Abstract
Ehrlichia chaffeensis is an obligately intracellular bacterium that exhibits tropism for mononuclear phagocytes and survives by evading host cell defense mechanisms. Recently, molecular interactions between E. chaffeensis 47-kDa tandem repeat (TR) protein (TRP47) and the eukaryotic host cell have been described. In this investigation, yeast (Saccharomyces cerevisiae) two-hybrid analysis demonstrated that E. chaffeensis-secreted tandem repeat protein 120 (TRP120) interacts with a diverse group of host cell proteins associated with major biological processes, including transcription and regulation, cell signaling, protein trafficking, and actin cytoskeleton organization. Twelve target proteins with the highest frequency of interaction with TRP120 were confirmed by cotransformation in yeast. Host targets, including human immunoglobulin lambda locus (IGL), cytochrome c oxidase subunit II (COX2), Golgi-associated gamma adaptin ear-containing ARF binding protein 1 (GGA1), polycomb group ring finger 5 (PCGF5), actin gamma 1 (ACTG1), and unc-13 homolog D (UNC13D; Caenorhabditis elegans), colocalized strongly with TRP120 in HeLa cells and with E. chaffeensis dense-cored morulae and areas adjacent to morulae in the host cytoplasm. The TR domain of TRP120 interacted only with PCGF5, indicating that distinct TRP120 domains contribute to specific host target interactions and that multiple domains are required to reconstitute TRP120 interactions with other host targets. Three previously defined molecular interactions between TRP47 and host proteins, PCGF5, IGLL1, and CAP1, were also associated with TRP120, demonstrating that molecular cross talk occurs between Ehrlichia TRPs and host targets. These findings further support the role of TRPs as effectors that reprogram the host cell.
INTRODUCTION
Ehrlichia chaffeensis is an obligately intracellular Gram-negative bacterium and is the etiologic agent of human monocytotropic ehrlichiosis (HME) (34). E. chaffeensis exhibits tropism for mononuclear phagocytes and enters the cell through receptor-mediated endocytosis, residing and multiplying within a cytoplasmic vacuole that phenotypically resembles an early endosome (34, 37). E. chaffeensis has one of the smallest bacterial genomes but has evolved molecular mechanisms that enable it to circumvent innate and adaptive host defense mechanisms, including killing by reactive oxygen species, lysosomal fusion, and gamma interferon signaling (8, 28, 37). The effector proteins involved in reprogramming the host cell have not been fully defined, but molecular interactions of tandem repeat (TR) protein (TRP) and ankyrin repeat protein (Ank) effectors of E. chaffeensis with the host cell have been associated with the cell nucleus and cytoplasmic vacuole containing the organism (42, 44, 51). Identified host targets suggest that there are numerous and complex interactions occurring in the nucleus, cytoplasm, cell membrane, and organelles, including mitochondria and endoplasmic reticulum.
E. chaffeensis proteins strongly recognized by the host immune response include the 120-kDa TRP (TRP120), TRP47, TRP32 (a variable-length PCR target [VLPT] protein), Ank200, and outer membrane protein 1 (OMP1; p28) (5, 7, 24–26). The TRPs are secreted, and two (TRP120 and TRP47) are differentially expressed on dense-cored (DC) ehrlichiae (7, 24, 26, 36, 43). TRP120 is involved in ehrlichial binding and internalization and directly binds host DNA, demonstrating that it has important and diverse roles in ehrlichial molecular pathobiology (20, 36, 50). Furthermore, new pathogen-host interactions have been defined for TRP47, which interacts with multiple host cell proteins associated with cell signaling, transcriptional regulation, and vesicle trafficking, including polycomb group ring finger 5 (PCGF5), Src protein tyrosine kinase FYN (FYN), protein tyrosine phosphatase nonreceptor type 2 (PTPN2), adenylate cyclase-associated protein 1 (CAP1), and immunoglobulin lambda-like polypeptide 1 (IGLL1) (42). In addition, the E. chaffeensis ankyrin repeat protein, Ank200, is translocated to the host cell nucleus, where it binds Alu elements located in the promoter region of many genes associated with ehrlichial pathobiology (51).
Ehrlichia replicates within membrane-bound cytoplasmic vacuoles, forming microcolonies called morulae. The ehrlichial morula membrane is a dynamic pathogen-host interface, and many secreted ehrlichial proteins associate with the morula membrane (7, 26, 36). The interaction between E. chaffeensis TRP47 and human CAP1 has been defined at this interface and is associated with vesicle trafficking (42). Similarly, the Anaplasma phagocytophilum-occupied vacuole selectively recruits Rab GTPases that are primarily associated with recycling endosomes (16). Pathogen-host interactions that occur at this interface are critically important for pathogen entry, vacuole trafficking within the host cell, avoiding host defense mechanisms, and, eventually, release from the host cell (28, 44). In addition, E. chaffeensis modulates host gene expression of numerous host cell processes to facilitate its intracellular survival and proliferation, including genes for cell cycle and differentiation, immune response, membrane trafficking and lysosomal fusion, apoptosis, and signal transduction (49), and modulation of host gene expression appears to be mediated in part by ehrlichial DNA-binding proteins such as Ank200 and TRP120.
A better understanding of the role of ehrlichial TRPs in molecular host interactions is an important step in defining ehrlichial effectors and molecular mechanisms involved in ehrlichial pathobiology. In order to define the role of TRP120, we used a yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) assay to identify molecular TRP120-host interactions. We determined that E. chaffeensis TRP120 interacts with a diverse group of eukaryotic proteins involved in multiple cellular processes and has molecular cross talk with the TRP47 network. Thus, TRP120 appears to be involved in the molecular reprogramming of the host cell that facilitates ehrlichial survival in mononuclear phagocytes.
MATERIALS AND METHODS
Cell culture and cultivation of E. chaffeensis.
Human cervical epithelial adenocarcinoma (HeLa) cells and human monocytic leukemia (THP-1) cells were propagated in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) as previously described (42). E. chaffeensis (Arkansas strain) was cultivated in THP-1 cells as previously described (42).
Antibodies.
Rabbit anti-TRP120 and anti-PCGF5 antibodies have been described previously (24, 42). Other antibodies used in this study were mouse anti-human immunoglobulin lambda locus (IGL), cytochrome c oxidase subunit II (COX2), Golgi-associated gamma adaptin ear-containing ARF binding protein 1 (GGA1), actin gamma 1 (ACTG1), and unc-13 homolog D (UNC13D; Caenorhabditis elegans) (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-green fluorescent protein (GFP), and mouse anti-GAL4 DNA-binding domain (GAL4 DNA-BD) tag (Clonetech, Mountain View, CA). All antibodies used for immunofluorescence were tested by the vendor to ensure the specificity, which was confirmed by Western blotting, immunofluorescence, or both (Santa Cruz).
Yeast two-hybrid system.
A Matchmaker Gold yeast two-hybrid system (Clontech), including all yeast strains, yeast media and supplements, vectors, and yeast transformation system, was used.
E. chaffeensis TRP120 cloning and expression in yeast.
The coding region of TRP120 (GenBank accession number U49426) was amplified by PCR from E. chaffeensis genomic DNA using forward (5′-GGCGAATTCATGGATATTGATAATAGTAACATAAG) and reverse (5′-GGCGTCGACTACAATATCATTTACTACATTGTG) (restriction enzyme sites are in boldface; Sigma-Genosys, Woodlands, TX) primers and cloned into the EcoRI-SalI site of the pGBKT7 vector containing the GAL4 DNA-BD. The resulting bait plasmid, pGBKT7-TRP120, was transformed into yeast (Saccharomyces cerevisiae) strain Y2HGold using Yeastmaker yeast transformation system 2, and the expression of bait protein TRP120 (GAL4 DNA-BD fused) in yeast was confirmed by Western immunoblotting using transformed yeast cell lysates probed with mouse anti-GAL4 DNA-BD antibody (1:2,000) or rabbit anti-TRP120 antibody (1:2,000). Yeast protein extracts were prepared by the urea/sodium dodecyl sulfate method described in Clontech's Yeast Protocols Handbook.
Yeast two-hybrid assay.
The Matchmaker human bone marrow library (Clontech), a high-complexity cDNA library cloned into the yeast GAL4 activation domain (GAL4-AD) vector pGADT7-Rec and pretransformed into S. cerevisiae host strain Y187, was screened by yeast mating with bait strain Y2HGold containing pGBKT7-TRP120 according to the manufacturer's protocol. A positive control was created by mating Y2HGold containing pGBKT7-p53 (murine p53 protein) with Y187 containing pGADT7-T (simian virus 40 large T antigen), and Y2HGold containing pGBKT7-Lam (human nuclear lamin) was mated with Y187 containing pGADT7-T as a negative control. Positive clones expressing prey proteins interacting with TRP120 (bait) were selected on minimal synthetically defined (SD) quadruple-dropout (QDO) medium (SD medium without Ade, His, Leu, and Trp [SD/−Ade/−His/−Leu/−Trp]) supplemented with 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-Gal) and aureobasidin A (QDO/X/A). Blue colonies with normal size were segregated three times on SD double-dropout (DDO; SD medium without Leu and Trp [SD/−Leu/−Trp]) plates containing X-Gal (DDO/X), and the prey cDNA inserts were amplified by colony PCR and then sequenced in the University of Texas Medical Branch Protein Chemistry Core Laboratory.
Confirmation of positive interactions by cotransformation.
The prey plasmid responsible for positive interactions was rescued from segregated colonies using an Easy yeast plasmid isolation kit (Clontech), transformed into Escherichia coli Fusion-Blue (Clontech), and isolated. To distinguish positive from false-positive interactions, Y2HGold yeast cells were cotransformed with bait (pGBKT7-TRP120) and prey plasmids or with empty pGBKT7 and the prey plasmids as the control. The positive interactions were confirmed by selection on QDO/X/A.
Cotransfection of mammalian cells and co-IP.
A Matchmaker chemiluminescent coimmunoprecipitation (co-IP) system (Clontech) was used to confirm interactions between bait and prey proteins identified by Y2H assay in mammalian cells. Briefly, two mammalian expression vectors, pAcGFP1-C and pProLabel-C, which encode a fluorescent Aequorea coerulescens green fluorescent protein (AcGFP1) and enzymatic ProLabel (PL; an ∼6-kDa fragment of β-galactosidase) tags, respectively, were used for generation of N-terminal AcGFP1-bait and PL-prey fusion proteins. The bait TRP120 was amplified from pGBK-TRP120 and cloned in-frame downstream of the AcGFP1 tag of pAcGFP1-C using In-Fusion PCR cloning (Clontech), while the prey gene was amplified from pGADT7-prey and cloned in-frame downstream of the PL tag of pProLabel-C.
An approach similar to that described above for the full-length TRP120 was used to generate amino-terminal TRP120 (TRP120N; amino acids 1 to 54), TRP120 tandem repeats (TRP120TRs; amino acids 48 to 220), and carboxy-terminal TRP120 (TRP120C; amino acids 401 to 548) fragments, except In-Fusion PCR cloning was used for cloning of TRP120 fragments into pGBKT7. Primers used for TRP120 fragment amplification from E. chaffeensis genomic DNA included the In-Fusion cloning leader sequence CATGGAGGCCGAATTC at the 5′ end of each primer. Fragment-specific primer sequences were TRP120N (forward, 5′-ATGGATATTGATAATAGTAACATAAGTAC; reverse, 5′-TGTGTCATCTTCTTGCTCTTG), TRP120TR (forward, 5′-CAAGAGCAAGAAGATGACAC; reverse, 5′-TGATGAAGGTTGAGATACTATTTC), and TRP120C (forward, 5′-ATTCTAGTAGAAGATTTGCCATTAG; reverse, 5′-TACAATATCATTTACTACATTGTGATT) (Sigma-Genosys).
Plasmids pAcGFP1-C (control without insert), pAcGFP1-TRP120, pAcGFP1-TRP120N, pAcGFP1-TRP120TR, or pAcGFP1-TRP120C and pProLabel-prey were cotransfected into HeLa cells using Lipofectamine 2000 (Invitrogen), and expression of GFP was confirmed under an inverted fluorescence microscope (IX71; Olympus, Japan) at 2 days posttransfection. Then, interacting proteins were immunoprecipitated according to the manufacturer's protocol, and PL activity (in relative light units [RLUs]) was measured at different time intervals after addition of substrate using a Veritas microplate luminometer (Turner Biosystems, Sunnyvale, CA). Statistical differences between experimental groups were assessed with the two-tailed Student's t test, and significance was indicated by a P value of <0.05.
Immunofluorescence microscopy.
Transfected (pAcGFP1-TRP120 or pAcGFP1-empty) HeLa cells were collected at 2 days posttransfection, washed once in phosphate-buffered saline (PBS), and fixed in 3% paraformaldehyde in PBS for 15 min. Cells were permeabilized and blocked with a mixture of 0.1% Triton X-100 and 1% bovine serum albumin (Sigma) in PBS for 1 h. Then, cells were incubated with mouse anti-IGL, -COX2, -GGA1, -PCGF5, -ACTG1, or -UNC13D antibodies (1:100) for 1 h, washed, and stained with Alexa Fluor 568 goat anti-mouse IgG (H+L) secondary antibodies (1:100; Molecular Probes, Eugene, OR) for 30 min. Uninfected or E. chaffeensis-infected THP-1 cells (3 days postinfection) were cytocentrifuged onto glass slides, fixed in 3% paraformaldehyde for 20 min, permeabilized, and blocked with a mixture of 0.3% Triton X-100 and 2% bovine serum albumin in PBS for 1 h. Cells were then incubated with mouse anti-IGL, -COX2, -GGA1, -ACTG1, or -UNC13D antibody (1:100) and rabbit anti-TRP120 antibody (1:40,000) for 1 h, washed, and dually stained with Alexa Fluor 568 goat anti-mouse IgG (H+L) and Alexa Fluor 488 goat anti-rabbit IgG (H+L) secondary antibodies (1:100; Molecular Probes) for 30 min. Slides were washed and mounted with ProLong Gold antifade reagent (Invitrogen), and fluorescence images were obtained using an Olympus BX61 epifluorescence microscope. The images were further processed, and fluorescence intensity profiles representing the pixel intensity values in a digitized section along a user-defined area were created using Slidebook (version 5.0) software (Intelligent Imaging Innovations, Denver, CO).
GO analysis.
Gene ontology (GO) analysis of TRP120 target proteins was performed by using the Babelomics FatiGO tool (http://babelomics3.bioinfo.cipf.es) (2).
RESULTS
Biochemical and functional characteristics of E. chaffeensis TRP120.
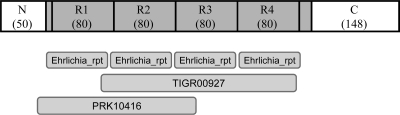
E. chaffeensis TRP120 is a 548-amino-acid, highly acidic (pI 3.91) protein containing four nearly identical 80-mer TRs flanked by N (50 amino acids) and C (148 amino acids) termini (Fig. 1). TRs constitute the majority of TRP120, and all four TRs were predicted to be surface accessible. TRP120 was predicted to be secreted by a nonclasssical and leaderless mechanism by the SecretomeP (version 2.0) server (www.cbs.dtu.dk/services/SecretomeP). Neither the full-length TRP120 nor any of three regions (N terminus, TRs, or C terminus) had substantial homology with any known protein except E. canis ortholog TRP140. However, a conserved domain search (http://www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi) identified four putative single domains of the Ehrlichia repeat superfamily and two multidomains, K+-dependent Na+/Ca+ exchanger (TIGR00927; bit score, 44.22; E value, 5.92e−05) and FtsY (PRK10416; bit score, 38.34; E value, 3.92e−03) (Fig. 1). FtsY is a cell division membrane protein and a signal recognition particle receptor involved in cell division and protein secretion of bacteria.
Fig. 1.
E. chaffeensis TRP120 regions and conserved domains. (Top) Schematic of E. chaffeensis TRP120 showing the regions of N terminus, TRs, and C terminus (numbers of amino acids are in parentheses; R, repeat). The TR region is shown in gray. There are two incomplete repeats (10 and 20 amino acids, respectively) preceding the first repeat and following the last repeat, respectively, which are homologous to tandem repeats and are also shown in gray. (Bottom) The Ehrlichia repeat (rpt), TIGR00927 (K+-dependent Na+/Ca+ exchanger), and PRK10416 (cell division and signal recognition particle-docking protein FtsY) conserved domains were identified from the NCBI Conserved Domain Database. The computationally detected Ehrlichia repeat multidomains are present in the same region of TRP120 that contains four TRs.
Analysis of E. chaffeensis TRP120 interactions with human proteins using a yeast two-hybrid system.
To confirm that the bait (TRP120) did not autonomously activate (autoactivate) the reporter genes in yeast strain Y2HGold in the absence of a prey protein, Y2HGold cells were transformed with bait plasmid pGBKT7-TRP120 and plated on SD without Trp (SD/−Trp), SD without Trp supplemented with X-Gal and aureobasidin A (SD/−Trp/X/A), and SD without Trp, His, and Ade supplemented with X-Gal (SD/−Trp/−His/−Ade/X-Gal; triple dropout [TDO/X]) plates. Many colonies were observed on the SD/−Trp plate after 3 to 5 days of incubation, but no growth or only a few tiny white/pale blue colonies were observed on SD/−Trp/X/A and TDO/X plates, confirming a lack of autoactivation by TRP120 (data not shown). To examine the toxicity of TRP120 in yeast cells, Y2HGold cells transformed with pGBKT7-empty or pGBKT7-TRP120 were grown in both solid and liquid media. No significant differences in yeast growth in solid or liquid medium containing bait and empty plasmids were observed, demonstrating that the bait was nontoxic to yeast.
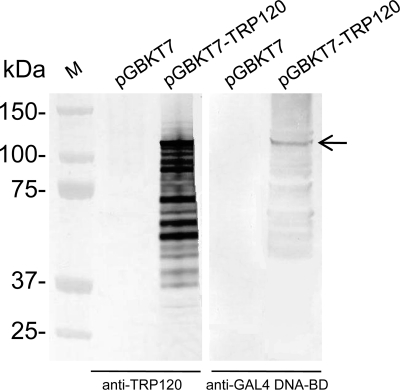
To confirm GAL4 DNA-BD-TRP120 fusion protein expression in yeast, yeast proteins were extracted from Y2HGold cells transformed with pGBKT7-empty or pGBKT7-TRP120, and the fusion protein was detected by Western immunoblotting with an anti-GAL4 DNA-BD monoclonal antibody or an anti-TRP120 antibody (Fig. 2). The fusion protein exhibited a molecular mass (∼125 kDa) larger than the predicted mass (∼83 kDa=61 kDa of TRP120 + 22 kDa of GAL4 DNA-BD tag) on the basis of the amino acid sequence, which was consistent with our previous report regarding the aberrant electrophoretic mobility of native and recombinant TRP120 (24). In addition, it has previously been demonstrated that TRP120 likely degraded, and multiple bands were detected by Western immunoblotting (24).
Fig. 2.
Detection of TRP120 expression in yeast. Expression of GAL4 DNA-BD-TRP120 fusion protein in yeast (arrow) was determined by immunoblotting of yeast cell extracts transformed with pGBKT7 (empty vector) or pGBKT7-TRP120 using the anti-TRP120 antibody or anti-GAL4 DNA-BD antibody. M, molecular mass marker.
By yeast two-hybrid screening using yeast mating, ∼19% mating efficiency was achieved and ∼21 million diploid clones were screened. In total, 271 yeast colonies with blue color and normal size were observed and selected from SD/−Trp/−Leu/−Ade/−His/X/A (QDO/X/A) plates for identification of potential positive clones that exhibited bait and prey protein-protein interactions. After segregation of the colony three times, yeast colony PCR was performed, followed by DNA sequencing to eliminate the duplicate clones and identify interacting targets. Ninety-eight different potentially interacting human proteins were identified (see Table S1 in the supplemental material), and the 12 most abundant clones in order of frequency were human immunoglobulin light-chain lambda (IGL), cytochrome c oxidase subunit II (COX2), immunoglobulin kappa constant (IGKC), immunoglobulin heavy constant alpha 1 (IGHA1), erythrocyte membrane protein band 4.2 (EPB42), Golgi-associated gamma adaptin ear-containing ARF binding protein 1 (GGA1), prosaposin (PSAP), solute carrier family 2, member 3 (SLC2A3), unc-13 homolog D (UNC13D; C. elegans), polycomb group ring finger 5 (PCGF5), actin gamma 1 (ACTG1), and F-box and WD repeat domain containing 7 (FBXW7) (Table 1).
Table 1.
Summary of 12 human proteins that interact with E. chaffeensis TRP120 determined by yeast two-hybrid assay and cotransformation
| Interacting human proteina | GenBank accession no. | Properties/functions |
|---|---|---|
| Immunoglobulin lambda locus (IGL) | XM_002348112.1 | Recognizes foreign antigens and initiates immune responses such as phagocytosis and the complement system |
| Cytochrome c oxidase subunit II (COX2) | XR_078889.1 | Terminal enzyme of the mitochondrial respiratory chain, catalyzing the transfer of electrons from reduced cytochrome c to molecular oxygen |
| Immunoglobulin kappa constant (IGKC) | NW_001838785.1 | Recognizes foreign antigens and initiates immune responses such as phagocytosis and the complement system |
| Immunoglobulin heavy constant alpha 1 (IGHA1) | NW_001838121.1 | Recognizes foreign antigens and initiates immune responses such as phagocytosis and the complement system |
| Erythrocyte membrane protein band 4.2 (EPB42) | NM_001114134.1 | An ATP-binding protein which may regulate the association of protein 3 with ankyrin; it probably has a role in erythrocyte shape and mechanical property regulation |
| Golgi-associated, gamma adaptin ear-containing, ARF binding protein 1 (GGA1) | NM_013365.3 | Ubiquitous coat proteins that regulate the trafficking of proteins between the trans-Golgi network and the lysosome |
| Prosaposin (PSAP) | NM_001042466.1 | A highly conserved glycoprotein which is a precursor for saposins; saposins localize primarily to the lysosomal compartment, where they facilitate the catabolism of glycosphingolipids |
| Solute carrier family 2, member 3 (SLC2A3) | NM_006931.2 | Glucose transmembrane transporter |
| unc-13 homolog D (C. elegans) (UNC13D) | NM_199242.2 | A member of the UNC13 family that appears to play a role in vesicle maturation during exocytosis and is involved in regulation of cytolytic granules secretion |
| Polycomb group ring finger 5 (PCGF5) | NM_032373.3 | DNA-dependent regulation of transcription, metal ion binding, and protein binding |
| Actin, gamma 1 (ACTG1) | NM_001614.2 | A cytoplasmic actin found in nonmuscle cells; highly conserved proteins that are involved in various types of cell motility and maintenance of the cytoskeleton |
| F-box and WD repeat domain containing 7 (FBXW7) | NT_016354.19 | The F-box proteins constitute one subunit of ubiquitin protein ligase complex SCFs (SKP1 [S-phase KimSC-associated protein 1] cullin-F-box) which function in phosphorylation-dependent ubiquitination; seven tandem WD40 repeats bind directly to cyclin E, probably for ubiquitin-mediated degradation |
Listed by the order of identification frequency.
In order to reveal general biological processes that may be affected by TRP120, a functional analysis was performed on the 98 Y2H assay-identified TRP120-interacting targets using the Babelomics FatiGO tool. Proteins were classified in three gene ontology databases (biological process, molecular function, and cellular component) (see Fig. S1 in the supplemental material). The most highly represented target proteins were associated with major biological processes, including transcription and transcription regulation, posttranslational protein modification, transcription, actin cytoskeleton organization and biogenesis, and regulation of nucleic acid metabolic processes, which were consistent with their biological functions, including zinc ion binding, ATP-binding, and pyrophosphatase activity and with their major structural associations with the nucleus (see Fig. S1 in the supplemental material).
Confirmation of interactions by cotransformation and coimmunoprecipitation.
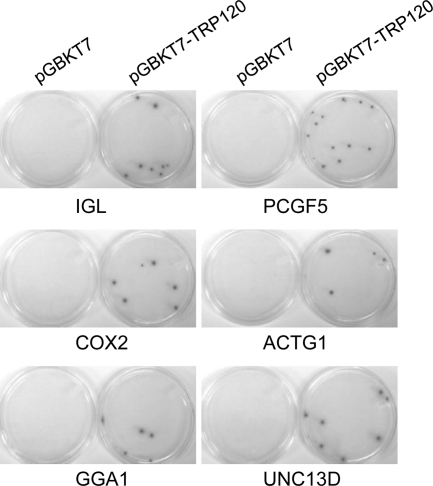
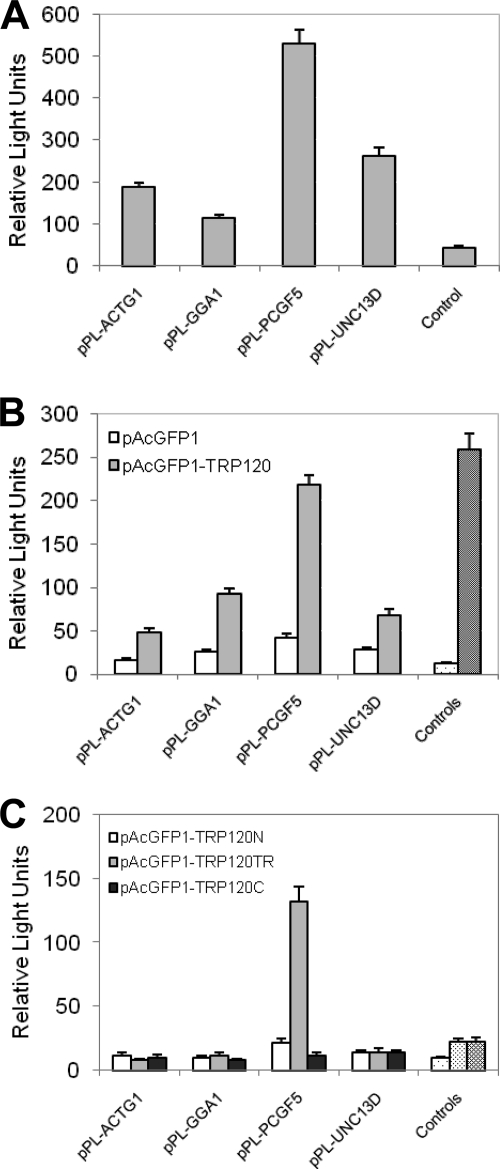
To confirm the true interactions, cotransformation assays were performed in yeast with candidate prey plasmids from the 12 most abundant clones. Interactions in yeast were confirmed with all these prey proteins (Fig. 3, showing interactions of TRP120 with six prey proteins: IGL, COX2, GGA1, PCGF5, ACTG1, and UNC13D). In order to further examine the interaction of TRP120 with these prey proteins in mammalian cells by co-IP, nine candidate preys were selected from the above-described 12 most abundant clones except IGKC, IGHA1, and SLC2A3, which had significant overlap in functions with another target IGL or were of low priority (SLC2A3). Nine prey genes were cloned into the mammalian expression vector pProLabel-C, but only four proteins (ACTG1, GGA1, PCGF5, and UNC13D) were expressed and detected by PL chemiluminescence (Fig. 4A). Co-IP confirmed a direct interaction between TRP120 and all four expressed prey proteins. The relative strengths of the interaction between TRP120 and PCGF5, GGA1, ACTG1, and UNC13D were 5.2, 3.5, 3.0, and 2.4 times higher than those of the respective controls (AcGFP1 without TRP120; P < 0.05) (Fig. 4B). The full-length TRP120 interacted with each prey protein specifically and differentially, as demonstrated by high relative physical interaction values (218 RLUs for PCGF5 to 48 RLUs for ACTG1) (Fig. 4B). To confirm the specificity of the protein-protein interaction, HeLa cells were cotransfected with pAcGFP1-TRP120 and pProLabel-C, and co-IP was performed. The relatively low RLU value (12) indicated that there was no protein-protein interaction and demonstrated that the presence of a specific prey protein was required for TRP120-prey protein interaction (Fig. 4B). This experiment confirmed the Y2H assay results and also confirmed that TRP120 interacted most strongly with PCGF5, followed by GGA1, ACTG1, and UNC13D.
Fig. 3.
Confirmation of positive interactions in yeast by cotransformation. Y2HGold yeast cells were cotransformed with bait plasmid pGBKT7-TRP120 and prey plasmid pGADT7-IGL, -COX2, -GGA1, -PCGF5, -ACTG1, or -UNC13D. As a control, pGBKT7 (empty vector) was used to cotransform with each prey plasmid. The positive interactions were confirmed by selection on QDO/X/A plates.
Fig. 4.
E. chaffeensis TRP120 interactions with multiple human proteins detected by chemiluminescent coimmunoprecipitation. AcGFP1 (without insert) or AcGFP1-TRP120 was coexpressed with the PL-ACTG1, -GGA1, -PCGF5, or -UNC13D fusion protein in HeLa cells. A positive control (B, controls, right bars in the pair of bars) was performed by coexpressing AcGFP1-p53 (human tumor suppressor p53 protein) and PL-T (simian virus 40 large T antigen), while AcGFP1-TRP120 (B, controls, left bars in the pair of bars), -TRP120N, -TRP120TR, or -TRP120C (C, controls, from left to right) was coexpressed with PL (without insert) as the negative control. ProLabel activity in relative light units (RLUs) was measured 1 h after addition of substrate. The results were from three independent experiments, and the values are means ± standard deviations. (A) The relative strength of expression of four proteins (ACTG1, GGA1, PCGF5, and UNC13D) was detected in HeLa cells by PL chemiluminescence activity. Control, pPL-C. (B) Relative strength and interaction of AcGFP1-TRP120 with PL-ACTG1, -GGA1, -PCGF5, or -UNC13D. (C) Relative strength and interaction of AcGFP1-TRP120N, -TRP120TR, and -TRP120C with PL-ACTG1, -GGA1, -PCGF5, or -UNC13D. TRP120N, amino-terminal TRP120; TRP120TR, tandem repeats of TRP120; TRP120C, carboxyl-terminal TRP120.
Tandem repeats of E. chaffeensis TRP120 interact with PCGF5.
To define the TRP120 domain that was involved in specific interactions with the identified host proteins, N-terminal (TRP120N; amino acids 1 to 54), tandem repeat (TRP120TR; amino acids 48 to 220), and C-terminal (TRP120C; amino acids 401 to 548) regions of TRP120 were cloned in-frame downstream of the pAcGFP1 coding sequence and expressed as a C-terminal AcGFP1 fusion protein. Plasmid pAcGFP1-TRP120N, pAcGFP1-TRP120TR, or pAcGFP1-TRP120C was cotransfected with a prey plasmid (pProLabel-ACTG1, -GGA1, -PCGF5, or -UNC13D) into HeLa cells. The co-IP results demonstrated that only TRP120TR containing the TRs interacted with PCGF5, but no interaction was observed with the other three proteins (Fig. 4C). The N- or C-terminal region of TRP120 did not exhibit any substantial interaction with ACTG1, GGA1, PCGF5, or UNC13D (Fig. 4C), although the full-length TRP120 interacted with these proteins (Fig. 4B). However, the relative interaction with the TRP120TR construct with PCGF5 was weaker (∼3 times the control) than the relative interaction of full-length TRP120 with PCGF5 (∼5 times the control) (Fig. 4B and C).
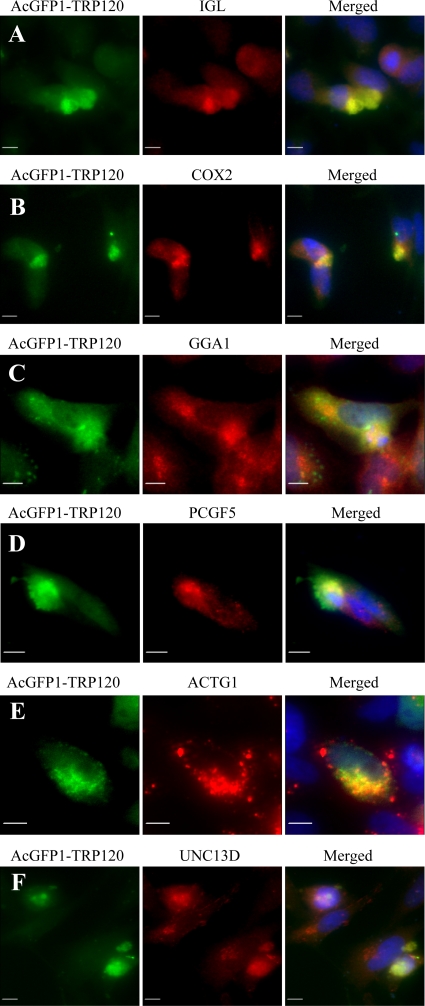
Recombinant E. chaffeensis TRP120 (AcGFP1-TRP120) colocalizes with human target proteins in HeLa cells.
Interactions between TRP120 and nine selected proteins were examined in mammalian (HeLa) cells transfected with the TRP120 expression vector. Immunofluorescence microscopy revealed that recombinant TRP120 colocalized with all nine host targets, but six proteins (IGL, COX2, GGA1, PCGF5, ACTG1, and UNC13D) exhibited the strongest colocalization (Fig. 5A to F, showing colocalization of six proteins). Recombinant E. chaffeensis TRP120 and host cell proteins exhibited primarily diffused cytoplasmic colocalization in the cell, although punctate colocalization of TRP120 and ACTG1 was observed (Fig. 5A to F).
Fig. 5.
Colocalization of IGL, COX2, GGA1, PCGF5, ACTG1, and UNC13D with AcGFP-TRP120 in HeLa cells. pAcGFP1-TRP120-transfected HeLa cells (2 days posttransfection) were labeled and observed by fluorescence microscopy. The AcGFP-TRP120 (green; A to F) and anti-IGL, -COX2, -GGA1, -PCGF5, -ACTG1, and -UNC13D (red; A to F, respectively) signals were merged with 4,6′-diamidino-2-phenylindole staining (blue) (A to F). Bars, 10 μm.
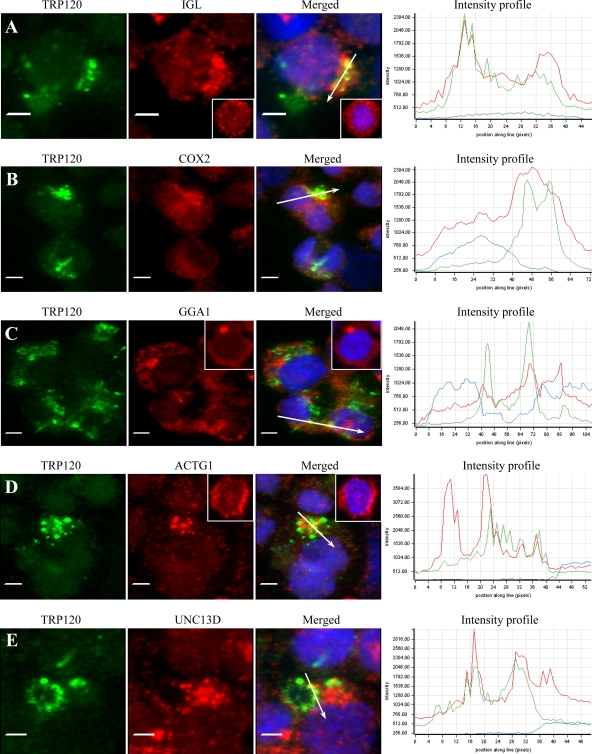
Differentially expressed TRP120 on DC ehrlichiae colocalizes with human target proteins in E. chaffeensis-infected THP-1 cells.
Consistent with our previous report (36), we observed that only dense-cored (DC) E. chaffeensis cells reacted with anti-TRP120 antibody, in contrast to anti-Dsb (a constitutively expressed pan-Ehrlichia marker) antibody, which reacted with all ehrlichiae (data not shown). Consistent with the colocalization in HeLa cells, double-immunofluorescence labeling of E. chaffeensis-infected THP-1 cells revealed that nine identified host targets colocalized with morulae (green) that stained with anti-TRP120 antibody, but six proteins, IGL, COX2, GGA1, PCGF5, ACTG1, and UNC13D (red), exhibited the strongest colocalization (Fig. 6A to E, showing colocalization of five proteins). Colocalization of E. chaffeensis DC with PCGF5 in THP-1 cells has been reported previously (42). Target proteins colocalized with the morulae or in the host cell cytoplasm adjacent to the morula membrane. To further examine the distribution and colocalization of host proteins with TRP120-expressing E. chaffeensis morulae, the fluorescence intensity profiles across the colocalized regions in the fluorescence images were analyzed (Fig. 6A to E, right). The intensity analysis demonstrated that TRP120 (green curve) and IGL, COX2, GGA1, ACTG1, or UNC13D (red curve) had consistent colocalization with morulae and revealed a similar pattern of elevated peaks across and adjacent to the morulae profile. Redistribution of TRP120 target proteins in E. chaffeensis-infected cells compared to uninfected cells was observed; for example, IGL, GGA1, and ACTG1 were mostly associated with morulae in infected THP-1 cells (Fig. 6A, C, and D), while IGL was diffusely distributed in uninfected THP-1 cells, mainly in the cytoplasm (Fig. 6A, inset); GGA1 mainly localized in a small area in the cytoplasm adjacent to the nucleus in uninfected cells, which was consistent with the location of the Golgi apparatus (Fig. 6C, inset); and ACTG1 was mainly distributed in the cytoplasm (Fig. 6D, insets).
Fig. 6.
Colocalization of IGL, COX2, GGA1, ACTG1, and UNC13D with E. chaffeensis TRP120 in E. chaffeensis-infected THP-1 cells. Fluorescence microscopy and intensity profiles of infected THP-1 cells stained with 4,6′-diamidino-2-phenylindole (blue), anti-TRP120 (green), and anti-IGL, -COX2, -GGA1, -ACTG1, or -UNC13D antibodies (red; A to E, respectively) show colocalization of E. chaffeensis TRP120-labeled morulae with IGL, COX2, GGA1, ACTG1, and UNC13D (A to E). The white arrows in the fluorescence images indicate the areas selected for fluorescence intensity profile analyses, which are displayed in graph form. The x axis shows the position along the line (pixels), and the y axis shows the fluorescence intensity. The insets (A, C, and D) show the distribution of IGL, GGA1, and ACTG1 in normal uninfected THP-1 cells. Bars, 5 μm.
DISCUSSION
Following entry into mononuclear phagocytes, E. chaffeensis subverts the host cell immune system and modulates cellular processes to create a supportive replicative microenvironment within phagocytes, involving, for the most part, complex molecularly undefined E. chaffeensis-host interactions. Some host cell responses during Ehrlichia infection have been identified and characterized, but little is known regarding the ehrlichial effector proteins and the host cell targets responsible for cellular permissiveness (37, 44).
Recently, we have determined for the first time that TRP47, an ehrlichial surface-exposed and secreted protein, is an effector protein that interacts with multiple host proteins associated with cell signaling, transcriptional regulation, and vesicle trafficking, including PCGF5, FYN, PTPN2, CAP1, IGLL1, and some others (42). In this study, we demonstrated that TRP120, another ehrlichial surface-exposed and secreted protein, appears to be an effector protein involved in the direct interactions with a diverse group of eukaryotic proteins, including the targets that are associated with TRP47. By yeast two-hybrid analysis, TRP120-interacting human targets which are involved in diverse cellular processes were identified, which suggests that TRP120 is a multifunctional protein intimately involved in a complex molecular strategy to alter molecular host cell processes. This study expands on our previous conclusion that an array of diverse interactions is occurring between ehrlichial effectors and host proteins in vivo. Recently, we also demonstrated that E. chaffeensis Ank200 and TRP120 can translocate to the host cell nucleus and directly bind host DNA associated with genes that regulate transcription, signal transduction, and apoptosis (50, 51). Therefore, ehrlichial effector proteins, such as TRP47, TRP120, and Ank200, appear to play important roles in ehrlichial modulation of expression of host genes associated with numerous host cellular processes through protein-protein or protein-DNA interactions.
Many target proteins with a high frequency of interaction with TRP120 were confirmed, including IGL, COX2, GGA1, PCGF5, ACTG1, and UNC13D, which exhibited strong colocalization with E. chaffeensis morulae. Consistent with findings from our recent study that the PCGF5 protein exhibited the strongest interaction with E. chaffeensis TRP47, we also detected the strongest interaction to be between PCGF5 and TRP120 in this study. The role of PCGF5 is not fully defined, but it appears to be involved in DNA-dependent regulation of transcription through metal ion binding, protein-protein interactions, and epigenetic modification (13), which is also consistent with our conclusion from gene ontology analysis of TRP120-interacting partners. Polycomb group (PcG) proteins are members of a set of transcriptional repressors that control expression of developmental regulator genes in animals and plants (19, 32). PCGF proteins are PcG proteins with a specialized ring finger motif characterized by a cysteine-rich Zn2+-binding domain. The ring finger domains are involved in protein-DNA and protein-protein interactions and E2-dependent ubiquitination and consequently function in a variety of fundamental cellular processes, including the regulation of gene expression, signal transduction, and proteolysis (18, 27). PCGF proteins, such as PCGF1 (NSPC1), PCGF2 (Mel-18), PCGF4 (Bmi-1), and PCGF6 (MBLR), play important roles in the regulation of embryogenesis, lymphocyte differentiation and migration, immune responses, and self-renewing cell divisions (1, 11, 14, 17, 30). Thus, both TRP47 and TRP120 may interact with PCGF5 in order to modulate host cell gene expression to favor ehrlichial survival. E. chaffeensis changes the global gene expression pattern of the host cell (49), but the mechanisms involved are largely unknown.
The yeast two-hybrid assay revealed that the IGL protein was the most frequently identified partner interacting with TRP120. Our data also identified other immunoglobulin loci, including IGKC and IGHA1, interacting with TRP120 at a high frequency. Thus, TRP120 appears to directly interact with a wide variety of regions of human immunoglobulins. Interestingly, our previous study also identified an immunoglobulin lambda-like polypeptide (IGLL1) to be a partner interacting with E. chaffeensis TRP47 (42). The IGLL1 gene encodes one of the surrogate light-chain subunits, which is a member of the immunoglobulin gene superfamily (6). IGLL1 is involved in transduction of signals for cellular proliferation, differentiation from the pro-B cell to the pre-B cell stage, allelic exclusion at the immunoglobulin heavy-chain gene locus, and promotion of immunoglobulin light-chain gene rearrangements (40). To the best of our knowledge, the role of immunoglobulin chains in macrophage differentiation or other cellular processes is undetermined. Hence, understanding of the role and significance of IGL and IGLL1 and their interactions with ehrlichial TRPs in the macrophage will require further study, but the association with the host immune system suggests convergence on defined cellular networks by Ehrlichia effectors.
COX2 was the second most frequently identified partner interacting with TRP120. Cytochrome c oxidase is a multimeric enzymatic complex which is the terminal enzyme of the mitochondrial respiratory chain and is involved in the transfer of electrons from cytochrome c to molecular oxygen. The human COX complex is located in the mitochondrial inner membrane and consists of 13 subunits. Subunits 1, 2, and 3 (COX1, -2, and -3) are large, highly hydrophobic, transmembrane proteins encoded in the mitochondrial genome and form the catalytic core of the enzyme. COX2 transfers the electrons from reduced cytochrome c to the catalytic subunit COX1, and this is one of the key functional and regulatory sites of mammalian energy metabolism (9). Gene mutations of COX2 have been reported to cause encephalopathy or myopathy in patients (38). Associations between Ehrlichia morulae and mitochondria have been consistently observed (31, 35, 47, 48), and recently, Ehrlichia morulae were found to interact with mitochondria and inhibit mitochondrial metabolism, including the transcriptional level of the mitochondrial gene NADPH2 (23). The association of TRP120 with the respiratory chain suggests that E. chaffeensis may inhibit the production of ATP and superoxide in the host cell through the interaction of TRP120 and COX2 and subsequently suppress some cellular processes that utilize ATP, including biosynthetic reactions, motility, cell division, and superoxide production. Downregulation of host cell superoxide generation by E. chaffeensis has previously been reported (22).
Mitochondria play a central role in programmed cell death through both apoptotic and necrotic signals (46). Proapoptotic signals that disrupt the mitochondrial membrane barrier cause the release of proapoptotic contents such as cytochrome c and apoptosis-inducing factor (AIF), which separately activates a cascade of caspases and promotes nuclear changes in a caspase-independent manner (41). Recently, we reported the interaction of E. chaffeensis TRP47 with CAP1, which is a highly conserved monomeric actin-binding protein involved in regulation of actin remodeling in response to cellular signals and functions as an actin shuttle to mitochondria independent of caspase activation to promote apoptosis (42, 45). Therefore, like TRP47 and CAP1, the association of TRP120 with COX2 may also promote apoptosis in the late stage of ehrlichial infection and serve multiple functions in the host cell.
Caveola-mediated endocytosis directs E. chaffeensis to an intracellular compartment or inclusion that retains characteristics of the early endosome and does not fuse with lysosomes, thus providing protection from nonoxidative and oxidative damage (31). However, how Ehrlichia modulates host cell vesicular trafficking to avoid morula delivery to lysosomes remains unclear. The strong TRP120-GGA1 interaction suggests that TRP120 may play a role in this process. GGA1 is a member of the Golgi-localized, gamma adaptin ear-containing, ADP-ribosylation factor (ARF)-binding protein (GGA) family. Members of this family are ubiquitous coat proteins that regulate the trafficking of proteins between the trans-Golgi network (TGN) and the endosome/lysosome system mediated by clathrin-coated vesicles (4). In eukaryotic cells, newly synthesized proteins in the endoplasmic reticulum are transported via the Golgi apparatus to the TGN, where they are sorted for delivery to various cellular destinations, such as endosomes, lysosomes, and the cell membrane domains. GGAs possess four conserved functional domains, each of which interacts with cargo proteins, including mannose 6-phosphate receptors and other proteins, the small GTPase ARF, clathrin, or accessory proteins (33). Cargo proteins are packaged into carrier vesicles that detach from the TGN, dock, and fuse at the appropriate acceptor compartment; for example, human acid hydrolases are targeted to lysosomes by the mannose 6-phosphate recognition system (10). Therefore, by the interaction between TRP120 and GGA1, E. chaffeensis may manipulate host cell protein transportation from the Golgi apparatus to the endosome, preventing early endosomal maturation into late endosomes, thereby avoiding the late endosome-lysosome pathway.
Among other TRP120-interacting proteins identified by Y2H assay, the identification of ACTG1 and UNC13D was of significant interest. Actins are highly conserved proteins that are involved in various types of cell motility and maintenance of the cytoskeleton (21). Actin gamma 1 is a cytoplasmic actin found in nonmuscle cells. We previously reported that E. chaffeensis TRP47 interacts with an actin-binding protein, CAP1 (42), and a recent report showed that inhibition of actin polymerization in Ehrlichia-infected cells prevented filopodia formation, followed by the localization of Ehrlichia in the periphery of macrophages (39). Thus, actins appear to play important roles in both ehrlichial entry into and exit from the host cell, although the full details of the exact mechanisms of ehrlichial entry and exit remain incompletely defined. Therefore, associations of both TRP120 and TRP47 with actins suggest their importance in E. chaffeensis pathobiology, although the effects of the TRP interactions are unknown. An actin-related protein 2/3 complex subunit 2 (ARPC2) which has been implicated in the control of actin polymerization in cells (29) was also found to be associated with TRP120 by the Y2H assay (see Table S1 in the supplemental material). UNC13D appears to play a role in vesicle maturation during exocytosis and is involved in regulation of the secretion of cytolytic granules (15). Thus, the interaction of TRP120 and UNC13D may also play a role in ehrlichial release.
E. chaffeensis TRP120 consists of three major regions: the N-terminal region (50 amino acids), TR-containing region (350 amino acids), and C-terminal region (148 amino acids). Homology between TRP120 and other known proteins was not identified, except for E. canis TRP140. However, in addition to the single Ehrlichia repeat domain, conserved multidomains in K+-dependent Na+/Ca+ exchanger and FtsY were identified in the TR region of TRP120. FtsY is involved in cell division and protein secretion signal recognition of bacteria (3, 12). The significance of these conserved domains in the TR region is not clear but implicates a functional significance of the TR region, consistent with our findings. A co-IP experiment using single-region mutants of TRP120 identified the TR-containing region of E. chaffeensis TRP120 to be the primary interaction domain with PCGF5. However, the interaction between PCGF5 and the TR region of TRP120 was weaker than the interaction between PCGF5 and full-length TRP120, demonstrating the contribution of the TRP120 N- and C-terminal regions in the interaction. Moreover, none of the examined full-TRP120-interacting partners, including PCGF5, GGA1, ACTG1, and UNC13D, interacted with the N- or C-terminal region of TRP120 devoid of TRs, further implicating the importance of all regions, especially the TR region, in these protein-protein interactions, which is consistent with our previous observations regarding TRP47 (42).
TRP120 is a DC ehrlichia-expressed and secreted protein, and detection of TRP120 in the culture supernatants of host cells indicates that it can be secreted extracellularly into the host cell (24). Immunofluorescent microscopy demonstrated that interactions between TRP120 and host targets occurred either on morula-containing DC ehrlichiae or in the host cell cytoplasm adjacent to morula, which was consistent with our previous report regarding interactions between TRP47 and host targets (42). The interactions between TRP120 and TRP47 and host targets cause the redistribution of some host proteins to the morula or cytoplasm around the morulae in E. chaffeensis-infected cells, suggesting that TRP120 and TRP47 have profound effects on host cell protein recruitment and cellular distribution.
It is becoming more and more evident that Ehrlichia major immunoreactive proteins, such as TRP47, TRP120, and Ank200, are important effectors in the pathobiology of ehrlichial infection. TRP120 and TRP47 have some common molecular host interactions, suggesting an increased importance of overlapping or function-associated targets such as PCGF5, IGL, and CAP1. E. chaffeensis TRP120 interactions with both host proteins and DNA indicate that it is an ehrlichial effector with complex and diverse functions. Further research is needed to understand the mechanisms underlying these host-pathogen molecular interactions and their importance in ehrlichial pathobiology. Identification of other ehrlichial effector proteins and host cell ligands will facilitate studies to elucidate and define the molecular mechanisms involved.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI 071145 and AI 0071145 and by funding from the Clayton Foundation for Research. Jeeba A. Kuriakose was supported by NIH Biodefense Training grant T32 AI060549.
We thank David Walker and Xue-jie Yu (University of Texas Medical Branch) for reviewing the manuscript and providing helpful suggestions.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 22 August 2011.
REFERENCES
- 1. Akasaka T., et al. 2002. MBLR, a new RING finger protein resembling mammalian polycomb gene products, is regulated by cell cycle-dependent phosphorylation. Genes Cells 7:835–850 [DOI] [PubMed] [Google Scholar]
- 2. Al-Shahrour F., Minguez P., Vaquerizas J. M., Conde L., Dopazo J. 2005. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 33:W460–W464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arends S. J., Kustusch R. J., Weiss D. S. 2009. ATP-binding site lesions in FtsE impair cell division. J. Bacteriol. 191:3772–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonifacino J. S. 2004. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 5:23–32 [DOI] [PubMed] [Google Scholar]
- 5. Chen S. M., Cullman L. C., Walker D. H. 1997. Western immunoblotting analysis of the antibody responses of patients with human monocytotropic ehrlichiosis to different strains of Ehrlichia chaffeensis and Ehrlichia canis. Clin. Diagn. Lab. Immunol. 4:731–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Donohoe M. E., Blomberg B. B. 1997. The 14.1 surrogate light chain promoter has lineage- and stage-restricted activity. J. Immunol. 158:1681–1691 [PubMed] [Google Scholar]
- 7. Doyle C. K., Nethery K. A., Popov V. L., McBride J. W. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74:711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dunning Hotopp J. C., et al. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fontanesi F., Soto I. C., Barrientos A. 2008. Cytochrome c oxidase biogenesis: new levels of regulation. IUBMB Life 60:557–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghosh P., Kornfeld S. 2004. The GGA proteins: key players in protein sorting at the trans-Golgi network. Eur. J. Cell Biol. 83:257–262 [DOI] [PubMed] [Google Scholar]
- 11. Gong Y., et al. 2006. NSPc1 is a cell growth regulator that acts as a transcriptional repressor of p21Waf1/Cip1 via the RARE element. Nucleic Acids Res. 34:6158–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grudnik P., Bange G., Sinning I. 2009. Protein targeting by the signal recognition particle. Biol. Chem. 390:775–782 [DOI] [PubMed] [Google Scholar]
- 13. Grupe A., et al. 2006. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am. J. Hum. Genet. 78:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo W. J., Datta S., Band V., Dimri G. P. 2007. Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol. Biol. Cell 18:536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holt O. J., Gallo F., Griffiths G. M. 2006. Regulating secretory lysosomes. J. Biochem. 140:7–12 [DOI] [PubMed] [Google Scholar]
- 16. Huang B., et al. 2010. The Anaplasma phagocytophilum-occupied vacuole selectively recruits Rab-GTPases that are predominantly associated with recycling endosomes. Cell. Microbiol. 12:1292–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Itahana K., et al. 2003. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol. Cell. Biol. 23:389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joazeiro C. A., Weissman A. M. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552 [DOI] [PubMed] [Google Scholar]
- 19. Kerppola T. K. 2009. Polycomb group complexes—many combinations, many functions. Trends Cell Biol. 19:692–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kumagai Y., Matsuo J., Hayakawa Y., Rikihisa Y. 2010. Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J. Bacteriol. 192:4122–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee S. H., Dominguez R. 2010. Regulation of actin cytoskeleton dynamics in cells. Mol. Cells 29:311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin M., Rikihisa Y. 2007. Degradation of p22phox and inhibition of superoxide generation by Ehrlichia chaffeensis in human monocytes. Cell. Microbiol. 9:861–874 [DOI] [PubMed] [Google Scholar]
- 23. Liu Y., et al. 2010. Obligate intracellular bacterium Ehrlichia inhibiting mitochondrial activity. Microbes Infect. 13:232–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo T., Zhang X., McBride J. W. 2009. Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin. Vaccine Immunol. 16:982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo T., Zhang X., Nicholson W. L., Zhu B., McBride J. W. 2010. Molecular characterization of antibody epitopes of Ehrlichia chaffeensis ankyrin protein 200 and tandem repeat protein 47 and evaluation of synthetic immunodeterminants for serodiagnosis of human monocytotropic ehrlichiosis. Clin. Vaccine Immunol. 17:87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo T., Zhang X., Wakeel A., Popov V. L., McBride J. W. 2008. A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infect. Immun. 76:1572–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matthews J. M., Sunde M. 2002. Zinc fingers—folds for many occasions. IUBMB Life 54:351–355 [DOI] [PubMed] [Google Scholar]
- 28. McBride J. W., Walker D. H. 2011. Molecular and cellular pathobiology of Ehrlichia infection: targets for new therapeutics and immunomodulation strategies. Expert Rev. Mol. Med. 13:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mitsushima M., et al. 2010. Revolving movement of a dynamic cluster of actin filaments during mitosis. J. Cell Biol. 191:453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morey L., Helin K. 2010. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 35:323–332 [DOI] [PubMed] [Google Scholar]
- 31. Mott J., Barnewall R. E., Rikihisa Y. 1999. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect. Immun. 67:1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muller J., Verrijzer P. 2009. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19:150–158 [DOI] [PubMed] [Google Scholar]
- 33. Nakayama K., Wakatsuki S. 2003. The structure and function of GGAs, the traffic controllers at the TGN sorting crossroads. Cell Struct. Funct. 28:431–442 [DOI] [PubMed] [Google Scholar]
- 34. Paddock C. D., Childs J. E. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16:37–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Popov V. L., Chen S. M., Feng H. M., Walker D. H. 1995. Ultrastructural variation of cultured Ehrlichia chaffeensis. J. Med. Microbiol. 43:411–421 [DOI] [PubMed] [Google Scholar]
- 36. Popov V. L., Yu X., Walker D. H. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb. Pathog. 28:71–80 [DOI] [PubMed] [Google Scholar]
- 37. Rikihisa Y. 2010. Molecular events involved in cellular invasion by Ehrlichia chaffeensis and Anaplasma phagocytophilum. Vet. Parasitol. 167:155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shoubridge E. A. 2001. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106:46–52 [DOI] [PubMed] [Google Scholar]
- 39. Thomas S., Popov V. L., Walker D. H. 2010. Exit mechanisms of the intracellular bacterium Ehrlichia. PLoS One 5:e15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thompson E. C., et al. 2007. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity 26:335–344 [DOI] [PubMed] [Google Scholar]
- 41. Vander Heiden M. G., Thompson C. B. 1999. Bcl-2 proteins: regulators of apoptosis or of mitochondrial homeostasis? Nat. Cell Biol. 1:E209–E216 [DOI] [PubMed] [Google Scholar]
- 42. Wakeel A., Kuriakose J. A., McBride J. W. 2009. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect. Immun. 77:1734–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wakeel A., Luo T., Zhang X., McBride J. W. 2011. Extracellular secretion of Ehrlichia chaffeensis tandem repeat and Ank proteins is reduced in the absence of Escherichia coli TolC protein, abstr. B-2092. Abstr. 111th Gen. Meet. Am. Soc. Microbiol American Society for Microbiology, Washington, DC [Google Scholar]
- 44. Wakeel A., Zhu B., Yu X. J., McBride J. W. 2010. New insights into molecular Ehrlichia chaffeensis-host interactions. Microbes Infect. 12:337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang C., Zhou G. L., Vedantam S., Li P., Field J. 2008. Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J. Cell Sci. 121:2913–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang X. 2001. The expanding role of mitochondria in apoptosis. Genes Dev. 15:2922–2933 [PubMed] [Google Scholar]
- 47. Xiong Q., Bao W., Ge Y., Rikihisa Y. 2008. Ehrlichia ewingii infection delays spontaneous neutrophil apoptosis through stabilization of mitochondria. J. Infect. Dis. 197:1110–1118 [DOI] [PubMed] [Google Scholar]
- 48. Zhang J. Z., Popov V. L., Gao S., Walker D. H., Yu X. J. 2007. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell Microbiol. 9:610–618 [DOI] [PubMed] [Google Scholar]
- 49. Zhang J. Z., Sinha M., Luxon B. A., Yu X. J. 2004. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect. Immun. 72:498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu B., et al. 2011. Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function. Infect. Immun. 79:4370–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu B., et al. 2009. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect. Immun. 77:4243–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.