a percentage scale. Cohesive clustering according to infection intensity is indicated by shading.
The midgut response to Plasmodium is infection intensity dependent.
Expression analyses were carried out using competitive two-dye hybridizations of DNA microarrays, in which infected midguts were hybridized against the respective noninfected controls (mosquitoes fed on inactivated gametocytes). A total of 6,384 genes that registered expression values in at least one of the three infection stages in at least one of the Plasmodium species were identified (see Table S2 in the supplemental material). Hierarchical clustering revealed that expression profiles clustered primarily on the basis of the infection stage and secondarily on the basis of infection intensity (Fig. 1C). This result indicated that midgut responses at each infection stage are infection intensity dependent, and it prompted an additional analysis where expression data were pooled into three new data sets per parasite: (i) all infections regardless of intensity, (ii) high-intensity infections (mean oocyst number, >15) (E1 to E3), and (iii) low-intensity infections (mean oocyst numbers, <15) (E4 to E6). A total of 2,203 genes exhibiting a >0.7-fold expression change (on a log2 scale) in infected versus noninfected mosquitoes or that were transiently and significantly regulated (P ≤ 0.025) during the course of an infection were used in downstream analyses (see Fig. S1B and C and Table S2 in the supplemental material).
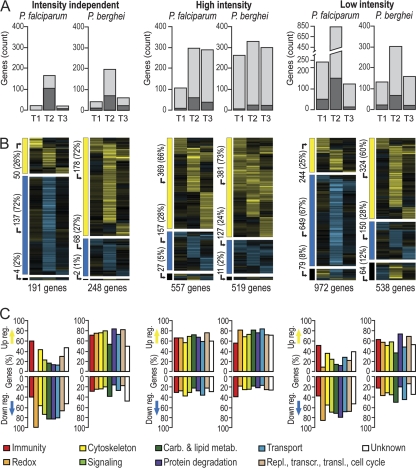
Intensity-independent analysis of P. falciparum infections identified 191 differentially regulated genes with expression values in all infection stages, while P. berghei elicited responses from 248 genes (Fig. 2A). Analysis of intensity-dependent data sets yielded a significantly higher number of genes, indicating fine-tuning of the transcriptional response depending on the infection intensity. The total numbers of genes in high-intensity infections were similar for the two parasites (557 and 519 in P. falciparum and P. berghei, respectively). Strikingly, low-intensity P. falciparum infections elicited a stronger response than low-intensity P. berghei infections (972 versus 538 genes, respectively). In intensity-independent and low-intensity infection analyses, the most prominent response occurred at T2. However, in high-intensity infections, the numbers of differentially regulated genes were similar for the three P. falciparum infection stages, as well as for the P. berghei T2 and T3 infection stages.
Fig. 2.
Differentially regulated genes in the Anopheles midgut upon Plasmodium infection. (A) Total numbers of differentially regulated genes per Plasmodium species and infection stage, grouped according to infection intensity. Light shaded bars, genes showing an expression change higher than 0.7 log2; dark shaded bars, genes showing smaller but significant changes (P ≤ 0.025) across infection stages. (B) Transcriptional profiles of genes for which data are presented in panel A. Eisengrams are divided into three parts: upregulation (yellow) in at least one infection stage (top), downregulation (blue) in at least one infection stage (middle), and upregulation and downregulation in at least one infection stage (bottom). The total numbers of genes in each eisengram and each eisengram part are given. (C) Up- and downregulated fractions of genes belonging to distinct functional classes. Carb., carbohydrate; metab., metabolism; Repl., replication; transcr., transcription; transl., translation.
Analysis of transcriptional bias of differentially regulated genes across all infection stages showed that 60 to 73% of genes were upregulated and 24 to 28% downregulated in all P. berghei data sets and in P. falciparum high-intensity infections, consistent with the findings of previous studies (30). However, in low-intensity P. falciparum infections, the transcriptional bias was strikingly different: ∼67% of genes were downregulated (Fig. 2B).
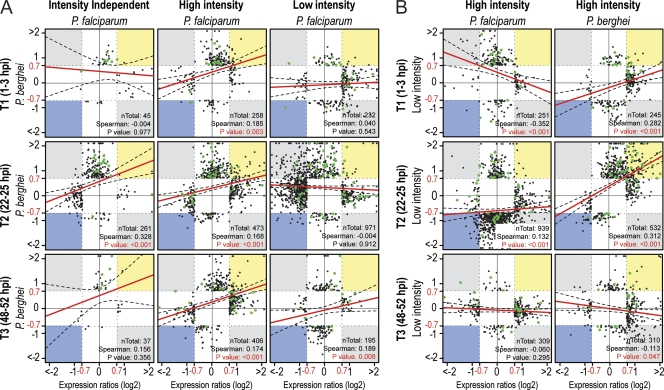
An overall similarity of responses to the two parasite species was detected in high-intensity but not in low-intensity infections, especially at T1 and T2 (Fig. 3A). Responses during early P. falciparum infection stages differed greatly depending on the infection intensity; however, responses during the P. berghei T1 and T2 infection stages were significantly similar for low- and high-intensity infections (Fig. 3B).
Fig. 3.
Comparison of A. gambiae transcriptional responses to the different Plasmodium species and infection intensities. (A) Scatter plot comparisons of expression values of genes differentially regulated (expression ratios greater than 0.7 log2) upon P. falciparum (x axis) or P. berghei (y axis) infection in the various data sets and infection stages. (B) Scatter plot comparisons of expression values of genes differentially regulated (expression ratios greater than 0.7 log2) upon high-intensity (x axis) or low-intensity (y axis) infection with P. falciparum (left) or P. berghei (right). The total number of genes present in each comparison and the significance of the Spearman correlation between the expression ratios of the two data sets are presented at the lower right of each panel. Red lines show the best linear distribution fit; black dashed lines indicate the 95% freedom interval. Green dots indicate immunity-related genes.
The distribution of functional gene classes in differentially regulated genes, as assigned through available databases (31) and manual annotation, showed clear differences between the responses to the two parasite species and between the responses at different infection intensities (Fig. 2C). The strong transcriptional suppression observed in low-intensity P. falciparum infections was evident for all functional classes except immunity, which was always biased toward transcriptional induction, regardless of infection intensity and parasite species (Fig. 3, green dots; see also Fig. S2 in the supplemental material). Importantly, genes involved in regulating the cellular oxidative status showed the most divergent transcriptional behavior for different infection intensities and parasite species. These genes were biased toward suppression in low-intensity P. falciparum infections but not in any other data set.
Phylogenetic analysis of Anopheles responses to Plasmodium.
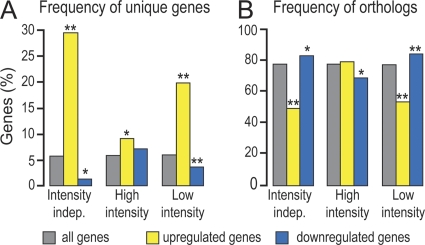
We investigated the phylogenetic relationships between differentially regulated A. gambiae genes during T2 and genes of Aedes aegypti and Drosophila melanogaster. A statistically significant enrichment of genes unique to A. gambiae was detected within the genes upregulated by P. falciparum infection (Fig. 4). This was especially evident in low-intensity infections and was accompanied by a significant depletion of genes with identifiable orthologs in the other two insect genomes. In contrast, the set of genes downregulated by P. falciparum was significantly depleted of unique genes and enriched in genes with identifiable orthologs. Interestingly, downregulation upon P. berghei infection showed an opposite pattern: significant underrepresentation of identifiable orthologs and overrepresentation of unique genes (see Fig. S3 in the supplemental material).
Fig. 4.
Phylogenetic analysis of genes regulated upon midgut invasion by P. falciparum. Percentages of genes either unique to A. gambiae (A) or with identifiable orthologs in Aedes aegypti and D. melanogaster (B) that are upregulated (yellow) or downregulated (blue) are shown. Significant differences from the overall transcriptome registered for each data set (gray) were analyzed statistically using hypergeometric distribution (single asterisks indicate a P value of <0.05; double asterisks indicate a P value of <0.05 after Bonferroni correction). indep., independent.
Functional gene classes involved in mosquito responses to Plasmodium.
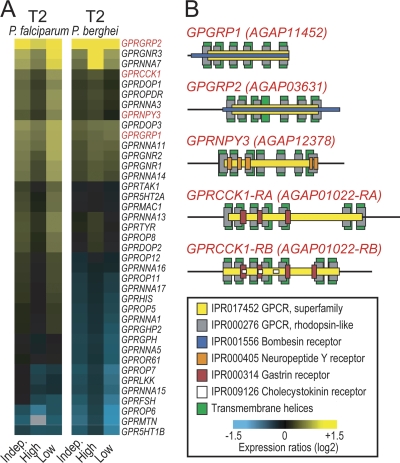
We investigated the putative functions of genes involved in mosquito responses during T2 regardless of infection intensity, using Gene Ontology (GO) (see Table S3 in the supplemental material) and InterPro (IPR) domain (see Table S4 in the supplemental material) annotations. Genes encoding proteins with signal transducer activity in particular G protein-coupled receptors (GPCRs) of the rhodopsin type (IPR000276) were significantly overrepresented during P. falciparum infections. Their directions of differential regulation were mostly similar for low- and high-intensity infections, as well as for P. falciparum and P. berghei infections, but of variable magnitude (Fig. 5). Additionally, various immunity-related GO terms and IPR domains were overrepresented independently of infection intensity. Genes involved in the recognition and catabolism of peptidoglycan, as well as genes containing leucine-rich repeats (LRRs), were overrepresented in P. berghei-infected midguts, while C-type lectins, which are known modulators of the mosquito defense against Plasmodium, were upregulated in P. falciparum infections (6, 22). Importantly, this analysis further highlighted the characteristic downregulation of genes related to oxidative metabolism in low-intensity infections.
Fig. 5.
Rhodopsin-like GPCRs involved in the A. gambiae response to Plasmodium invasion. (A) Expression heat maps showing differential expression of rhodopsin-like GPCR-encoding genes upon invasion of the A. gambiae midgut by P. falciparum or P. berghei. Yellow and blue indicate upregulation and downregulation, respectively. Different color intensities represent different levels of regulation. Indep., independent of infection intensity; High or Low, high- or low-intensity infection. (B) Predicted protein domain structures of GPCR-encoding genes selected for phenotypic characterization. GRPCCK1 is predicted to encode two isoforms via alternative splicing (RA and RB).
Intensity-dependent effects of GPCR signaling on Plasmodium infections.
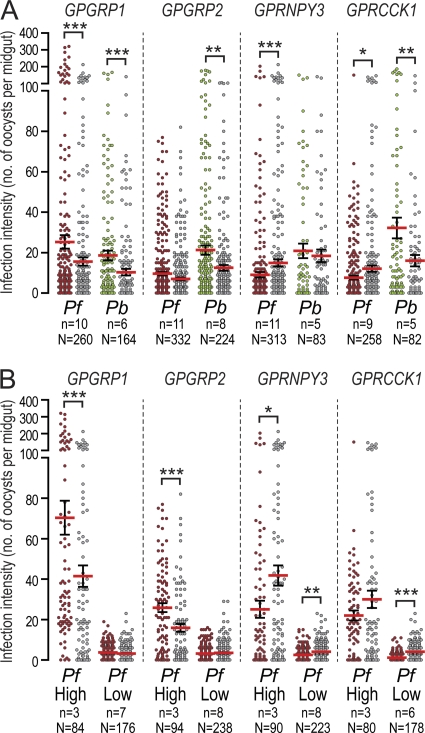
We investigated the role of rhodopsin-type GPCRs in A. gambiae infections with field isolates of P. falciparum and laboratory strains of P. berghei by means of RNAi-based gene silencing following injection of double-stranded RNA (dsRNA) into adult female mosquitoes. This analysis included two putative gastrin/bombesin receptors (GPGRP1 and GPGRP2), a putative gastrin/cholecystokinin receptor (GPRCCK1), and a putative neuropeptide Y receptor (GPRNPY3), all of which were upregulated at various levels during infections (Fig. 5). Mosquitoes injected with dsRNA of the Escherichia coli lacZ gene were used as controls. Five to 11 independent replicate infections, each involving a paired GPCR knockdown and lacZ control, were carried out for each gene and parasite.
Infection intensity-independent analysis revealed antagonistic effects of GPGRP1 on both P. falciparum and P. berghei and of GPGRP2 on P. berghei (Fig. 6A; see also Fig. S4 and Table S5 in the supplemental material). In contrast, GPRNPY3 depletion resulted in a significant reduction in P. falciparum infection intensities, while GPRCCK1 silencing had opposite effects on the two parasites: a reduction in P. falciparum and an increase in P. berghei oocyst numbers.
Fig. 6.
Effect of A. gambiae rhodopsin-like GPCR silencing on Plasmodium infection. The numbers of oocysts in the midguts of A. gambiae mosquitoes with GPCR knockdown, infected with P. falciparum (Pf) (red) or P. berghei (Pb) (green), were plotted against the numbers of oocysts in corresponding lacZ dsRNA-injected controls (gray). Data were analyzed either independently of infection intensity (A) or after grouping of infections into high- or low-intensity infections (B) as assessed in lacZ dsRNA-injected controls. Infection intensity analyses were possible only for P. falciparum. Arithmetic means (red horizontal lines) and standard errors (error bars) of pooled data from replicate experiments (n) are shown. The total number of midguts (N) in each data set is given at the bottom. P values for the meta-analysis are indicated above each data set as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Independent analysis of high and low P. falciparum infection intensities (arithmetic means of oocyst numbers in double-stranded lacZ controls, >15 and <5, respectively) revealed intensity-dependent effects for three of the four genes (Fig. 6B; see also Fig. S5 and Table S6 in the supplemental material); only GPRNPY3 silencing led to a significant reduction in oocyst numbers in both high- and low-intensity P. falciparum infections. The effect of GPGRP1 observed in the intensity-independent analysis was restricted to high-intensity P. falciparum infections, while GPGRP2, which had no knockdown phenotype in the intensity-independent analysis, showed a strongly increased number of P. falciparum oocysts in high-intensity infections. Similarly, the effect of GPRCCK1 silencing was restricted to low-intensity infections.
DISCUSSION
Renewed efforts to eliminate malaria transmission rely on deep biological understanding of vector-parasite interactions. Since genetic diversity in mosquito and parasite populations affects these interactions, research in environments as close as possible to natural settings is essential. The mosquito infections with human parasites presented in this study were carried out in an African equatorial region with continuous malaria transmission by using blood sampled from local P. falciparum gametocyte carriers and an A. gambiae colony created from local mosquitoes.
Our research has revealed that mosquito responses to malaria parasites are infection intensity dependent, especially during ookinete invasion of the mosquito midgut. An overall correlation was observed between responses to high-intensity P. falciparum infections and responses to infections with the rodent parasite P. berghei; however, responses to low-intensity P. falciparum infections are unique and encompass a striking downregulation of genes. Such a unique A. gambiae response to P. falciparum was not detected in either of two earlier studies that investigated the transcriptional responses of genes to infections with human and rodent malaria parasites. This is likely due to the high infection intensities obtained with the laboratory P. falciparum parasite strain that was used in these studies, which do not reflect the low infection intensities most commonly observed in natural P. falciparum infections in Africa (8, 27).
Suppression of immune responses has been demonstrated previously in Aedes aegypti infections with the avian Plasmodium gallinaceum parasite (3) but not in A. gambiae infections with African P. falciparum isolates (17). This is consistent with our data showing that transcriptional suppression in low-intensity P. falciparum infections does not encompass classical immunity-related processes. Most of the immunity genes were transcriptionally induced independently of the infection intensity, which is consistent with the finding that induction involves genes that are phylogenetically unique to A. gambiae. While it is possible that P. falciparum is recognized by mosquito immune surveillance, recent studies suggest that induction of immunity genes is due partly to an increase in the number of bacteria in the mosquito gut upon blood feeding rather than representing a bona fide anti-Plasmodium defense (9, 19). Such a response is therefore expected to be independent of the initial parasite density and infection intensity.
Transcriptional suppression upon low-intensity P. falciparum infection is strongly associated with evolutionarily conserved genes, most likely involved in midgut homeostasis, and is most prominent for genes involved in modulation of the cellular oxidative metabolism. Oxidative responses are known to be important in the host defense against invaders and have been implicated in mosquito midgut defense against Plasmodium (15). However, these reactions are also among the costliest with regard to energy and are thus expected to be intensity dependent. Oxidative responses are also involved in apoptosis of invaded midgut cells, the level of which is proportional to the number of invading parasites (14, 15, 23). Thus, genes involved in these processes are upregulated mostly in high-intensity infections.
Our study identifies GPCR genes as representing one of the most prominent differentially regulated gene classes. GPCR genes constitute as much as 2% of all animal genomes and encode receptors that function in cell communication by bridging extracellular ligands (hormones, neurotransmitters, light and odor sensors, chemokines, etc.) with downstream intracellular effectors, thereby playing an essential role in important physiological processes such as development, reproduction, feeding behavior, diuresis, and immunity (4, 18).
A. gambiae encodes 276 putative GPCRs grouped into five families (13). We provide evidence that signaling through rhodopsin-like GPCRs (family A) regulates A. gambiae responses to Plasmodium infection in a parasite species-dependent and infection intensity-dependent manner. The gastrin/bombesin receptors GPGRP1 and GPGRP2 act as infection antagonists; their depletion increases the number of oocysts in A gambiae infections by Plasmodium, but the effect on P. falciparum is limited to high infection intensities. In contrast, the putative neuropeptide Y receptor GPRNPY3 and the gastrin/cholecystokinin receptor GPRCCK1 are P. falciparum agonists but either are dispensable (GPRNPY3) in P. berghei infections or, surprisingly, play an opposite role, acting as P. berghei antagonists (GPRCCK1). These data highlight the importance of GPCRs in controlling A. gambiae infections with malaria parasites and also show that analyzing the effect of gene silencing on a sum of data from infections of different intensities can skew the results by either hiding a significant phenotype or extending its specific effect beyond particular infection intensities.
Further research is needed to understand the mechanisms by which GPCRs affect Plasmodium infections. Recent studies reveal a role for GPCRs in fine-tuning the immune response, leading to the negative regulation of immune responses, mainly, but not exclusively, in a p38 mitogen-activated protein kinase (MAPK) pathway-dependent manner (1, 25). Such an immunosuppressive effect would be consistent with the reduction in P. falciparum oocyst numbers upon silencing of GPCRs, such as GPRNYP3. The possible roles of these receptors in activating the A. gambiae MAPK pathway in a dose-dependent manner, as demonstrated previously for this pathway (26), remain to be investigated.
Overall, our data concur with the concept that hosts and parasites are coadapted and that no universal mechanism of host response exists against all infections and infection intensities. By directly analyzing the A. gambiae response to sympatric P. falciparum parasites circulating among children in Africa, we show that the often-low infection intensities elicit a distinct response in the mosquito midgut, characterized by suppression of genes regulating midgut homeostasis and oxidative metabolism. The mechanism that causes this suppression is unknown and could involve signaling through GPCRs. In contrast, high-intensity infections, which are rare in nature, lead to a robust transcriptional response that includes genes regulating homeostasis, e.g., cellular oxidative status, cytoskeleton reorganization, and immunity. This response is similar to that observed in P. berghei infections, indicating that high infection levels cause a potent response that involves both infection resistance and endurance processes.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Mfou primary school pupils and their parents or guardians for making this study possible. We thank Sylvie Kemleu-Zebaze and Rose Nyambam for blood smear readings, Etienne Onana and Isaac Tchikangwa for mosquito rearing and dissections, and the medical team of Mfou Hospital, Engelbert Manga, Emmanuel Bodzwan, and Constance Efemba for medical assistance. We thank Fanny Turlure for assistance with parasitological surveys, RNAi experiments, mosquito infections, and dissections.
This work was funded by Wellcome Trust Programme grant GR077229/Z/05/Z, a European Commission FP7 Collaborative project grant (TransMalariaBloc, HEALTH-F3-2008-223736), and an ANR project grant (ANR-06-MIME-001-01). The research leading to these results has also received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement 242095. A.M.M. was partly supported by a BioMalPar FP6 European Network of Excellence Ph.D. fellowship (LSHP-CT-2004-503578).
A.M.M. designed and performed research, analyzed data, and cowrote the report. S.E.N. and A.C. participated in parasitological and mosquito surveys. D.F. contributed new materials and reagents. P.H.A.-A. organized parasitological surveys in the Mfou area. F.C.K. contributed new materials and reagents. G.K.C. designed research and cowrote the report. I.M. designed research, coordinated the field work, and contributed new materials and reagents. D.V. designed research, analyzed data, contributed new materials and reagents, oversaw the project, and cowrote the report.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Aballay A. 2009. Neural regulation of immunity: role of NPR-1 in pathogen avoidance and regulation of innate immunity. Cell Cycle 8:966–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Annan Z., et al. 2007. Population genetic structure of Plasmodium falciparum in the two main African vectors, Anopheles gambiae and Anopheles funestus. Proc. Natl. Acad. Sci. U. S. A. 104:7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boete C., Paul R. E., Koella J. C. 2004. Direct and indirect immunosuppression by a malaria parasite in its mosquito vector. Proc. Biol. Sci. 271:1611–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Broeck J. V. 2001. Insect G protein-coupled receptors and signal transduction. Arch. Insect Biochem. Physiol. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 5. Castillo-Davis C. I., Hartl D. L. 2003. GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19:891–892 [DOI] [PubMed] [Google Scholar]
- 6. Cohuet A., et al. 2006. Anopheles and Plasmodium: from laboratory models to natural systems in the field. EMBO Rep. 7:1285–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dawes E. J., Churcher T. S., Zhuang S., Sinden R. E., Basanez M. G. 2009. Anopheles mortality is both age- and Plasmodium-density dependent: implications for malaria transmission. Malar. J. 8:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong Y., et al. 2006. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong Y., Manfredini F., Dimopoulos G. 2009. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 5:e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garver L. S., Dong Y., Dimopoulos G. 2009. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog. 5:e1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris C., et al. 2010. Polymorphisms in Anopheles gambiae immune genes associated with natural resistance to Plasmodium falciparum. PLoS Pathog. 6:e1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hill C. A., et al. 2002. G protein-coupled receptors in Anopheles gambiae. Science 298:176–178 [DOI] [PubMed] [Google Scholar]
- 14. Kumar S., Barillas-Mury C. 2005. Ookinete-induced midgut peroxidases detonate the time bomb in anopheline mosquitoes. Insect Biochem. Mol. Biol. 35:721–727 [DOI] [PubMed] [Google Scholar]
- 15. Kumar S., et al. 2003. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 100:14139–14144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambrechts L., Halbert J., Durand P., Gouagna L. C., Koella J. C. 2005. Host genotype by parasite genotype interactions underlying the resistance of anopheline mosquitoes to Plasmodium falciparum. Malar. J. 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lambrechts L., et al. 2007. Effect of infection by Plasmodium falciparum on the melanization immune response of Anopheles gambiae. Am. J. Trop. Med. Hyg. 76:475–480 [PubMed] [Google Scholar]
- 18. Liebmann C. 2004. G protein-coupled receptors and their signaling pathways: classical therapeutical targets susceptible to novel therapeutic concepts. Curr. Pharm. Des. 10:1937–1958 [DOI] [PubMed] [Google Scholar]
- 19. Meister S., et al. 2009. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 5:e1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mendes A. M., et al. 2008. Conserved mosquito/parasite interactions affect development of Plasmodium falciparum in Africa. PLoS Pathog. 4:e1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Michel K., et al. 2006. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 103:16858–16863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osta M. A., Christophides G. K., Kafatos F. C. 2004. Effects of mosquito genes on Plasmodium development. Science 303:2030–2032 [DOI] [PubMed] [Google Scholar]
- 23. Peterson T. M., Gow A. J., Luckhart S. 2007. Nitric oxide metabolites induced in Anopheles stephensi control malaria parasite infection. Free Radic. Biol. Med. 42:132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sinden R. E., et al. 2007. Progression of Plasmodium berghei through Anopheles stephensi is density-dependent. PLoS Pathog. 3:e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Styer K. L., et al. 2008. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Surachetpong W., Singh N., Cheung K. W., Luckhart S. 2009. MAPK ERK signaling regulates the TGF-β1-dependent mosquito response to Plasmodium falciparum. PLoS Pathog. 5:e1000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tahar R., Boudin C., Thiery I., Bourgouin C. 2002. Immune response of Anopheles gambiae to the early sporogonic stages of the human malaria parasite Plasmodium falciparum. EMBO J. 21:6673–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tchuinkam T., et al. 1993. Experimental infections of Anopheles gambiae with Plasmodium falciparum of naturally infected gametocyte carriers in Cameroon: factors influencing the infectivity to mosquitoes. Trop. Med. Parasitol. 44:271–276 [PubMed] [Google Scholar]
- 29. Vernick K. D., et al. 2005. Molecular genetics of mosquito resistance to malaria parasites. Curr. Top. Microbiol. Immunol. 295:383–415 [DOI] [PubMed] [Google Scholar]
- 30. Vlachou D., Schlegelmilch T., Christophides G. K., Kafatos F. C. 2005. Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr. Biol. 15:1185–1195 [DOI] [PubMed] [Google Scholar]
- 31. Waterhouse R. M., et al. 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316:1738–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.